Background: Although ribosomal protein L11 (RPL11) inhibits c-Myc activity, it remains unclear if other RPs can do so.
Results: RPS14 negates c-Myc functions by reducing its transcriptional activity and level.
Conclusion: RPS14 suppresses c-Myc activity and cell proliferation independently of p53.
Significance: RPS14 may serve as a tumor suppressor that not only activates p53, but also inhibits c-Myc.
Keywords: Oncogene, p53, Proliferation, Signal Transduction, Transcription Regulation, 5-q Syndrome, c-Myc, Ribosomal Protein L11, Ribosomal Protein S14
Abstract
The ribosomal gene RPS14 is associated with the cancer-prone 5q-syndrome, which is caused by an interstitial deletion of the long arm of human chromosome 5. Previously, we found that ribosomal protein S14 (RPS14) binds to and inactivates MDM2, consequently leading to p53-dependent cell-cycle arrest and growth inhibition. However, it remains elusive whether RPS14 regulates cell proliferation in a p53-independent manner. Here, we show that RPS14 interacts with the Myc homology box II (MBII) and the C-terminal basic helix-loop-helix leucine zipper (bHLH-LZ) domains of the oncoprotein c-Myc. Further, RPS14 inhibited c-Myc transcriptional activity by preventing the recruitment of c-Myc and its cofactor, TRRAP, to the target gene promoters, as thus suppressing c-Myc-induced cell proliferation. Also, siRNA-mediated RPS14 depletion elevated c-Myc transcriptional activity determined by its target gene, Nucleolin, expression. Interestingly, RPS14 depletion also resulted in the induction of c-Myc mRNA and subsequent protein levels. Consistent with this, RPS14 promoted c-Myc mRNA turnover through an Argonaute 2 (Ago2)- and microRNA-mediated pathway. Taken together, our study demonstrates that RPS14 negates c-Myc functions by directly inhibiting its transcriptional activity and mediating its mRNA degradation via miRNA.
Introduction
Tumorigenesis is a complex process by which normal cells are transformed into malignant cells that undergo uncontrolled growth and division. This process is promoted by overexpression of numerous oncogenes, but prevented by activated tumor suppressor proteins. The c-Myc oncoprotein is a transcriptional factor that regulates expression of a large number of genes involved in cell cycle progression, cell proliferation and differentiation, angiogenesis, and metabolism, leading to neoplastic transformation (1). Hyper-expression of c-Myc was observed in multiple human cancers (2, 3). The Myc overexpression was shown to boost oncogenesis in cells (4) and in transgenic mouse models (5–7). Additionally, an appropriate c-Myc level is required for normal cell growth and mammalian development, as c-Myc deficiency causes cells to exit the cell cycle and blocks cell proliferation (8, 9), and homozygous deletion of the c-Myc gene resulted in embryonic lethality in mice (10).
c-Myc is a transcriptional factor with several domains critical for cell transformation. The N-terminal transactivation domain (TAD)2 confers both activation and repression of gene transcription. Two highly conserved domains within TAD are Myc homology box I (MBI) and Myc homology box II (MBII) domains, both of which have been shown to regulate c-Myc stability and activity (11). The MBI domain contains two phosphorylation sites, threonine 58 and serine 62 (12, 13). Phosphorylation of Thr-58 and dephosphorylation of Ser-62 elicit the subsequent ubiquitination and degradation of c-Myc by the SCFFbw7 complex, which also binds to the MBI domain (11, 14–17). The MBII domain is crucial for the recruitment of several co-activators to the target promoters of c-Myc. For instance, the GCN5 and TIP60 histone acetyltransferase complexes are recruited to these promoters through the MBII-mediated TRRAP association (18–21). These complexes promote histone acetylation, leading to a loose chromatin structure and engaging the transcription of the c-Myc target genes. Although the phosphorylation sites in the MBII domain remain to be determined, this domain is targeted by the ubiquitin ligase, Skp2, which mediates c-Myc ubiquitination and activation (22, 23). The other essential domain of c-Myc is the C-terminal basic helix-loop-helix leucine zipper (bHLH-LZ) domain (24, 25), which is responsible for its interaction with Max, a close partner of c-Myc. c-Myc-Max heterodimers bind to a CACGTG E-box sequence with high affinity (26, 27). Furthermore, the bHLH-LZ domain is also targeted by the ubiquitin ligase, Skp2, leading to c-Myc activation (22, 23).
The c-Myc activity is subjected to multiple regulations, because of its critical role in cell growth and transformation. In addition to the aforementioned post-translational modifications, several nucleolar proteins have also been shown to modulate the c-Myc transcriptional activity through direct contact with c-Myc. The tumor suppressor ARF, which is well-known for its key role in activating the p53 pathway (28–31), has also been shown to interact with c-Myc and to block its transcriptional activity, thus inhibiting c-Myc-promoted hyperproliferation and transformation (32–34). Another nucleolar protein, Nucleophosmin (NPM or B23), enhances c-Myc oncogenic activity by elevating the expression of its target genes (35). Interestingly, NPM has also been found to be required for the nucleolar localization and stabilization of Fbw7γ, a component of the E3 ligase complex mediating c-Myc ubiquitination and degradation (36). Thus, NPM controls c-Myc activity by both bolstering c-Myc transcriptional activity and improving c-Myc stabilization.
Recently, our group unveiled that ribosomal protein L11 (RPL11) can bind to the MBII domain of c-Myc and prevent the recruitment of the co-activator, TRRAP, to the target gene promoters of c-Myc, therefore hampering its transcriptional activity (37). Later, RPL11 was also found to control c-Myc mRNA turnover by recruiting miRISC/miR-24 complex to the 3′-UTR and thus leading to c-Myc mRNA degradation upon nucleolar stress (38). Furthermore, treatment of cells with the nucleolar stress-inducing chemicals, actinomycin D (Act D) or 5-fluorouracil (5-FU), drastically reduced c-Myc mRNA level in cells (38). The finding of the nucleolar stress-RPL11-c-Myc pathway prompted us to explore whether there are additional ribosomal proteins that are involved in the regulation of c-Myc.
RPS14, encoding the ribosomal protein S14 (RPS14), was identified as the human 5q-syndrome gene (39, 40). Our group has recently shown that RPS14 activates p53 upon nucleolar stress via binding to MDM2 and inhibiting its E3-ligase activity toward p53 (41). To explore the p53-independent tumor suppressive function of RPS14, we investigated whether RPS14 play a role in the c-Myc regulation. Indeed, we found that RPS14 could bind to the MBII and bHLH-LZ domains of c-Myc and inhibit its transcriptional activity by impeding the recruitment of c-Myc and its cofactor, TRRAP, to the promoter of its target gene, Nucleolin. Consistently, overexpression of RPS14 suppressed c-Myc-induced E2F2 and Nucleolin mRNA expression and cell proliferation. Conversely, ablation of RPS14 by siRNAs elevated both c-Myc and Nucleolin levels. Finally, we found that RPS14 could interact with Ago2 and triggered Ago2/miR-145 mediated c-Myc mRNA decay. Hence, our study as presented here demonstrates that RPS14 can also regulate c-Myc activity.
EXPERIMENTAL PROCEDURES
Cell Lines, Plasmids, and Antibodies
Human H1299 and HCT116p53−/− cells were cultured in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum, 50 units/ml penicillin and 0.1 mg/ml streptomycin at 37 °C in a 5% CO2 humidified atmosphere. Flag-RPS14, Flag-RPL30, V5-c-Myc, and deletion mutants of V5-c-Myc-coding plasmids have been described previously (37, 41). The pEGFP-Max plasmid was generated by inserting the human Max full-length cDNAs amplified by RT-PCR from H1299 cells into the pEGFP-C1 vector. The primers used for PCR amplification were 5′-CCGGAATTCAATGAGCGATAACGATGACA-3′ and 5′- CGCGGATCCTTAGCTGGCCTCCATCCGG-3′. Anti-HA (12CA5) has been described previously (41). Anti-Flag (Sigma-Aldrich), anti-V5 (Invitrogen and Thermo Scientific), anti-RPS14 (H-130, Santa Cruz Biotechnology), anti-c-Myc (N262 and 9E10, Santa Cruz Biotechnology), anti-GFP (B2, Santa Cruz Biotechnology), anti-BrdU (IIB5, Santa Cruz Biotechnology), anti-Ago2 (Abcam) were commercially purchased.
Transient Transfection, Immunoblotting, and Co-immunoprecipitation Analyses
Cells were transfected with plasmids as indicated in figure legends using METAFECTENE transfection reagent following the manufacturer's protocol (Biontex Laboratories GmbH). The cells were harvested at 30–48 h post transfection and lysed in lysis buffer consisting of 50 mm Tris/HCl (pH 8.0), 0.5% Nonidet P-40 (Nonidet P-40), 1 mm EDTA, 150 mm NaCl, 1 mm phenylmethylsulfonyl fluoride, 1 mm, 0.25 μg/ml pepstatin A, and 1 mm leupeptin. Equal amounts of clear cell lysate (20–50 μg) were used for immune-blotting (IB) analysis. Immunoprecipitation (IP) was performed using antibodies as indicated in the figure legends and described previously (41). Protein G or A beads (Santa Cruz Biotechnology) were washed twice with lysis buffer, and once with RIPA buffer (50 mm Tris-HCl (pH 8.0), 5 mm EDTA, 1% Nonidet P-40, 0.5% deoxycholate, 0.1% SDS, 150 mm NaCl). Dynabeads Protein G (Invitrogen) was also used in IP assays according to the manufacturer's protocol. The immunoprecipitated proteins were detected by IB with antibodies as indicated in the figure legends.
Chromatin Immunoprecipitation (ChIP) Assay
ChIP assay was performed as described previously (42) using anti-c-Myc (N262, Santa Cruz Biotechnology) and anti-TRRAP (H300, Santa Cruz Biotechnology). The reverse cross-linked immunoprecipitated DNA fragments were purified using GeneJET gel extraction kit (Thermo Scientific) followed by qPCR analysis for the Nucleolin promoter using primers as previously described (37).
BrdU Incorporation Assay
The BrdU incorporation assay was performed as described previously (37). Briefly, cells were incubated with 10 μm BrdU for 2 to 3 h followed by being fixed with 95% ethanol and 5% acetic acid, treated with 2 m HCl containing 1% Triton X-100 and stained with the monoclonal anti-BrdU antibody (IIB5, Santa Cruz Biotechnology). Cells were then stained with Alexa Fluor 546 (red) goat anti-mouse antibodies and DAPI and analyzed under a Zeiss Axiovert 25 fluorescent microscope.
Reverse Transcription (RT) and Quantitative (q) PCR Analysis
Total RNA was isolated from cells using Trizol (Invitrogen) following the manufacturer's protocol. Total RNAs of 50 ng were used as templates for reverse transcription using poly-(T)20 primers and M-MLV reverse transcriptase (Promega). Quantitative PCR (qPCR) was conducted using SYBR Green Mix according to the manufacturer's protocol (Bio-Rad). Primers for E2F2, Nucleolin, c-Myc, and GAPDH mRNAs were described previously (37).
RNA Interference
The siRNA pool against RPS14 and Ago2 (Santa Cruz Biotechnology) were commercially purchased. 40∼100 nm of siRNAs were introduced into cells using METAFECTENE siRNA transfection reagent following the manufacturer's protocol (Biontex Laboratories GmbH). Cells were harvested 48 to 72 h after transfection for immunoblot and quantitative RT-PCR.
RESULTS
RPS14 Interacts wtih c-Myc
The identification of RPL11 as a dual MDM2- and c-Myc-interacting protein made us wonder if other MDM2-binding proteins, such as RPS14, may also play a similar role in the c-Myc regulation. We thus tested whether RPS14 could bind to c-Myc by conducting a set of reciprocal co-IP-IB experiments. Indeed, V5-c-Myc could be specifically co-immunoprecipitated with Flag-S14 when the anti-Flag was used for the IP assay (Fig. 1A). Conversely, Flag-S14 was also co-immunoprecipitated with V5-c-Myc by the anti-V5 (Fig. 1B). To determine which domain(s) of c-Myc RPS14 might bind to, we performed a series of co-IP-IB assays by co-expressing V5-tagged c-Myc deletion mutants together with Flag-S14. Interestingly, we found that RPS14 bound to both N terminus (amino acids 1–144) and C-terminal bHLH-LZ domain (amino acids 355–439) (Fig. 1C). The exact RPS14 binding site in the N terminus (amino acids 1–144) of c-Myc was further examined. Similar to RPL11, RPS14 also bound to the MBII domain as the MBII-deleting N-terminal fragment (amino acids 1–144/ΔMBII) did not co-immunoprecipitate with RPS14 (Fig. 1D). Furthermore, we confirmed the endogenous interaction between RPS14 and c-Myc in H1299 cells by employing anti-c-Myc to co-immunoprecipitate RPS14-c-Myc complex (Fig. 1F). Taken together, these results indicate that RPS14 associates with c-Myc at both of its MBII and bHLH-LZ domains (Fig. 1E). This implies that RPS14 might regulate c-Myc activity partially through a different mechanism than that by RPL11.
FIGURE 1.
RPS14 associates with c-Myc at the MBII and bHLH-LZ domains. A and B, exogenous RPS14 binds to exogenous c-Myc in H1299 cells. Cells were transfected with V5-c-Myc and/or Flag-S14 plasmids as indicated and harvested at 48 h post-transfection for IP assays (0.3 mg total proteins per sample) using anti-Flag, anti-c-Myc (N262), or mouse/rabbit IgG, followed by IB assays with anti-c-Myc (N262) or anti-Flag. C and D, mapping the RPS14-binding domains of c-Myc. H1299 cells were transfected with Flag-S14 plasmid together with full-length c-Myc or c-Myc fragment-expressing plasmids and harvested at 48 h post-transfection for a set of IP assays using anti-V5 followed by IB assays with the antibodies as indicated. Arrows indicate the V5-c-Myc or V5-c-Myc-fragments. E, schematic representation of the RPS14-binding domains of c-Myc. F, endogenous RPS14 interacts with endogenous c-Myc in H1299 cells. Cell lysates (1 mg total proteins per sample) were prepared, and IP assays were performed using anti-c-Myc (N262) or rabbit IgG, followed by IB assays with anti-c-Myc (9E10) or anti-RPS14.
RPS14 Reduces the Recruitment of Both c-Myc and Its Co-factor TRRAP to the Target Promoter
To determine whether RPS14 affects the association of c-Myc with its target promoter, we performed a set of ChIP assays. In the representative result with the Nucleolin promoter, we could show that the association of c-Myc with this promoter was dampened when RPS14 was overexpressed in cells (Fig. 2A). Consistent with this data, we also found that RPS14 could impair the recruitment of exogenous expressing c-Myc to the target promoter (Fig. 2B). Conversely, siRNA-mediated knockdown of endogenous RPS14 enhanced the binding of c-Myc to the Nucleolin promoter (Fig. 2C). Given that RPS14 also bound to the MBII domain of c-Myc, which is important for TRRAP binding (18, 37), we tested whether RPS14 could inhibit the recruitment of TRRAP to the Nucleolin gene promoter. As expected, overexpression of RPS14 decreased the TRRAP recruitment (Fig. 2A) while knockdown of RPS14 elevated the association of TRRAP to the promoter (Fig. 2C). These results indicate that RPS14 might be able to inhibit c-Myc transcriptional activity by preventing the recruitment of c-Myc and TRRAP to the target promoters.
FIGURE 2.
RPS14 influences the recruitment of c-Myc and TRRAP to c-Myc target promoters. A, RPS14 overexpression reduces the recruitment of c-Myc and TRRAP to c-Myc target promoters. H1299 cells were transfected with control or Flag-S14 plasmid followed by serum starvation for overnight and stimulation for 3 h before harvesting for ChIP assay using antibodies or rabbit IgG as indicated. B, RPS14 overexpression reduces exogenous c-Myc recruitment to the target gene promoter. V5-c-Myc together with or without Flag-S14 was transfected into H1299 cells. Cells were harvested and subjected to ChIP assay using anti-c-Myc. C, siRNA-mediated knockdown of endogenous RPS14 increases the recruitment of c-Myc and TRRAP to the c-Myc target promoters. H1299 cells were transfected with control or RPS14 siRNAs for 48–72 h before harvesting for ChIP assays using antibodies or rabbit IgG and for IB (inset) as indicated. The co-immunoprecipitated DNAs in A, B, and C were subjected to qPCR analysis of the Nucleolin gene promoter. * indicates p < 0.05. D, RPS14 overexpression affects the c-Myc-Max interaction in H1299 cells. Cells were transfected with combinations of V5-c-Myc, pEGFP-Max, and Flag-S14 plasmids as indicated and were harvested for IP assays using anti-c-Myc (N262). The immunoprecipitated proteins were detected by IB assays with anti-V5, anti-GFP, or anti-Flag. E, RPS14 overexpression affects the interaction between the amino acids 355–439 of c-Myc and Max. The same experiments were performed as shown in D, except for using the V5-c-Myc/355–439 plasmid for transfection and anti-V5 for IP assays.
Next, we wondered how RPS14 could regulate the association of c-Myc with its target promoter. The c-Myc partner, Max, has been shown to improve the c-Myc binding affinity to the E-Box sequence and consequently, enhance its transcriptional activity via forming a heterodimer with c-Myc (26, 27). Since RPS14 bound to the bHLH-LZ domain which is required for the c-Myc-Max dimerization, we examined whether RPS14 could impair c-Myc-Max interaction. To test this idea, we introduced V5-c-Myc, pEGFP-Max, and Flag-S14 into H1299 cells in different combinations and performed co-IP-IB assays as shown in Fig. 2D. We found that overexpression of RPS14 could impair the c-Myc-Max interaction. Additionally, we also showed that overexpression of RPS14 could reduce the interaction between Max and amino acids 355–439 of c-Myc including the bHLH-LZ domain (Fig. 2E). Hence, these data suggest that perturbation of the interaction between c-Myc and Max may be one of the mechanisms underlying RPS14 inhibition of c-Myc association with its target promoter.
Overexpression of RPS14 Inhibits c-Myc Transcriptional Activity and Cell Proliferation
Since RPS14 impeded the association of c-Myc and TRRAP to the promoter, RPS14 might suppress c-Myc transcriptional activity. To test this possibility, we introduced RPS14 and/or c-Myc into H1299 cells followed by detecting the expression of endogenous E2F2 and Nucleolin mRNAs. While overexpression of RPS14 alone slightly reduced the E2F2 and Nucleolin expression, RPS14, when co-expressed with c-Myc, markedly negated the elevated expression of these genes induced by c-Myc (Fig. 3, A and B). Furthermore, RPS14 restrained the c-Myc-induced cell proliferation as shown by the BrdU incorporation assay (Fig. 3, C and D), which was consistent with our previous study showing that RPS14 inhibits cell growth through a p53-independent manner (41). Together, these results suggest that RPS14 can suppress c-Myc activity and c-Myc-induced cell proliferation.
FIGURE 3.

RPS14 inhibits c-Myc transcriptional activity and subsequent cell proliferation in H1299 cells. A and B, overexpression of RPS14 down-regulates c-Myc transcriptional activity determined by E2F2 and Nucleolin expression. Cells were transfected with Flag-S14 and V5-c-Myc plasmids individually or together for 30 h and harvested for RT-qPCR analysis to detect the expression of c-Myc target genes, E2F2 and Nucleolin. C and D, RPS14 suppresses c-Myc-induced cell proliferation as determined by BrdU incorporation assays. Cells were transfected with Flag-S14 and V5-c-Myc plasmids individually or together for 30 h and supplemented with 10 μm BrdU for 2 to 3 h before subjected to BrdU staining (C). Percentage of BrdU-positive cells is shown in panel D. * indicates p < 0.05, ** indicates p < 0.005.
Endogenous RPS14 Is Required for Down-regulation of c-Myc Activity and Level
To further determine whether endogenous RPS14 is essential for down-regulation of c-Myc transcriptional activity, we conducted siRNA-mediated RPS14 depletion and detected the mRNA level of c-Myc target gene, Nucleolin. To exclude the influence of p53, as RPS14 depletion can activate p53 pathway (41), we employed p53-deficient H1299 cells and HCT116p53−/− cells in this experiment. Interestingly, we found that these two cells differentially responded to RPS14 depletion. In H1299 cells, knockdown of RPS14 showed significant elevation of Nucleolin gene expression no matter whether cells were treated with 5-FU (Fig. 4A). Nevertheless, RPS14 depletion could only increase Nucleolin gene expression in response to 5-FU treatment in HCT116p53−/− cells (Fig. 4D).
FIGURE 4.
Knockdown of RPS14 leads to the elevation of c-Myc activity and level. A and D, siRNA-mediated RPS14 depletion elevates the expression of Nucleolin in both H1299 and HCT116p53−/− cells. Cells were transfected with control or RPS14 siRNAs for 48–72 h and supplemented with or without 5-FU overnight before harvesting for RT-qPCR. B and E, RPS14 knockdown increases c-Myc mRNA level as determined by RT-qPCR in both H1299 and HCT116p53−/− cells. * indicates p < 0.05, ** indicates p < 0.005. C and F, the representative results for the expression of endogenous c-Myc and RPS14, which were detected by IB assays using antibodies as indicated.
Moreover, we examined whether c-Myc level is affected upon RPS14 depletion and found that shortage of RPS14 increased c-Myc mRNA level (Fig. 4B) and subsequent protein level (Fig. 4C) in H1299 cells. Although RPS14 knockdown had undetectable impact on c-Myc level in HCT116p53−/− cells under normal culture condition, 5-FU treatment sensitized c-Myc expression upon RPS14 deprivation. When cells were supplemented with 5-FU, c-Myc expression was reduced (Fig. 4, E and F), which was consistent with previously reported data (38). By further knocking down RPS14, both mRNA and protein levels of c-Myc were induced HCT116p53−/− cells (Fig. 4, E and F). It appeared probable that less ribosome-free forms of RPS14 dwell in the nucleoplasm of the HCT116p53−/− cells compared with H1299 cells under non-stressful circumstance, while nucleolar stress, like 5-FU treatment, would promote the release of RPS14 to the nucleoplasm and enhance the connection between RPS14 and c-Myc, hence, RPS14 ablation in this situation could induce the c-Myc activity and level.
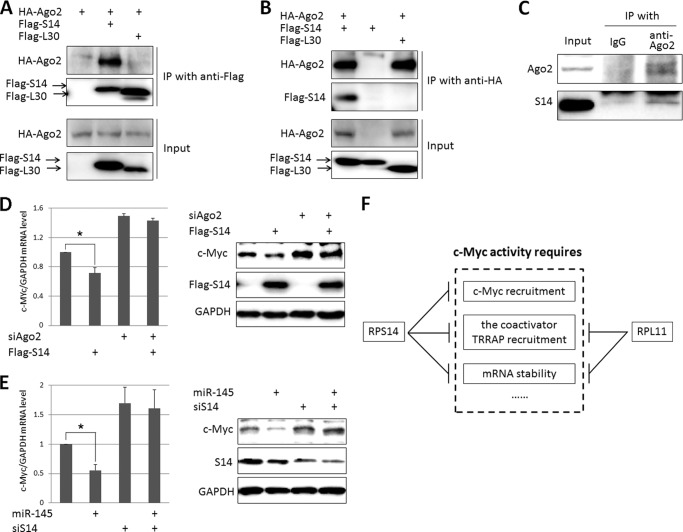
RPS14 Interacts with Ago2 and Prompts Ago2/miR-145-mediated c-Myc mRNA Turnover
Next, we sought to figure out how RPS14 could regulate c-Myc mRNA level. It has been shown that RPL11 binds to Ago2, the core component of microRNA induced silencing complex (miRISC), and leads to miRISC/miR-24-dependent c-Myc mRNA reduction (38). To test whether RPS14 also binds to Ago2, we transfected a combination of Flag-S14, Flag-L30, and HA-Ago2 into H1299 cells followed by reciprocal co-IP-IB assays. HA-Ago2 could be specifically co-immunoprecipitated with Flag-S14, but not Flag-L30 using the anti-Flag antibody (Fig. 5A). Consistently, Flag-S14, but not Flag-L30, could be co-immunoprecipitated with HA-Ago2 by the anti-HA antibody (Fig. 5B). In addition, we also detected the endogenous binding of RPS14 and Ago2 (Fig. 5C). We further explored whether Ago2 was required for RPS14 triggered c-Myc mRNA turnover. As shown in the left panel of Fig. 5D, overexpression of RPS14 could reduce c-Myc mRNA level as expected, whereas c-Myc mRNA level was elevated, but not repressed by RPS14 when Ago2 was knocked down in these cells (Fig. 5D). The representative protein expression was shown in the right panel of Fig. 5D. These results suggest that the reduction of c-Myc mRNA by RPS14 is through a miRISC/microRNA-mediated pathway. We thereafter asked whether RPS14 control c-Myc mRNA decay through specific miRNA activity. Several microRNAs, including let-7 (43), miR-24 (44), and miR-145 (45), have been reported to down-regulate c-Myc mRNA level. In this study, we introduced miR-145 together with or without RPS14 siRNAs in H1299 cells (Fig. 5E). In consistence with previous data, we also showed that miR-145 supplementation reduced c-Myc mRNA level. However, this reduction was blocked when RPS14 was knocked down (Fig. 5E). Together, these results indicate that c-Myc mRNA turnover is dependent on both RPS14 and miRISC/miR-145.
FIGURE 5.
RPS14 interacts with Ago2 and leads to miR-145/Ago2-mediated c-Myc mRNA turnover. A and B, exogenous RPS14, but not RPL30, binds to exogenous Ago2 in H1299 cells. Cells were transfected with a combination of HA-Ago2, Flag-S14, and Flag-L30 plasmids as indicated and harvested at 48 h post-transfection for IP assays using anti-Flag, anti-HA, or mouse IgG, followed by IB assays with anti-HA or anti-Flag. C, the endogenous binding of RPS14 and Ago2 in H1299 cells. IP assays were performed using anti-Ago2 followed by IB assay using antibodies as indicated. D, Ago2 is required for suppression of c-Myc mRNA expression by RPS14. H1299 cells were transfected with plasmids or siRNA as indicated. Cells were harvested 48–72 h post-transfection and subjected to qPCR analysis (left panel) and IB assay (right panel). E, RPS14 knockdown impairs miR-145-mediated c-Myc mRNA decay. H1299 cells were transfected with miR-145 or siRNA as indicated. Cells were harvested 48–72 h post-transfection and subjected to qPCR analysis (left panel) and IB assay (right panel). * indicates p < 0.05. F, schematic for c-Myc regulation by ribosomal proteins via multiple mechanisms. Both RPS14 and RPL11 can negatively regulate c-Myc activity by preventing the recruitment of TRRAP to the c-Myc target promoters and prompting c-Myc mRNA degradation. Additionally, RPS14 can also hinder the binding of c-Myc to its target gene promoter.
DISCUSSION
Ribosomal proteins have emerged as the activators of p53 in response to nucleolar stress (46). However, it remains intriguing to explore their extra-ribosomal functions in the p53-indepentent pathway. RPL11 was recently found to not only inhibit c-Myc transcriptional activity by directly binding to its MBII domain (37, 47), but also suppress c-Myc mRNA expression via a miRNA-mediated mechanism (38). It remains possible that RPL11 might not be the only ribosomal protein that regulates c-Myc activity and stability, given that nearly a dozen of ribosomal proteins, including RPL11, RPL5 RPL23, RPS7, and RPS14, have been shown to play a role in regulation of the MDM2-p53 pathway (48–55). In this study, we revealed that RPS14 is also capable of inhibiting c-Myc activity through multiple mechanisms (Fig. 5F). First, RPS14 impaired the association of c-Myc with the target gene promoter probably via binding to the bHLH-LZ domain of c-Myc. Second, RPS14, like RPL11, bound to the MBII domain of c-Myc and impeded the recruitment of its co-factor, TRRAP, to its target promoter. Lastly, we found that RPS14 cooperated with Ago2 and elicited miR-145-induced c-Myc mRNA turnover.
It is interesting and surprising to learn that RPS14, different from RPL11 (37), also associates with the bHLH-LZ domain (Fig. 1C), which is critical for c-Myc to bind to DNA and Max. This finding indicates that RPS14 can modulate c-Myc transcriptional activity via an additional mechanism compared with RPL11 (37). Even though the c-Myc-Max interaction is so robust that several proteins binding to the bHLH-LZ domain have not been shown to disrupt this heterodimerization (56), our results showed that overexpression of RPS14 in H1299 cells could reduce the formation of the c-Myc-Max complex (Fig. 2D) as well as the interaction between the bHLH-LZ domain (amino acids 355–439) of c-Myc and Max (Fig. 2E). These findings suggest that RPS14 might also inhibit c-Myc transcriptional activity by negatively influencing the c-Myc-Max interaction, which could be further enhanced by associating with other ribosomal proteins.
As mentioned above, RPS14 reduces the association of c-Myc to its target promoter (Fig. 2, A, B, and C). This could be the result from the following three possible mechanisms. First, it could be associated with the reduction of the c-Myc-Max interaction by RPS14 (Fig. 2, D and E). Alternatively, the RPS14 association with the bHLH-LZ domain may directly conceal this DNA-binding domain from contacting the E-box sequence of its target promoter. Finally, RPS14, like RPL11, also suppresses c-Myc mRNA expression (Figs. 4 and 5), thus decreases c-Myc protein accumulation on the target gene promoter.
The ARF tumor suppressor has been previously found to suppress c-Myc oncogenic activity (33, 34, 57). Interestingly, ARF also binds to both MBII and bHLH-LZ domains (33, 34, 57), which overlaps with the RPS14 binding regions. Recently, we reported that RPL11 and ARF bind to each other and enhance p53 activity (58). It is thus possible that RPS14 might crosstalk with ARF in regulating c-Myc activity as well.
Ribosomal proteins are able to regulate c-Myc mRNA levels (Figs. 4 and 5) (38). RPL11 has been reported to bind to the 3′-UTR of c-Myc mRNA and to recruit the miR-24/RISC complex for the c-Myc mRNA degradation (38), which might also be the molecular basis for the suppression of c-Myc mRNA by RPS14. Indeed, we found that RPS14, but not RPL30, specifically interacted with Ago2 and promoted c-Myc RNA degradation through a miR-145/RISC mediated pathway. In addition to these observations, increasing evidence has shown that ribosomal proteins may play an important role in microRNA-mediated mRNA decay or translation inhibition. First, several eukaryotic ribosomal proteins, including RPS13, RPS14, RPL12, and RPL30, have been shown to inhibit their own mRNA production by binding to the mRNA (59), although it remained unclear whether such inhibition is through microRNAs. Also, Rps14 was found to interact with the nematode Argonaute homolog, ALG-1, and modulate the microRNA function in Caenorhabditis elegans (60). Recently, it has been reported that ribosomal proteins are essential for microRNA-mediated gene repression, as reduced ribosomal protein gene expression dissociated microRNA/RISC complex from the target mRNAs, thus enhanced target mRNA stability and translation, which is consistent with our current study. Therefore, additional ribosomal proteins are expected to be found to regulate c-Myc mRNA turnover.
It has been an open question of whether each ribosomal protein acts in a complex with other MDM2-binding ribosomal proteins or independently to execute their extra-ribosomal functions since several ribosomal proteins were found to interact with MDM2 (48–52). MDM2 was shown to form a complex with RPL5, RPL11, and RPL23 (51). Later, RPL5 and RPL11 were found to bind to each other through 5S rRNA and enhance p53 activity synergistically (61). Surprisingly, we recently found that RPS14 competed with RPL5 for the MDM2 binding (55), suggesting that these two ribosomal proteins might not be able to cooperate with each other in activating p53. But, it remains possible that RPS14 and RPL11 may cooperate with each other to suppress MDM2 and c-Myc activity since they have different binding patterns on MDM2 or c-Myc (Fig. 1, C and D) (37, 48, 49, 55).
The rps14 gene has been linked with 5q-syndrome, a subtype of myelodysplastic syndrome (MDS) that is characterized by defects in erythroid differentiation, due to an interstitial deletion of the long arm of chromosome 5 (62). Recently, an RNAi-based screening demonstrated that partial loss of function of RPS14 is responsible for this genetic disease (39), which was later verified by an Rps14-haploinsufficient mouse model (40). It has been shown that Rps14 deficiency results in the accumulation of p53, which in turn causes increased bone marrow cell apoptosis (40). Even though with elevated p53 level in their erythroid lineage (63), the 5q-syndrome patients have high risk of developing acute myeloid leukemia (AML) (64). This suggests that other mechanisms may contribute to this high incidence of cancer bypassing the p53 supervision. The overexpression or amplification of c-Myc has been observed in AML patients (65). Moreover, the increased c-Myc mRNA level has also been reported in some AML patients (66). Hence, the identification of RPS14 as a negative regulator of c-Myc may provide a clue as to why 5q-syndrome patients with impaired function of RPS14 are susceptible to AML.
Acknowledgments
We thank Shelya X. Zeng for technical assistance and all of our colleagues in the Lu laboratory for active discussion and help.
This work was supported in whole or in part by National Institutes of Health-NCI Grants CA095441, CA 079721, CA129828, and CA 172468 (to H. L.).
- TAD
- N-terminal transactivation domain
- Act D
- actinomycin D
- 5-FU
- 5-fluorouracil
- MBI
- Myc homology box I
- MBII
- Myc homology box II
- bHLH-LZ
- basic helix-loop-helix leucine zipper
- MDS
- myelodysplastic syndrome
- AML
- acute myeloid leukemia.
REFERENCES
- 1. Meyer N., Penn L. Z. (2008) Nature Rev. 8, 976–990 [DOI] [PubMed] [Google Scholar]
- 2. Nesbit C. E., Tersak J. M., Prochownik E. V. (1999) MYC oncogenes and human neoplastic disease. Oncogene 18, 3004–3016 [DOI] [PubMed] [Google Scholar]
- 3. Pelengaris S., Khan M., Evan G. (2002) c-MYC: more than just a matter of life and death. Nature Rev. 2, 764–776 [DOI] [PubMed] [Google Scholar]
- 4. Land H., Parada L. F., Weinberg R. A. (1983) Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304, 596–602 [DOI] [PubMed] [Google Scholar]
- 5. Adams J. M., Harris A. W., Pinkert C. A., Corcoran L. M., Alexander W. S., Cory S., Palmiter R. D., Brinster R. L. (1985) The c-myc oncogene driven by immunoglobulin enhancers induces lymphoid malignancy in transgenic mice. Nature 318, 533–538 [DOI] [PubMed] [Google Scholar]
- 6. Stewart T. A., Pattengale P. K., Leder P. (1984) Spontaneous mammary adenocarcinomas in transgenic mice that carry and express MTV/myc fusion genes. Cell 38, 627–637 [DOI] [PubMed] [Google Scholar]
- 7. Schoenenberger C. A., Andres A. C., Groner B., van der Valk M., LeMeur M., Gerlinger P. (1988) Targeted c-myc gene expression in mammary glands of transgenic mice induces mammary tumours with constitutive milk protein gene transcription. EMBO J. 7, 169–175 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Alboran I. M., O'Hagan R. C., Gärtner F., Malynn B., Davidson L., Rickert R., Rajewsky K., DePinho R. A., Alt F. W. (2001) Analysis of C-MYC function in normal cells via conditional gene-targeted mutation. Immunity 14, 45–55 [DOI] [PubMed] [Google Scholar]
- 9. Trumpp A., Refaeli Y., Oskarsson T., Gasser S., Murphy M., Martin G. R., Bishop J. M. (2001) c-Myc regulates mammalian body size by controlling cell number but not cell size. Nature 414, 768–773 [DOI] [PubMed] [Google Scholar]
- 10. Davis A. C., Wims M., Spotts G. D., Hann S. R., Bradley A. (1993) A null c-myc mutation causes lethality before 10.5 days of gestation in homozygotes and reduced fertility in heterozygous female mice. Genes Dev. 7, 671–682 [DOI] [PubMed] [Google Scholar]
- 11. Dai M. S., Jin Y., Gallegos J. R., Lu H. (2006) Balance of Yin and Yang: ubiquitylation-mediated regulation of p53 and c-Myc. Neoplasia 8, 630–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Sears R., Nuckolls F., Haura E., Taya Y., Tamai K., Nevins J. R. (2000) Multiple Ras-dependent phosphorylation pathways regulate Myc protein stability. Genes Dev. 14, 2501–2514 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Lutterbach B., Hann S. R. (1994) Hierarchical phosphorylation at N-terminal transformation-sensitive sites in c-Myc protein is regulated by mitogens and in mitosis. Mol. Cell. Biol. 14, 5510–5522 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Yeh E., Cunningham M., Arnold H., Chasse D., Monteith T., Ivaldi G., Hahn W. C., Stukenberg P. T., Shenolikar S., Uchida T., Counter C. M., Nevins J. R., Means A. R., Sears R. (2004) A signalling pathway controlling c-Myc degradation that impacts oncogenic transformation of human cells. Nature Cell Biol. 6, 308–318 [DOI] [PubMed] [Google Scholar]
- 15. Welcker M., Orian A., Jin J., Grim J. A., Harper J. W., Eisenman R. N., Clurman B. E. (2004) The Fbw7 tumor suppressor regulates glycogen synthase kinase 3 phosphorylation-dependent c-Myc protein degradation. Proc. Natl. Acad. Sci.U.S.A. 101, 9085–9090 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Moberg K. H., Mukherjee A., Veraksa A., Artavanis-Tsakonas S., Hariharan I. K. (2004) The Drosophila F box protein archipelago regulates dMyc protein levels in vivo. Curr. Biol. 14, 965–974 [DOI] [PubMed] [Google Scholar]
- 17. Yada M., Hatakeyama S., Kamura T., Nishiyama M., Tsunematsu R., Imaki H., Ishida N., Okumura F., Nakayama K., Nakayama K. I. (2004) Phosphorylation-dependent degradation of c-Myc is mediated by the F-box protein Fbw7. EMBO J. 23, 2116–2125 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. McMahon S. B., Van Buskirk H. A., Dugan K. A., Copeland T. D., Cole M. D. (1998) The novel ATM-related protein TRRAP is an essential cofactor for the c-Myc and E2F oncoproteins. Cell 94, 363–374 [DOI] [PubMed] [Google Scholar]
- 19. McMahon S. B., Wood M. A., Cole M. D. (2000) The essential cofactor TRRAP recruits the histone acetyltransferase hGCN5 to c-Myc. Mol. Cell. Biol. 20, 556–562 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Park J., Kunjibettu S., McMahon S. B., Cole M. D. (2001) The ATM-related domain of TRRAP is required for histone acetyltransferase recruitment and Myc-dependent oncogenesis. Genes Dev. 15, 1619–1624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Frank S. R., Parisi T., Taubert S., Fernandez P., Fuchs M., Chan H. M., Livingston D. M., Amati B. (2003) MYC recruits the TIP60 histone acetyltransferase complex to chromatin. EMBO Rep. 4, 575–580 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. von der Lehr N., Johansson S., Wu S., Bahram F., Castell A., Cetinkaya C., Hydbring P., Weidung I., Nakayama K., Nakayama K. I., Söderberg O., Kerppola T. K., Larsson L. G. (2003) The F-box protein Skp2 participates in c-Myc proteosomal degradation and acts as a cofactor for c-Myc-regulated transcription. Mol. Cell 11, 1189–1200 [DOI] [PubMed] [Google Scholar]
- 23. Kim S. Y., Herbst A., Tworkowski K. A., Salghetti S. E., Tansey W. P. (2003) Skp2 regulates Myc protein stability and activity. Mol. Cell 11, 1177–1188 [DOI] [PubMed] [Google Scholar]
- 24. Landschulz W. H., Johnson P. F., McKnight S. L. (1988) The leucine zipper: a hypothetical structure common to a new class of DNA binding proteins. Science 240, 1759–1764 [DOI] [PubMed] [Google Scholar]
- 25. Murre C., McCaw P. S., Baltimore D. (1989) A new DNA binding and dimerization motif in immunoglobulin enhancer binding, daughterless, MyoD, and myc proteins. Cell 56, 777–783 [DOI] [PubMed] [Google Scholar]
- 26. Blackwood E. M., Eisenman R. N. (1991) Max: a helix-loop-helix zipper protein that forms a sequence-specific DNA-binding complex with Myc. Science 251, 1211–1217 [DOI] [PubMed] [Google Scholar]
- 27. Amati B., Brooks M. W., Levy N., Littlewood T. D., Evan G. I., Land H. (1993) Oncogenic activity of the c-Myc protein requires dimerization with Max. Cell 72, 233–245 [DOI] [PubMed] [Google Scholar]
- 28. Honda R., Yasuda H. (1999) Association of p19(ARF) with Mdm2 inhibits ubiquitin ligase activity of Mdm2 for tumor suppressor p53. EMBO J. 18, 22–27 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Weber J. D., Taylor L. J., Roussel M. F., Sherr C. J., Bar-Sagi D. (1999) Nucleolar Arf sequesters Mdm2 and activates p53. Nat. Cell Biol. 1, 20–26 [DOI] [PubMed] [Google Scholar]
- 30. Zhang Y., Xiong Y. (1999) Mutations in human ARF exon 2 disrupt its nucleolar localization and impair its ability to block nuclear export of MDM2 and p53. Mol. Cell 3, 579–591 [DOI] [PubMed] [Google Scholar]
- 31. Tao W., Levine A. J. (1999) P19(ARF) stabilizes p53 by blocking nucleo-cytoplasmic shuttling of Mdm2. Proc. Natl. Acad. Sci. U.S.A. 96, 6937–6941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Cleveland J. L., Sherr C. J. (2004) Antagonism of Myc functions by Arf. Cancer Cell 6, 309–311 [DOI] [PubMed] [Google Scholar]
- 33. Datta A., Nag A., Pan W., Hay N., Gartel A. L., Colamonici O., Mori Y., Raychaudhuri P. (2004) Myc-ARF (alternate reading frame) interaction inhibits the functions of Myc. J. Biol. Chem. 279, 36698–36707 [DOI] [PubMed] [Google Scholar]
- 34. Qi Y., Gregory M. A., Li Z., Brousal J. P., West K., Hann S. R. (2004) p19ARF directly and differentially controls the functions of c-Myc independently of p53. Nature 431, 712–717 [DOI] [PubMed] [Google Scholar]
- 35. Li Z., Boone D., Hann S. R. (2008) Nucleophosmin interacts directly with c-Myc and controls c-Myc-induced hyperproliferation and transformation. Proc. Natl. Acad. Sci.U.S.A. 105, 18794–18799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Bonetti P., Davoli T., Sironi C., Amati B., Pelicci P. G., Colombo E. (2008) Nucleophosmin and its AML-associated mutant regulate c-Myc turnover through Fbw7 gamma. J. Cell Biol. 182, 19–26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dai M. S., Arnold H., Sun X. X., Sears R., Lu H. (2007) Inhibition of c-Myc activity by ribosomal protein L11. EMBO J. 26, 3332–3345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Challagundla K. B., Sun X. X., Zhang X., DeVine T., Zhang Q., Sears R. C., Dai M. S. (2011) Mol. Cell. Biol. 31, 4007–4021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Ebert B. L., Pretz J., Bosco J., Chang C. Y., Tamayo P., Galili N., Raza A., Root D. E., Attar E., Ellis S. R., Golub T. R. (2008) Identification of RPS14 as a 5q- syndrome gene by RNA interference screen. Nature 451, 335–339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Barlow J. L., Drynan L. F., Hewett D. R., Holmes L. R., Lorenzo-Abalde S., Lane A. L., Jolin H. E., Pannell R., Middleton A. J., Wong S. H., Warren A. J., Wainscoat J. S., Boultwood J., McKenzie A. N. (2010) Nat. Med. 16, 59–66 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Zhou X., Hao Q., Liao J., Zhang Q., Lu H. Oncogene 32, 388–396 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Zeng S. X., Dai M. S., Keller D. M., Lu H. (2002) SSRP1 functions as a co-activator of the transcriptional activator p63. EMBO J. 21, 5487–5497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Kim H. H., Kuwano Y., Srikantan S., Lee E. K., Martindale J. L., Gorospe M. (2009) HuR recruits let-7/RISC to repress c-Myc expression. Genes Dev. 23, 1743–1748 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Lal A., Navarro F., Maher C. A., Maliszewski L. E., Yan N., O'Day E., Chowdhury D., Dykxhoorn D. M., Tsai P., Hofmann O., Becker K. G., Gorospe M., Hide W., Lieberman J. (2009) miR-24 Inhibits cell proliferation by targeting E2F2, MYC, and other cell-cycle genes via binding to “seedless” 3'UTR microRNA recognition elements. Mol. Cell 35, 610–625 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Sachdeva M., Zhu S., Wu F., Wu H., Walia V., Kumar S., Elble R., Watabe K., Mo Y. Y. (2009) p53 represses c-Myc through induction of the tumor suppressor miR-145. Proc. Natl. Acad. Sci. U.S.A. 106, 3207–3212 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang Y., Lu H. (2009) Signaling to p53: ribosomal proteins find their way. Cancer Cell 16, 369–377 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Dai M. S., Sears R., Lu H. (2007) Feedback regulation of c-Myc by ribosomal protein L11. Cell Cycle 6, 2735–2741 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Zhang Y., Wolf G. W., Bhat K., Jin A., Allio T., Burkhart W. A., Xiong Y. (2003) Ribosomal protein L11 negatively regulates oncoprotein MDM2 and mediates a p53-dependent ribosomal-stress checkpoint pathway. Mol. Cell. Biol. 23, 8902–8912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Lohrum M. A., Ludwig R. L., Kubbutat M. H., Hanlon M., Vousden K. H. (2003) Regulation of HDM2 activity by the ribosomal protein L11. Cancer Cell 3, 577–587 [DOI] [PubMed] [Google Scholar]
- 50. Jin A., Itahana K., O'Keefe K., Zhang Y. (2004) Inhibition of HDM2 and activation of p53 by ribosomal protein L23. Mol. Cell. Biol. 24, 7669–7680 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Dai M. S., Zeng S. X., Jin Y., Sun X. X., David L., Lu H. (2004) Ribosomal protein L23 activates p53 by inhibiting MDM2 function in response to ribosomal perturbation but not to translation inhibition. Mol. Cell. Biol. 24, 7654–7668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Dai M. S., Lu H. (2004) Inhibition of MDM2-mediated p53 ubiquitination and degradation by ribosomal protein L5. J. Biol. Chem. 279, 44475–44482 [DOI] [PubMed] [Google Scholar]
- 53. Chen D., Zhang Z., Li M., Wang W., Li Y., Rayburn E. R., Hill D. L., Wang H., Zhang R. (2007) Ribosomal protein S7 as a novel modulator of p53-MDM2 interaction: binding to MDM2, stabilization of p53 protein, and activation of p53 function. Oncogene 26, 5029–5037 [DOI] [PubMed] [Google Scholar]
- 54. Zhu Y., Poyurovsky M. V., Li Y., Biderman L., Stahl J., Jacq X., Prives C. (2009) Ribosomal protein S7 is both a regulator and a substrate of MDM2. Mol. Cell 35, 316–326 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Deleted in proof
- 56. Escamilla-Powers J. R., Daniel C. J., Farrell A., Taylor K., Zhang X., Byers S., Sears R. (2010) The tumor suppressor protein HBP1 is a novel c-myc-binding protein that negatively regulates c-myc transcriptional activity. J. Biol. Chem. 285, 4847–4858 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Amente S., Gargano B., Varrone F., Ruggiero L., Dominguez-Sola D., Lania L., Majello B. (2006) p14ARF directly interacts with Myc through the Myc BoxII domain. Cancer Biology Therapy 5, 287–291 [DOI] [PubMed] [Google Scholar]
- 58. Dai M. S., Challagundla K. B., Sun X. X., Palam L. R., Zeng S. X., Wek R. C., Lu H. (2012) Physical and functional interaction between ribosomal protein L11 and the tumor suppressor ARF. J. Biol. Chem. 287, 17120–17129 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Warner J. R., McIntosh K. B. (2009) How common are extraribosomal functions of ribosomal proteins? Mol. Cell 34, 3–11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Chan S. P., Slack F. J. (2009) Ribosomal protein RPS-14 modulates let-7 microRNA function in Caenorhabditis elegans. Dev. Biol. 334, 152–160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Horn H. F., Vousden K. H. (2008) Cooperation between the ribosomal proteins L5 and L11 in the p53 pathway. Oncogene 27, 5774–5784 [DOI] [PubMed] [Google Scholar]
- 62. Van den Berghe H., Cassiman J. J., David G., Fryns J. P., Michaux J. L., Sokal G. (1974) Distinct haematological disorder with deletion of long arm of no. 5 chromosome. Nature 251, 437–438 [DOI] [PubMed] [Google Scholar]
- 63. Dutt S., Narla A., Lin K., Mullally A., Abayasekara N., Megerdichian C., Wilson F. H., Currie T., Khanna-Gupta A., Berliner N., Kutok J. L., Ebert B. L. (2011) Blood 117, 2567–2576 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Narla A., Ebert B. L. (2010) Blood 115, 3196–3205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Delgado M. D., Leon J. (2010) Genes & cancer 1, 605–616 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Court E. L., Smith M. A., Avent N. D., Hancock J. T., Morgan L. M., Gray A. G., Smith J. G. (2004) DNA microarray screening of differential gene expression in bone marrow samples from AML, non-AML patients and AML cell lines. Leukemia Res. 28, 743–753 [DOI] [PubMed] [Google Scholar]