Background: Intracellular trafficking of RGS proteins determines their function as G-protein inhibitors.
Results: The function and trafficking of RGS4 through intracellular endosomal pools are highly regulated by different Rab proteins.
Conclusion: Rab5 and Rab11 regulate the intracellular trafficking and function of RGS4.
Significance: Identifying novel mechanisms whereby RGS4 localization and function may be regulated might lead to new strategies for the selective regulation of G-protein signaling.
Keywords: Cell Biology, Endocytosis, G-protein-coupled Receptors (GPCR), G-proteins, RGS Proteins, Signaling
Abstract
RGS4, a heterotrimeric G-protein inhibitor, localizes to plasma membrane (PM) and endosomal compartments. Here, we examined Rab-mediated control of RGS4 internalization and recycling. Wild type and constitutively active Rab5 decreased RGS4 PM levels while increasing its endosomal targeting. Rab5, however, did not appreciably affect the PM localization or function of the M1 muscarinic receptor (M1R)/Gq signaling cascade. RGS4-containing endosomes co-localized with subsets of Rab5-, transferrin receptor-, and Lamp1/Lysotracker-marked compartments suggesting RGS4 traffics through PM recycling or acidified endosome pathways. Rab7 activity promoted TGN association, whereas Rab7(dominant negative) trapped RGS4 in late endosomes. Furthermore, RGS4 was found to co-localize with an endosomal pool marked by Rab11, the protein that mediates recycling/sorting of proteins to the PM. The Cys-12 residue in RGS4 appeared important for its Rab11-mediated trafficking to the PM. Rab11(dominant negative) decreased RGS4 PM levels and increased the number of RGS4-containing endosomes. Inhibition of Rab11 activity decreased RGS4 function as an inhibitor of M1R activity without affecting localization and function of the M1R/Gq signaling complex. Thus, both Rab5 activation and Rab11 inhibition decreased RGS4 function in a manner that is independent from their effects on the localization and function of the M1R/Gq signaling complex. This is the first study to implicate Rab GTPases in the intracellular trafficking of an RGS protein. Thus, Rab GTPases may be novel molecular targets for the selective regulation of M1R-mediated signaling via their specific effects on RGS4 trafficking and function.
Introduction
The regulator of G-protein signaling (RGS)2 proteins modulate cellular signaling mediated by G-protein-coupled receptors. The superfamily of RGS proteins (1, 2) contains >35 members, many of which function as GTPase-activating proteins for Gα subunits (3, 4). RGS4 is one of the most widely studied members of this family. RGS4 has been shown to be an effective inhibitor of M1 muscarinic and other receptors coupled to both Gq and Gi. Indeed, genetic deletion models have been used to show that RGS4 regulates a wide variety of GPCR-mediated physiological pathways in the brain (5, 6), sinoatrial node (7), pancreas (8, 9), and metastatic tumor cells (10). Indeed, changes in RGS4 expression have been identified in numerous pathogenic conditions, including schizophrenia (11, 12), Alzheimer disease (13, 14), and heart failure (15), linking its altered function to disease pathogenesis and progression. Thus, understanding the cellular mechanisms by which RGS4 function is regulated at the intracellular level may be an important step toward understanding its role in various tissues under normal and pathophysiological conditions. Specifically, we are interested in uncovering novel intracellular pathways that regulate the function of RGS4 as an inhibitor of G-protein-coupled receptor signaling in mammalian cells as a first step to the design of therapeutic strategies for the treatment and prevention of disease.
Previously, our group showed that efficient localization of RGS4 to the plasma membrane was an important determinant of its G-protein inhibitory function. Somewhat unexpectedly, RGS4 was also observed to traffic through an endosomal compartment in a manner that was highly dependent on Cys-2 palmitoylation (16). Thus, we set out to examine the cellular mechanisms that mediate RGS4 localization to the endosomal compartment and to characterize the cellular pathways that control RGS4 trafficking between endosomal pools and the plasma membrane.
The Rab superfamily of small GTPase proteins contains a large number of key regulators of endosome trafficking and remodeling. In their GTP-bound (activated) state, Rabs have a number of physiological roles, including the following: (i) binding specific tethering proteins (i.e. EEA1, Fip) that are critical for endosome membrane recognition (17–19); (ii) interacting with SNARE proteins that are important for membrane fusion (20, 21); and (iii) coupling endosomes to microtubules to allow anterograde and retrograde trafficking throughout the cytoplasm (22, 23). Some Rab proteins are known to specifically reside within distinct endosomal pools. For example, Rab5, Rab7, and Rab11 are widely established markers for early/sorting, late, and recycling endosomal pools, respectively (24–26). Specifically, Rab5 regulates clathrin-mediated endocytosis from the plasma membrane to the early/sorting endosome pool (27). The early/sorting endosome is an intersection point for many proteins where they can either be sorted to undergo degradation via a Rab7-dependent late endosome/lysosomal route or be sorted for recycling back to the plasma membrane via a Rab11-dependent recycling endosome pathway (28). Like the Rabs, the small GTPases such as Arf6 also can direct endosomal internalization and trafficking. Arf6 is located at the plasma membrane and mediates clathrin-independent endocytosis from the plasma membrane Rab5-containing early/sorting endosomes (29, 30).
Here, we used a variety of wild type and mutant constructs for the Rab and Arf family of proteins, discussed above, to study the effect of altered small GTPase activity on the intracellular trafficking and function of RGS4. Our data show for the first time that Rab-mediated internalization and intracellular trafficking of an RGS protein are important components of the intracellular mechanisms that regulate its plasma membrane targeting and function as an inhibitor of GPCR signaling.
EXPERIMENTAL PROCEDURES
Materials
HEK293 cells (tsA-201 derivative) were a kind gift from Zhong-Ping Feng (University of Toronto). All tissue culture media and transfection reagents were purchased from Invitrogen and Roche Applied Science, respectively. The pEYFP-C1 plasmid was originally purchased from Clontech/BD Biosciences. The pIRESpuro-GLUE plasmid, used to generate the hemagglutinin (HA)-tagged RGS4-HA, RGS4-HA, was a kind gift from Stephane Anger (University of Toronto). Fluorescent-tagged versions of the trans-Golgi network marker protein TGN38 were from J. Lippincott-Schwartz (National Institutes of Health, Bethesda), and CFP-Rab5a, CFP-Rab5SN, and CFP-Rab11 were generously provided by Marino Zerial (Max Planck Institute). CFP-Arf6, CFP-Arf6TN, RFP-Lamp1, and RFP-Rab7 were obtained from addgene.org. Texas Red®-conjugated transferrin and Lysotracker® Red were purchased from Invitrogen. Rhodamine B-labeled phosphatidylethanolamine was from Avanti Polar Lipids (Alabaster, AL). Anti-HA antibody was from Roche Applied Science, and the horseradish peroxidase-coupled anti-mouse secondary antibody was from GE Healthcare (catalog no. NXA931). Unless otherwise stated, all other reagents and chemicals were from Sigma.
Cell Culture
HEK293 cells were grown in Dulbecco's modified Eagle's medium (DMEM)/Ham's F-12 medium (1:1), supplemented with 10% (v/v) heat-inactivated fetal bovine serum, 2 mm glutamine, 10 μg/ml streptomycin, 100 units/ml penicillin at 37 °C in a humidified atmosphere with 5% CO2.
RGS4 Expression Constructs
For subcellular localization studies, RGS4-YFP expression plasmids were generated in the pEYFP-C1 vector by cloning the human RGS4 cDNA into the NheI/AgeI sites to generate a carboxyl-terminal YFP fusion. Robust expression was ensured by inclusion of an optimized translation initiation signal in the context of the first methionine codon (GCCACCATGGCG). For analysis of RGS4 expression levels, RGS4-HA was generated by cloning RGS4 coding sequences into the NotI/AscI polylinker sites of the pIRESpuro-GLUE plasmid. Cysteine point mutations were introduced by site-directed mutagenesis as described previously (16). All plasmid constructs were purified using the Endofree Maxi kit (Qiagen) and verified by sequencing of the complete protein-coding region. Protein expression was analyzed by Western blotting on nitrocellulose membranes using anti-HA antibody and the ECL detection.
Generation of Mutant Rab Constructs
The dominant negative Rab11 construct, CFP-Rab11-S25N, was generated from the wild type CFP-Rab11 construct using the following primer pair: 5′-agattctggtgttgggaagaataatctcttgtctcg-3′ and its reverse complement. The constitutively active construct, CFP-Rab5-Q79L, was similarly created using the following primer: 5′-ata tgg gat aca gct ggt ctc gag cga tac cat agc cta gca-3′ and its reverse complement. Dominant negative Rab7, mRFP-Rab7-T22N, was likewise created from mRFP-Rab7 using 5′-gat tct gga gtt ggt aag aat tca ctc atg aac cag tat-3′.
Assays of Fura 2 Calcium Assay
The function of RGS4 as an inhibitor of Gq-coupled signaling was studied by ratiometric calcium imaging in cells selected for similar RGS4-YFP protein expression levels as described previously (31). In brief, M1-HEK cells were seeded on poly-l-lysine-coated number 1 glass coverslips and transiently transfected with the indicated constructs using Xtremegene HPTM transfection reagents. Twenty four hours after transfection, cells were loaded with fura-2AM in Ca1 buffer (11 mm glucose, 130 mm NaCl, 4.8 mm KCl, 1.2 mm MgCl2, 17 mm HEPES, and 1 mm CaCl2, pH 7.3), and coverslips were washed and loaded into a modified Leyden chamber. Cells were perfused at 37 °C with Ca1 buffer for 5 min. Base-line fluorescent ratio (FR) values were collected for 10–20 s before the addition of carbachol for a final concentration of 200 μm. Peak relative percent FR increase above base-line = ((peak stimulated FR/unstimulated baseline FR) − 1) × 100%.
Confocal Microscopy
HEK293 cells were plated at 50% in tissue culture-treated microscopy dishes (Ibidi, catalog no. 81156) and transfected overnight with 1 μg of each construct to be tested using 2.5 μl of Xtremegene HP transfection reagent according to the manufacturer's instructions. At the outset of each series of experiments, the experimenter was blinded to the identity of the transfectants immediately following the transfection step and unblinded following data collection and analysis (as detailed below). Following 24 h of incubation, dishes were examined by confocal microscopy to determine their plasma membrane/cytosol localization ratio containing transfected cells. Confocal microscopy was performed on live cells at 37 °C in an environmental chamber maintained at 5% CO2 that was built onto an Olympus FluoViewTM FV1000 laser-scanning confocal microscope. Images represent a single equatorial plane on the basal side of the cell obtained with a ×60 oil objective, 1.42 numerical aperture. Confocal images were processed with Microsoft Office 2010. Membrane/cytosol ratios were measured using the ImageJ (freeware) software package. For movies and co-localization data, the cells were visualized on a WaveFX spinning-disk confocal microscope (Quorum Technologies, Guelph, Canada), composed of an Olympus IX81 microscope stand, a Yokogawa CSU10 spinning-disk unit, and a Hamamatsu C9100–13 EM-CCD camera, controlled by the Volocity software. Imaging was performed using a ×60/1.42NA oil immersion objective, using 405-, 488-, and 561-nm solid-state lasers for the excitation of CFP, FP, and mRFP/rhodamine/Texas Red, respectively. Emission wavelength parameters of each were matched to the appropriate bandpass emission filters, and where more than one fluorescent channel was examined in a single cell, the possibility of bleed through fluorescence was excluded prior to evaluation of the co-localization of different proteins.
Supplemental Movie Acquisition
All movies were generated on a WaveFX Spinning-Disk confocal microscope (Quorum Technologies, Guelph, Canada as detailed above. In supplemental movie 1, RGS4WT traffics via intracellular endosome pools under basal conditions. HEK293 cells transfected with RGS4WT-YFP (green channel) was imaged 24 h post-transfection. Images were collected every 800 ms for 60 s. In supplemental movie 2, increased Rab5 activity increases the extent of RGS4 endosomal trafficking. HEK293 cell co-transfected with RGS4WT-YFP (green channel) and Rab5Q79L were analyzed 24 h post-transfection. Images were collected every 500 ms for 70 s. In supplemental movie 3, RGS4WT co-localizes with transferrin-Texas Red, HEK293 cells transfected with RGS4WT-YFP (green channel) were imaged 40 min after transferrin-Texas Red (red channel) addition, for a final concentration of 25 μg/ml. Merged two-color images were collected every 1.5 s for 60 s.
Preparation of Rhodamine B Phosphatidylethanolamine
The lipid was received in chloroform/methanol (2:1; v/v) and stored at −20 °C. The day of use, aliquots were dried under a N2 stream and resuspended in the same volume using 100% ethanol. After addition of 10 μl of the lipids to cells and incubation for 1 h at 4 °C, cells were washed with cold PBS and incubated at 37 °C, 5% CO2 for 3 h prior to observation.
Western Blotting
Proteins were transferred to Trans-Blot (Bio-Rad) nitrocellulose membrane. Membranes were blocked for 1 h in 0.1% Tween 20 and 5% bovine serum albumin. Primary antibodies were added to 5% BSA at concentrations provided by the vendor's instructions and incubated with membranes overnight at 4 °C before removing by washing. Horseradish peroxidase-linked secondary mouse antibody in 5% BSA was added for 2 h, before washing and signal detection using Super Signal West Pico Chemiluminescent Substrate (Thermo Scientific).
Statistical Analysis
One-way ANOVA with Tukey's post hoc analysis was used to analyze the experimental results. p < 0.05 was considered significant. Where indicated in the text, summarization of the raw data (including means ± S.E. and p values) appear in the data supplement in the form of supplemental tables.
RESULTS
RGS4 localizes to the plasma membrane in mammalian cells (53) where it inhibits GPCR signaling and function. Previous work using immunogold labeling and electron microscopic analysis of RGS4 in the macaque cerebral cortex showed that it can be localized to intracellular endosomal structures (32). Consistent with this observation, our data showed that exogenously expressed RGS4 is localized to different endosomal pools and that its trafficking through this compartment is highly dependent on its palmitoylation status (16). To investigate the pathways controlling the intracellular trafficking and function of RGS4, we here examined its cellular distribution and function in the presence of several co-expressed markers and mediators of intracellular endosomal trafficking. Under basal conditions, when RGS4-YFP is co-expressed with the pECFP control vector, it localizes efficiently to the plasma membrane and to a variety of endosomal structures of different size, speed, and fluorescence intensity that are visible against the background fluorescence of RGS4-YFP in the cytosol (Fig. 1, A and B, and supplemental movie 1). In the absence of perturbations in membrane trafficking, RGS4-containing endosomes were clearly identified in 43% of RGS4-transfected cells (Fig. 1C); however, this number is likely an underestimate based on our observation that cytosolic RGS4-YFP can sometimes interfere with the detection of RGS4 on smaller endosomes. Indeed, as demonstrated previously in our original description of this intracellular pool (16), the majority of the RGS4-containing endosomes observed under the basal state are small in size and relatively motile. Given that RGS4 may be degraded via ubiquitin-dependent pathways (10, 33, 34), we examined whether some of the larger and more fluorescent endosomal structures may represent deposition of RGS4 in aggresomes, centrosome-associated sites of protein accumulation/degradation that had been previously described for some overexpressed proteins (35). Notably, the large RGS4-containing endosome structures were neither co-localized with PLK1 (a centrosomal marker) nor were they affected in size or number by addition of the proteasomal inhibitor MG132 (supplemental Fig. 1, A and B). Moreover, these structures are regularly shaped with an obvious lumen when we use confocal image collection and image processing parameters (lower hυ values and deconvolution) that allow us to study them in isolation (supplemental Fig. 1C). Together, these data suggested that the larger RGS4-containing endosomes were not strictly aggresome-type sites of misfolded RGS4 deposition.
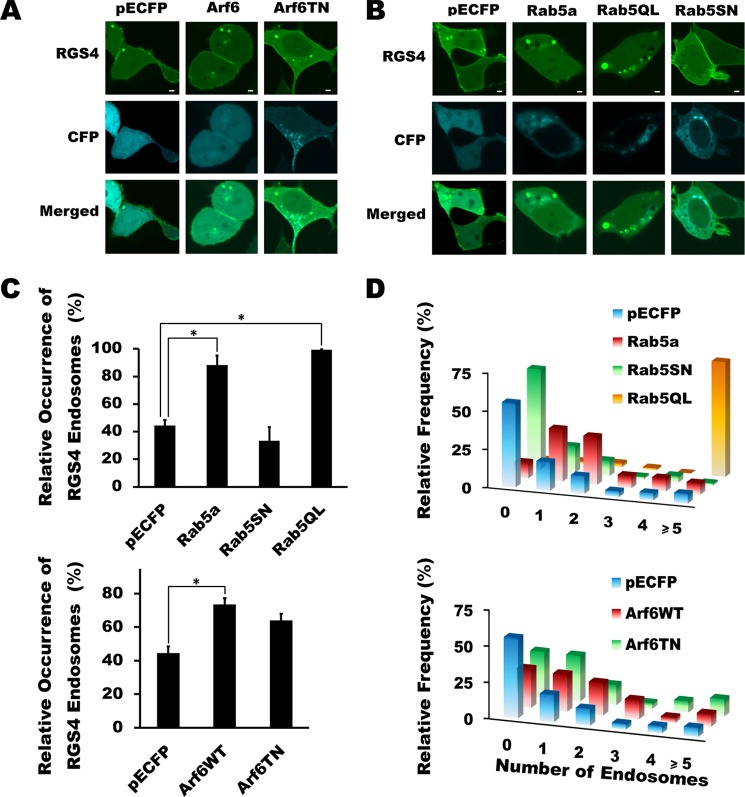
FIGURE 1.
Rab5 and Arf6 activity promote altered endosomal trafficking of RGS4. A and B, RGS4-YFP expression was imaged using a spinning disc confocal microscope following a 24-h co-transfection with the indicated constructs. Images are taken at the equatorial plane of the cells and are representative of >30 cells viewed in each of three independent experiments carried out on separate days. Scale bars represent 1 μm. C, for each experimental condition, cells with low to intermediate RGS4 fluorescence intensity were scored for the presence or absence of endosome structures. Shown are the mean values for the percentage of cells with visible RGS4-containing endosomes. (n > 30 cells/experiment/condition.) One-way ANOVA was used to determine differences between groups (*, p < 0.05). S.E. is indicated by error bars. D, histogram plots for the relative frequency of RGS4 visible endosomes per cell when co-expressed with the indicated construct. Mean ± S.E. values are presented in supplemental Table 1.
We next examined the relationship between the plasma membrane and endosomal pools of RGS4. Two members of the small GTPase superfamily, Arf6 and Rab5, are known to promote endocytosis of a wide number of plasma membrane proteins, including GPCRs (36–39). Manipulation of Arf6 function via expression of either wild type or dominant negative Arf6 (T27N) resulted in a modest increase in the percentage of transfected cells containing visible endosomes and the number of endosomes/cell (Fig. 1, C and D). By contrast, manipulation of Rab5a function had pronounced Rab5 activity-dependent effects on the endocytotic activity and endosome distribution profiles. Specifically, increased Rab5a activity (WT and Q79L) markedly increased both the percentage of cells showing endosomes and the number of endosomes/cell, with the Q79L clone having the greatest effect (Fig. 1, B–D, supplemental movie 2, and supplemental Table 1), whereas dominant negative Rab5(S17N) showed a trend toward reduced cellular endosome content.
Consistent with the notion that endocytotic activity can affect the plasma membrane targeting of RGS4, co-expression with both Rab5a and Rab5a(Q79L) decreased the plasma membrane/cytosol YFP fluorescence ratio, whereas dominant negative Rab5(S17N) showed a trend toward increased plasma membrane targeting (Fig. 2A). Notably, manipulation of Arf6 activity had very little effect on plasma membrane targeting of RGS4. Because plasma membrane targeting is a determinant of the RGS4 Gq inhibitory function (16), we examined whether changing Rab5 activity could affect the ability of RGS4 to inhibit M1 muscarinic receptor signaling. Using fura-2-loaded cells stably expressing the M1 receptor, we examined the function of RGS4 as an inhibitor of receptor/Gq-mediated calcium release (Fig. 2B). In the absence of Rab5 co-expression and in the presence of dominant negative Rab5S17N (supplemental Fig. 2), RGS4 potently inhibited peak intracellular calcium release following stimulation of the M1 muscarinic receptor when compared with the RGS-inactive mutant control (EN-AA). Notably, Rab5 co-expression reduced the function of RGS4 as an inhibitor of M1 receptor-dependent calcium release by 29.3% relative to the controls. The effect of Rab5 in this setting appeared to be selective for its effect on RGS4 localization, because Rab5 did not alter the base-line M1 receptor activity (compared with CFP controls) in cells expressing the inactive RGS4 mutant (EN-AA). Consistent with this observation, neither the M1R nor the Gq protein showed altered levels of plasma membrane localization when co-expressed with Rab5 compared with the empty vector controls (Fig. 2C).
FIGURE 2.
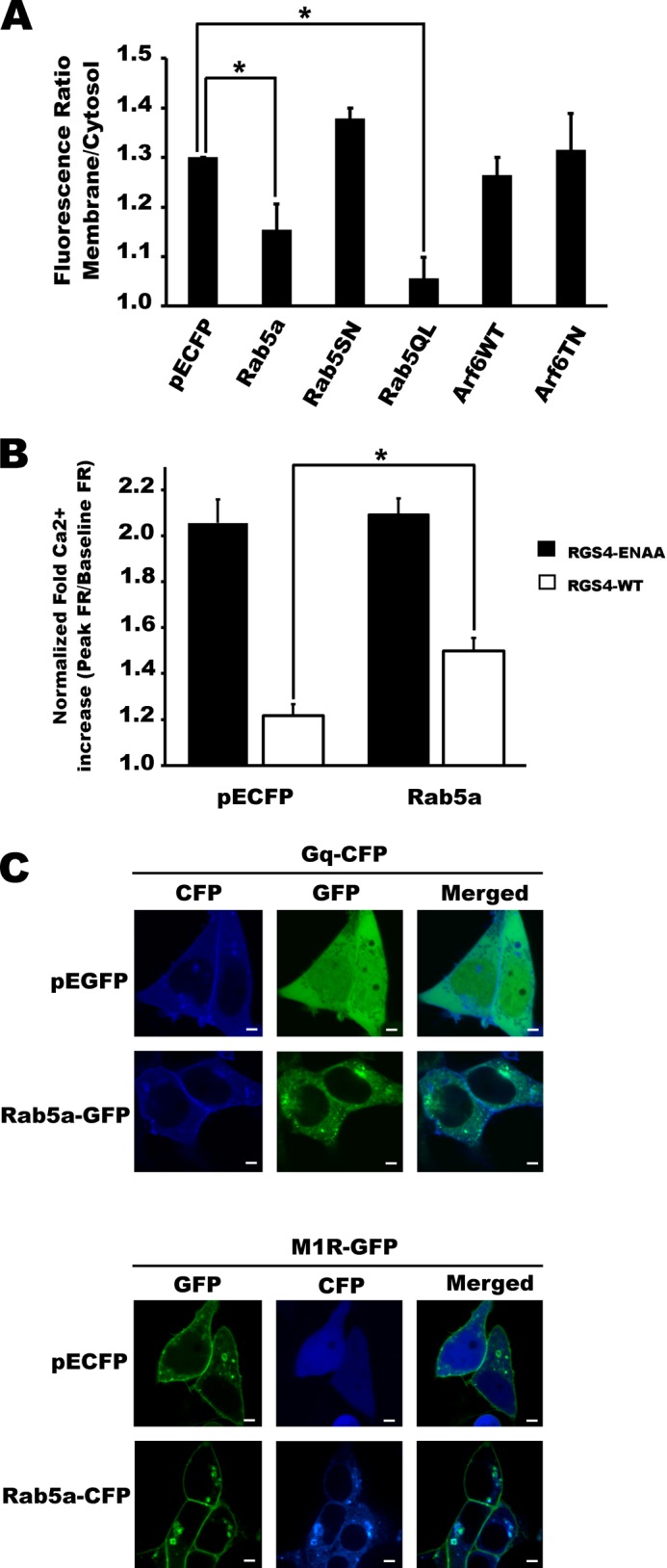
Rab5 activity alters the plasma membrane localization and function of RGS4 as an inhibitor of M1 muscarinic signaling. A, using images collected as in Fig. 1 above, the ratio of the RGS4-YFP fluorescent signal between the plasma membrane and cytosol was analyzed by densitometry using the ImageJ software package. Shown are the mean plasma membrane/cytosol ratio data of n > 80 cells recorded over at least three independent experiments carried out on separate days. p values are shown in supplemental Table 3. B, M1 muscarinic receptor signaling was examined in M1-HEK cells co-expressing RGS4-YFP and the indicated constructs. Changes in intracellular calcium levels were recorded as changes in fluorescence ratio (FR = (emission at 510 nm upon excitation at 355 nm)/(emission at 510 nm upon excitation at 396 nm)). Peak relative changes in intracellular calcium (percentage of FR increase above base line = ((peak stimulated FR/unstimulated base-line FR) −1) × 100%) following 200 μm carbachol stimulation was used as an index of M1 receptor activity for each condition (n >100 cell from at least five independent experiments). Similar data were observed following 1 μm carbachol addition (see supplemental Fig. 3) S.E. are indicated by error bars. One-way ANOVA was used to determine differences between groups (*, p < 0.05 was considered as significant). p values are shown in supplemental Table 4. C, laser-scanning confocal images showing localization of Gq and M1R under conditions of co-expression with Rab5 or control vector.
It is widely appreciated that proteins endocytosed via Rab5a-dependent pathways may be recycled back to the plasma membrane or sent to the lysosomal compartment for degradation (40–42). Consistent with the intracellular trafficking of RGS4 via a Rab5-dependent pathway, we observed that RGS4 was localized to the large Rab5-containing endosomes either as spheroid accessory compartments decorating the cytosolic surface of the rings (analogous to gemstones; Fig. 3A, arrows) or to the ring structures themselves (Fig. 3A, arrowheads) as well as to the zones of focal Rab5 accumulation. Corroborating evidence that RGS4 associated with a Rab5-dependent sorting endosome pool was the observation that these rings were also labeled by the sorting endosome marker RFP-2×FYVE (Fig. 3B) that binds selectively to phosphatidylinositol 3-phosphate enriched on sorting endosomes. The current paradigm for Rab5-mediated sorting of membrane proteins suggests that the appearance of RGS4 in accessory domains decorating the sorting endosome may represent a collection/departure point for membranes and trafficking proteins from the sorting endosome before their trafficking to another endosomal compartment. Indeed, we occasionally observe a transfer of RGS4 from these structures to other intracellular domains (as seen in the lower cell of supplemental movie 2). Finally, Lamp1, a protein that traffics through the PM-sorting endosome-late endosome-lysosome pathway, can also co-localize with RGS4 in the same accessory compartment (Fig. 3C). Lamp1 and RGS4 co-localization also occurred in numerous endosomes that are not associated with the Rab5-containing pool (Fig. 3D) indicating that RGS4 may also target the late endosome or lysosome pools.
FIGURE 3.
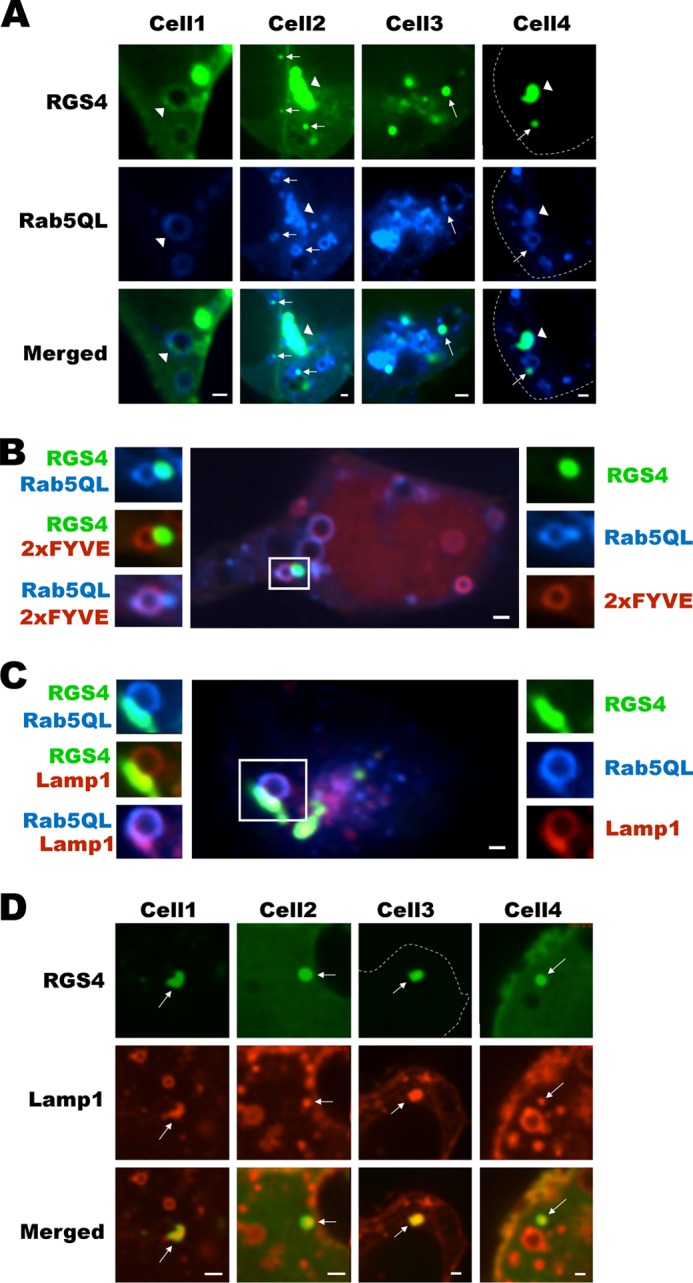
RGS4 traffics via Rab5-containing sorting endosomes and targets a Lamp1-positive intracellular pool. A, confocal images of cells co-expressing RGS4 and Rab5QL. Arrows indicate domains of co-localization between RGS4 and Rab5QL associated with individual sorting endosomes (rings), and arrowheads denote regions of RGS4 and Rab5 co-localization associated with focal clusters of Rab5QL-containing endosomes. B, large image (center) is a three-channel composite view of cells marked with RGS4-YFP (green), CFP-Rab5QL (blue), and RFP-2×FYVE (red). Small images on the right show single channel views, and left side images represent overlap of the different combinations of two channels. C, as in B, the center and surrounding images represent different channel views of cells marked in three colors with RGS4-YFP (green), CFP-Rab5QL (blue), and RFP-Lamp1 (red). D, shown are single confocal plane views of four cells co-transfected with RGS4 and RFP-Lamp1. Arrows indicate endosomes containing RGS4 and Lamp1. All images were captured 24 h post-transfection using a spinning disc microscope with ×60 oil objective. Scale bars represent 1 μm.
The ESCORT-Gs protein complex within multivesicular bodies (MVBs) is a step downstream of the sorting endosome that directs many GPCRs to either lysosomal degradation or plasma membrane recycling (43, 44). We labeled the MVBs with the phosphatidylethanolamine tracer; RGS4-YFP did not localize to MVB under basal conditions or following manipulation of Rab5 function (Fig. 4, A and B). By contrast, RGS4 showed marked co-localization with LysoTrackerTM, a marker of acidified compartments, including late endosomes and lysosomes. Consistent with the notion of Rab5 mediating increased endocytic flux of RGS4-YFP, the extent of co-localization between RGS4 and LysoTrackerTM was much greater when Rab5a was co-expressed relative to the dominant negative Rab5SN (Fig. 5). Together, these data suggest that RGS4 is endocytosed from the plasma membrane and is internalized/sorted in a Rab5-dependent fashion en route to an acidic/Lamp1-positive compartment such as late endosomes or lysosomes.
FIGURE 4.
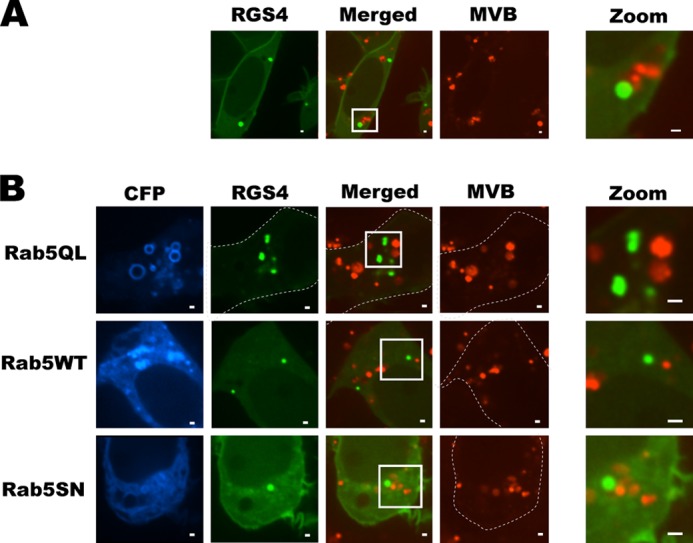
RGS4 does not target MVBs. Rhodamine ethanolamine was used to mark MVBs in cells expressing RGS4-YFP without (A) or with (B) the indicated Rab5 constructs. Virtually no co-localization between RGS4 and MVBs was observed by spinning disc confocal microscopy, 24 h post-transfection under any of the conditions tested. Scale bars represent 1 μm.
FIGURE 5.
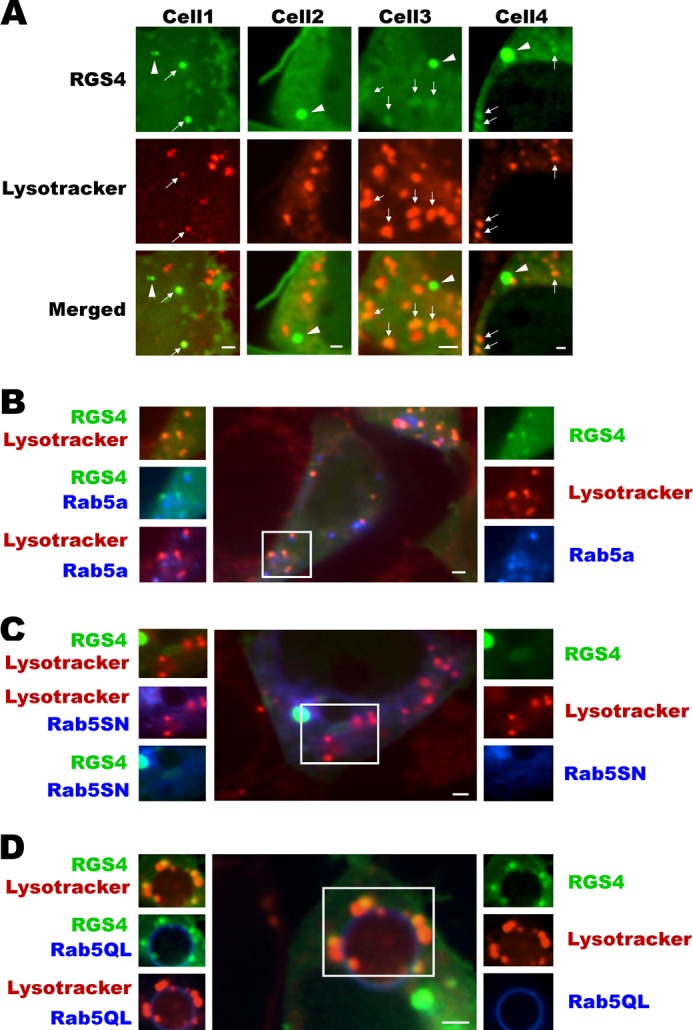
Rab5 promotes increased RGS4 co-localization with a Lysotracker® Red-labeled pool of endosomes. A, shown are four different cells transfected with RGS4-YFP and subsequently labeled with Lysotracker® Red under base-line conditions. Arrows indicate co-localized endosomes between RGS4 and Lysotracker® Red, and arrowheads indicate non-co-localized endosomes. B–D, as in the legend of Fig. 3 above, the center and surrounding images represent different channel views of cells that are marked in three colors with RGS4-YFP (green), the indicated CFP-Rab5 construct (blue), and Lysotracker® Red. For each panel, the right side images are single channel, and left side images represent overlap of two channels. All images were captured 24 h post-transfection using a spinning disc microscope with ×60 oil objective. Scale bars, 1 μm.
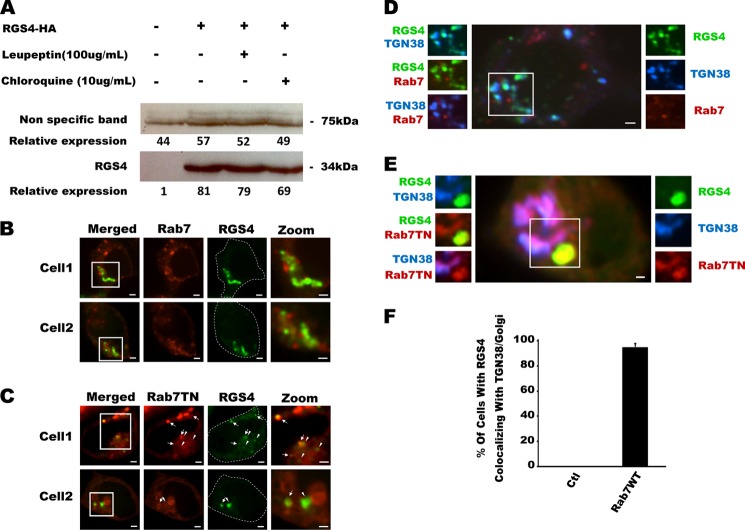
It is widely appreciated that altered stability of the RGS4 protein may lead to increased susceptibility to diseases such as cancer (10, 45, 46). Although the primary mode of degradation appears to be proteasomal degradation via the N-end rule pathway (10, 33, 34), there have been reports that suggest RGS4 may also be degraded via the lysosomal compartment (47). Based on its localization to these acidified compartments, we here tested whether RGS4 expression levels were altered by leupeptin or chloroquine, two inhibitors of lysosomal degradative activity. No changes in RGS4 protein levels were observed with chloroquine (10 μg/ml for 24 h) or leupeptin (100 μg/ml for 24 h) (Fig. 6A). Accordingly we considered the possibility that the RGS4 localization to Lysotracker/Lamp1 compartments was instead part of its intracellular trafficking to another intracellular compartment. Because Rab7 mediates the interaction of many different membranes with the late endosomal pool (25, 48), we studied the localization of RGS4-YFP when different functional variants of Rab7-RFP were co-expressed. Surprisingly, when wild type Rab7 was expressed, RGS4-YFP displayed extensive co-localization with large perinuclear structures in the vicinity of the Golgi (Fig. 6B), and when a dominant negative Rab7 (T22N) was introduced, RGS4 localized to irregularly shaped endosome structures (Fig. 6C). Triple-labeling cells with the trans-Golgi network (TGN) as marker TGN38-CFP revealed that RGS4 was localized to the TGN in the presence of wild type Rab7 (Fig. 6D) but not the dominant negative construct (Fig. 6E). Compared with 0% of cells showing TGN targeting under basal conditions, we observed almost 100% of cells showing RGS4 at the TGN when it was co-transfected with Rab7WT (Fig. 6F). These data are consistent with RGS4 trafficking from the late endosome compartment to the Golgi via a Rab7-dependent mechanism.
FIGURE 6.
Rab7 activity promotes RGS4 trafficking to the TGN. A, effect of two lysosomal inhibitors, leupeptin and chloroquine, on the stability of RGS4 protein stability were tested by Western blotting after 24 h post-transfection. The displayed Western blot is representative of three independent experiments. Densitometric analysis of RGS4 expression levels, relative to a nonspecific band from the same gel used as protein loading control, is shown. B and C, confocal images of two representative cells co-transfected with RGS4-YFP and the indicated RFP-Rab7 construct. Arrows indicate co-localization between RGS4 and Rab7TN, and the arrowheads indicate RGS4 endosomes that do not overlap Rab7TN. D and E, as in the legend of Fig. 3, the center and surrounding images represent different channel views of cells that are marked in three colors with RGS4-YFP (green), the indicated CFP-Rab5 construct (blue), and the indicated RFP-Rab7 construct (red). For each panel, right-sided images are single channel, and left-sided images represent overlap of two channels. F, cells co-transfected with RGS4 and co-empty vector or Rab7WT were scored for the presence or absence of Golgi-like perinuclear localization of RGS4-YFP. Shown are the mean of the percentage of cells with visible RGS4-endosomes (n > 20 cells evaluated/experiment, experiments performed on at least 3 separate days for total of >60 cells evaluated). All images were captured 24 h post-transfection using a spinning disc microscope with a ×60 oil objective. Scale bars, 1 μm.
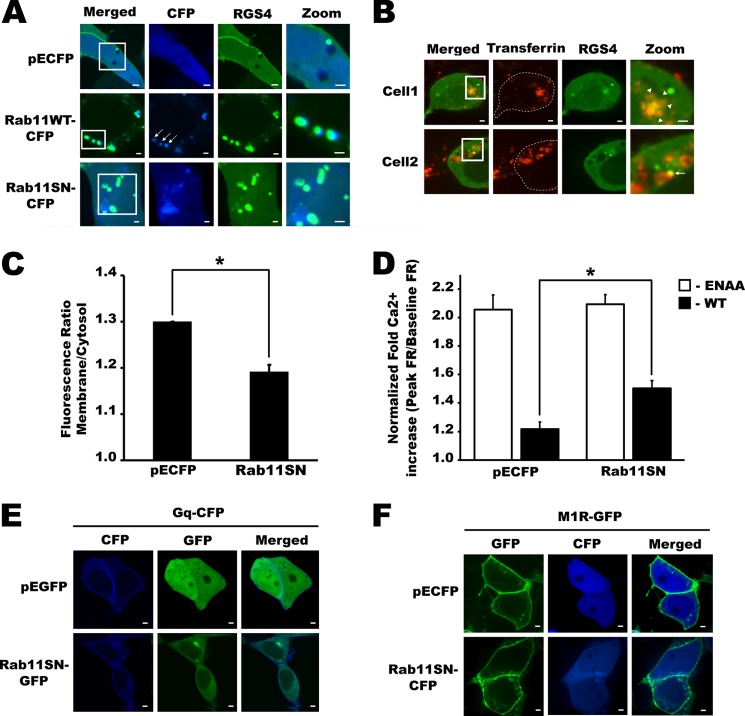
The recycling endosome pool is an important pathway for protein trafficking to the plasma membrane through different intracellular membrane compartments (19, 49, 50). We routinely observed co-localization of RGS4-containing endosomes with wild type Rab11, a marker for the recycling endosome pool, whereas expression of dominant negative Rab 11(S25N) appeared to increase the number of RGS4-labeled endosome structures in the perinuclear region (Figs. 7A and 8, A and C). Together, these data suggested that RGS4 may traffic through the recycling endosome center in a Rab11-dependent manner. To explore this possibility, we examined RGS4-YFP localization following treatment of cells with transferrin-Texas Red, a fluorescent transferrin receptor ligand that accumulates in perinuclear endosomes before it is recycled back to the plasma membrane via a Rab11-dependent pathway (51). After 30–40 min of incubation, ∼30% of RGS4-transfected cells displayed increased accumulation of RGS4-YFP and transferrin in the perinuclear region, and RGS4-YFP could also be observed in a subset of fast-moving endosomes that labeled with the transferrin-Texas Red ligand (Fig. 7B and supplemental movie 3). These data suggested that RGS4 may traffic via the recycling endosomal pool toward the plasma membrane. Therefore, we tested the role of Rab11 function as a determinant of RGS4 targeting to the plasma membrane. Notably, plasma membrane targeting was markedly impaired when RGS4 was co-transfected with dominant negative Rab11 (Rab11(S25N)) (Fig. 7C). Together these data support the trafficking of RGS4 through the Rab11/transferrin receptor positive recycling endosome toward the plasma membrane in a Rab11 activity-dependent manner. Because Rab11(S25N) reduced the levels of RGS4 at the plasma membrane, we finally asked whether Rab11(S25N) affected the ability of RGS4 to inhibit of the M1 muscarinic receptor signaling. Consistent with its ability to decrease RGS4 targeting to the plasma membrane, Rab11(S25N) also decreased the Gq inhibitory activity of RGS4. In calcium signaling assays, Rab11(S25N) co-expression reduced the function of RGS4 as an inhibitor of M1 receptor-dependent calcium release by 29.8% relative to CFP controls (Fig. 7D). The effect of Rab11(S25N) appeared to be selective for its effect on RGS4 mislocalization, because Rab11(S25N) did not alter the base-line M1 receptor activity compared with CFP in cells expressing the inactive RGS4 mutant (EN-AA). Consistent with these data, neither the Gq protein (Fig. 7E) nor the M1R (Fig. 7F) showed altered levels of plasma membrane localization when co-expressed with Rab11(S25N) compared with the empty vector controls. Taken together, these data indicate that Rab11 activity is a selective regulator of RGS4 plasma membrane localization and function relative to the other components of the M1R/Gq signaling pathway.
FIGURE 7.
Rab11DN impaired RGS4-mediated inhibition of the M1 muscarinic receptor via decreasing its plasma membrane targeting. A, shown are confocal images layer of cells co-transfected with RGS4-YFP and the indicated control or Rab11 construct (CFP-labeled). Arrows show RGS4-containing endosomes that co-localize with Rab11. B, confocal images of two representative RGS4-YFP-transfected cells labeled with transferrin-Texas Red (25 μg/ml) at different time points following transferring addition. Arrows indicate RGS4-containing endosomes co-localized with Texas Red dye after 30 min of transferrin treatment. Arrowheads indicate RGS4 co-localization with Texas Red at the recycling endosomal center after 40 min of transferrin treatment. C, confocal images of RGS4-YFP-expressing cells were used to determine the ratio of RGS4-YFP fluorescence between the plasma membrane and cytosol. Shown are the mean plasma membrane/cytosol ratio data of n >80 cells recorded over at least three independent experiments carried out on separate days. p values are shown in supplemental Table 3. D, M1 muscarinic receptor signaling was examined in M1-HEK cells co-expressing RGS4-YFP and the indicated constructs. Changes in intracellular calcium levels were recorded as changes in fluorescence ratio (FR = (emission at 510 nm upon excitation at 355 nm)/(emission at 510 nm upon excitation at 396 nm)). Peak relative changes in intracellular calcium (percentage of FR increase above base line = ((peak stimulated FR/unstimulated base-line FR) −1) × 100%) following 200 μm carbachol stimulation was used as an index of M1 receptor activity for each condition (n >100 cell from at least five independent experiments). Similar data were observed following 1 μm carbachol addition (see supplemental Fig. 3). S.E. are indicated by error bars. One-way ANOVA was used to determine differences between groups (*, p < 0.05 was considered as significant). p values are shown in supplemental Table 4. Scale bars represent 1 μm. E and F, laser-scanning confocal images showing membrane localization of Gq (E) and M1R (F) under conditions of co-expression with dominant negative Rab11 or control vector.
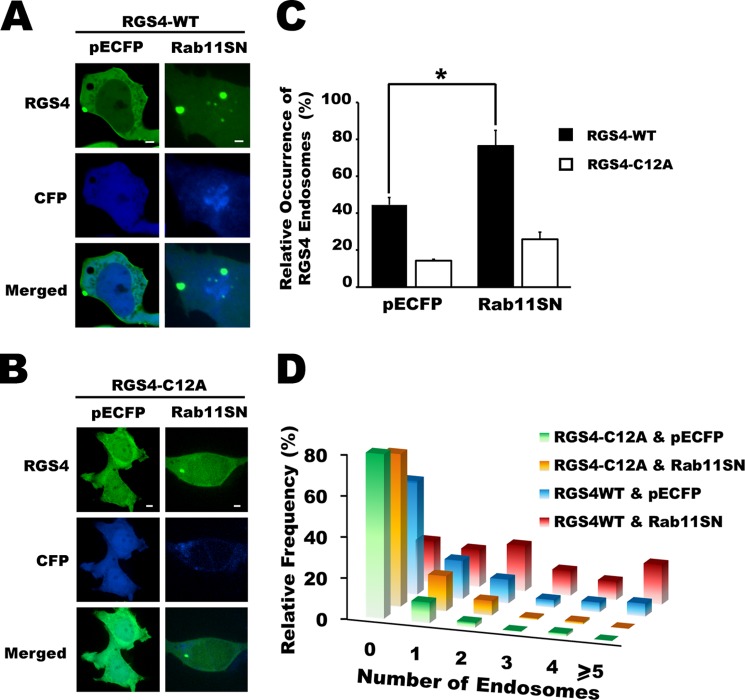
FIGURE 8.
RGS4 Cys-12 mutation impaired Rab11 dominant negative retention of RGS4 in intracellular compartments. A and B, confocal images of cells expressing YFP-tagged RGS4 WT(A) and C12A (B) cells following 24 h of co-transfection with dominant negative Rab11. All images were captured using a spinning disc microscope, ×60 oil objective. Images are representative of three independent experiments and are taken at the equatorial plan of the cells. Scale bars, 1 μm. C, shown are the mean values for the percentage of cells with visible RGS4-WT or C12A endosomes. (n > 30 cells/experiment/condition, where each condition was studied at least three times for a total of n > 90 total cells/condition). One-way ANOVA was used to determine differences between groups (*, p < 0.05 was considered as significant). S.E. is indicated by error bars. D, histogram plots for the relative frequency of visible RGS4-containing endosomes when RGS4 WT- and C12A-expressing cells were co-transfected with a Rab11SN or control vector as indicated. A summary of the mean and S.E. values for all conditions tested is presented in supplemental Table 2.
Previously, our group showed that palmitoylation on Cys-2 and Cys-12 mediated RGS4 trafficking and function in mammalian cells (16). Unlike wild type RGS4, the RGS4(C12A) mutant appeared unable to associate with certain specific pools of intracellular endosomes and, in addition, showed markedly decreased plasma membrane targeting. We therefore asked whether the Cys-12 residue was required by RGS4 to access the Rab11-mediated recycling endosomal pool. As noted above, wild type RGS4-containing endosomes were observed in 43% of the cells; however, co-expression with the dominant negative Rab11(S25N) construct increased the relative incidence of these endosomes to 76% (Fig. 8, A and B) and altered the population distribution of endosome number/cell to higher values (Fig. 8D). By contrast, RGS4(C12A)-containing endosomes were generally observed at lower frequency and numbers/cell and appeared to be much less sensitive to the effects of the dominant negative Rab11SN construct (Fig. 8, C and D). Together, these data were consistent with the notion that Cys-12 is required for RGS4 to target the plasma membrane in a Rab11 activity-dependent manner.
DISCUSSION
The function of heterotrimeric G-protein α subunits is regulated by palmitoylation (52). As a consequence, α subunits have been shown to traffic between the plasma membrane, where they exert their functional effects, and the Golgi, where a set of specific palmitoyl-CoA transferase enzymes are located (52). Likewise, we have shown that a regulator of heterotrimeric G-protein signaling, RGS4, is palmitoylated on two amino-terminal residues and that its palmitoylation is associated with its ability to traffic between intracellular membrane compartments. Specifically, palmitoylation on Cys-2 is required for trafficking of RGS4 within endosomal pathways, whereas palmitoylation on Cys-12 modulates its targeting with both the plasma membrane and a specific subset of endosomal pools. Accordingly, we proposed that the internalization of RGS4 may allow for its sequential palmitoylation on Cys-2 and Cys-12 to target and exit the endosomal pool, respectively. Indeed, RGS4 was shown localize to distinct endosomal pools, and prevention of its endosomal trafficking had marked effects on its function as an inhibitor of Gq (16). Here, we set out to identify the pathways mediating RGS4 trafficking through various endosomal compartments and the consequences of altered trafficking on its function as a GPCR inhibitor.
In general, relatively little information is available regarding the intracellular trafficking of RGS proteins (53–57), particularly when compared with that available for their biological targets, the GPCRs and heterotrimeric G-proteins. We show here for the first time that the Rab family of small GTPases plays a critical role in the intracellular trafficking of RGS4. Based on its ability to regulate Gq signaling in mammalian cells, we anticipated that Arf6 would play an important role in the internalization of RGS4 from the plasma membrane. Indeed, Gαq has been shown to interact with ARNO, an Arf6 GEF activator (58–60), and both ARNO and Arf6 are required for the phosphatidylinositol 2,4-bisphosphate synthesis that selectively mediates vesicle fission at the plasma membrane (61). Finally, Arf6 has been directly implicated in the internalization of a number of GPCRs (62–65). We were thus surprised that, in our hands, manipulation of Arf6 activity did not appreciably alter RGS4 plasma membrane levels despite the cells showing a modest increase in its endosomal targeting. It may be that RGS4 activity as a Gq inhibitor is sufficient to prevent increased endosome internalization via the Arf6-dependent pathways. In this regard it would be interesting to examine whether RGS4 activity can modulate Arf6-dependent internalization of specific GPCRs. Perhaps these data are consistent with other known functions of Arf6 involving endosome recycling (66, 67) or cytoskeletal reorganization because (68, 69) we did observe a significant change in the morphology of the cells (specifically to a more rounded cell shape) when Arf6 was co-transfected.
Rab5 is another small GTPase known to regulate the internalization of plasma membrane proteins (70, 71). Rab5 activity promotes endocytosis of numerous GPCRs and other cellular signaling components via stimulation of the clathrin-mediated endocytic pathway (38, 72). Rab5 is also known to promote homotypic vesicle fusion and is involved in protein sorting at the level of the sorting/early endosome compartment (73–75). The observation that increased Rab5 activity promotes increased RGS4 internalization and reduces its plasma membrane levels is consistent with the notion that RGS4 is endocytosed via a similar clathrin-dependent pathway. Indeed, we can detect RGS4 at the level of the sorting endosome, a compartment that requires Rab5 for its normal protein sorting function. At this stage, however, we cannot rule out the possibility that RGS4 may also be internalized via clathrin-independent pathways (e.g. caveolae) as Rab5 has been reported to intersect with and regulate several other membrane internalization pathways (76, 77). In any case, the observation that Rab5 activity promotes internalization and decreases Gq inhibitory function of RGS4 is a novel observation that will likely inform the design of new experiments to test whether activators and inhibitors of Rab5 function may be used to modulate RGS4 expression and function at the plasma membrane.
The subsequent transfer of RGS4 from the sorting endosome to a Rab7/Lamp1 compartment is reminiscent of the intracellular fate of some GPCRs (72, 78) that undergo internalization following agonist stimulation and are targeted for degradation at the lysosomal compartment. However, in our expression model, RGS4 protein levels were not increased by the lysosomal inhibitors chloroquine or leupeptin, suggesting that RGS4 is localized to the cytosolic (rather than the lumenal) surface of the late endosome/lysosomal lipid bilayer. Indeed, most of the proteins targeted to late endosomes and lysosomes are degraded for recycling of energy/ATP (79) and amino acids (80), or alternatively, they are secreted as protein aggregates (81). Further evidence that RGS4 is not found within the lumen of the degradative endosomes comes from the observation that RGS4 endosomes are never found co-localized with MVBs, a common route for several GPCRs undergoing degradation. Thus, it appears that RGS4 may undergo intracellular trafficking in a manner that is distinct from some of its targets, the GPCRs and G-proteins. As was demonstrated by our M1 muscarinic receptor signaling data, this knowledge may provide unique opportunities to fine tune GPCR signaling levels via RGS proteins without dramatically affecting the trafficking and function of the core signaling pathway receptors, G-proteins or effector molecules. This notion is further supported by the previous report that Gq, the G-protein specifically coupled to the M1 muscarinic receptor, does not appear to traffic via a Rab11-mediated pathway (82).
Although the endocytic trafficking of proteins via the TGN has been reported for numerous proteins, including furin and Pseudomonas exotoxin A (83, 84), it was somewhat surprising to observe that wild type Rab7 promoted this pathway for RGS4. Additional evidence for this late endosome-TGN trafficking pathway for RGS4 was the observation that dominant negative Rab7TN, prevented TGN targeting and trapped RGS4 irregularly shaped endosome structures reminiscent of late endosomes. Indeed, Rab9 is the commonly described GTPase that mediates TGN targeting of endocytosed proteins (85), so it will be interesting to determine whether Rab7 and Rab9 will have overlapping functions with respect to the late endosome-TGN trafficking of RGS4 and other proteins. The notion that RGS4 may traffic through the Golgi was not completely unexpected because previous reports have shown that RGS4 can interact with βCOP-I coatomer protein (86), and our own data showed co-localization of RGS4-containing endosomes with the trans-Golgi marker protein, TGN38, suggesting the two proteins traffic through similar compartments. Notably, however, RGS4 was never observed in high concentrations at the trans-Golgi compartment unless Rab7 was co-expressed suggesting its time within the TGN may be very transient in the absence of increased membrane/protein flux to the TGN such as may be the case when Rab7 activity is increased.
Proteins that are targeted to the plasma membrane often traffic via the Rab11 recycling endosome pool (19, 49, 87). In our cell model, we observed extensive co-localization between RGS4 and Rab11WT suggesting that Rab11 was also involved in delivering RGS4 to the plasma membrane. However, Rab11 labels several types of vesicles in addition to its marking of the recycling endosome center. For example, Rab11 mediates retrograde trafficking between the recycling endosomal center and the TGN (88). The identification of RGS4 within highly motile transferrin-labeled endosomes after 30 min of incubation is a strong indication that RGS4 normally traffics via the recycling endosome pathway that is regulated by Rab11. We rarely observe such rapidly moving endosomes under basal (non-transferrin treated) conditions suggesting that the Rab11 compartment may serve as a storage pool of RGS4 that can be released by agonist treatments that activate Rab11. This is the first report that Rab11 and recycling endosome activity is an important step in the trafficking of an RGS protein to the plasma membrane. As such, it will be interesting to determine whether extracellular signaling pathways that stimulate the Rab11 activity can modulate the trafficking of RGS4 to the plasma membrane. For instance, PI3K/AKT signaling pathway has been described to modulate the Rab11 activity and may thus be expected to have a marked effect on RGS4 function (89, 90).
Finally, we have extended our understanding of the role that palmitoylation plays in the trafficking of RGS4 (16). In our previous publication, we observed that the Cys-12 mutants of RGS4 showed markedly decreased association with the plasma membrane. We proposed that one explanation for this observation was that palmitoylation on Cys-12 would extend the length of the hydrophobic face of an amino-terminal amphipathic helix that is known to be required for the plasma membrane targeting of RGS4. The data herein suggest another possible complementary function of Cys-12 palmitoylation. Previously, we reported that the mutation of Cys-12 prevents the association of RGS4 and TGN38 in a specific pool of relatively small sized endosomes. We considered the possibility that this pool of endosomes may represent the RGS4 as it traffics to the plasma membrane from the TGN or recycling endosome center. Indeed, the dominant negative Rab11(S25N) clone markedly alters the incidence and number of wild type RGS4-containing endosomes/cell, the majority of which are clustered in the vicinity of the putative recycling endosome center as marked by Rab11 accumulation. These data are consistent with the notion that RGS4 may be trapped in the recycling endosome pool when Rab11 activity is decreased. With Cys-12 mutated, however, we observe very little increase in either the relative incidence or number of RGS4-containing endosomes suggesting that palmitoylation of RGS4 on Cys-12 may be a critical step for exit from the TGN or recycling endosome center. It will be interesting to determine the extent to which the loss of plasma membrane targeting by the Cys-12 mutant of RGS4 is due to its altered affinity for lipid bilayers versus its improper trafficking through different intracellular endosome pools. In any case, these data should help inform the future search for Golgi- and recycling endosome-resident palmitoyl-CoA transferases that can selectively palmitoylate RGS4 on Cys-12 to regulate its cell trafficking and function.
In summary, we have identified multiple key steps regulating the intracellular trafficking and function of the regulator of the G-protein signaling protein, RGS4. Our summary of the proposed intracellular trafficking pathway for RGS4 is presented in Fig. 9. The data herein suggest that RGS4 is internalized from the plasma membrane in a Rab5-dependent manner to sorting endosomes. From there, it appears that RGS4 is collected and trafficked to an acidic/Rab7/Lamp1-containing late endosomal or lysosomal compartment. Finally, the internalized RGS4 may enter the Rab11-mediated exocytic pathway, either directly or via a Rab7-mediated circuit that involves trafficking through the Golgi. It may be that there still remain additional pathways through which RGS4 may traffic to because manipulation of Rab5 or Rab11 did not completely disrupt the localization and function of the RGS4 protein; however, given that there is a measurable level of constitutive cytosolic expression, this observation may not be surprising.
FIGURE 9.
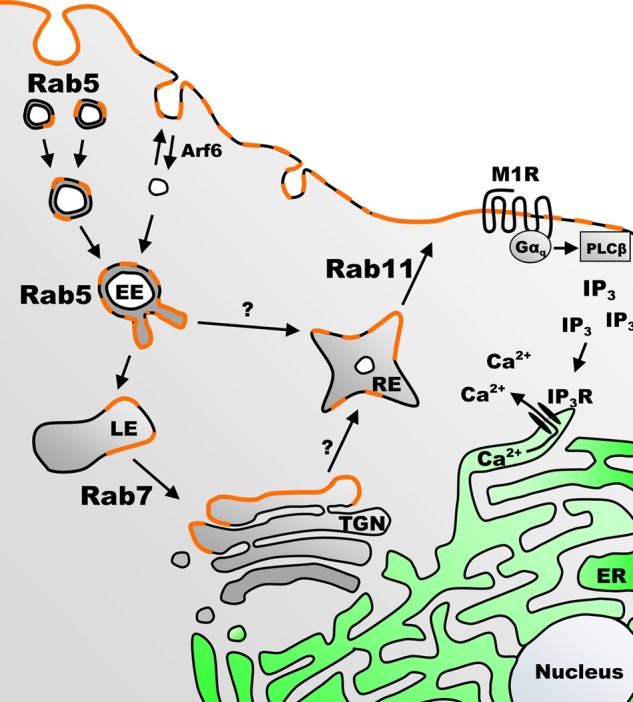
Proposed model for Rab-mediated trafficking of RGS4 between the plasma membrane and various endosomal compartments in mammalian cells. Shown is the proposed cycle of intracellular trafficking of RGS4 (orange color). The cycle begins at the plasma membrane, where RGS4 acts as an inhibitor of M1 muscarinic receptor-mediated phospholipase Cβ (PLCβ) activation, inositol phosphate (IP3)/inositol phosphate receptor activation (IP3R) and intracellular calcium release (Ca2+). The model predicts that RGS4 is internalized and sorted in early endosomes (EE) in a Rab5-dependent manner, and then it is either recycled back to the plasma membrane or transferred to a late endosome (LE) pool. Trafficking from the late endosome pool may direct RGS4 to the trans-Golgi network (TGN) via Rab7 and other sorting proteins, where it is sorted via unknown mechanisms (perhaps involving compartment-specific palmitoylation on Cys-2 and Cys-12) before it re-enters the Rab11 recycling endosome (RE) pool for trafficking back to the plasma membrane. The cycle is completed when the Rab11 recycling pathway returns RGS4 to the plasma membrane where it may once again function as a G-protein inhibitor. ER, endoplasmic reticulum.
By understanding the mechanisms that control RGS4 trafficking, we may uncover new models for the modulation of GPCR signaling. For example, the observation that RGS4 function may be selectively regulated by Rab5 and Rab11 in a manner that is independent from the M1R-Gq signaling complexes provides new insight into mechanisms whereby manipulating Rab activity may allow for differential control of GPCR pathways. Although we focused our attention on the regulation of the M1 muscarinic receptor by RGS4, we expect that these same paradigms will be relevant to the regulation of other GPCRs with which it is known to interact (6, 9, 47, 53, 91–94). These data may also help us to uncover additional pathophysiological mechanisms that contribute to altered disease susceptibility in the face of decreased RGS4 function (5–7, 10, 46, 93, 95). Finally, because the key determinants modulating its intracellular localization and function are in the amino-terminal domain of RGS4, and this domain is highly conserved within RGS5 and RGS16, these observations may indicate novel mechanisms for the regulation of other RGS protein family members. Together, this work represents an advance in our understanding of RGS4 trafficking and highlights new mechanistic insights for the intracellular regulation of GPCR signaling.
Supplementary Material
Acknowledgments
We acknowledge Drs. Christian Rolando and Caroline Tokarski at “La Plateforme Proteomique de Lille,” University of Lille 1, France, for the generous support, mentorship (to G. B.), and critical evaluation of this work. We are also grateful for technical support from Kevin Singh.
This work was supported by Canadian Institutes of Health Research Operating Grants Program Grant MOP-106670 (to S. P. H.).

This article contains supplemental movies 1–3, Figs. 1–3, and Tables 1–4.
- RGS
- regulator of G-protein signaling
- GPCR
- G-protein-coupled receptor
- Arf
- adenosine diphosphate ribosylation factor
- M1R
- M1 muscarinic receptor
- Lamp1
- lysosomal associated membrane protein 1
- MVB
- multivesicular body
- CFP
- cyan fluorescent protein
- RFP
- red fluorescent
- TGN
- trans-Golgi network
- HEK
- human embryonic kidney
- PM
- plasma membrane
- G-protein
- guanine nucleotide-binding protein
- ANOVA
- analysis of variance
- FR
- fluorescence ratio.
REFERENCES
- 1. Ross E. M., Wilkie T. M. (2000) GTPase-activating proteins for heterotrimeric G proteins: regulators of G protein signaling (RGS) and RGS-like proteins. Annu. Rev. Biochem. 69, 795–827 [DOI] [PubMed] [Google Scholar]
- 2. Hollinger S., Hepler J. R. (2002) Cellular regulation of RGS proteins: modulators and integrators of G protein signaling. Pharmacol. Rev. 54, 527–559 [DOI] [PubMed] [Google Scholar]
- 3. Berman D. M., Kozasa T., Gilman A. G. (1996) The GTPase-activating protein RGS4 stabilizes the transition state for nucleotide hydrolysis. J. Biol. Chem. 271, 27209–27212 [DOI] [PubMed] [Google Scholar]
- 4. Watson N., Linder M. E., Druey K. M., Kehrl J. H., Blumer K. J. (1996) RGS family members: GTPase-activating proteins for heterotrimeric G-protein α-subunits. Nature 383, 172–175 [DOI] [PubMed] [Google Scholar]
- 5. Pacey L. K., Heximer S. P., Hampson D. R. (2009) Increased GABAB receptor-mediated signaling reduces the susceptibility of fragile X knockout mice to audiogenic seizures. Mol. Pharmacol. 76, 18–24 [DOI] [PubMed] [Google Scholar]
- 6. Lerner T. N., Kreitzer A. C. (2012) RGS4 is required for dopaminergic control of striatal LTD and susceptibility to parkinsonian motor deficits. Neuron 73, 347–359 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Cifelli C., Rose R. A., Zhang H., Voigtlaender-Bolz J., Bolz S. S., Backx P. H., Heximer S. P. (2008) RGS4 regulates parasympathetic signaling and heart rate control in the sinoatrial node. Circ. Res. 103, 527–535 [DOI] [PubMed] [Google Scholar]
- 8. Ruiz de Azua I., Scarselli M., Rosemond E., Gautam D., Jou W., Gavrilova O., Ebert P. J., Levitt P., Wess J. (2010) RGS4 is a negative regulator of insulin release from pancreatic β-cells in vitro and in vivo. Proc. Natl. Acad. Sci. U.S.A. 107, 7999–8004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Ruiz de Azua I., Gautam D., Guettier J. M., Wess J. (2011) Novel insights into the function of β-cell M3 muscarinic acetylcholine receptors: therapeutic implications. Trends Endocrinol. Metab. 22, 74–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Xie Y., Wolff D. W., Wei T., Wang B., Deng C., Kirui J. K., Jiang H., Qin J., Abel P. W., Tu Y. (2009) Breast cancer migration and invasion depend on proteasome degradation of regulator of G-protein signaling 4. Cancer Res. 69, 5743–5751 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Ding L., Hegde A. N. (2009) Expression of RGS4 splice variants in dorsolateral prefrontal cortex of schizophrenic and bipolar disorder patients. Biol. Psychiatry 65, 541–545 [DOI] [PubMed] [Google Scholar]
- 12. Mirnics K., Middleton F. A., Stanwood G. D., Lewis D. A., Levitt P. (2001) Disease-specific changes in regulator of G-protein signaling 4 (RGS4) expression in schizophrenia. Mol. Psychiatry 6, 293–301 [DOI] [PubMed] [Google Scholar]
- 13. Muma N. A., Mariyappa R., Williams K., Lee J. M. (2003) Differences in regional and subcellular localization of Gq/11 and RGS4 protein levels in Alzheimer's disease: correlation with muscarinic M1 receptor binding parameters. Synapse 47, 58–65 [DOI] [PubMed] [Google Scholar]
- 14. Emilsson L., Saetre P., Jazin E. (2006) Low mRNA levels of RGS4 splice variants in Alzheimer's disease: association between a rare haplotype and decreased mRNA expression. Synapse 59, 173–176 [DOI] [PubMed] [Google Scholar]
- 15. Mittmann C., Chung C. H., Höppner G., Michalek C., Nose M., Schüler C., Schuh A., Eschenhagen T., Weil J., Pieske B., Hirt S., Wieland T. (2002) Expression of 10 RGS proteins in human myocardium: functional characterization of an up-regulation of RGS4 in heart failure. Cardiovasc. Res. 55, 778–786 [DOI] [PubMed] [Google Scholar]
- 16. Bastin G., Singh K., Dissanayake K., Mighiu A. S., Nurmohamed A., Heximer S. P. (2012) Amino-terminal cysteine residues differentially influence RGS4 protein plasma membrane targeting, intracellular trafficking, and function. J. Biol. Chem. 287, 28966–28974 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Christoforidis S., McBride H. M., Burgoyne R. D., Zerial M. (1999) The Rab5 effector EEA1 is a core component of endosome docking. Nature 397, 621–625 [DOI] [PubMed] [Google Scholar]
- 18. Simonsen A., Lippé R., Christoforidis S., Gaullier J. M., Brech A., Callaghan J., Toh B. H., Murphy C., Zerial M., Stenmark H. (1998) EEA1 links PI(3)K function to Rab5 regulation of endosome fusion. Nature 394, 494–498 [DOI] [PubMed] [Google Scholar]
- 19. Baetz N. W., Goldenring J. R. (2013) Rab11-family interacting proteins define spatially and temporally distinct regions within the dynamic Rab11a-dependent recycling system. Mol. Biol. Cell 24, 643–658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sørensen J. B., Wiederhold K., Müller E. M., Milosevic I., Nagy G., de Groot B. L., Grubmüller H., Fasshauer D. (2006) Sequential N- to C-terminal SNARE complex assembly drives priming and fusion of secretory vesicles. EMBO J. 25, 955–966 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Luzio J. P., Gray S. R., Bright N. A. (2010) Endosome-lysosome fusion. Biochem. Soc. Trans. 38, 1413–1416 [DOI] [PubMed] [Google Scholar]
- 22. Ishida M., Ohbayashi N., Maruta Y., Ebata Y., Fukuda M. (2012) Functional involvement of Rab1A in microtubule-dependent anterograde melanosome transport in melanocytes. J. Cell Sci. 125, 5177–5187 [DOI] [PubMed] [Google Scholar]
- 23. Horgan C. P., McCaffrey M. W. (2011) Rab GTPases and microtubule motors. Biochem. Soc. Trans. 39, 1202–1206 [DOI] [PubMed] [Google Scholar]
- 24. Feng Y., Press B., Wandinger-Ness A. (1995) Rab 7: an important regulator of late endocytic membrane traffic. J. Cell Biol. 131, 1435–1452 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Chavrier P., Parton R. G., Hauri H. P., Simons K., Zerial M. (1990) Localization of low molecular weight GTP-binding proteins to exocytic and endocytic compartments. Cell 62, 317–329 [DOI] [PubMed] [Google Scholar]
- 26. Urbé S., Huber L. A., Zerial M., Tooze S. A., Parton R. G. (1993) Rab11, a small GTPase associated with both constitutive and regulated secretory pathways in PC12 cells. FEBS Lett. 334, 175–182 [DOI] [PubMed] [Google Scholar]
- 27. Cavalli V., Corti M., Gruenberg J. (2001) Endocytosis and signaling cascades: a close encounter. FEBS Lett. 498, 190–196 [DOI] [PubMed] [Google Scholar]
- 28. Burd C. G. (2011) Physiology and pathology of endosome-to-Golgi retrograde sorting. Traffic 12, 948–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lau A. W., Chou M. M. (2008) The adaptor complex AP-2 regulates post-endocytic trafficking through the non-clathrin Arf6-dependent endocytic pathway. J. Cell Sci. 121, 4008–4017 [DOI] [PubMed] [Google Scholar]
- 30. Tanabe K., Torii T., Natsume W., Braesch-Andersen S., Watanabe T., Satake M. (2005) A novel GTPase-activating protein for ARF6 directly interacts with clathrin and regulates clathrin-dependent endocytosis. Mol. Biol. Cell 16, 1617–1628 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Gu S., He J., Ho W. T., Ramineni S., Thal D. M., Natesh R., Tesmer J. J., Hepler J. R., Heximer S. P. (2007) Unique hydrophobic extension of the RGS2 amphipathic helix domain imparts increased plasma membrane binding and function relative to other RGS R4/B subfamily members. J. Biol. Chem. 282, 33064–33075 [DOI] [PubMed] [Google Scholar]
- 32. Paspalas C. D., Selemon L. D., Arnsten A. F. (2009) Mapping the regulator of G protein signaling 4 (RGS4): presynaptic and postsynaptic substrates for neuroregulation in prefrontal cortex. Cereb. Cortex 19, 2145–2155 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Lee M. J., Tasaki T., Moroi K., An J. Y., Kimura S., Davydov I. V., Kwon Y. T. (2005) RGS4 and RGS5 are in vivo substrates of the N-end rule pathway. Proc. Natl. Acad. Sci. U.S.A. 102, 15030–15035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Davydov I. V., Varshavsky A. (2000) RGS4 is arginylated and degraded by the N-end rule pathway in vitro. J. Biol. Chem. 275, 22931–22941 [DOI] [PubMed] [Google Scholar]
- 35. García-Mata R., Bebök Z., Sorscher E. J., Sztul E. S. (1999) Characterization and dynamics of aggresome formation by a cytosolic GFP-chimera. J. Cell Biol. 146, 1239–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Grimsey N. L., Goodfellow C. E., Dragunow M., Glass M. (2011) Cannabinoid receptor 2 undergoes Rab5-mediated internalization and recycles via a Rab11-dependent pathway. Biochim. Biophys. Acta 1813, 1554–1560 [DOI] [PubMed] [Google Scholar]
- 37. Esseltine J. L., Dale L. B., Ferguson S. S. (2011) Rab GTPases bind at a common site within the angiotensin II type I receptor carboxyl-terminal tail: evidence that Rab4 regulates receptor phosphorylation, desensitization, and resensitization. Mol. Pharmacol. 79, 175–184 [DOI] [PubMed] [Google Scholar]
- 38. Hinkle P. M., Gehret A. U., Jones B. W. (2012) Desensitization, trafficking, and resensitization of the pituitary thyrotropin-releasing hormone receptor. Front. Neurosci. 6, 180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Claing A., Chen W., Miller W. E., Vitale N., Moss J., Premont R. T., Lefkowitz R. J. (2001) β-Arrestin-mediated ADP-ribosylation factor 6 activation and β2-adrenergic receptor endocytosis. J. Biol. Chem. 276, 42509–42513 [DOI] [PubMed] [Google Scholar]
- 40. Fan G. H., Lapierre L. A., Goldenring J. R., Richmond A. (2003) Differential regulation of CXCR2 trafficking by Rab GTPases. Blood 101, 2115–2124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bouley R., Lin H. Y., Raychowdhury M. K., Marshansky V., Brown D., Ausiello D. A. (2005) Down-regulation of the vasopressin type 2 receptor after vasopressin-induced internalization: involvement of a lysosomal degradation pathway. Am. J. Physiol. Cell Physiol. 288, C1390–C1401 [DOI] [PubMed] [Google Scholar]
- 42. Anborgh P. H., Seachrist J. L., Dale L. B., Ferguson S. S. (2000) Receptor/β-arrestin complex formation and the differential trafficking and resensitization of β2-adrenergic and angiotensin II type 1A receptors. Mol. Endocrinol. 14, 2040–2053 [DOI] [PubMed] [Google Scholar]
- 43. Kantamneni S., Holman D., Wilkinson K. A., Nishimune A., Henley J. M. (2009) GISP increases neurotransmitter receptor stability by down-regulating ESCRT-mediated lysosomal degradation. Neurosci. Lett. 452, 106–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Dores M. R., Chen B., Lin H., Soh U. J., Paing M. M., Montagne W. A., Meerloo T., Trejo J. (2012) ALIX binds a YPX(3)L motif of the GPCR PAR1 and mediates ubiquitin-independent ESCRT-III/MVB sorting. J. Cell Biol. 197, 407–419 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Tatenhorst L., Senner V., Püttmann S., Paulus W. (2004) Regulators of G-protein signaling 3 and 4 (RGS3, RGS4) are associated with glioma cell motility. J. Neuropathol. Exp. Neurol. 63, 210–222 [DOI] [PubMed] [Google Scholar]
- 46. Hurst J. H., Mendpara N., Hooks S. B. (2009) Regulator of G-protein signalling expression and function in ovarian cancer cell lines. Cell. Mol. Biol. Lett. 14, 153–174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Wang Q., Traynor J. R. (2011) Opioid-induced down-regulation of RGS4: role of ubiquitination and implications for receptor cross-talk. J. Biol. Chem. 286, 7854–7864 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Bucci C., Thomsen P., Nicoziani P., McCarthy J., van Deurs B. (2000) Rab7: a key to lysosome biogenesis. Mol. Biol. Cell 11, 467–480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Takahashi S., Kubo K., Waguri S., Yabashi A., Shin H. W., Katoh Y., Nakayama K. (2012) Rab11 regulates exocytosis of recycling vesicles at the plasma membrane. J. Cell Sci. 125, 4049–4057 [DOI] [PubMed] [Google Scholar]
- 50. Gidon A., Bardin S., Cinquin B., Boulanger J., Waharte F., Heliot L., de la Salle H., Hanau D., Kervrann C., Goud B., Salamero J. (2012) A Rab11A/myosin Vb/Rab11-FIP2 complex frames two late recycling steps of langerin from the ERC to the plasma membrane. Traffic 13, 815–833 [DOI] [PubMed] [Google Scholar]
- 51. Maxfield F. R., McGraw T. E. (2004) Endocytic recycling. Nat. Rev. Mol. Cell Biol. 5, 121–132 [DOI] [PubMed] [Google Scholar]
- 52. Tsutsumi R., Fukata Y., Noritake J., Iwanaga T., Perez F., Fukata M. (2009) Identification of G protein α subunit-palmitoylating enzyme. Mol. Cell. Biol. 29, 435–447 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Roy A. A., Lemberg K. E., Chidiac P. (2003) Recruitment of RGS2 and RGS4 to the plasma membrane by G proteins and receptors reflects functional interactions. Mol. Pharmacol. 64, 587–593 [DOI] [PubMed] [Google Scholar]
- 54. Huang J., Fisher R. A. (2009) Nuclear trafficking of regulator of G protein signaling proteins and their roles in the nucleus. Prog. Mol. Biol. Transl. Sci. 86, 115–156 [DOI] [PubMed] [Google Scholar]
- 55. Chatterjee T. K., Liu Z., Fisher R. A. (2003) Human RGS6 gene structure, complex alternative splicing, and role of N terminus and G protein γ-subunit-like (GGL) domain in subcellular localization of RGS6 splice variants. J. Biol. Chem. 278, 30261–30271 [DOI] [PubMed] [Google Scholar]
- 56. Grabowska D., Jayaraman M., Kaltenbronn K. M., Sandiford S. L., Wang Q., Jenkins S., Slepak V. Z., Smith Y., Blumer K. J. (2008) Postnatal induction and localization of R7BP, a membrane-anchoring protein for regulator of G protein signaling 7 family-Gβ5 complexes in brain. Neuroscience 151, 969–982 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Bernstein L. S., Grillo A. A., Loranger S. S., Linder M. E. (2000) RGS4 binds to membranes through an amphipathic α-helix. J. Biol. Chem. 275, 18520–18526 [DOI] [PubMed] [Google Scholar]
- 58. Delaney K. A., Murph M. M., Brown L. M., Radhakrishna H. (2002) Transfer of M2 muscarinic acetylcholine receptors to clathrin-derived early endosomes following clathrin-independent endocytosis. J. Biol. Chem. 277, 33439–33446 [DOI] [PubMed] [Google Scholar]
- 59. Giguère P., Rochdi M. D., Laroche G., Dupré E., Whorton M. R., Sunahara R. K., Claing A., Dupuis G., Parent J. L. (2006) ARF6 activation by Gαq signaling: Gαq forms molecular complexes with ARNO and ARF6. Cell. Signal. 18, 1988–1994 [DOI] [PubMed] [Google Scholar]
- 60. Laroche G., Giguère P. M., Dupré E., Dupuis G., Parent J. L. (2007) The N-terminal coiled-coil domain of the cytohesin/ARNO family of guanine nucleotide exchange factors interacts with Gαq. Mol. Cell. Biochem. 306, 141–152 [DOI] [PubMed] [Google Scholar]
- 61. Moreau K., Ravikumar B., Puri C., Rubinsztein D. C. (2012) Arf6 promotes autophagosome formation via effects on phosphatidylinositol 4,5-bisphosphate and phospholipase D. J. Cell Biol. 196, 483–496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Macia E., Partisani M., Paleotti O., Luton F., Franco M. (2012) Arf6 negatively controls the rapid recycling of the β2 adrenergic receptor. J. Cell Sci. 125, 4026–4035 [DOI] [PubMed] [Google Scholar]
- 63. Reiner C., Nathanson N. M. (2008) The internalization of the M2 and M4 muscarinic acetylcholine receptors involves distinct subsets of small G-proteins. Life Sci. 82, 718–727 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Kanamarlapudi V., Owens S. E., Saha K., Pope R. J., Mundell S. J. (2012) ARF6-dependent regulation of P2Y receptor traffic and function in human platelets. PLoS One 7, e43532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Borroni V., Barrantes F. J. (2011) Cholesterol modulates the rate and mechanism of acetylcholine receptor internalization. J. Biol. Chem. 286, 17122–17132 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Fielding A. B., Schonteich E., Matheson J., Wilson G., Yu X., Hickson G. R., Srivastava S., Baldwin S. A., Prekeris R., Gould G. W. (2005) Rab11-FIP3 and -FIP4 interact with Arf6 and the exocyst to control membrane traffic in cytokinesis. EMBO J. 24, 3389–3399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Prigent M., Dubois T., Raposo G., Derrien V., Tenza D., Rossé C., Camonis J., Chavrier P. (2003) ARF6 controls post-endocytic recycling through its downstream exocyst complex effector. J. Cell Biol. 163, 1111–1121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Boshans R. L., Szanto S., van Aelst L., D'souza-Schorey C. (2000) ADP-ribosylation factor 6 regulates actin cytoskeleton remodeling in coordination with Rac1 and RhoA. Mol. Cell. Biol. 20, 3685–3694 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Bose A., Cherniack A. D., Langille S. E., Nicoloro S. M., Buxton J. M., Park J. G., Chawla A., Czech M. P. (2001) G(α)11 signaling through ARF6 regulates F-actin mobilization and GLUT4 glucose transporter translocation to the plasma membrane. Mol. Cell. Biol. 21, 5262–5275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Zeigerer A., Gilleron J., Bogorad R. L., Marsico G., Nonaka H., Seifert S., Epstein-Barash H., Kuchimanchi S., Peng C. G., Ruda V. M., Del Conte-Zerial P., Hengstler J. G., Kalaidzidis Y., Koteliansky V., Zerial M. (2012) Rab5 is necessary for the biogenesis of the endolysosomal system in vivo. Nature 485, 465–470 [DOI] [PubMed] [Google Scholar]
- 71. Stokes L. (2013) Rab5 regulates internalisation of P2X4 receptors and potentiation by ivermectin. Purinergic Signal. 9, 113–121 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Dale L. B., Seachrist J. L., Babwah A. V., Ferguson S. S. (2004) Regulation of angiotensin II type 1A receptor intracellular retention, degradation, and recycling by Rab5, Rab7, and Rab11 GTPases. J. Biol. Chem. 279, 13110–13118 [DOI] [PubMed] [Google Scholar]
- 73. Shin H. W., Hayashi M., Christoforidis S., Lacas-Gervais S., Hoepfner S., Wenk M. R., Modregger J., Uttenweiler-Joseph S., Wilm M., Nystuen A., Frankel W. N., Solimena M., De Camilli P., Zerial M. (2005) An enzymatic cascade of Rab5 effectors regulates phosphoinositide turnover in the endocytic pathway. J. Cell Biol. 170, 607–618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Huotari J., Helenius A. (2011) Endosome maturation. EMBO J. 30, 3481–3500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Rink J., Ghigo E., Kalaidzidis Y., Zerial M. (2005) Rab conversion as a mechanism of progression from early to late endosomes. Cell 122, 735–749 [DOI] [PubMed] [Google Scholar]
- 76. Hagiwara M., Shirai Y., Nomura R., Sasaki M., Kobayashi K., Tadokoro T., Yamamoto Y. (2009) Caveolin-1 activates Rab5 and enhances endocytosis through direct interaction. Biochem. Biophys. Res. Commun. 378, 73–78 [DOI] [PubMed] [Google Scholar]
- 77. Pelkmans L., Bürli T., Zerial M., Helenius A. (2004) Caveolin-stabilized membrane domains as multifunctional transport and sorting devices in endocytic membrane traffic. Cell 118, 767–780 [DOI] [PubMed] [Google Scholar]
- 78. Seachrist J. L., Ferguson S. S. (2003) Regulation of G protein-coupled receptor endocytosis and trafficking by Rab GTPases. Life Sci. 74, 225–235 [DOI] [PubMed] [Google Scholar]
- 79. Hubbard V. M., Valdor R., Patel B., Singh R., Cuervo A. M., Macian F. (2010) Macroautophagy regulates energy metabolism during effector T cell activation. J. Immunol. 185, 7349–7357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Korolchuk V. I., Saiki S., Lichtenberg M., Siddiqi F. H., Roberts E. A., Imarisio S., Jahreiss L., Sarkar S., Futter M., Menzies F. M., O'Kane C. J., Deretic V., Rubinsztein D. C. (2011) Lysosomal positioning coordinates cellular nutrient responses. Nat. Cell Biol. 13, 453–460 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Ioannou Y. A., Bishop D. F., Desnick R. J. (1992) Overexpression of human α-galactosidase A results in its intracellular aggregation, crystallization in lysosomes, and selective secretion. J. Cell Biol. 119, 1137–1150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Svensson L., Stanley P., Willenbrock F., Hogg N. (2012) The Gαq/11 proteins contribute to T lymphocyte migration by promoting turnover of integrin LFA-1 through recycling. PLoS One 7, e38517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Chia P. Z., Gasnereau I., Lieu Z. Z., Gleeson P. A. (2011) Rab9-dependent retrograde transport and endosomal sorting of the endopeptidase furin. J. Cell Sci. 124, 2401–2413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Smith D. C., Spooner R. A., Watson P. D., Murray J. L., Hodge T. W., Amessou M., Johannes L., Lord J. M., Roberts L. M. (2006) Internalized Pseudomonas exotoxin A can exploit multiple pathways to reach the endoplasmic reticulum. Traffic 7, 379–393 [DOI] [PubMed] [Google Scholar]
- 85. Ng E. L., Gan B. Q., Ng F., Tang B. L. (2012) Rab GTPases regulating receptor trafficking at the late endosome-lysosome membranes. Cell Biochem. Funct. 30, 515–523 [DOI] [PubMed] [Google Scholar]
- 86. Sullivan B. M., Harrison-Lavoie K. J., Marshansky V., Lin H. Y., Kehrl J. H., Ausiello D. A., Brown D., Druey K. M. (2000) RGS4 and RGS2 bind coatomer and inhibit COPI association with Golgi membranes and intracellular transport. Mol. Biol. Cell 11, 3155–3168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Takata K. (2006) Aquaporin-2 (AQP2): its intracellular compartment and trafficking. Cell. Mol. Biol. 52, 34–39 [PubMed] [Google Scholar]
- 88. Jing J., Junutula J. R., Wu C., Burden J., Matern H., Peden A. A., Prekeris R. (2010) FIP1/RCP binding to Golgin-97 regulates retrograde transport from recycling endosomes to the trans-Golgi network. Mol. Biol. Cell 21, 3041–3053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89. Dugani C. B., Klip A. (2005) Glucose transporter 4: cycling, compartments and controversies. EMBO Rep. 6, 1137–1142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Wieman H. L., Wofford J. A., Rathmell J. C. (2007) Cytokine stimulation promotes glucose uptake via phosphatidylinositol-3 kinase/Akt regulation of Glut1 activity and trafficking. Mol. Biol. Cell 18, 1437–1446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Jaén C., Doupnik C. A. (2006) RGS3 and RGS4 differentially associate with G protein-coupled receptor-Kir3 channel signaling complexes revealing two modes of RGS modulation. Precoupling and collision coupling. J. Biol. Chem. 281, 34549–34560 [DOI] [PubMed] [Google Scholar]
- 92. Zhang Q., Pacheco M. A., Doupnik C. A. (2002) Gating properties of GIRK channels activated by Gα(o)- and Gα(i)-coupled muscarinic m2 receptors in Xenopus oocytes: the role of receptor precoupling in RGS modulation. J. Physiol. 545, 355–373 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Chen Y., Liu Y., Cottingham C., McMahon L., Jiao K., Greengard P., Wang Q. (2012) Neurabin scaffolding of adenosine receptor and RGS4 regulates anti-seizure effect of endogenous adenosine. J. Neurosci. 32, 2683–2695 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Schwendt M., Sigmon S. A., McGinty J. F. (2012) RGS4 overexpression in the rat dorsal striatum modulates mGluR5- and amphetamine-mediated behavior and signaling. Psychopharmacology 221, 621–635 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Harris I. S., Treskov I., Rowley M. W., Heximer S., Kaltenbronn K., Finck B. N., Gross R. W., Kelly D. P., Blumer K. J., Muslin A. J. (2004) G-protein signaling participates in the development of diabetic cardiomyopathy. Diabetes 53, 3082–3090 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.