Background: Neuronally LRP1-deficient mice show severe neurological signs and symptoms.
Results: Neuronal LRP1 is cleaved by γ-secretase and regulates NMDA-dependent signaling and protein turnover.
Conclusion: LRP1 modulates postsynaptic protein complexes and thus has the potential to influence synaptic transmission.
Significance: This might explain why neuronal LRP1 is essential in vivo and shed light on its genetic association with neurodegenerative disease (i.e. Alzheimer disease).
Keywords: Glutamate Receptors, Lipoprotein Receptor, Secretases, Signaling, Synapses
Abstract
The lipoprotein receptor LRP1 is essential in neurons of the central nervous system, as was revealed by the analysis of conditional Lrp1-deficient mouse models. The molecular basis of its neuronal functions, however, is still incompletely understood. Here we show by immunocytochemistry, electron microscopy, and postsynaptic density preparation that LRP1 is located postsynaptically. Basal and NMDA-induced phosphorylation of the transcription factor cAMP-response element-binding protein (CREB) as well as NMDA target gene transcription are reduced in LRP1-deficient neurons. In control neurons, NMDA promotes γ-secretase-dependent release of the LRP1 intracellular domain (LRP1-ICD). However, pull-down and chromatin immunoprecipitation (ChIP) assays showed no direct interaction between the LRP1-ICD and either CREB or target gene promoters. On the other hand, NMDA-induced degradation of the postsynaptic scaffold protein PSD-95 was impaired in the absence of LRP1, whereas its ubiquitination was increased, indicating that LRP1 influences the composition of postsynaptic protein complexes. Accordingly, NMDA-induced internalization of the AMPA receptor subunit GluA1 was impaired in LRP1-deficient neurons. These results show a role of LRP1 in the regulation and turnover of synaptic proteins, which may contribute to the reduced dendritic branching and to the neurological phenotype observed in the absence of LRP1.
Introduction
The LDL receptor-related protein 1 (LRP1) is a ubiquitously expressed transmembrane protein that functions both as a cargo transporter and as a signaling receptor (1). Its essential functions in the central nervous system were demonstrated by conditional gene targeting in mice using the CRE-loxP system (2, 3). Both expression of CRE recombinase under the control of the synapsin promoter in differentiated neurons and expression driven by the αCaMKII promoter in adult forebrain neurons of mice with loxP-marked Lrp1 alleles led to behavioral and motor deficits in animals with recombined genes (2, 3). The molecular basis of these phenotypical consequences of LRP1 deficiency in neurons is only incompletely understood, partly because Lrp1-deficient neurons could not be cultivated from these mouse models, in which recombination mainly takes place in adult animals.
The neuronal functions of LRP1 that have been elucidated so far include the regulation of brain lipid metabolism (3), presumably via its endocytic function, the modulation of trafficking and processing of the amyloid β precursor protein (4), as well as modulating toxicity of the resulting amyloid β fragment, in part by mediating its cellular uptake (5). In addition, LRP1 has been demonstrated to modulate NMDA receptor function by regulating tissue plasminogen activator-dependent calcium influx via the NMDA receptor ion channel through the interaction with the adaptor protein PSD-95 (6) and to mediate the transactivation of Trk receptors by an Src family kinase-dependent pathway (7). These distinct functions seem to play a role in securing neuronal viability and differentiation as well as preventing neural inflammation (3). It is unclear whether proteolytical processing and release of the LRP1 intracellular domain (LRP1-ICD), which limit inflammatory signaling in macrophages (8), play a role in the aforementioned actions of LRP1. It has been found in cerebral artery occlusion models, however, that there is an increased amount and nuclear translocation of LRP1-ICD in the penumbra of ischemic brain tissue (9), indicating that the proteolytical processing of LRP1 is of potential importance in the CNS.
Here, we use conditionally LRP1-deficient mice, where recombination of loxP-marked Lrp1 alleles takes place in neuronal precursors to isolate Lrp1-deficient primary cortical neurons. We find that these cells are of normal viability but exhibit reduced dendritic branching. Immunocytochemical and electron microscopic examination of LRP1-expressing control neurons as well as biochemical purification of proteins of the postsynaptic density demonstrate that LRP1 is located on dendritic processes, where it is partly localized on dendritic spines and in the postsynaptic density. NMDA-dependent signaling is altered in the absence of LRP1 with reduced target gene transcription and reduced phosphorylation of the transcription factor cAMP-response element (CRE)5-binding protein (CREB). In addition, we show that NMDA treatment induces proteolytical processing of LRP1 in neurons, possibly implying a role of the free LRP1-ICD in regulating these NMDA-induced intracellular events. In the absence of direct transcriptional regulation by the LRP1-ICD, its role could be to regulate postsynaptic protein complexes, because in the absence of LRP1, NMDA-induced degradation of PSD-95 is reduced, its ubiquitination is increased, and internalization of the AMPA receptor subunit GluA1 is impaired.
EXPERIMENTAL PROCEDURES
Chemicals
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT), NMDA, N-methyl-l-aspartate ifenprodil (+)-tartrate salt, and AP5 were purchased from Sigma-Aldrich. DAPT was obtained from Merck, MG-132 from Enzo Life Sciences (Lörrach, Germany), L-685,458 from Tocris Bioscience (Germany), and epoxomicin from Biomol (Hamburg, Germany).
Animals
Mice carrying a loxP-marked Lrp1 allele were described previously (10). To obtain recombination of the floxed allele in neuronal precursor cells, these mice were bred to animals transgenic for the viral CRE recombinase under control of the Nestin promoter. These mice (B6.Cg-Tg(Nes-cre)1Kln/J) were obtained from JAX (Bar Harbor, ME). Additionally, mice homozygous for a genetically engineered disruption of the very low density lipoprotein receptor (VLDLR) gene (11) were bred to NesCre/LRP1wt/lox mice to generate animals deficient in VLDLR in addition to neuronal LRP1.
Animals were kept under standard laboratory conditions. Experiments were carried out according to the principles of good laboratory animal care and were approved by local authorities (Regierungspräsidium Freiburg).
Neuron and Astrocyte Preparation
Primary cortical neurons were prepared from E15–E16 mouse embryos as described (12) and seeded at 500,000 cells/well of a poly-d-lysine (0.05 mg/ml)-coated 6-well plate or at 5000 cells/10-mm glass coverslip. Neurons were cultured in an H2O-saturated atmosphere containing 5% CO2 at 37 °C.
Astrocytes were prepared in the same way, except that they were seeded in Dulbecco's modified Eagle's medium (DMEM) with 1 g/liter d-glucose supplemented with l-glutamine (Invitrogen), 10% FCS (Sigma), and 10 units/ml penicillin/streptomycin (Lonza). Astrocytes were trypsinized and dissociated when they formed monolayers of 80% confluence to obtain pure cultures, which were then further incubated for 7 days.
Quantitative Real-time PCR
RNA was extracted from primary cortical neuronal cultures with TRIzol (Invitrogen) and was treated with RNase-free DNase I (Fermentas). For cDNA synthesis, random primers (Promega), Moloney murine leukemia virus reverse transcriptase (Promega), RNase inhibitor (Promega), and dNTPs (Genaxxon) were used.
The real-time PCR was set up with 2× AbsoluteTM QPCR SYBR® Green mix with fluorescein (Abgene) on a single-color real-time PCR detection system (Bio-Rad MyiQ with MyIQTM optical system software version 1.0). The sequences of the primers were as follows: Gapdh, 5′-TGTGTCCGTCGTGGATCTGA-3′ (forward) and 5′-CCTGCTTCACCACCTTCTTGAT-3′ (reverse); Arc, 5′-AGCAGCAGACCTGACATCCT-3′ (forward) and 5′-AGCTCTGCTCTTCTTCACTGGT-3′ (reverse); BDNF, 5′-AGGCTGCAGGGGCATAGAC-3′ (forward) and 5′-AATCGCCAGCCAATTCTCTTT-3′ (reverse); c-Fos, 5′-GGGACAGCCTTTCCTACTACC-3′ (forward) and 5′-GATCTGCGCAAAAGTCCTGT-3′ (reverse); Homer1a, 5′-CGGCTTGCAAAGGAGAAGT-3′ (forward) and 5′-TCTCCTCCTGCTGATTCCTG-3′ (reverse). To compare expression levels, the ΔΔCt method was used (8). Ct values were standardized with respect to Gapdh. For all experiments, samples were assayed in duplicate and the mean of Ct was used for further calculations. Experiments were repeated independently at least seven times. Data were averaged and statistically analyzed as described in the legend for Fig. 3B.
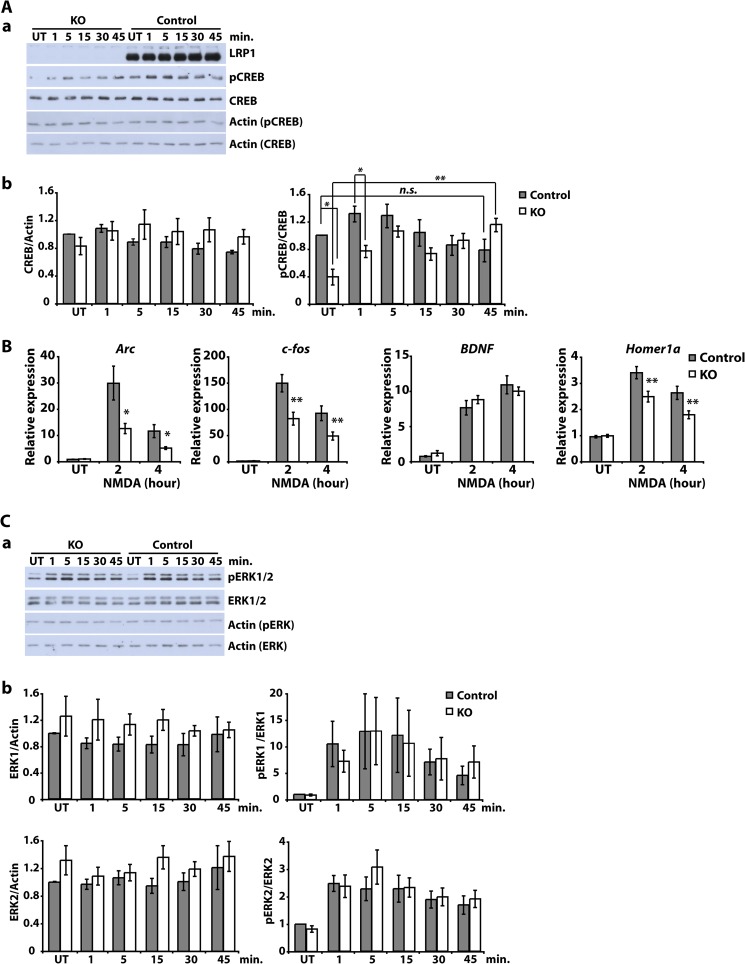
FIGURE 3.
NMDA-induced intracellular signaling and gene transcription are altered in LRP1-deficient neurons. A, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15 LRP1wt/lox (Control) mouse embryos. On DIV 7–8, neurons were treated with 50 μm NMDA for the time indicated or left untreated (UT). Then whole cell lysates were prepared and analyzed by immunoblotting with antibodies directed against LRP1, CREB, pCREB, and actin, which served as a loading control. Representative blots (a) and quantitative analysis (b) of four independent experiments are shown. Error bars, S.E. Statistical analysis was done by Student's t test (*, p < 0.05; **, p < 0.01, n = 4). B, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15 LRP1lox/lox (Control) mouse embryos. On DIV 5, neurons were treated with 50 μm NMDA for the time indicated. Then total RNA was isolated and analyzed by quantitative real-time PCR for the expression of NMDA target genes Arc, BDNF, c-fos, and Homer 1a. Expression levels were normalized to GAPDH and are represented as -fold induction over untreated control. Bars, means ± S.E. from at least seven independent experiments. Statistical analysis was done by Student's t test (*, p < 0.05; **, p < 0.01, n ≥ 7). C, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15 LRP1wt/lox (Control) mouse embryos. On DIV 7–8, neurons were treated with 50 μm NMDA for the time indicated or left untreated (UT). Then whole cell lysates were prepared and analyzed by immunoblotting with antibodies directed against LRP1, ERK1/2, phospho-ERK1/2 (pErk 1/2), and actin, which served as a loading control. Representative blots (a) and quantitative analysis (b) of four independent experiments are shown. Error bars, S.E. Statistical analysis was done by Student's t test (*, p < 0.05; **, p < 0.01, n = 4).
Immunocytochemistry
Primary cortical neurons cultured on glass coverslips were fixed with 4% paraformaldehyde, permeabilized, and blocked with 5% donkey serum, 0.5% BSA, 0.1% Triton X-100 in phosphate-buffered saline (PBS). Incubation with the first antibody (affinity-purified rabbit polyclonal α-LRP1 (13) and mouse monoclonal α-synaptophysin (SY38, catalog no. 61012, Progen) or α-PSD-95 (6G6-1C9, catalog no. MAB 1596, Millipore) was done overnight at 4 °C. Alexa 488- or Alexa 555-labeled donkey anti-rabbit or anti-mouse antibody (Invitrogen) was used diluted 1:1000 in blocking solution as secondary antibody. Coverslips were mounted in aqueous mounting medium (ThermoScientific), and double-immunofluoresence images were acquired with an inverted LSM-I-NL02 confocal microscope using a Plan-Apochromat ×63/1.4 oil objective (Zeiss, Oberkochern, Germany) and appropriate filter sets.
Preparation of Whole Cell Lysates and Western Blot Analysis
Neurons were briefly washed with ice-cold PBS and lysed in RIPA buffer containing protease inhibitor mixture (Sigma) and phosphatase inhibitor mixtures 1 and 2 (Sigma). Lysates were subjected to SDS-PAGE and Western blotting according to standard procedures. The following primary antibodies were used: a rabbit polyclonal antibody against a C-terminal epitope of LRP1 (13), a polyclonal rabbit antibody against actin (catalog no. A5060, Sigma-Aldrich), α-ApoER2 (a rabbit polyclonal antibody against a C-terminal epitope of ApoER2 (ApoE receptor 2) (14)), α-Dab1 (catalog no. AB5840, Millipore), α-VLDLR (catalog no. sc18824, Santa Cruz Biotechnology, Inc.), α-PSD-95 (7E3-1B8, catalog no. MAB1598, Millipore), α-CREB (48H2) (catalog no. 9197, Cell Signaling), α-pCREB (Ser-133) (catalog no. 9198, Cell Signaling), α-ERK1/2 (catalog no. 9102, Cell Signaling), α-phospho-ERK1/2 Thr-202/Tyr-204 (catalog no. 4376, Cell Signaling), α-NR1 (catalog no. 556308, BD Pharmingen), α-NR2A (NMDAe1 (c-17), catalog no. sc-1468, Santa Cruz Biotechnology, Inc., Santa Cruz Biotechnology, Inc.), α-NR2B (NMDAe2 (c-20), catalog no. sc-1469, Santa Cruz Biotechnology, Inc.), α-phosphotyrosine (clone 4G10, catalog no. 05-321, Millipore), α-GluR1 (catalog no. AB1504, Millipore), and α-mono- and poly-ubiquitinated protein (catalog no. PW8801, clone FK2, Enzo). After incubation with an HRP-conjugated secondary antibody, bound antibodies were visualized by enhanced chemiluminescence (ECL).
Quantification of results was done by densitometry with the help of the program ImageJ version 1.42q (National Institutes of Health, Bethesda, MD). Statistical analysis was done with Student's t test or ANOVA, and the threshold of significance was defined as p < 0.05.
MTT Assay
Cell viability was measured according to MTT reduction capability. Briefly, neurons (DIV 6 and 14) were incubated with MTT at a final concentration of 500 μg/ml for 4 h under normal culture conditions. The amount of the blue formazan reduction product was measured photometrically at 595 nm after lysis with 10% SDS in 0.01 n HCl (reference wavelength: A690 nm).
Lactate Dehydrogenase (LDH) Cytotoxicity Assay
The cytotoxicity of NMDA treatment (50 μm, 2 h) of neurons was examined by quantitative analysis of LDH release using the Cytotoxicity Detection Kit (Roche Applied Science). Conditions for neuronal culture were the same as for the MTT assay. LDH was measured according to the manufacturer's instructions at A490 nm (reference wavelength was A690 nm).
Spine Density
LRP1-deficient and control cortical neurons were fixed on DIV 24 with 4% PFA, 4% sucrose in 0.1 m phosphate buffer (PB) for 10 min. Neuronal processes were stained with 0.2 μm phalloidin-TRITC (Fluka) in 0.1 m PB for 30 min. Images were acquired using a Zeiss Axioplan-2 imaging microscope equipped with Plan-Apochromat ×63/1.40 oil objective (Zeiss). Dendritic spine density was determined with the help of ImageJ version 1.42q (National Institutes of Health). The number of spines on dendritic branches (starting 30 μm away from the soma) was determined by manual counting from black and white images by using the “threshold” and “particle analyzing” functions. 20 dendrites from five independent cultures were analyzed for both genotypes.
Neurite Outgrowth
Primary cortical neurons were cultured for 4 or 9 DIV, fixed, and stained with an α-MAP-2 antibody (clone AP20, Leinco, Germany, catalog no. M119). 120 LRP1-deficient and control neurons from four independent cultures were analyzed with METAMORPH software (Universal Imaging Corp., Downington, PA) for neurite length and branch number. Images were acquired using a Zeiss Axioplan-2 imaging microscope equipped with a Plan-Neofluar ×40/1.30 oil-differential interference contrast objective (Zeiss).
Preparation of Postsynaptic Density Fractions
Proteins of the postsynaptic density were prepared as described (15, 16). Briefly, cortices (∼400 mg of tissue/preparation) from adult mice were homogenized in sucrose buffer. The homogenate was subjected to sucrose gradient centrifugation, and the synaptosomal fraction was collected. Three rounds of solubilization with detergent and centrifugation led to extraction of PSD fractions I–III (I, Triton X-100; II, SDS; III, Sarkosyl).
Electron Microscopy
The pre-embedding immunogold method was used as described previously (17). Briefly, adult mice (n = 3/group) were perfused with 4% PFA, 15% saturated picric acid, and 0.05% glutaraldehyde in 0.1 m PB. Vibratome brain sections (50 μm) were blocked in 20% normal goat serum in 50 mm Tris-buffered saline (TBS) for 1 h followed by incubation with the primary antibody (affinity-purified α-LRP1, final concentration 2.0 μg/ml), diluted in 50 mm TBS containing 3% normal goat serum at 4 °C overnight. After washing with 50 mm TBS, the sections were incubated with 1.4-nm gold-coupled goat anti-rabbit secondary antibody (Fab fragment; 1:100 in 50 mm TBS with 2% normal goat serum; Nanogold; Nanoprobes, Stony Brook, NY) at 4 °C overnight. The sections were washed with 50 mm TBS and 25 mm PBS and postfixed with 1% glutaraldehyde in 25 mm PBS for 10 min. After washing in PBS, the HQ Silver kit (Nanoprobes) was used for intensifying gold particles. Sections were washed with 0.1 m PB for 5 min and then postfixed with 1% OsO4 for 40 min. The sections were dehydrated once with 50% ethanol and stained with uranyl acetate (1% in 70% ethanol) for 35 min in the dark, followed by further dehydration with 90, 95, and 100% ethanol and then propylene oxide for 2 × 10 min. They were then soaked in epoxy resin (Durcupan, Fluka) overnight at room temperature. The tissues were flat-embedded in epoxy resin on glass slides and polymerized for 2 days at 56 °C. They were then trimmed for the hippocampal stratum lacunosum-moleculare (CA1 region) and were re-embedded into resin. This area was chosen because intense immunostaining for LRP1 was observed in apical dendrites of CA1 pyramidal cells, particularly on distal dendritic segments, located in the stratum lacunosum-moleculare. 70-nm ultrathin sections were cut on a Leica EM UC6 ultramicrotome. Analysis and documentation were performed on a Philips CM100 electron microscope equipped with a digital camera device (Gatan, Orius SC600).
Generation of GST Fusion Proteins
A pGEX plasmid containing the receptor-associated protein (RAP) cDNA was employed. Fusion proteins were expressed in BL21 codon+ bacteria (Stratagene) following induction by 1 mm isopropyl-thio-d-galactopyranoside for 5 h. Proteins were recovered by Triton lysis (PBS with 1% Triton X-100 and protease inhibitors (Roche Applied Science)) and purified using glutathione-agarose beads (Sigma-Aldrich).
Chromatin Immunoprecipitation Assays
ChIP analysis was performed using the ChIP assay kit (catalog no. 17-295, Millipore, Schwalbach, Germany). Briefly, E15–E16 mouse embryonic cortical neurons were prepared, and 5 × 106 cells were used per condition. On DIV 7, cells were either left untreated or stimulated with 50 μm NMDA for 4 h. Then they were harvested, and DNA was sheared by sonification with a Branson Digital Sonifier (model 450) and a 3-mm diameter microtip probe (4 cycles of 15-s pulse/20-s interval with an amplitude of 10%). For the immunoprecipitation, α-CREB (in a 1:50 dilution; catalog no. 9197, Cell Signaling), α-LRP1 (1:30 dilution), or 5 μg of control IgG was used. The following primers were employed for the PCR of the c-fos promoter: CRE (−64 and −294), 5′-TCCATATTAGGACATCTGCGTCA-3′ (forward) and 5′-CGGCTCTATCCAGTCTTCTCAGT-3′ (reverse); CRE (−342), 5′-GGGTCCACATTGAATCAGGT-3′ (forward) and 5′-TGTAAAGGAGGGAGGGATTG (reverse); serum-response element, 5′-GCGAGCTGTTCCCGTCAATC-3′ (forward) and 5′-GGATGGACTTCCTACGTCAC-3′ (reverse). PCR conditions were as follows: 94 °C for 30 s, 60 °C for 30 s, 67 °C for 2 min, 30 cycles.
Biotinylation Studies
Cortical neurons were seeded on 60-mm dishes (1.5 million cells/dish) and cultured to 10–12 DIV. For surface biotinylation, they were washed with ice-cold PBS, and all further steps were performed on ice with ice-cold buffers to prevent endocytosis. Neurons were then incubated with 1 mg/ml EZ-Link Sulfo-NHS-SS-Biotin (Thermo Scientific) in PBS for 20 min, washed in TBS, and lysed in 300 μl of RIPA buffer with protease inhibitors. The homogenates were centrifuged at 14,000 × g for 15 min at 4 °C. Protein concentration of the supernatant was adjusted to 1 mg/ml by adding RIPA buffer. 10% of the supernatant was boiled with 4× SDS loading buffer for determination of total cellular levels of the respective protein of interest. The remaining supernatant was incubated with 50% immobilized NeutrAvidin (Thermo Scientific) in a 2:1 volume ratio for 6 h at 4 °C. After washing, proteins bound to the beads were resuspended in 2× SDS loading buffer and boiled at 95 °C for 5 min. Western blots were performed to determine both total and surface (biotinylated) levels of GluN1, GluN2A, GluN2B, and LRP1. For the analysis of surface GluA1 turnover, neurons were treated with 20 μm NMDA for 3 min and then incubated in fresh medium for a total length of time as indicated before biotin labeling.
For the internalization assays, neurons were biotinylated for 2 min at 37 °C with 0.3 mg/ml EZ-Link Sulfo-NHS-SS-Biotin solution and were then washed in TBS. Then neurons were incubated in medium containing 20 μm NMDA or in medium alone for 3 min, washed with fresh medium, and further incubated to allow endocytosis of labeled surface proteins. To strip biotin off of the cell surface, the cells were placed on ice, incubated with glutathione solution (150 mm glutathione, 150 mm NaCl, pH 8.75) for 20 min, washed with iodoacetamide solution (50 mm iodoacetamide in PBS), and further processed as described above. Total and internalized GluA1 and LRP1 were determined by Western blotting.
Immunoprecipitation
If not indicated otherwise, DIV 10–12 cortical neurons were lysed in lysis buffer (50 mm Tric-HCl (pH 7.5), 150 mm NaCl, 1 mm MgCl2, 1 mm CaCl2, 1% Triton X-100, containing protease and phosphatase inhibitors). Alternatively, cortical neurons were fractionated to obtain detergent-soluble synaptosomal membrane fractions. Cells were scraped in HEPES-buffered sucrose (0.32 m sucrose, 4 mm HEPES, pH 7.4) and homogenized in a glass-Teflon homogenizer. Pooled samples were centrifuged at 1000 × g for 10 min. Then supernatants were centrifuged at 10,000 × g for 15 min. The pellet was washed in HEPES-buffered sucrose, resuspended in HEPES buffer (4 mm HEPES, pH 7.4), mixed for 30 min on the rotor, and centrifuged at 25,000 × g for 20 min. The pellet was suspended in lysis buffer or in RIPA buffer. RIPA buffer was used for detecting the interaction between LRP1 and GluN1 or PSD-95, respectively. The suspension was ultracentrifuged at 100,000 × g for 30 min, and the resulting supernatant was taken as the detergent-soluble synaptosomal membrane fraction.
Neuronal lysates or soluble synaptosomal membrane fractions were incubated on ice for 1 h with the first antibody for immunoprecipitation (anti-GluN2A or anti-GluN2B for phosphotyrosine detection; anti-PSD-95 (clone 6G6–1C9, Millipore, catalog no. MAB1596) for detection of ubiquitinated PSD-95; and anti-GluN1, anti-GluN2A, anti-GluN2B, anti-PSD-95, or anti-GluA1 (Millipore, catalog no. AB1504) for detection of LRP1). Immune complexes were precipitated with Protein A/G Plus-agarose (Santa Cruz Biotechnology, Inc.). After washing, bound proteins were extracted by boiling with SDS loading buffer and further analyzed by Western blotting.
Metabolic Turnover of PSD-95
DIV 14–16 cortical neurons were incubated with methionine- and cysteine-free DMEM (Invitrogen) for 15 min and then in medium containing 300 μCi/ml EXPRE35S35S [35S] protein labeling mix (PerkinElmer Life Sciences) for 1 h. The labeled cultures were chased for the times indicated in methionine- and cysteine-free DMEM with 2 mm methionine. Cells were lysed, and PSD-95 was immunoprecipitated as described above. Immunoprecipitated samples were subjected to SDS-PAGE. The gels were dried, and autoradiographs were obtained by exposure to x-ray films for 5–9 days at −80 °C.
RESULTS
LRP1 Is Completely Absent from NestinCreLRP1lox/lox Neurons
The LDL receptor-related protein 1 (LRP1) is critically important for the normal functioning of the central nervous system, as shown by the analysis of different conditionally LRP1-deficient mouse models (2, 3). To pinpoint its role in neuronal cells, several studies with primary cultured neurons have been done, but so far the direct comparison of Lrp1-deficient and wild type neurons has not been accomplished because Lrp1-deficient neurons could not be cultivated from the available mouse models. We therefore bred mice with floxed Lrp1 alleles (10) to a line transgenic for the CRE recombinase under the control of the Nestin promoter, which is active in neuronal precursor cells, among others (18, 19). Thus, contrary to the models studied so far, recombination of the floxed Lrp1 alleles already takes place during embryonic development. We therefore examined whether Lrp1-deficient embryonic cortical neurons could be prepared and cultured from this line.
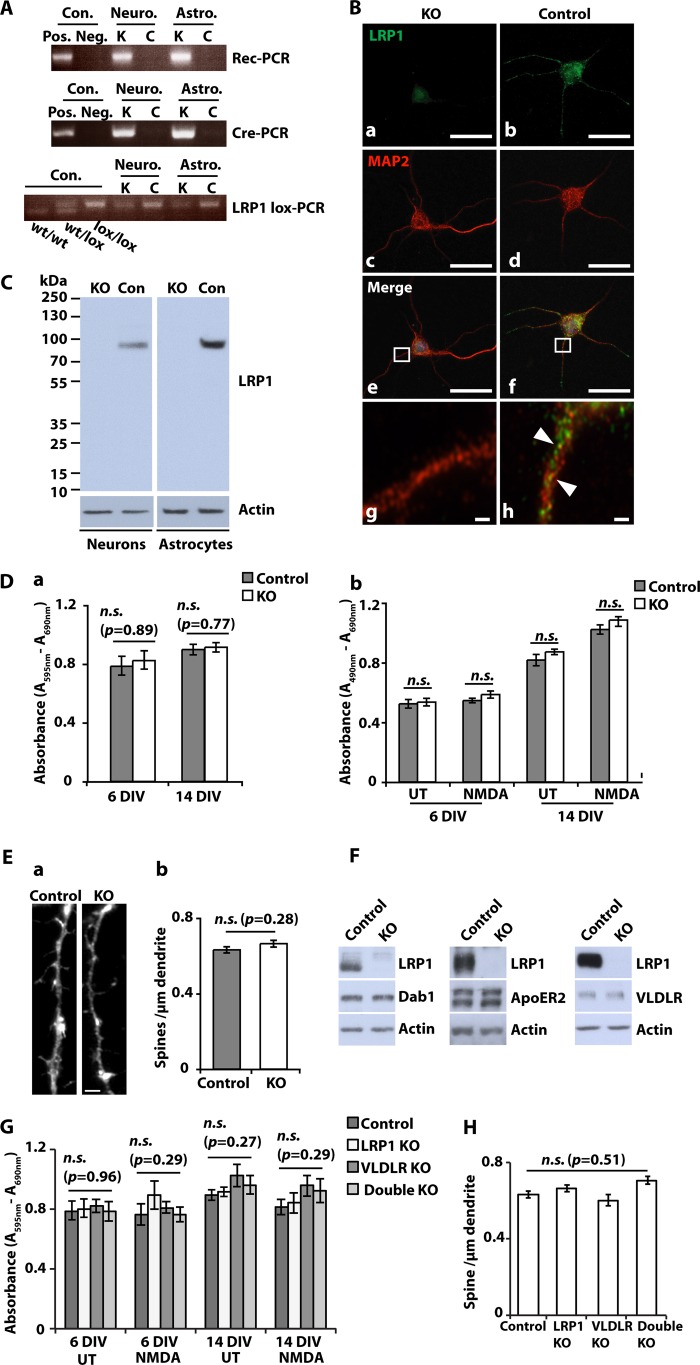
NestinCreLRP1lox/lox embryos were dissected from time-mated female mice on day E15–E16 for cortical neuron preparation. The isolated neuronal cells were examined for recombination of Lrp1 by a PCR specific for the recombined allele (8) and for the presence of LRP1 protein by immunocytochemistry and by immunoblotting with a C-terminal anti-LRP1 antibody. Fig. 1A shows recombination of the floxed Lrp1 alleles in NestinCre-positive neurons. LRP1 protein was completely absent from DIV 5 and 10 NestinCreLRP1lox/lox cultures (Fig. 1, B and C). Because recombination takes place in precursors from which also glial cells develop, LRP1 was lost from astrocytes too (Fig. 1, A and C).
FIGURE 1.
Complete loss of LRP1 in NesCreLRPlox/lox neurons and astrocytes. A, primary cultured cortical neurons (Neuro.) and astrocytes (Astro.) were prepared from E15–E16 NesCreLRP1lox/lox (K) and E15–E16 LRP1lox/lox (C) mouse embryos. Genomic DNA was prepared from the cultures and examined by PCRs specific for the recombined LRP1 allele (Rec-PCR), the CRE transgene (Cre-PCR), and the non-recombined loxP-marked Lrp1 allele (LRP1lox-PCR), respectively. B, primary cultured cortical neurons were prepared from E15 NesCreLRP1lox/lox (KO) and E15 LRP1lox/lox (Control) mouse embryos and were seeded on glass coverslips. On DIV 5, immunocytochemical staining with antibodies directed against LRP1 and MAP-2 neuronal protein was performed. Scale bar, 25 μm. Inset, partial overlap of the LRP1 signal (green) with the MAP-2 signal (red) on dendrites of control neurons (arrowheads in h). Scale bars, 25 μm (a–f) and 1 μm (g and h). C, primary cultured cortical neurons and astrocytes were prepared from E15–E16 NesCreLRP1lox/lox (KO) and LRP1lox/lox (control (Con)) mouse embryos. Whole cell lysates were prepared on DIV 10 and analyzed by SDS-PAGE and immunoblotting with an antiserum directed against LRP1. Actin served as a loading control. D, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15–E16 LRP1lox/lox (control) mouse embryos. On DIV 6 and 14, MTT cell viability assays and LDH release cytotoxicity assays were performed. a, the results of MTT cell viability assays are shown as mean values. Error bars, S.E. Statistical analysis was done by Student's t test. n.s., not significant (p > 0.05, n = 4). b, LDH release was measured without (UT) and after 2 h of treatment with 50 μm NMDA. The results are shown as mean values ± S.E. Statistical analysis was done by Student's t test. n.s., not significant (p > 0.05, n = 4). E, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15-E16 LRP1lox/lox (Control) mouse embryos. On DIV 24, neurons were stained with phalloidin, and spine density was assessed on 20 dendrites/culture. a, representative spines and dendrite from KO and control. Scale bar, 5 μm. b, bars represent mean values ± S.E. from five independent cultures. Spine density did not differ significantly between KO and control neurons (statistical analysis was done by Student's t test). F, primary cultured cortical neurons from NesCreLRP1lox/lox (KO) and LRP1lox/lox (Control) embryos were prepared and lysed on 10–16 DIV with RIPA buffer. Whole cell lysates were subjected to SDS-PAGE, and expression levels of Dab1, ApoER2, and VLDLR were examined by Western blotting. Actin served as a loading control. Representative results of at least three independent experiments are shown. G, primary cultured cortical neurons were prepared from NesCreLRP1lox/lox (LRP1 KO), LRP1lox/lox (Control), LRP1lox/lox/VLDLR−/− (VLDLR KO), and NesCreLRP1lox/lox/VLDLR−/− (Double KO) mouse embryos. On DIV 6 and 14, MTT cell viability assays were performed. The cells were kept untreated or treated with 50 μm NMDA for 2 h before assays. Bars, means ± S.E. (Student's t test; n.s., not significant (p > 0.05), n = 4). H, primary cultured cortical neurons were prepared from NesCreLRP1lox/lox (LRP1 KO), LRP1lox/lox (Control), LRP1lox/lox/VLDLR−/− (VLDLR KO), and NesCreLRP1lox/lox/VLDLR−/− (Double KO) mouse embryos. Spine density on 24 DIV neurons was assessed as described above. Bars, means ± S.E. (error bars) (ANOVA; n.s., not significant (p > 0.05), n = 5).
Normal Viability and Maturation but Reduced Dendritic Branching of LRP1-deficient Cortical Neurons
LRP1 is an endocytic receptor that internalizes a vast number of ligands, including lipoproteins, and could therefore play a critical role in the nutritional supply of neurons. Thus, we next tested whether LRP1-deficient neurons were normally viable. Comparing cortical neuronal cultures from NestinCreLRP1lox/lox embryos with control cultures from LRP1lox/lox offspring, no differences in viability were recorded, as measured by MTT (Fig. 1D, a) and LDH release assays (Fig. 1D, b).
Because LRP1 is lost already in neuronal precursors of the NestinCreLRP1lox/lox genotype, we examined whether LRP1-defiecient neurons mature regularly. As a measurement for maturity, we assessed the spine density of NestinCreLRP1lox/lox and LRP1lox/lox neurons on DIV 24 and found no significant difference between the two groups (Fig. 1E, a and b).
To analyze whether the loss of LRP1 is compensated for by other lipoprotein receptors, we examined the expression of ApoER2 and VLDLR, respectively, in NestinCreLRP1lox/lox and LRP1lox/lox neurons. There was no difference in expression levels between the two genotypes (Fig. 1F), nor was there any change in the amount of the ApoER2 and VLDLR adaptor protein Dab1 in Lrp1-deficient cells.
Moreover, the analysis by MTT assay showed no change in viability in cortical neurons lacking both LRP1 and VLDLR compared with Lrp1-deficient or control neurons (Fig. 1G). Likewise, there was no difference in spine density between neurons of either genotype (Fig. 1H), indicating no prominent role for compensation by other lipoprotein receptors in Lrp1-deficient neurons.
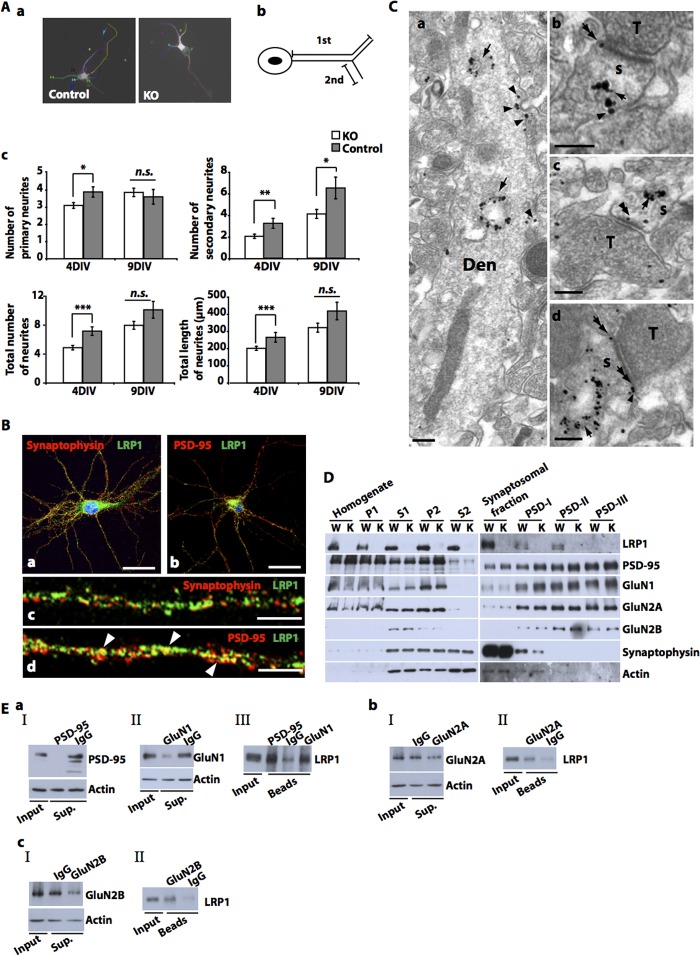
We next processed neuronal cultures for immunostaining with the dendritic marker MAP-2. Morphological analysis of neuronal cultures after 4 DIV revealed a reduction in the formation, length, and branching of MAP-2-positive neurites (Fig. 2A, a–c). These overall abnormalities were not observed after 9 DIV, but the number of secondary neurites was reduced in the absence of LRP1 (Fig. 2A, c).
FIGURE 2.
Impaired neurite outgrowth of LRP1-deficient neurons and postsynaptic localization of LRP1. A, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15–E16 LRP1lox/lox (Control) mouse embryos and were seeded on glass coverslips. The total number of primary neurites, the total number of branches (secondary neurites), and the total number and length of neurites were assessed on DIV 4 and 9 after immunocytochemical staining with a MAP-2 antibody. a, representative micrographs of control and KO neurons. b, schematic representation of primary and secondary neurites. c, quantitative representation of the number of primary and secondary neurites per cell, average total number of neurites (primary neurites plus branches) per cell, and average total length of neurites. The average number of neurites/cell was obtained by dividing the total number of neurites by the number of analyzed cells (120 randomly chosen neurons from four independent experiments). Bars, means ± S.E. (error bars). Statistical analysis was done by Student's t test (*, p < 0.05; **, p < 0.01; ***, p < 0.001). B, primary cultured cortical neurons were prepared from E15–E16 wild type mouse embryos and were seeded on glass coverslips. On DIV 21, immunocytochemical analysis was performed with antibodies directed against LRP1 and synaptophysin (a and c) or PSD-95 (b and d), respectively, and documented by confocal microscopy. Co-localization of LRP1 and PSD-95 was observed (arrowheads in d). Scale bar, 25 μm (a and b) and 10 μm (c and d). C, electron microscopic analysis of the CA1 region (stratum lacunosum-moleculare) of 2–5-month-old LRP1lox/lox mice was performed by the pre-embedding immunogold method with an LRP1 antibody. Clusters of immunogold particles were found on intracellular membranes, such as lysosomes and endosomes (arrows in a–d) of dendritic compartments (Den, s) of presumed pyramidal cells. Immunoparticles were also found on the extrasynaptic (arrowheads in a, b, and d) and perisynaptic membrane (double arrows in b and d) of dendritic shafts (Den) and dendritic spines (s) and also appeared occasionally over the postsynaptic specialization at synapses between spines and axon terminals (T) (double arrowheads in c). No membrane-bound particles were detected in tissues obtained from NesCreLRP1lox/lox animals (not shown). Scale bars, 0.2 μm. D, synaptosomes were prepared from the brains of 3-month-old LRP1wt/wt (WT, W) and NesCreLRP1lox/lox (KO, K) mice, and postsynaptic proteins were extracted. Subsequent immunoblot analysis was done with antibodies against the proteins indicated. A representative photograph of three independent preparations is shown. P1, first pellet (nuclei and large debris); S1, supernatant 1 (cortex homogenate); P2, pellet 2 (crude synaptosomal membranes); S2, supernatant 2 (cytosol and light membranes); PSDI-III, postsynaptic fractions I–III (I, extracted with Triton X-100; II, additional extraction with SDS; III, additional extraction with Sarkosyl). E, primary cultured cortical neurons were prepared from E15 wild type mouse embryos. Detergent-soluble synaptosomal membrane fractions were prepared on DIV 10. Co-immunoprecipitation was performed using primary antibodies against PSD-95 (a), GluN1 (a), GluN2A (b), or GluN2B (c). After separation of precipitates by SDS-PAGE, immunoblotting with an antibody against LRP1 was performed. IgG was used as a negative control. Blots of IP supernatants (Sup.) (a, I and II; b, I; c, I) and of proteins extracted from beads (a, III; b, II; c, II) are represented. Representative blots of two independent experiments are shown.
Subcellular Localization of LRP1 in Neurons
To better understand the role of LRP1 in neurons and for dendritic branching in particular, the subcellular localization of LRP1 was determined. As shown previously (2), confocal laser microscopy revealed the presence of LRP1 on the cell soma and on dendritic shafts, partly co-localizing with the postsynaptic protein PSD-95 but not with the presynaptic marker synaptophysin (Fig. 2B, a–d). Electron microscopic examination of sections obtained from the hippocampal CA1 region of adult and LRP1lox/lox control mice confirmed the presence of LRP1 on the dendritic shafts and spines of putative pyramidal cells (Fig. 2C, a–d). It was found both on intracellular membrane structures, such as lysosomes, and on the perisynaptic and extrasynaptic plasma membrane (Fig. 2C, a, b, and d) as well as occasionally over the synaptic membrane specialization between presynaptic terminals and postsynaptic dendritic spines of principal cells (Fig. 2C, c). In contrast, no immunogold labeling was found on the presynaptic axon terminals (Fig. 2C, a and d). Biochemical analysis of synaptosomes and PSD fractions prepared from cortices of adult LRP1-expressing mice showed the presence of LRP1 in the postsynaptic density, from where it was partly co-purified with PSD-95 and NMDA receptor subunits (Fig. 2D). Moreover, in concordance with our previous findings (2), LRP1 could be co-precipitated with PSD-95 and the NMDA receptor subunits GluN1, GluN2A, and GluN2B (Fig. 2E, a–c) in immunoprecipitation experiments from soluble synaptosomal membrane preparations.
LRP1 Modulates NMDA Receptor-dependent Intracellular Signaling
The dendritic localization of LRP1 strongly suggests that it can modify the postsynaptic changes induced by synaptic transmission. Indeed, earlier studies have implied LRP1 in the modulation of NMDA receptor-dependent signaling, although evidence was indirect and did not involve the study of Lrp1-deficient neurons (4, 20, 21). In addition, some results obtained were contradictory (20, 21). We thus employed primary cultured cortical neurons from NestinCreLRP1lox/lox and LRP1lox/lox control embryos to compare downstream signaling events in NMDA-treated cultures of both genotypes. Immunoblot analysis of lysates from neuronal cultures after stimulation with 50 μm NMDA showed considerably reduced basal levels of phosphorylated CREB and consequently delayed increase in phospho-CREB (pCREB) after NMDA treatment (Fig. 3A, a; quantitative analysis in Fig. 3A, b). Subsequently, qRT-PCR analysis of NMDA-induced gene transcription revealed reduced expression of certain NMDA-responsive genes in Lrp1-deficient neurons (Fig. 3B). The reduced phosphorylation of CREB and decreased transcription of NMDA-dependent genes in the Lrp1-deficient neurons were not mediated by a decrease in upstream ERK activation as measured by ERK phosphorylation after NMDA treatment of Lrp1-deficient and control neurons (Fig. 3C; quantitative analysis in Fig. 3C, b). Measurement of LDH release by LRP1-deficient and control neurons after treatment with NMDA excluded cytotoxicity as a reason for the observed difference in intracellular signaling (Fig. 1D, b).
NMDA Induces Proteolytical Processing of LRP1
LRP1 has been shown to release its ICD by intramembranous proteolysis (22). The free LRP1-ICD has a direct role in transcriptional regulation in the context of the inflammatory response (8). To test whether the rate of LRP1-ICD processing is regulated by activation of NMDA receptors and whether the LRP1-ICD could thus play a role in the regulation of pCREB availability and NMDA target gene transcription, cultured primary neurons were treated with NMDA, and neuronal lysates were examined for LRP1 cleavage products. Shedding of the LRP1 extracellular domain is the regulated step during the proteolytical processing of LRP1, and cleavage by the γ-secretase complex to release the LRP1-ICD follows constitutively. Neuronal cultures were therefore pretreated with the γ-secretase inhibitor DAPT, and neuronal lysates were examined for the presence of the still membrane-bound γ-secretase substrate (i.e. the LRP1 C-terminal fragment of 25 kDa (LRP1-CTF)), which is generated by ectodomain shedding. After NMDA treatment, LRP1-CTF increased in a time-dependent manner in wild type cortical neuronal cultures but not in Lrp1-deficient ones, indicating that activation of NMDA receptors accelerates the rate of LRP1 proteolytical processing (Fig. 4A, a, c, and d). This effect was abolished by the addition of the NMDA receptor antagonist AP5 (Fig. 4A, b, c, and e). Treatment with the inactive stereoisomer N-methyl-l-aspartate, on the other hand, did not increase the rate of proteolytical processing of LRP1 (Fig. 4A, b and c). In addition, the GluN2B-specific antagonist ifenprodil did not prevent NMDA-induced generation of the LRP1-CTF, indicating that GluN2B-containing NMDA receptors are not essential for increased LRP1 processing (Fig. 4A, e). The NMDA-induced increase of LRP1 cleavage does not involve direct binding of NMDA to LRP1. This could be shown by co-incubation of cultured primary neurons with NMDA and the lipoprotein receptor chaperone RAP (receptor-associated protein), which blocks ligand binding to LRP1. NMDA treatment increased proteolytical processing of LRP1 regardless of the presence of RAP (Fig. 4B, a and b).
FIGURE 4.
NMDA induces proteolytical processing of LRP1 in primary cultured cortical neurons. A (a), primary cultured cortical neurons were prepared from E15–E16 wild type mouse embryos. On DIV 5, neurons were pretreated with 1 μm DAPT for 2 h, and then 50 μm NMDA was added for the time indicated. DMSO was used as control (vehicle treatment). Whole cell lysates were prepared and analyzed by immunoblotting with antiserum directed against LRP1. Actin served as a loading control. b, primary cultured cortical neurons were prepared from E15–E16 wild type mouse embryos. On DIV 5, neurons were pretreated with 1 μm DAPT for 2 h, and then 50 μm NMDA, 50 μm AP5, 50 μm NMDA plus 50 μm AP5, or 50 μm N-methyl-l-aspartate was added for 12 h (UT, untreated control). Whole cell lysates were prepared and analyzed by immunoblotting with α-LRP1 antiserum. Actin served as a loading control. c, densitometric quantification of LRP1-CTF from b. Data are means ± S.E. (error bars) of five independent experiments. Statistical analysis was done by ANOVA (*, p < 0.05; **, p < 0.01; n.s., not significant). d, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15-E16 LRP1lox/lox (Control) mouse embryos. On DIV 5, control neurons were pretreated with 1 μm DAPT for 2 h, and then 50 μm NMDA was added for 12 h. KO neurons were pretreated with 1 μm DAPT for 2 h, and then 50 μm NMDA, 50 μm AP5, or 50 μm NMDA plus 50 μm AP5 was added for 12 h (UT, untreated control). Whole cell lysates were prepared and analyzed by immunoblotting with α-LRP1 antiserum. Actin served as a loading control. e, primary cultured cortical neurons were prepared from E15–E16 wild type mouse embryos. On DIV 5, neurons were pretreated with 1 μm DAPT for 2 h, and then 50 μm NMDA, 50 μm AP5, or 50 μm NMDA plus 50 μm AP5, 50 μm NMDA plus 1 μm ifenprodil, or 50 μm NMDA plus 10 μm ifenprodil was added for 12 h. DMSO was used as control (vehicle treatment). Whole cell lysates were prepared and analyzed by immunoblotting with α-LRP1 antiserum. Actin served as a loading control. B, primary cultured cortical neurons were prepared from E15 wild type mouse embryos. On DIV 5, neurons were pretreated with 10 μg/ml GST or 10 μg/ml GST-RAP for 30 min, and then 1 μm DAPT was added, followed by 50 μm NMDA for 12 h. Whole cell lysates were prepared and analyzed by immunoblotting with α-LRP1 antiserum. Actin served as a loading control. A representative blot (a) and quantitative analysis of four independent experiments (b) are shown. Error bars in b, S.E. Statistical analysis was done by ANOVA (*, p < 0.05). C, primary cultured cortical neurons were prepared from E15 wild type mouse embryos. On DIV 7, neurons were treated with 50 μm NMDA (N) for 4 h or left untreated (UT). After cross-linking with 1% paraformaldehyde, cell lysates were prepared, and DNA was sheared by sonification. Subsequently, chromatin immunoprecipitation was performed with an α-LRP1 antibody, rabbit IgG as negative control, or α-CREB as positive control. PCR amplification of c-fos promoter fragments was done with different primer sets to include transcription factor binding sites CRE −64, CRE −294, and CRE −342, and the serum-response element (SRE). The numbering refers to the fos gene in the Ensembl database (ENSMUSG00000021250).
The LRP1-ICD Does Not Bind to the Promoter of the NMDA Target Gene c-fos
Theoretically, cleavage of the LRP1-ICD from the membrane allows the cytoplasmic tail of the receptor to directly modulate NMDA-dependent intracellular signaling, including transcriptional regulation. We therefore tested whether the LRP1-ICD directly interacts with the promoter of the NMDA target gene c-fos, which was differentially expressed in Lrp1-deficient neurons. In chromatin immunoprecipitation assays with subsequent PCR employing primers that covered the c-fos promoter with its CREs and its serum-response element (SRE), no direct interaction with the LRP1-ICD could be demonstrated (Fig. 4C). This indicates that modulation of NMDA-dependent signaling processes by LRP1 does not occur on the level of target gene promoters but is likely to take place higher upstream.
LRP1 Facilitates NMDA-induced Degradation of PSD-95, and in the Absence of LRP1, Ubiquitinated PSD-95 Is Increased, whereas NMDA-induced GluA1 Internalization Is Reduced
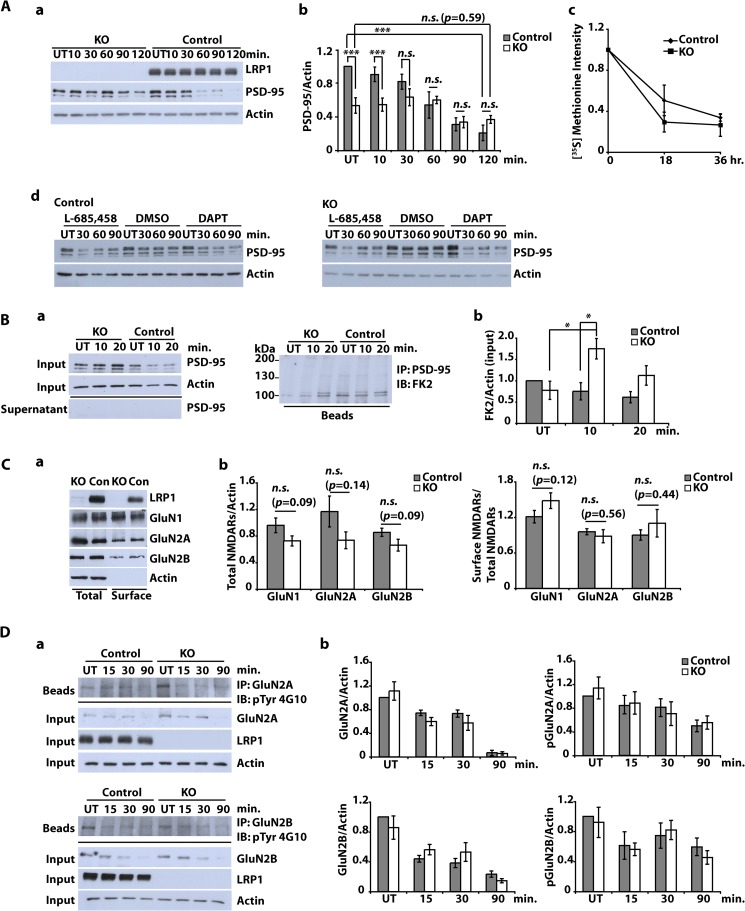
In addition to transcriptional regulation, γ-secretase-generated free ICDs are known to regulate the degradation of other proteins (23). Because the LRP1-ICD binds PSD-95 (24) and because PSD-95 is degraded in response to NMDA treatment (25), we next examined whether NMDA-induced loss of PSD-95 was LRP1-dependent and LRP1 could thus regulate the composition of postsynaptic protein complexes. Indeed, NMDA-induced degradation of PSD-95 was reduced in LRP1-deficient cortical neurons compared with control cells (Fig. 5A, a and b). Basal levels of PSD-95 were also reduced in the absence of LRP1, although turnover of PSD-95 in untreated cells was not affected by the lack of LRP1 in pulse-chase experiments (Fig. 5A, c).
FIGURE 5.
Subcellular localization of NMDA receptor subunits and NMDA-dependent PSD-95 degradation in LRP1-deficient neurons. A (a and b), primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15 LRP1wt/lox (Control) mouse embryos. On DIV 10–12, neurons were treated with NMDA (20 μm, 3 min), and whole cell lysates were prepared after the time indicated. Immunoblot analysis was performed with α-LRP1 and α-PSD-95 antibodies. Actin served as a loading control. Representative blot (a) and quantitative analysis (b) of six independent experiments are shown. Error bars, S.E. Statistical analysis was done by Student's t test (***, p < 0.001). c, cortical neurons were cultured from NesCreLRP1lox/lox (KO) and LRP1wt/lox (Control) mouse embryos and pulse-labeled with [35S]methionine on DIV 14–16. The cultures were chased with unlabeled methionine for 0, 18, or 36 h. PSD-95 was immunoprecipitated from the lysates, and samples were analyzed by SDS-PAGE and subsequent autoradiography. Densitometric analysis of the results is shown. Error bars, S.E. No statistical significance between KO and control was found by Student's t test (n = 3). d, cortical neurons from E15 LRP1lox/lox (Control) and NesCre/LRP1lox/lox (KO) animals were prepared. On DIV 12, neurons were pretreated with 1 μm DAPT, 10 μm L-685,458, or DMSO (1:1000; vehicle) for 2 h and were then further treated with NMDA (20 μm, 3 min). After the NMDA treatment, the cells were briefly washed with fresh medium and then cultured in fresh medium for the time indicated. Whole cell lysates were subjected to Western blotting to detect PSD-95 and actin. Actin served as a loading control. A representative blot of two independent experiments is shown. B, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15 LRP1wt/lox (Control) mouse embryos. On DIV 10–12, neurons were treated with NMDA (20 μm, 3 min). After further incubation for 10 or 20 min, whole cell lysates were prepared and subjected to immunoprecipitation (IP) with a PSD-95 antibody. Lysates, supernatants, and beads were analyzed by immunoblotting (IB) with the antibodies indicated. FK2, an antibody against mono- and polyubiquitin. Representative blots (a) and quantitative analysis (b) of five independent experiments are shown. Statistical analysis was done by Student's t test (*, p < 0.05). C, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15 LRP1wt/lox (Con) mouse embryos. On DIV 10–12, cell surface proteins were biotinylated. Subsequently, whole cell lysates were prepared, and biotinylated proteins were precipitated with Neutravidin. Aliquots from the lysates and precipitated proteins were analyzed by immunoblotting with anti-LRP1, -GluN1, -GluN2A, and -GluN2B antibodies. Actin served as a loading control. Representative blot (a) and quantitative analysis (b) of at least seven independent experiments are shown. Proteins in the lysate (total) were normalized to actin, and cell surface proteins were measured as biotinylated protein/total protein. Statistical analysis was done by Student's t test. n.s., not significant (p ≥ 0.05). D, primary cultured cortical neurons were prepared from E15–E16 NesCreLRP1lox/lox (KO) and E15 LRP1wt/lox (Control) mouse embryos. On DIV 10–12, cultures were treated with 50 μm NMDA for the time indicated or left untreated (UT). Whole cell lysates were prepared and subjected to immunoprecipitation with either α-GluN2A or α-GluN2B antibody. Input, supernatants, and precipitated proteins (beads) were analyzed by immunoblotting with the antibodies indicated. Actin served as a loading control. Representative blots (a) and quantitative analysis (b) of three independent experiments are shown. Statistical analysis was done by Student's t test (p ≥ 0.05).
To examine whether γ-secretase-dependent processing of LRP1 plays a role in regulating PSD-95 degradation, control and Lrp1-deficient neurons were pretreated with γ-secretase inhibitors DAPT or L-685,458 before exposure to NMDA (Fig. 5A, d). Interestingly, treatment of Lrp1-deficient cells with either inhibitor normalized NMDA-induced degradation in Lrp1-deficient cells, indicating that processing of proteins other than LRP1 influences NMDA-induced PSD-95 degradation. The fact that pretreatment with γ-secretase inhibitors did not alter PSD-95 turnover in control cells, however, still argues for a role of LRP1 cleavage in regulating PSD-95 degradation because inhibition of LRP1 processing might counterbalance the effect of other γ-secretase substrates so that PSD-95 degradation remains unchanged instead of enhanced as in Lrp1-deficient neurons.
Furthermore, NMDA treatment led to the accumulation of ubiquitinated PSD-95 in LRP1-deficient cortical neurons (Fig. 5B, a and b). Interestingly, treatment with the proteasome inhibitors MG-132 and epoxomicin blocked NMDA-induced degradation of PSD-95 neither in LRP1-deficient nor in control cells (data not shown). This indicates that degradation was non-proteasomal and that ubiquitination served a purpose other than delivery of PSD-95 to the proteasome, as found in an earlier work by Bianchetta et al. (26), who linked ubiquitination of PSD-95 to its interaction with the endocytosis machinery of neurons.
To test whether this interaction between LRP1 and PSD-95 would modulate the subcellular distribution of NMDA receptor subunits, we determined their distribution in LRP1-deficient and control neurons. Biotinylation experiments revealed no difference in NMDA receptor subunits with regard to both cell surface and total levels (Fig. 5C, a and b), indicating that LRP1 has no role in regulating NMDA receptor trafficking. In concordance with these results, there was no difference in degradation of NMDA receptor subunits and in the levels of phosphorylated NMDA receptor subunits between LRP1-deficient and control cells (Fig. 5D, a and b).
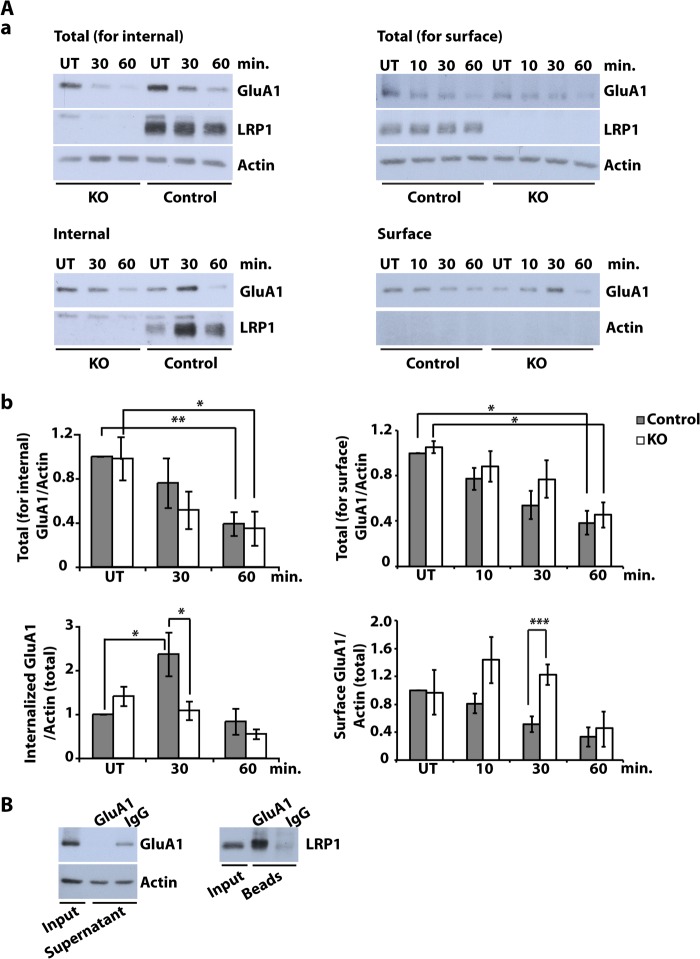
Examination of the AMPA receptor subunit GluA1, on the other hand, demonstrated greatly reduced NMDA-induced internalization of this receptor in the absence of LRP1 both by measurement of internalized protein and by biotin labeling of surface-localized subunits (Fig. 6A, a and b). Endocytosis of GluA1 and LRP1 followed a similar time course (Fig. 6A, a and c). Furthermore, we could show by co-immunoprecipitation from neuronal lysates that there is a physical interaction between LRP1 and GluA1 (Fig. 6B) that might play a role, directly or through other interacting proteins, in the effect of LRP1 on NMDA-induced GluA1 endocytosis.
FIGURE 6.
NMDA-induced GluA1 turnover is altered in LRP1-deficient neurons. A, primary cultured cortical neurons were prepared from E15 NesCreLRP1lox/lox (KO) and E15 LRP1wt/lox (Control) mouse embryos. Internal and surface GluA1 were analyzed under NMDA treatment. To measure internal GluA1, on DIV 10–12, cell surface proteins were biotinylated, and then neurons were treated with NMDA (20 μm, 3 min). Neurons were washed and incubated in fresh medium for the time indicated to allow endocytosis of GluA1. After stripping off cell surface biotin, whole cell lysates were prepared, and biotinylated proteins were precipitated with Neutravidin. To measure surface GluA1, on DIV 10–12, neurons were first treated with NMDA (20 μm, 3 min). Then neurons were washed and incubated in fresh medium for the time indicated. Subsequently, cell surface proteins were biotinylated. Whole cell lysates were prepared, and biotinylated proteins were precipitated with Neutravidin. Aliquots from the lysates and supernatants were analyzed by immunoblotting with anti-LRP1 and anti-GluA1 antibodies. Actin served as a loading control and as an indicator of purity for surface samples. Shown are representative Western blots of total and internalized or surface GluA1, respectively (a), and quantitative analysis of total (for internalized analysis) GluA1 (b), internalized GluA1 (b), total (for surface analysis) GluA1 (b), and surface GluA1 (b). All values are normalized to actin from total lysate. Bars, means ± S.E. (error bars) (Student's t test; *, p < 0.05; **, p < 0.01; ***, p < 0.001, n = 4 for internal GluA1, n = 3 for surface GluA1). B, detergent-soluble synaptosomal membrane fractions from DIV 10–12 wild type cortical neurons were prepared and subjected to immunoprecipitation with an anti-GluA1 antibody or rabbit-IgG as a negative control. Precipitated proteins were examined with an antibody against LRP1 by immunoblotting. Representative blots of five independent experiments are shown.
In summary, we found by comparison of LRP1-deficient and control neurons that LRP1 predominantly localized to dendrites of hippocampal pyramidal cells, where it regulates NMDA-dependent intracellular signaling processes. This concerns both the activation of NMDA-regulated intracellular messengers like CREB and the regulation of target genes as well as NMDA-dependent changes of the composition of postsynaptic protein complexes, including the surface localization of the AMPA receptor subunit GluA1. In addition, treatment with NMDA induces the proteolytical processing of LRP1, suggesting that release of the LRP1-ICD has a role in these regulatory functions. On a phenotypic level, the LRP1-dependent changes lead to reduced neurite outgrowth despite normal viability of LRP1-deficient neurons.
DISCUSSION
The LDL receptor gene family member LRP1 plays an essential role in the central nervous system of mammals. Conditionally Lrp1-deficient mice exhibit severe neurological phenotypes (2, 3), which are still incompletely understood on the molecular level despite recent advances in our understanding of neuronal LRP1 functions.
Here we employed a novel mouse model, where recombination of loxP-marked Lrp1 alleles takes place in neuronal precursors, which allowed us to isolate and cultivate LRP1-deficient primary cortical neurons from this line. These neurons were of normal viability and maturity but exhibited reduced branching of neurites. Basal levels of pCREB, NMDA-dependent accumulation of pCREB, and NMDA-dependent target gene transcription were reduced in LRP1-deficient neurons. LRP1, which was found to be partly located on dendritic spines in the postsynaptic density by immunocytochemistry, biochemical PSD preparation, and electron microscopy, was proteolytically processed in response to NMDA treatment, allowing the free LRP1-ICD to directly regulate intracellular signaling events. This includes NMDA-dependent degradation and ubiquitination of PSD-95, both of which were altered in LRP1-deficient neurons, and subsequent reduction in NMDA-induced endocytosis of AMPA receptor subunits, suggesting that LRP1 has a role in the regulation of synaptic transmission.
Subcellular Localization of LRP1
LRP1 is expressed throughout the central nervous system (27). Earlier studies carried out by in situ hybridization and immunohistochemistry demonstrated its presence on neurons (28). At the electron microscopic level, the subcellular localization was described as on the plasma membrane and intracellularly on ribosomes and multivesicular bodies of both neurons and glial cells (29). Our own previous immunocytochemical studies had shown LRP1 on dendritic spines co-localizing with postsynaptic proteins (2). Our current electron microscopic studies of the CA1 hippocampal area of mice confirmed the presence of LRP1 on the plasma membrane of neurons as well as on intracellular membranaceous structures like lysosomes (Fig. 2C). In addition, we could specify that LRP1 is partly located in and around postsynaptic densities on dendritic spines (Fig. 2C). The synaptic localization of LRP1 was confirmed biochemically by purification of proteins of the postsynaptic density, which showed that LRP1 could be partly retrieved with postsynaptic proteins like PSD-95 and NMDA receptor subunits (Fig. 2D), with which it co-precipitated in immunoprecipitation experiments (Fig. 2E), whereas our current immunocytochemical and confocal microscopic examinations confirmed the co-localization (Fig. 2B).
This comprehensive analysis of the subcellular localization of LRP1 shows that a fraction of the receptor is found in the vicinity or within the main body of synapses, where it is suitably located to modulate signaling events following synaptic transmission and to influence the composition of postsynaptic protein complexes. Indeed, it has been described by other investigators that LRP1 is necessary for tissue plasminogen activator-induced NMDA receptor activation and fulfills its role via its direct interaction with PSD-95 (6). In addition, earlier studies also showed that calcium influx through NMDA receptor channels is modulated by LRP1 (20, 21). These findings are especially interesting because NMDA receptor activity has been shown to be essential for normal dendritic branching (30), which we found to be defective in LRP1-deficient primary neurons (Fig. 2A), indicating that the postsynaptic fraction of LRP1 fulfills physiologically important tasks.
NMDA-dependent Intracellular Signaling Is Compromised in LRP1-deficient Neurons
LRP1 is known to modulate NMDA receptor-mediated calcium currents (6, 20). The molecular nature of the interaction is incompletely understood, although it was found to depend on LRP1 binding to PSD-95 (6). Here, we examined downstream intracellular signaling events in NMDA-treated LRP1-deficient primary neurons and found that phosphorylation of the transcription factor CREB was reduced in resting cells and that the NMDA-induced increase in phosphorylation was delayed in comparison with wild type control neurons (Fig. 3A). Subsequently, induction of NMDA target genes was reduced in the absence of LRP1 (Fig. 3B). ERK phosphorylation, on the other hand, followed a constant time course, regardless whether LRP1 was present or not (Fig. 3C). These findings indicate that LRP1 either selectively influences the fast calmodulin kinase-dependent activation of CREB with preservation of the slower ERK-mediated pathway or directly modulates the availability of pCREB. GST pull-down assays did not show a direct interaction between the LRP1-ICD and CREB (data not shown), and from chromatin immunoprecipitation with an LRP1 antibody, no association with the promoter of the NMDA target gene c-fos could be demonstrated (Fig. 4C). These findings indicate that LRP1 intervenes upstream of the pCREB nuclear action and that it selectively modulates fast CREB activation.
In contrast to our findings, Martin et al. (6) described the lack of NMDA receptor-dependent ERK activation in cells where LRP1 was reduced by shRNA-mediated gene silencing. It has to be noted, however, that this study examined tissue plasminogen activator-induced NMDA receptor gating, whereas we investigated NMDA-dependent signaling events. In the former constellation, LRP1 functions as a receptor for tissue plasminogen activator and thus relates extracellular signals via PSD-95 to the NMDA receptor channel (6). In the latter, direct NMDA receptor activation is modulated by LRP1, apparently by a feed forward mechanism that includes NMDA-induced proteolytical cleavage of LRP1.
NMDA-dependent Degradation of PSD-95 Is Delayed and Reduced in LRP1-deficient Neurons with Subsequent Reduction in AMPA Receptor Endocytosis
In addition to alteration of downstream mediators of NMDA receptor signaling, basal levels of PSD-95 and NMDA-induced degradation of PSD-95 were reduced and delayed, respectively, in LRP1-deficient cortical neurons. Furthermore, ubiquitinated PSD-95 accumulated after NMDA treatment in LRP1-deficient cells.
Ubiquitination of PSD-95 has been linked to NMDA-dependent proteasomal degradation (25), but alternative routes of PSD-95 proteolysis have also been described (31). In contrast to Colledge et al. (25), we found that, at least in our experimental system, NMDA-induced PSD-95 degradation was not proteasome-dependent because it could not be prevented by a variety of proteasome inhibitors. This is in line with the findings of Bianchetta et al. (26), who did not find proteasomal degradation of PSD-95 and suggested a non-proteolytical regulatory function for ubiquitinated PSD-95 in the regulation of NMDA-induced AMPA receptor endocytosis. Still, NMDA-induced degradation of PSD-95 was impaired in the absence of LRP1 (Fig. 5A), indicating that LRP1 modulates the composition of postsynaptic protein complexes. Specifically, the ubiquitinated fraction of PSD-95 was increased in the absence of LRP1 (Fig. 5B), suggesting dysregulation of AMPA receptor trafficking and subsequently synaptic transmission. Indeed, our experiments demonstrated that NMDA-induced endocytosis of the AMPA receptor subunit GluA1 was greatly reduced in the absence of LRP1 (Fig. 6A). These findings are especially interesting in light of recent findings that there is synaptic autoregulation by NMDA-induced metalloprotease- and γ-secretase-dependent proteolysis (32). γ-Secretase-dependent release of the LRP1-ICD is increased by NMDA (Fig. 4A), and the LRP1-ICD interacts with PSD-95 (24), indicating that LRP1 may play a central role in the γ-secretase-dependent regulatory function proposed by Restituito et al. (32). This modulation of the postsynaptic structure may also underlie the differential NMDA-induced activation of CREB and NMDA target genes in the LRP1-deficient cells, although an additional direct role for the LRP1-ICD in facilitating CREB phosphorylation is also conceivable.
Both functions of LRP1 would be compatible with the finding that it modulates long term potentiation in hippocampal slices (3) and with its presumed role in synaptic transmission underlying the severe motor and behavioral abnormalities in structurally normal conditional Lrp1-deficient mice that lack the receptor in differentiated neurons (2).
This work was supported by Deutsche Forschungsgemeinschaft Grants MA 2410/1-3 and 1-4 (to P. M.), SFB 780/TP5 (to J. H., H. B., and M. F.) and BO1806/3-1 (to H. B.).
- CRE
- cAMP-response element
- CREB
- CRE-binding protein
- pCREB
- phospho-CREB
- DAPT
- N-(N-(3,5-difluorophenacetyl)-l-alanyl)-S-phenylglycine t-butyl ester
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- ICD
- intracellular domain
- PSD
- postsynaptic density
- VLDLR
- very low density lipoprotein receptor
- E15 and E16
- embryonic day 15 and 16, respectively
- RIPA
- radioimmune precipitation assay
- ANOVA
- analysis of variance
- DIV
- day(s) in vitro
- LDH
- lactate dehydrogenase
- PB
- phosphate buffer
- TRITC
- tetramethylrhodamine isothiocyanate
- CTF
- C-terminal fragment.
REFERENCES
- 1. Lillis A. P., Van Duyn L. B., Murphy-Ullrich J. E., Strickland D. K. (2008) LDL receptor-related protein 1. Unique tissue-specific functions revealed by selective gene knockout studies. Physiol. Rev. 88, 887–918 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. May P., Rohlmann A., Bock H. H., Zurhove K., Marth J. D., Schomburg E. D., Noebels J. L., Beffert U., Sweatt J. D., Weeber E. J., Herz J. (2004) Neuronal LRP1 functionally associates with postsynaptic proteins and is required for normal motor function in mice. Mol. Cell. Biol. 24, 8872–8883 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Liu Q., Trotter J., Zhang J., Peters M. M., Cheng H., Bao J., Han X., Weeber E. J., Bu G. (2010) Neuronal LRP1 knockout in adult mice leads to impaired brain lipid metabolism and progressive, age-dependent synapse loss and neurodegeneration. J. Neurosci. 30, 17068–17078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Pietrzik C. U., Busse T., Merriam D. E., Weggen S., Koo E. H. (2002) The cytoplasmic domain of the LDL receptor-related protein regulates multiple steps in APP processing. EMBO J. 21, 5691–5700 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Kang D. E., Pietrzik C. U., Baum L., Chevallier N., Merriam D. E., Kounnas M. Z., Wagner S. L., Troncoso J. C., Kawas C. H., Katzman R., Koo E. H. (2000) Modulation of amyloid β-protein clearance and Alzheimer's disease susceptibility by the LDL receptor-related protein pathway. J. Clin. Invest. 106, 1159–1166 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Martin A. M., Kuhlmann C., Trossbach S., Jaeger S., Waldron E., Roebroek A., Luhmann H. J., Laatsch A., Weggen S., Lessmann V., Pietrzik C. U. (2008) The functional role of the second NPXY motif of the LRP1 β-chain in tissue-type plasminogen activator-mediated activation of N-methyl-d-aspartate receptors. J. Biol. Chem. 283, 12004–12013 [DOI] [PubMed] [Google Scholar]
- 7. Shi Y., Mantuano E., Inoue G., Campana W. M., Gonias S. L. (2009) Ligand binding to LRP1 transactivates Trk receptors by a Src family kinase-dependent pathway. Sci. Signal. 2, ra18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Zurhove K., Nakajima C., Herz J., Bock H. H., May P. (2008) γ-Secretase limits the inflammatory response through the processing of LRP1. Sci. Signal. 1, ra15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Zhang C., An J., Strickland D. K., Yepes M. (2009) The low-density lipoprotein receptor-related protein 1 mediates tissue-type plasminogen activator-induced microglial activation in the ischemic brain. Am. J. Pathol. 174, 586–594 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Rohlmann A., Gotthardt M., Willnow T. E., Hammer R. E., Herz J. (1996) Sustained somatic gene inactivation by viral transfer of Cre recombinase. Nat. Biotechnol. 14, 1562–1565 [DOI] [PubMed] [Google Scholar]
- 11. Frykman P. K., Brown M. S., Yamamoto T., Goldstein J. L., Herz J. (1995) Normal plasma lipoproteins and fertility in gene-targeted mice homozygous for a disruption in the gene encoding very low density lipoprotein receptor. Proc. Natl. Acad. Sci. U.S.A. 92, 8453–8457 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Bock H. H., Herz J. (2003) Reelin activates SRC family tyrosine kinases in neurons. Curr. Biol. 13, 18–26 [DOI] [PubMed] [Google Scholar]
- 13. Herz J., Hamann U., Rogne S., Myklebost O., Gausepohl H., Stanley K. K. (1988) Surface location and high affinity for calcium of a 500-kd liver membrane protein closely related to the LDL-receptor suggest a physiological role as lipoprotein receptor. EMBO J. 7, 4119–4127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Trommsdorff M., Gotthardt M., Hiesberger T., Shelton J., Stockinger W., Nimpf J., Hammer R. E., Richardson J. A., Herz J. (1999) Reeler/Disabled-like disruption of neuronal migration in knockout mice lacking the VLDL receptor and ApoE receptor 2. Cell 97, 689–701 [DOI] [PubMed] [Google Scholar]
- 15. Carlin R. K., Grab D. J., Cohen R. S., Siekevitz P. (1980) Isolation and characterization of postsynaptic densities from various brain regions. Enrichment of different types of postsynaptic densities. J. Cell Biol. 86, 831–845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cho K. O., Hunt C. A., Kennedy M. B. (1992) The rat brain postsynaptic density fraction contains a homolog of the Drosophila discs-large tumor suppressor protein. Neuron 9, 929–942 [DOI] [PubMed] [Google Scholar]
- 17. Kulik A., Nakadate K., Nyíri G., Notomi T., Malitschek B., Bettler B., Shigemoto R. (2002) Distinct localization of GABAB receptors relative to synaptic sites in the rat cerebellum and ventrobasal thalamus. Eur. J. Neurosci. 15, 291–307 [DOI] [PubMed] [Google Scholar]
- 18. Dubois N. C., Hofmann D., Kaloulis K., Bishop J. M., Trumpp A. (2006) Nestin-Cre transgenic mouse line Nes-Cre1 mediates highly efficient Cre/loxP mediated recombination in the nervous system, kidney, and somite-derived tissues. Genesis 44, 355–360 [DOI] [PubMed] [Google Scholar]
- 19. Tronche F., Kellendonk C., Kretz O., Gass P., Anlag K., Orban P. C., Bock R., Klein R., Schütz G. (1999) Disruption of the glucocorticoid receptor gene in the nervous system results in reduced anxiety. Nat. Genet. 23, 99–103 [DOI] [PubMed] [Google Scholar]
- 20. Bacskai B. J., Xia M. Q., Strickland D. K., Rebeck G. W., Hyman B. T. (2000) The endocytic receptor protein LRP also mediates neuronal calcium signaling via N-methyl-d-aspartate receptors. Proc. Natl. Acad. Sci. U.S.A. 97, 11551–11556 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Qiu Z., Strickland D. K., Hyman B. T., Rebeck G. W. (2002) α-2-Macroglobulin exposure reduces calcium responses to N-methyl-d-aspartate via low density lipoprotein receptor-related protein in cultured hippocampal neurons. J. Biol. Chem. 277, 14458–14466 [DOI] [PubMed] [Google Scholar]
- 22. May P., Reddy Y. K., Herz J. (2002) Proteolytic processing of low density lipoprotein receptor-related protein mediates regulated release of its intracellular domain. J. Biol. Chem. 277, 18736–18743 [DOI] [PubMed] [Google Scholar]
- 23. Marambaud P., Wen P. H., Dutt A., Shioi J., Takashima A., Siman R., Robakis N. K. (2003) A CBP binding transcriptional repressor produced by the PS1/ϵ-cleavage of N-cadherin is inhibited by PS1 FAD mutations. Cell 114, 635–645 [DOI] [PubMed] [Google Scholar]
- 24. Gotthardt M., Trommsdorff M., Nevitt M. F., Shelton J., Richardson J. A., Stockinger W., Nimpf J., Herz J. (2000) Interactions of the low density lipoprotein receptor gene family with cytosolic adaptor and scaffold proteins suggest diverse biological functions in cellular communication and signal transduction. J. Biol. Chem. 275, 25616–25624 [DOI] [PubMed] [Google Scholar]
- 25. Colledge M., Snyder E. M., Crozier R. A., Soderling J. A., Jin Y., Langeberg L. K., Lu H., Bear M. F., Scott J. D. (2003) Ubiquitination regulates PSD-95 degradation and AMPA receptor surface expression. Neuron 40, 595–607 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Bianchetta M. J., Lam T. T., Jones S. N., Morabito M. A. (2011) Cyclin-dependent kinase 5 regulates PSD-95 ubiquitination in neurons. J. Neurosci. 31, 12029–12035 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Bu G., Maksymovitch E. A., Nerbonne J. M., Schwartz A. L. (1994) Expression and function of the low density lipoprotein receptor-related protein (LRP) in mammalian central neurons. J. Biol. Chem. 269, 18521–18528 [PubMed] [Google Scholar]
- 28. Wolf B. B., Lopes M. B., VandenBerg S. R., Gonias S. L. (1992) Characterization and immunohistochemical localization of α2-macroglobulin receptor (low-density lipoprotein receptor-related protein) in human brain. Am. J. Pathol. 141, 37–42 [PMC free article] [PubMed] [Google Scholar]
- 29. Tooyama I., Kawamata T., Akiyama H., Kimura H., Moestrup S. K., Gliemann J., Matsuo A., McGeer P. L. (1995) Subcellular localization of the low density lipoprotein receptor-related protein (α2-macroglobulin receptor) in human brain. Brain Res. 691, 235–238 [DOI] [PubMed] [Google Scholar]
- 30. Kalb R. G. (1994) Regulation of motor neuron dendrite growth by NMDA receptor activation. Development 120, 3063–3071 [DOI] [PubMed] [Google Scholar]
- 31. Lu X., Rong Y., Baudry M. (2000) Calpain-mediated degradation of PSD-95 in developing and adult rat brain. Neurosci. Lett. 286, 149–153 [DOI] [PubMed] [Google Scholar]
- 32. Restituito S., Khatri L., Ninan I., Mathews P. M., Liu X., Weinberg R. J., Ziff E. B. (2011) Synaptic autoregulation by metalloproteases and γ-secretase. J. Neurosci. 31, 12083–12093 [DOI] [PMC free article] [PubMed] [Google Scholar]