Background: PP2A is a serine/threonine phosphatase with a central role in the pathogenesis of SLE.
Results: Suppression of PP2A mediates increased DNA methylation through the MEK/ERK/DNMT1 pathway in normal and SLE T-cells.
Conclusion: PP2A overexpression accounts for DNA hypomethylation in SLE T-cells.
Significance: Here, we propose a link between increased PP2A expression and reduced DNA methylation in SLE.
Keywords: DNA Methylation, Epigenetics, ERK, Protein Phosphatase, T-cell, SLE
Abstract
DNA hypomethylation is a characteristic feature of systemic lupus erythematosus (SLE) immune cells. Numerous reports have implicated the involvement of the MEK/ERK pathway in the reduction of DNA methyltransferase (DNMT) expression, hence inducing the transcription of methylation-sensitive genes in SLE patients. However, the molecular mechanisms involved remain unclear. Here, we investigated whether the catalytic subunit of protein phosphatase 2A (PP2Ac), which is overexpressed in SLE T-cells, contributes to reduced DNA methylation. We show that both chemical suppression and siRNA silencing of PP2Ac in T-cells resulted in sustained phosphorylation of MEK and ERK following stimulation with phorbol 12-myristate 13-acetate and ionomycin. Furthermore, PP2Ac suppression resulted in increased DNMT enzyme activity, DNA hypermethylation, and decreased expression of methylation-sensitive genes. Similarly, in SLE T-cells, suppression of PP2Ac resulted in increased MEK/ERK phosphorylation, enhanced DNMT1 expression and suppressed expression of the methylation-sensitive CD70 gene. Our results demonstrate that PP2A regulates DNA methylation by influencing the phosphorylation of MEK/ERK. We propose that enhanced PP2Ac in SLE T-cells may dephosphorylate and activate the signaling pathway upstream of DNMT1, thus disturbing the tight control of methylation-sensitive genes, which are involved in SLE pathogenesis.
Introduction
Systemic lupus erythematosus (SLE)4 is an autoimmune disease that affects mainly women. It is complex and multifaceted in its pathogenesis and affects various organs, including kidneys, skin, joints, brain, heart, and lungs (1). Underlying abnormalities of the immune system are the leading cause of the clinical presentation in SLE (2). In particular, signaling defects in T-cells have been implicated in the pathogenesis of this disease (3).
Protein phosphatase 2A (PP2A) is a ubiquitously expressed and highly conserved serine/threonine phosphatase that plays an essential role in multiple cellular processes, including cell division, cytoskeletal dynamics, and various signaling pathways (4). The PP2A core enzyme consists of the scaffold subunit A and the catalytic subunit C. To form a functional holoenzyme, the core enzyme interacts with one of the many regulatory subunits, thus defining its specificity (5, 6). We have previously demonstrated that PP2A mRNA and protein levels, as well as the activity of the catalytic subunit (PP2Ac), are increased in T-cells isolated from SLE patients compared with healthy controls (7, 8). This enhanced expression and activity of PP2Ac have been shown to be responsible for some of the key signaling defects seen in SLE T-cells such as reduced expression of the T-cell receptor-associated CD3ζ chain and diminished production of the cytokine IL-2 (9). Furthermore, it dephosphorylates and activates the transcriptional enhancer of CREM, the specificity protein SP-1 (10). Thus, PP2Ac is a central component of the molecular mechanisms contributing to SLE.
Another hallmark of SLE T-cells is the general hypomethylated state of genomic DNA. The degree of hypomethylation has been shown to correlate with disease activity (11). Several genes, including ICAM1, CD70, and CD11a, are hypomethylated and overexpressed in SLE T-cells (12). DNMT1, the key methyltransferase responsible for DNA remethylation during cell division, has been demonstrated to be decreased in SLE T-cells, thus contributing to the hypomethylated state of DNA. Although the exact mechanisms leading to hypomethylation in SLE remain unknown, several studies have implicated a defective MAPK/ERK signaling pathway to contribute to reduced DNMT1 expression and function (13, 14).
In this study, we provide a link between two hallmarks of SLE T-cells that contribute to autoimmune pathology: the increased expression of PP2Ac and the general hypomethylated state of DNA. We show for the first time that the activity/expression of PP2A controls the expression of DNMT1 through the MEK/ERK signaling pathway and thus influences the expression of methylation-sensitive genes in normal and SLE T-cells.
EXPERIMENTAL PROCEDURES
Patients Enrolled in the Study and T-cell Isolation
A total of 27 patients were enrolled in this study. The patient demographics and treatment of 6 of the 27 patients are provided in Table 1. The rest of the 21 patients have been described elsewhere (15).
TABLE 1.
Patient demographics and treatment
SLEDAI, SLE disease activity index; F, female; H, hydroxychloroquine (milligrams); MTX, methotrexate (milligrams); A, azathioprine (milligrams); M, mycophenolate mofetil (milligrams).
| Age | SLEDAI | Race | Gender | Prednisone | Others |
|---|---|---|---|---|---|
| mg/day | |||||
| 28 years old | 0 | White | F | 0 | 0 |
| 51 years old | 0 | African American | F | 0 | H400 + MTX25 |
| 24 years old | 2 | African American | F | 10 | H400 + A100 |
| 42 years old | 0 | African American | F | 0 | H400 |
| 45 years old | 4 | White | F | 40 | M1000 |
| 29 years old | 2 | African American | F | 0 | H400 + M1000 |
Primary T-cells were purified from peripheral venous blood obtained from healthy volunteers as well as from patients. The blood was incubated for 30 min with a rosette T-cell purification kit (STEMCELL Technologies) that contained a tetrameric Ab mixture against CD14, CD16, CD19, CD56, and glycophorin A that attaches non-T-cells to erythrocytes. Ficoll-containing Lymphoprep gradient (Mediatech Inc., Manassas, VA) was subsequently used to separate these complexes from T-cells. T-cells were cultured in RPMI 1640 medium (Mediatech Inc.) supplemented with 2 mm l-glutamine, 10% fetal calf serum (Quality Biological), 50 units/ml penicillin (Sigma-Aldrich), and 50 μg/ml streptomycin (Sigma-Aldrich) and maintained in a humidified incubator (37 °C, 5% CO2). Studies were approved by the Institutional Review Board of the Beth Israel Deaconess Medical Center. Unless noted otherwise, the experiments were carried out with multiple donors (n = 6).
Inhibition/Knockdown of PP2A and Sample Preparations
We studied the effect of PP2A in T-cells using two different approaches. The first approach involved the pretreatment of T-cells with a chemical inhibitor of PP2A, okadaic acid (OA). Briefly, 5 × 106 T-cells were treated overnight with 5 nm OA (Upstate) before stimulation with phorbol 12-myristate 13-acetate (PMA; 5 ng/ml; Sigma-Aldrich) and ionomycin (1 μm; Sigma-Aldrich). The second approach was siRNA-mediated knockdown of PP2Ac. A combination of PP2Aca siRNA (Ambion s10957, s10958, and s10959) and PP2Acb siRNA (Ambion s10961) or the corresponding negative control siRNA-1 (Ambion AM4635) was transfected into 1 × 107 T-cells using an Amaxa Nucleoporator according to the manufacturer's instructions. For both PP2Ac siRNAs as well as the control siRNA, three individual transfections with 1 × 107 T-cells each were done. The cells were then cultured in 6-well plates in RPMI 1640 medium for 64 h. A total 3 × 107 cells from each group were collected, washed once with PBS, and then plated into 12-well plates (5 × 106 T-cells in 1 ml of fresh medium/well). Cells were stimulated with 5 ng/ml PMA and 1 μm ionomycin for 8 h. An equivalent volume of dimethyl sulfoxide was added to the unstimulated control. Cells were harvested for DNA and RNA purification using an AllPrep RNA/DNA/protein mini kit (Qiagen). Nuclear protein extracts were made using an EpiQuik nuclear extraction kit (Epigentek), and whole cell lysates were made using lysis buffer (1 mm EDTA (Boston BioProducts), 0.5% Triton X-100, 5 mm NaF (Sigma-Aldrich), 6 m urea, 25 μg/ml leupeptin (Sigma-Aldrich), 25 μg/ml pepstatin (Sigma-Aldrich), 3 μg/ml aprotinin (Sigma-Aldrich), 100 μm PMSF, and 1 mm activated sodium orthovanadate (Sigma-Aldrich) in PBS) for the different assays.
Quantification of Protein Levels
Whole cell lysates were used for Western blotting to assess the protein levels. Briefly, equal amounts of protein were loaded onto 4–12% Bis-Tris NuPAGE precast gels (Invitrogen) and then transferred to PVDF membrane. The membrane was blotted with the appropriate primary antibodies, which were purchased from Cell Signaling (phospho-MEK1/2 rabbit Ab (9154), MEK1/2 rabbit Ab (9122), phospho-ERK1/2 rabbit Ab (4370), ERK1/2 rabbit Ab (4695), and PP2Ac rabbit Ab (2038)), Abcam (DNMT1 rabbit Ab (ab16632)), and Sigma-Aldrich (β-actin rabbit Ab (A5060)). After incubation with the corresponding HRP-conjugated secondary antibody (Santa Cruz Biotechnology), protein bands were detected by enhanced chemiluminescence reagents (Amersham Biosciences). The digital images were scanned using a Fuji LAS-3000 scanner, and the density of each band was measured using Quantity One software. The densitometric ratio of phosphorylated protein to total protein or target protein to housekeeping protein was calculated for the semiquantification. Furthermore, the phospho-ERK/ERK ratio was also determined by ELISA (phospho-ERK1 and total ERK1 DuoSet IC, R&D Systems) according to the manufacturer's protocol.
Measurement of DNMT Enzyme Activity
DNMT enzyme activity in the nuclear extract described above was assessed using an EpiQuik DNMT activity/inhibition assay ultra kit (Epigentek) according to the manufacturer's protocol. The concentration of nuclear proteins was measured by the BCA method (Thermo Scientific), and an equal amount of nuclear extract was reacted with enzyme substrate (S-adenosylmethionine) for 90 min. Subsequently, capture antibody against 5-methylcytosine (5-mC) and then detection antibody were added. After the final washing step, the reaction was developed. Absorbance values were measured and quantified using a standard curve. All samples were run in duplicates, and the average was used for analysis.
Global DNA Methylation Analysis
DNA purified from T-cells described above was used for quantification of methylated DNA using a MethylFlash methylated DNA quantification kit (Epigentek) according to the manufacturer's instructions. To determine the relative methylation status of each DNA sample, the percentage of 5-mC in sample DNA was quantified using the absorbance value of the positive control. All samples were run in duplicates, and the average was used for analysis.
Methylated DNA Immunoprecipitation
The methylated DNA immunoprecipitation assay was carried out according to the recommendations of Zymo Research Corp. (16). Briefly, genomic DNA from T-cells of patients or healthy controls and from T-cells treated with various stimuli was isolated using the AllPrep RNA/DNA/protein mini kit. The DNA thus obtained was sheared to ∼200-bp fragments using DNA Shearase (Zymo Research Corp.), and 100 ng of this sheared DNA was used for immunoprecipitation of methylated DNA. Real-time PCR was performed with the methylated DNA using an ABI OneStepPlus system. Equal amounts of completely (100%) methylated human DNA and demethylated human DNA (Zymo Research Corp.) acted as input and negative controls, respectively.
Reverse Transcription and Real-time PCR
Total RNA (300 ng) was transcribed into cDNA in a conventional thermocycler using High Yield PCR EcoDry PremixTM (Clontech). Real-time PCR was performed in duplicate for every sample with a LightCycler® 480 system by adding SYBR Green (Roche Applied Science) to the reaction mixture. The following primers were used: human PP2Aca, 5′-TCCGAGTCCCAGGTCAAGAG-3′ (forward) and 5′-GCTACAAGCAGTGTAACTGTTTCA-3′ (reverse); human PP2Acb, 5′-AACGAGAACCAAGTGCGGAC-3′ (forward) and 5′-TAATGCTACAAGAAGAGTCACAGTC-3′ (reverse); human CD70, 5′-TACGTATCCATCGTGATG-3′ (forward) and 5′-GTTGGTGCAGAGTGTGTC-3′ (reverse); human ITGAL, 5′-GTCAGCTCATCATCCGAAACTG-3′ (forward) and 5′-AGACTGCAAGGTGCAGACACA-3′; and GAPDH, 5′-CAACTACATGGTTTACATGTTCC-3′ (forward) and 5′-GGACTGTGGTCATGAGTCCT-3′ (reverse). The averaged cycle threshold values of each reaction derived from the target gene, determined with LightCycler® 480 system software, were normalized to GAPDH levels. Cycle threshold values were used to calculate relative mRNA expression by the ΔCt relative quantification method.
Statistics
Data are presented as means ± S.E. Paired two-tailed Student's t tests were used for statistical analysis. Statistical significance was defined as p < 0.05.
RESULTS
Silencing of PP2A or Inhibition of Its Activity Results in Enhanced MEK/ERK Activation
To study the effect of PP2A in T-cells, we employed a two-pronged approach. First, we knocked down PP2Ac using siRNAs targeting both the PP2Aca and PP2Acb isoforms of PP2Ac. As shown in Fig. 1A, siRNA treatment of T-cells resulted in a 90% reduction in PP2Aca and PP2Acb mRNA levels. The reduction in PP2A protein levels was assessed by Western blotting of T-cell lysates (Fig. 1B). Densitometric analysis showed a 60% decrease in protein levels. In addition to siRNA-mediated knockdown of PP2A protein levels, we treated the cells with OA, a specific chemical inhibitor of PP2A (7).
FIGURE 1.
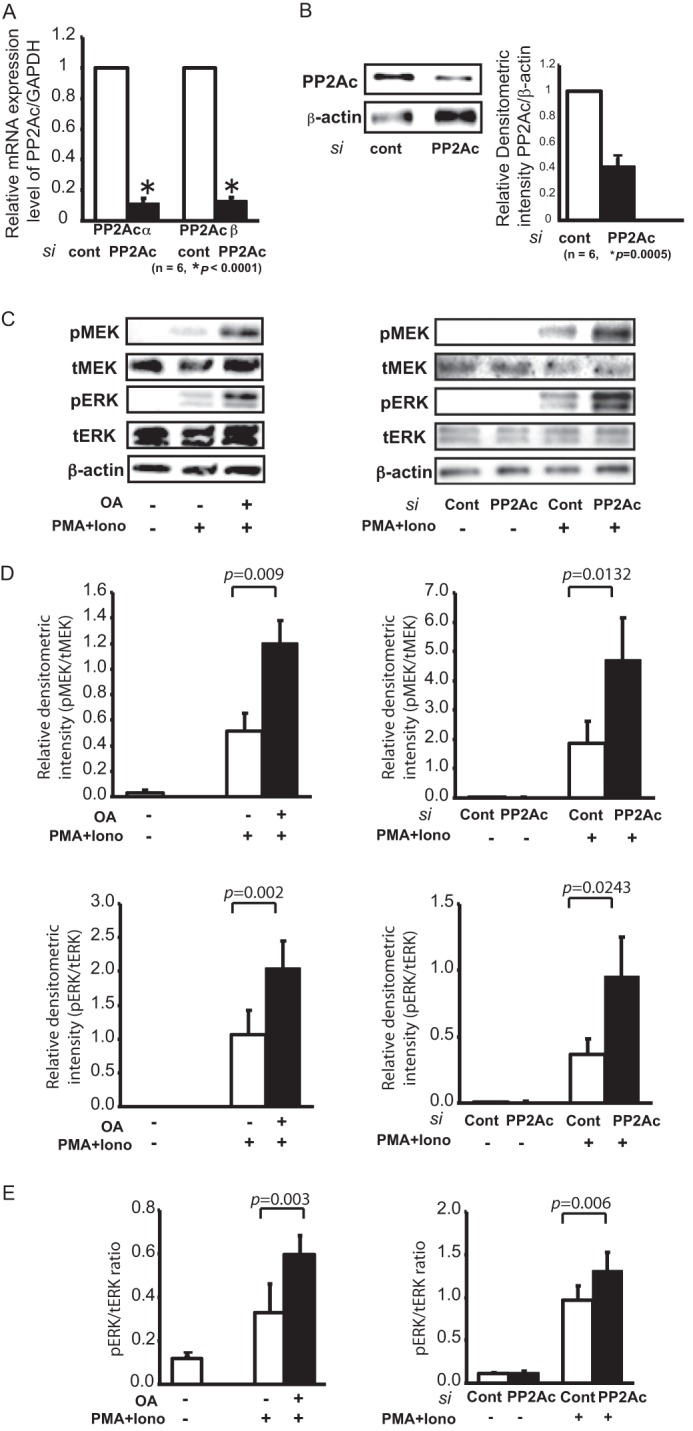
Silencing of PP2A or inhibition of its activity leads to enhanced MEK/ERK activation. A, efficacy of PP2Ac suppression by siRNA (si). A 300 nm combination of PP2Aca and PP2Acb siRNAs or the corresponding negative control (cont) siRNA was transfected into 1 × 107 T-cells by electroporation and incubated on 6-well plates for 64 h before stimulation. Both isoforms of PP2Ac transcripts were measured and normalized to GAPDH by real-time RT-PCR. We confirmed significant suppression of PP2Ac compared with control samples. The results represent the mean ± S.E. of six independent experiments. B, the suppression of PP2Ac protein was also determined by Western blotting. The densitometric ratio of PP2Ac to β-actin was calculated for the semiquantification. The results represent the mean ± S.E. of six independent experiments. C, lysates prepared from T-cells either treated overnight with OA (left panel) or transfected with PP2Ac siRNA (right panel) were used for immunoblotting to compare the relative phosphorylation levels of MEK (pMEK) and ERK (pERK) upon stimulation with 5 ng/ml PMA and 1 μm ionomycin (Iono) for 8 h. The basal level of each protein is also shown. Total ERK (tERK) and MEK (tMEK) proteins and β-actin served as controls. The blots represent one of six individual experiments. D, the densitometric ratio of phosphorylated MEK and ERK to the corresponding total protein was calculated for the semiquantification of OA-treated (left panels) and PP2Ac siRNA-treated (right panels) samples. The results represent the mean ± S.E. of six independent experiments. E, quantification of ERK phosphorylation by ELISA. The absorbance values of samples were measured using a phospho-ERK and total ERK ELISA system and converted to the concentration using a standard curve. The ratio of phospho-ERK to total ERK is shown. Left panel, OA treatment; right panel, PP2Ac siRNA transfection. The results represent the mean ± S.E. of six independent experiments.
T-cells isolated from SLE patients have been reported to exhibit impaired MEK/ERK signaling. To study the role of PP2A in MEK/ERK signaling, T-cells were isolated from healthy donors and either treated with OA or subjected to PP2A-specific siRNA. T-cells were stimulated with PMA and ionomycin, and activation of the MEK/ERK pathway was assessed. When activated, MAP2K/MEK and MAPK/ERK undergo phosphorylation. Phosphorylation was assessed by Western blotting (Fig. 1, C and D) and an ELISA-based assay (Fig. 1E). As shown in Fig. 1C, both OA treatment (left panel) and siRNA-mediated PP2A knockdown (right panel) resulted in an increase of MEK and ERK phosphorylation. Densitometric analysis was used to quantify the differences in the band intensity of phosphorylated MEK and ERK (Fig. 1D). Phosphorylation of ERK was also quantified using ELISA and confirmed the observations made by immunoblotting. As shown in Fig. 1E, cells treated with OA or PP2A siRNA exhibited higher levels of phosphorylated ERK. Total ERK was used for normalization in all samples. Thus, knockdown of PP2A or inhibition of its activity leads to amplified MEK/ERK signaling.
Suppression of PP2A Enhances DNMT1 Expression and Activity
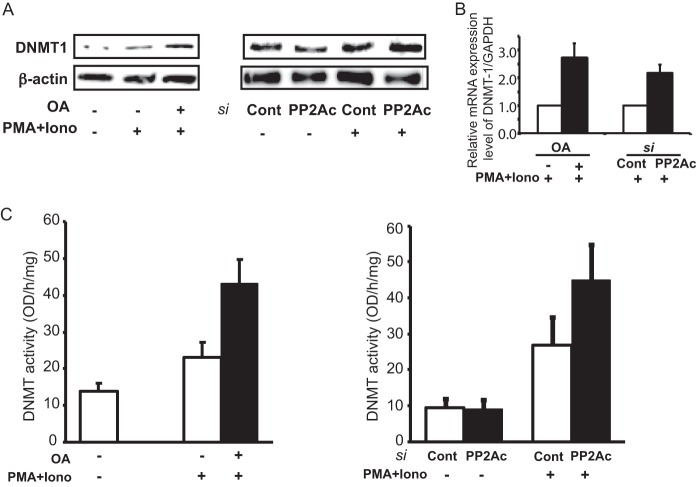
MEK/ERK activation is known to affect DNMT1 expression. Because the suppression of PP2A results in an enhanced MEK/ERK response, we tested its effects on DNMT1 expression and activity. T-cells treated with OA or PP2Ac-directed siRNA were stimulated with PMA and ionomycin. The expression levels of DNMT1 were then evaluated by immunoblotting. As shown in Fig. 2A, both OA treatment (left panel) and siRNA knockdown (right panel) resulted in increased DNMT1 expression. We also assessed the effect of PP2A suppression on DNMT1 mRNA and found that both OA treatment and siRNA-mediated knockdown of PP2Ac led to an increase in DNMT1 message (Fig. 2B).
FIGURE 2.
Suppression of PP2Ac enhances DNMT1 expression levels and enzyme activity. A, the relative expression level of DNMT1 protein against β-actin from whole cell extracts was assessed by Western blotting. Left panel, OA treatment; right panel, PP2Ac siRNA transfection. Iono, ionomycin; si, siRNA; Cont, control. B, the transcript levels of DNMT1 were measured and normalized to GAPDH by real-time PCR. The results represent the mean ± S.E. of six independent experiments. C, for the quantification of enzyme activity of DNMTs, nuclear extract was reacted with enzyme substrate and the ELISA system using 5-mC antibody. Left panel, OA treatment; right panel, PP2Ac siRNA transfection. The results represent the mean ± S.E. of six independent experiments.
Next, we aimed to determine the effect of PP2A suppression on the enzyme activity of DNMT1. Nuclear extracts from all samples were used in an ELISA utilizing 5-mC antibodies to quantify the methyltransferase activity of DNMT1. The samples in which PP2A had been suppressed had higher enzyme activity than the control samples (Fig. 2C, left (OA treatment) and right (PP2Ac siRNA transfection) panels). Thus, inhibition of PP2A activity mediates an increase in DNMT1 expression levels and enzyme activity.
PP2A Suppression Mediates Global DNA Methylation
The enhanced DNMT1 activity and expression we observed as a consequence of PP2A suppression led us to investigate its effect on global DNA methylation. DNMT1 is the key maintenance methyltransferase in mammals, and as such, a change in its expression or activity affects the levels of global DNA methylation (12). Nuclear extracts were prepared from T-cells treated with OA or subjected to siRNA-mediated knockdown of PP2A and subsequently stimulated with PMA and ionomycin. All samples were analyzed for the percentage of methylated DNA (5-mC percent) compared with total DNA using ELISA. Cells treated with vehicle control or control siRNA served as internal controls. As shown in Fig. 3, both OA treatment (left panel) and transfection with PP2A siRNA (right panel) resulted in an increase in the percentage of methylated DNA, as reflected in the 5-mC percent. Thus, PP2A suppression, either by chemical inhibition of its activity or by siRNA-mediated knockdown, mediates reduced DNA methylation through its effects on DNMT1.
FIGURE 3.
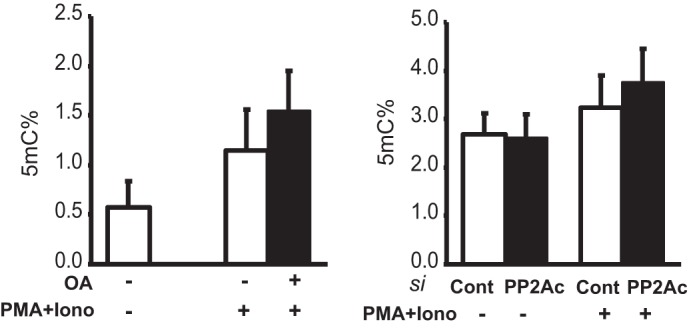
PP2Ac suppression promotes global DNA methylation. Each DNA extract was incubated with 5-mC antibody, and ELISA was used to calculate the percentage of 5-mC to total DNA in the case of both OA treatment (left panel) and PP2Ac siRNA transfection (right panel). The results represent the mean ± S.E. of six independent experiments. Iono, ionomycin; si, siRNA; Cont, control.
Chemical Inhibition of PP2A or siRNA-enabled Silencing Reduces Expression of Methylation-sensitive Genes
Increased DNA methylation within regulatory regions of methylation-sensitive genes results in reduced gene expression, whereas hypomethylation activates gene expression. Several genes known to be involved in the pathogenesis of SLE are hypomethylated and overexpressed (17, 18). Notable examples are perforin, CD11a (ITGAL), and CD70 (19, 20). We analyzed the methylation patterns of CD70 and ITGAL, as both of these genes have conserved CpG-rich regions (Fig. 4A) (21, 22). Using methylated DNA immunoprecipitation, we assessed the methylation patterns of CD70 and ITGAL in genomic DNA isolated from 21 SLE patients and 11 healthy controls. The methylation of CpG DNA at the CD70_ROI2 (where ROI is region of interest) locus was negligible in both patients and controls. However, in the case of both CD70_ROI1 and ITGAL_ROI1, patients displayed less DNA methylation compared with healthy controls, and this effect was more pronounced in patients with active disease (SLE disease activity index > 8) (Fig. 4B). Thus, as reported previously, both CD70 and ITGAL are hypomethylated in active SLE patients (19, 20). We also looked at the effect of PP2A suppression on the methylation patterns of the same regulatory regions in CD70 and ITGAL. As shown in Fig. 4C, both OA treatment and siRNA-mediated knockdown of PP2A resulted in significantly increased DNA methylation across all of the tested regions. Thus, the enhanced expression and activity of DNMT1 demonstrated above translate into increased methylation of methylation-sensitive genes such as CD70 and ITGAL.
FIGURE 4.
Suppression of PP2Ac reduces expression of methylation-sensitive genes CD70 and ITGAL. A, CpG sites within the regulatory regions of CD70 and ITGAL are shown. ROI, region of interest. B, DNA from T-cells of 21 SLE patients and 11 healthy individuals was subjected to methylated CpG DNA immunoprecipitation. Methylated DNA was recovered, and the indicated regions were amplified by real-time PCR. Completely methylated (input, 100%) and unmethylated human DNA samples were included as controls. Values are given as means ± S.D. SLEDAI, SLE disease activity index. C, OA-treated (left panel) or PP2Ac siRNA-treated (right panel) T-cells were stimulated with PMA/ionomycin for 8 h and subjected to methylated CpG DNA immunoprecipitation. Methylated DNA was recovered, and the indicated regions were amplified by real-time PCR. Completely methylated (input, 100%) and unmethylated human DNA samples were included as controls. Values are given as means ± S.D. of six independent experiments. si, siRNA; Cont, control. D, the transcript levels of CD70 and ITGAL were measured and normalized to GAPDH by real-time PCR. Upon PMA and ionomycin (Iono) stimulation, the expression level of CD70 in T-cells treated with OA (left black bar) or transfected with PP2Ac siRNA (right black bar) was decreased compared with each corresponding control (white bars). The results represent the mean ± S.E. of six independent experiments.
Next, we determined the expression of CD70 and ITGAL to confirm the downstream effect of enhanced methylation. As expected, in cells treated with OA or transfected with PP2Ac-directed siRNA, the mRNA expression of both CD70 (Fig. 4D, left panel) and ITGAL (Fig. 4D, right panel) was significantly lower compared with cells treated with vehicle control or scrambled control siRNA. This suggests that the increased DNMT1 expression and activity that we recorded contribute to reduced expression of methylation-sensitive genes such as CD70 and ITGAL. Conversely, in SLE patients, enhanced expression of PP2Ac may contribute to hypomethylation of DNA and increased expression of methylation-sensitive genes involved in the pathogenesis of SLE. However, we would like to note that DNMT1 has been known to suppress gene expression via mechanisms independent of its catalytic activity (23), and whether such mechanisms contribute to the effects we observed in this study merits further investigation.
Suppression of PP2A in SLE T-cells Enhances MEK/ERK Signaling and Reduces Expression of Methylation-sensitive Genes
All of the experiments described above were carried out in T-cells from healthy controls. Thus, we aimed to extend our key findings to T-cells isolated from SLE patients. To determine the effect of PP2Ac suppression on SLE T-cells, we either treated the cells with OA or transfected them with PP2Ac siRNA. Cells were then stimulated with PMA and ionomycin, and the protein and RNA samples were prepared for various analyses. Similar to healthy control T-cells, PP2A suppression enhanced MEK/ERK signaling in SLE T-cells (Fig. 5A). Moreover, cells treated with OA or transfected with PP2A siRNA displayed increased levels of DNMT1 mRNA (Fig. 5B) and consequently lower expression of the methylation-sensitive gene CD70 (Fig. 5C). Thus, as in healthy control T-cells, suppression of PP2A in SLE T-cells enhances MEK/ERK signaling and DNMT1 expression.
FIGURE 5.
Suppression of PP2A in SLE T-cells enhances MEK/ERK signaling and reduces expression of methylation-sensitive genes. A, T-cells from three SLE patients were treated with OA (left panel) or transfected with PP2Ac siRNA (right panel), followed by stimulation with PMA and ionomycin (Iono), and the lysates thus obtained were analyzed for ERK and MEK phosphorylation (p) via Western blotting. Total ERK and MEK protein levels and β-actin served as controls. The blots represent one of three independent experiments. si, siRNA; Cont, control. B and C, RNA samples from the same experiment were used to analyze the expression levels of DNMT1 (B) and CD70 (C) using real-time PCR. The transcript levels were normalized to GAPDH.
DISCUSSION
In this study, we have presented, for the first time, a link between DNA hypomethylation (one of the hallmarks of SLE) and the increased expression of PP2Ac seen in SLE patients. We demonstrated that suppression of PP2Ac results in increased MEK/ERK signaling. Furthermore, we showed that the suppression of PP2Ac, through its effect on the MEK/ERK pathways, contributes to enhanced expression and activity of DNMT1, which lead to an increase in the amount of methylated DNA. Thus, we have demonstrated that PP2A may affect DNA methylation in SLE T-cells through its effects on the MEK/ERK pathway.
SLE is a multifactorial autoimmune disease characterized by inflammatory damage to various organs (24). At the molecular level, there are a number of immune irregularities that are the basis of disease pathogenesis. One of the distinctive features of SLE T-cells is their general state of DNA hypomethylation (25). Numerous genes are hypomethylated, which results in increased gene expression in immune cells from SLE patients. These include cytokine genes such as IL6, IL4, IL10, and IL17A; costimulatory molecules such as ITGAL and CD70; and proinflammatory genes such as IFNGR2 (interferon-gamma receptor 2) and MMP14 (matrix metalloproteinase 14), to name a few (19, 20, 26–28). Increased expression of these and other genes contributes to the aberrant immune responses seen in SLE. Environmental and host factors that affect the methylation status of immune cells may thus play a role in the pathogenesis of SLE. Indeed, in vitro and in vivo studies using DNA-demethylating drugs, including hydralazine and procainamide, support the role of DNA methylation in the pathophysiology of SLE (29, 30).
A number of studies have documented impaired MEK/ERK signaling in SLE T-cells as one of the factors affecting the expression of the major maintenance methyltransferase DNMT1, thus affecting DNA methylation (13, 14). Furthermore, a defect in PKCδ in T-cells from SLE patients has been shown to affect DNMT1 activation (31, 32). Here, we have demonstrated that the levels of PP2A in T-cells can affect DNA methylation through its effects on MEK/ERK signaling. PP2Ac has been shown to be expressed at higher levels and to possess greater activity in SLE T-cells. However, to our knowledge, there has been no report establishing the connection between increased PP2Ac expression and reduced DNA methylation.
PP2A suppression using chemical inhibitors and siRNAs consistently demonstrated that lower expression or activity of PP2Ac enhances MEK/ERK signaling. Increased ERK signaling results in increased DNMT1 expression and activity. Furthermore, PP2A suppression mediates increased global DNA methylation and subsequently reduced expression of methylation-sensitive genes, including CD70 and ITGAL.
Extending our findings to T-cells from SLE patients, we documented that PP2A suppression leads to a similar pattern of signaling modulation in SLE T-cells compared with healthy donor T-cells. We propose that the higher expression of PP2A seen in SLE patients inhibits MEK/ERK signaling, which in turn inactivates DNMT1 and results in hypomethylation of genomic DNA. The aberrant methylation pattern of genes such as CD70 and ITGAL contributes to their overexpression, adding to the pathophysiology of SLE. Our findings are summarized in Fig. 6, in which, in addition to outlining the known effects of PP2A, we also propose a model for the link between PP2A and DNA hypomethylation. We have previously reported that PP2Ac itself is regulated through DNA methylation around a cAMP response element-binding site located in the proximal promoter (33), and therefore, PP2A may represent a potent accelerator of DNA demethylation through a positive feedback mechanism.
FIGURE 6.
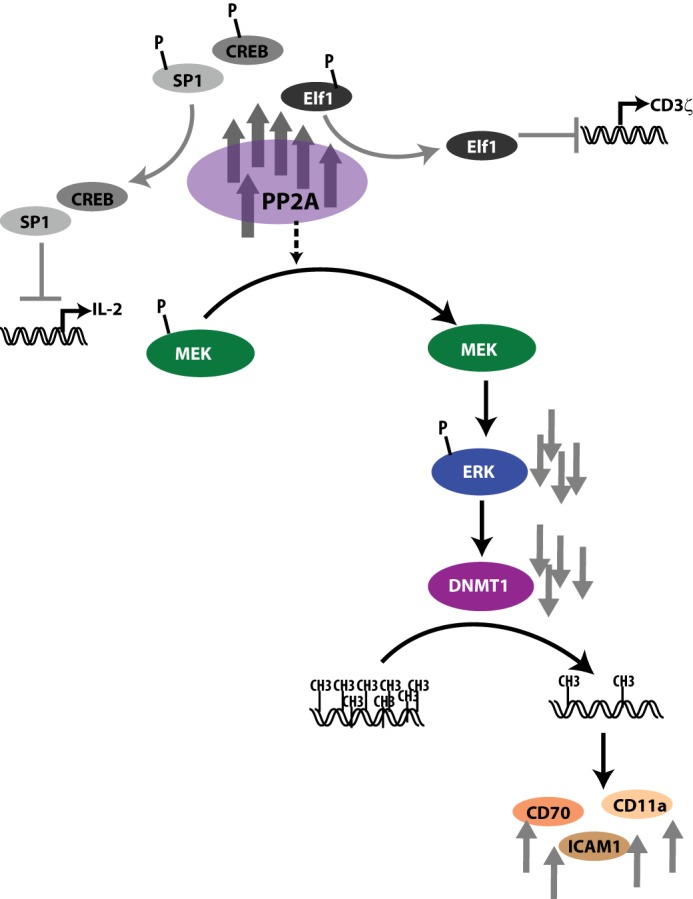
Model showing the effects of PP2A in SLE T-cells. In addition to dephosphorylating and activating SP-1 (resulting in increased CREM activity) and dephosphorylating Elf1 (resulting in decreased CD3ζ transcription), increased PP2A expression and activity dephosphorylate MEK, resulting in decreased ERK phosphorylation and decreased DNMT1 activity and DNA methylation. CREB, cAMP response element-binding protein.
Footnotes
- SLE
- systemic lupus erythematosus
- PP2A
- protein phosphatase 2A
- PP2Ac
- PP2A catalytic subunit
- DNMT
- DNA methyltransferase
- OA
- okadaic acid
- PMA
- phorbol 12-myristate 13-acetate
- Bis-Tris
- 2-[bis(2-hydroxyethyl)amino]-2-(hydroxymethyl)propane-1,3-diol
- 5-mC
- 5-methylcytosine.
REFERENCES
- 1. Tsokos G. C. (2011) Systemic lupus erythematosus. N. Engl. J. Med. 365, 2110–2121 [DOI] [PubMed] [Google Scholar]
- 2. Crispín J. C., Kyttaris V., Juang Y. T., Tsokos G. C. (2007) Systemic lupus erythematosus: new molecular targets. Ann. Rheum. Dis. 66, iii65–iii69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Crispín J. C., Kyttaris V. C., Terhorst C., Tsokos G. C. (2010) T cells as therapeutic targets in SLE. Nat. Rev. Rheumatol. 6, 317–325 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Virshup D. M. (2000) Protein phosphatase 2A: a panoply of enzymes. Curr. Opin. Cell Biol. 12, 180–185 [DOI] [PubMed] [Google Scholar]
- 5. Crispín J. C., Apostolidis S. A., Finnell M. I., Tsokos G. C. (2011) Induction of PP2A Bβ, a regulator of IL-2 deprivation-induced T-cell apoptosis, is deficient in systemic lupus erythematosus. Proc. Natl. Acad. Sci. U.S.A. 108, 12443–12448 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Janssens V., Goris J. (2001) Protein phosphatase 2A: a highly regulated family of serine/threonine phosphatases implicated in cell growth and signalling. Biochem. J. 353, 417–439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Katsiari C. G., Kyttaris V. C., Juang Y. T., Tsokos G. C. (2005) Protein phosphatase 2A is a negative regulator of IL-2 production in patients with systemic lupus erythematosus. J. Clin. Invest. 115, 3193–3204 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Sunahori K., Juang Y. T., Kyttaris V. C., Tsokos G. C. (2011) Promoter hypomethylation results in increased expression of protein phosphatase 2A in T cells from patients with systemic lupus erythematosus. J. Immunol. 186, 4508–4517 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Juang Y. T., Wang Y., Jiang G., Peng H. B., Ergin S., Finnell M., Magilavy A., Kyttaris V. C., Tsokos G. C. (2008) PP2A dephosphorylates Elf-1 and determines the expression of CD3ζ and FcRγ in human systemic lupus erythematosus T cells. J. Immunol. 181, 3658–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Juang Y. T., Rauen T., Wang Y., Ichinose K., Benedyk K., Tenbrock K., Tsokos G. C. (2011) Transcriptional activation of the cAMP-responsive modulator promoter in human T cells is regulated by protein phosphatase 2A-mediated dephosphorylation of SP-1 and reflects disease activity in patients with systemic lupus erythematosus. J. Biol. Chem. 286, 1795–1801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Richardson B., Scheinbart L., Strahler J., Gross L., Hanash S., Johnson M. (1990) Evidence for impaired T cell DNA methylation in systemic lupus erythematosus and rheumatoid arthritis. Arthritis Rheum. 33, 1665–1673 [DOI] [PubMed] [Google Scholar]
- 12. Liu Y., Chen Y., Richardson B. (2009) Decreased DNA methyltransferase levels contribute to abnormal gene expression in “senescent” CD4+CD28− T cells. Clin. Immunol. 132, 257–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Deng C., Kaplan M. J., Yang J., Ray D., Zhang Z., McCune W. J., Hanash S. M., Richardson B. C. (2001) Decreased Ras-mitogen-activated protein kinase signaling may cause DNA hypomethylation in T lymphocytes from lupus patients. Arthritis Rheum. 44, 397–407 [DOI] [PubMed] [Google Scholar]
- 14. Gorelik G., Richardson B. (2009) Aberrant T cell ERK pathway signaling and chromatin structure in lupus. Autoimmun. Rev. 8, 196–198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Hedrich C. M., Crispin J. C., Rauen T., Ioannidis C., Apostolidis S. A., Lo M. S., Kyttaris V. C., Tsokos G. C. (2012) cAMP response element modulator α controls IL2 and IL17A expression during CD4 lineage commitment and subset distribution in lupus. Proc. Natl. Acad. Sci. U.S.A. 109, 16606–16611 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Hedrich C. M., Rauen T., Kis-Toth K., Kyttaris V. C., Tsokos G. C. (2012) cAMP-responsive element modulator α (CREMα) suppresses IL-17F protein expression in T lymphocytes from patients with systemic lupus erythematosus (SLE). J. Biol. Chem. 287, 4715–4725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Richardson B. (2007) Primer: epigenetics of autoimmunity. Nat. Clin. Pract. Rheumatol. 3, 521–527 [DOI] [PubMed] [Google Scholar]
- 18. Hedrich C. M., Tsokos G. C. (2011) Epigenetic mechanisms in systemic lupus erythematosus and other autoimmune diseases. Trends Mol. Med. 17, 714–724 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Lu Q., Kaplan M., Ray D., Ray D., Zacharek S., Gutsch D., Richardson B. (2002) Demethylation of ITGAL (CD11a) regulatory sequences in systemic lupus erythematosus. Arthritis Rheum. 46, 1282–1291 [DOI] [PubMed] [Google Scholar]
- 20. Lu Q., Wu A., Richardson B. C. (2005) Demethylation of the same promoter sequence increases CD70 expression in lupus T cells and T cells treated with lupus-inducing drugs. J. Immunol. 174, 6212–6219 [DOI] [PubMed] [Google Scholar]
- 21. Lu Q., Ray D., Gutsch D., Richardson B. (2002) Effect of DNA methylation and chromatin structure on ITGAL expression. Blood 99, 4503–4508 [DOI] [PubMed] [Google Scholar]
- 22. Zhao M., Wu X., Zhang Q., Luo S., Liang G., Su Y., Tan Y., Lu Q. (2010) RFX1 regulates CD70 and CD11a expression in lupus T cells by recruiting the histone methyltransferase SUV39H1. Arthritis Res. Ther. 12, R227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Clements E. G., Mohammad H. P., Leadem B. R., Easwaran H., Cai Y., Van Neste L., Baylin S. B. (2012) DNMT1 modulates gene expression without its catalytic activity partially through its interactions with histone-modifying enzymes. Nucleic Acids Res. 40, 4334–4346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Moulton V. R., Tsokos G. C. (2011) Abnormalities of T cell signaling in systemic lupus erythematosus. Arthritis Res. Ther. 13, 207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Patel D. R., Richardson B. C. (2010) Epigenetic mechanisms in lupus. Curr. Opin. Rheumatol. 22, 478–482 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Lu Q., Wu A., Tesmer L., Ray D., Yousif N., Richardson B. (2007) Demethylation of CD40LG on the inactive X in T cells from women with lupus. J. Immunol. 179, 6352–6358 [DOI] [PubMed] [Google Scholar]
- 27. Singer N. G., Richardson B. C., Powers D., Hooper F., Lialios F., Endres J., Bott C. M., Fox D. A. (1996) Role of the CD6 glycoprotein in antigen-specific and autoreactive responses of cloned human T lymphocytes. Immunology 88, 537–543 [PMC free article] [PubMed] [Google Scholar]
- 28. Zhao M., Tang J., Gao F., Wu X., Liang Y., Yin H., Lu Q. (2010) Hypomethylation of IL10 and IL13 promoters in CD4+ T cells of patients with systemic lupus erythematosus. J. Biomed. Biotechnol. 2010, 931018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Ballestar E., Esteller M., Richardson B. C. (2006) The epigenetic face of systemic lupus erythematosus. J. Immunol. 176, 7143–7147 [DOI] [PubMed] [Google Scholar]
- 30. Richardson B. (2003) DNA methylation and autoimmune disease. Clin. Immunol. 109, 72–79 [DOI] [PubMed] [Google Scholar]
- 31. Gorelik G., Fang J. Y., Wu A., Sawalha A. H., Richardson B. (2007) Impaired T cell protein kinase Cδ activation decreases ERK pathway signaling in idiopathic and hydralazine-induced lupus. J. Immunol. 179, 5553–5563 [DOI] [PubMed] [Google Scholar]
- 32. Lavoie G., Estève P. O., Laulan N. B., Pradhan S., St-Pierre Y. (2011) PKC isoforms interact with and phosphorylate DNMT1. BMC Biol. 9, 31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Sunahori K., Juang Y. T., Tsokos G. C. (2009) Methylation status of CpG islands flanking a cAMP response element motif on the protein phosphatase 2Ac α promoter determines CREB binding and activity. J. Immunol. 182, 1500–1508 [DOI] [PMC free article] [PubMed] [Google Scholar]