Background: Pgp3 is an immunogenic protein secreted by Chlamydia trachomatis.
Results: The trimeric Pgp3 structure reveals globular domains connected by a triple helical coiled-coil.
Conclusion: The C-terminal domains resemble tumor necrosis factor, the helical coiled-coil has an unusual twist, and the N-terminal domain is a fusion of virus-like structural motifs.
Significance: The Pgp3 structure provides insight into its role in chlamydial pathogenesis.
Keywords: Bacterial Pathogenesis, Chlamydia, Immunology, Tumor Necrosis Factor (TNF), X-ray Crystallography, Beta-Helix, Beta-Propeller, Helical Coiled-coil, Pgp3, Sexually Transmitted Disease
Abstract
Chlamydia trachomatis infection is the most common sexually transmitted bacterial disease. Left untreated, it can lead to ectopic pregnancy, pelvic inflammatory disease, and infertility. Here we present the structure of the secreted C. trachomatis protein Pgp3, an immunodominant antigen and putative virulence factor. The ∼84-kDa Pgp3 homotrimer, encoded on a cryptic plasmid, consists of globular N- and C-terminal assemblies connected by a triple-helical coiled-coil. The C-terminal domains possess folds similar to members of the TNF family of cytokines. The closest Pgp3 C-terminal domain structural homologs include a lectin from Burkholderia cenocepacia, the C1q component of complement, and a portion of the Bacillus anthracis spore surface protein BclA, all of which play roles in bioadhesion. The N-terminal domain consists of a concatenation of structural motifs typically found in trimeric viral proteins. The central parallel triple-helical coiled-coil contains an unusual alternating pattern of apolar and polar residue pairs that generate a rare right-handed superhelical twist. The unique architecture of Pgp3 provides the basis for understanding its role in chlamydial pathogenesis and serves as the platform for its optimization as a potential vaccine antigen candidate.
Introduction
Chlamydia trachomatis is an obligate intracellular Gram-negative bacterial pathogen of different serovars that infect human ocular and urogenital tract epithelial cells. C. trachomatis infection of the urogenital tract is the most common cause of bacterial sexually transmitted disease in the world (1), and the Centers for Disease Control and Prevention estimate 50 million new cases occur every year. Left untreated, the infection can lead to complications such as ectopic pregnancy, pelvic inflammatory disease, and infertility (2). Ocular infection by C. trachomatis can lead to blindness (3). Host inflammatory responses induced by C. trachomatis infection are thought to contribute to pathogenicity (4). The infection starts with the entry of an infectious elementary body into an epithelial cell via pathogen-induced endocytosis (5, 6). The internalized elementary body rapidly differentiates to become a noninfectious but metabolically active reticulate body. After replication, the progeny reticulate bodies differentiate back to elementary bodies before exiting to invade adjacent cells (7).
All C. trachomatis organisms possess a highly conserved cryptic plasmid encoding eight open reading frames, the protein products of which are designated Pgp1–8 (8–10). Plasmid gene products are also known to regulate the expression of more than 20 genome-encoded genes (11). When depleted of the cryptic plasmid, C. trachomatis ocular serovar A is unable to induce pathology in monkey ocular tissues (12), consistent with an earlier report that plasmid-free Chlamydia muridarum fails to cause hydrosalpinx after inoculation of mouse genital tracts (13). Together, these findings suggest that the plasmid-encoded factors act in chlamydial pathogenesis.
A recently developed chlamydial plasmid transformation system (14) permits investigation of plasmid-encoded protein function (15). Pgp1, 2, 6, and 8 are required for plasmid maintenance. Pgp4 deletion transformants demonstrate reduced expression of Pgp3 and other plasmid-encoded genes as well as glgA in the chlamydial genome, resulting in a lack of glycogen accumulation in the inclusion. Pgp4 therefore appears to be a regulator of chlamydial gene expression (11). However, the roles of Pgp3, 5, and 7 in chlamydial biology remain unknown.
Pgp3 is a ∼84-kDa homotrimeric protein (16) both associated with the outer membrane (17) and secreted into the inclusion lumen and the host cell cytosol (8). It is one of the most immunodominant antigens in mammals infected by chlamydial organisms (18, 19). Human antibody recognition of Pgp3 is dependent on its trimeric quaternary structure (16). Purified Pgp3 is known to stimulate macrophages to release inflammatory cytokines (8), and vaccination with Pgp3 provides partial protection against challenge infection with chlamydial organisms (20). Together these observations suggest that the protein plays a prominent role in chlamydial pathogenesis and as such could be a promising vaccine antigen candidate. However, the precise role of Pgp3 in chlamydial pathogenesis and immunity remains unknown despite being the subject of extensive microbiological, immunological, and biochemical studies.
To better understand its action, we determined the structure of Pgp3 using the established tools of single crystal x-ray diffraction. The structure determination was a challenge, however, because crystals of the full-length protein suffer from diffuse scattering (21), limiting the resolution of the experimental electron density map. Although the protein backbone of the C-terminal domain (CTD)4 could be traced, the density for most of the triple helical coiled-coil and the N-terminal domain (NTD) was too weak and/or convoluted to permit interpretation.
A “divide-and-conquer” approach yielded high resolution structures of two Pgp3 truncation variants, providing detailed visualizations of the globular NTD and the trimeric arrangement of independently folded CTDs. The refined structures of these globular domains were positioned into the experimental electron density map of the full-length protein. Phase combination substantially improved the electron density and permitted the determination and refinement of the full-length molecule.
The structure reveals that Pgp3 is an elongated baton-like molecule with CTDs similar to members of the TNF family of cytokines. The NTD possesses a previously unobserved fold with internal pseudo-3-fold symmetry in which the three polypeptide chains intertwine and swap structural elements. The globular domains are connected by a parallel, triple-helical coiled-coil (THCC) with an unusual right-handed twist. The recent development of a chlamydial plasmid transformation system (14), combined with knowledge of the Pgp3 structure presented here, provides powerful tools to probe the role of the molecule in chlamydial pathogenesis and may assist in vaccine development.
EXPERIMENTAL PROCEDURES
Cloning, Expression, and Purification of Full-length Pgp3
DNA encoding full-length C. trachomatis Pgp3 from serovar D was PCR-amplified, subcloned into a pGEX vector, and transformed into Escherichia coli strain B834 (DE3). This construct encodes GST fused to the N terminus of Pgp3 separated by a protease cleavage site. Six liters of cells were grown at 37 °C for 20 h in minimal medium lacking methionine but containing selenomethionine (22). The temperature was decreased to 16 °C when the cells reached an A600 of 0.6, and expression was induced by adding isopropyl β-d-thiogalactoside to a final concentration of 0.5 mm. The cells were shaken overnight, harvested by centrifugation, and frozen at −20 °C.
Thawed cells were resuspended in ∼50 ml of 50 mm Tris, pH 8.0, 1% (v/v) Triton X-100, 400 mm NaCl, and Sigma protease inhibitor mixture, disrupted by sonication on ice, and centrifuged to remove cellular debris. The supernatant was incubated with glutathione-conjugated agarose beads (Pharmacia) in batch mode, and after washing with column buffer, the fusion protein was cleaved with GST-tagged PreScission Protease (GE Healthcare). Cleaved Pgp3 was released into the supernatant, dialyzed against 50 mm Tris (pH 8.0), loaded onto a mono Q column (Pharmacia), washed with five column volumes of buffer, and eluted in a single step using 0.5 m NaCl in 50 mm Tris buffer (pH 8.0). All of the solutions used in this process (as well as the purification processes below) contained the reducing agent tris(carboxyethyl)phosphine at a concentration of 2 mm. The full-length Pgp3 protein, estimated to be ∼98% pure by SDS-PAGE, was dialyzed into crystallization buffer consisting of 50 mm Tris buffer (pH 8.0) and concentrated to 17 mg/ml using the calculated extinction coefficient of 14,440 m−1 cm−1.
Cloning, Protein Expression, and Purification of Pgp3 Truncation Variants
Truncation variant constructs were generated by PCR with C. trachomatis serovar D plasmid DNA and the appropriate primers. The first encoded the CTD alone (residues 113–264). The second construct was engineered to encode Pgp3 lacking the THCC (residues 72–116), resulting in a NTD-CTD fusion (hereafter referred to as the NCD fusion). DNA fragments encoding the Pgp3 truncation variants were subcloned into pAG8H, a modified pET19d vector with a tobacco etch virus-cleavable His8 tag fused to the N terminus of the target protein (23).
Both variants were expressed in E. coli B834 (DE3) cells as described previously, except frozen cells were resuspended in 50 mm HEPES (pH 7.4), 400 mm NaCl, and Sigma protease inhibitor mixture. Cleared supernatants were loaded onto a GE Healthcare Life Sciences His-Trap nickel column, washed with buffer made 0.1 m in imidazole, and eluted with a linear imidazole gradient (0.1–0.5 m). The Pgp3-containing fractions, identified by SDS-PAGE, were pooled, dialyzed against 50 mm HEPES (pH 7.4), and incubated with His-tagged tobacco etch virus protease overnight at 20 °C. The solution was again passed over the His-Trap nickel column, and the His tag-free proteins were collected. Pgp3 truncation variants were dialyzed against 25 mm HEPES (pH 7.4), loaded onto a mono Q column, and eluted with a linear NaCl gradient (0.1–0.5 m) in 25 mm HEPES (pH 7.4). The proteins were dialyzed into crystallization buffer consisting of 25 mm HEPES (pH 7.4) and concentrated to 15 mg/ml using extinction coefficients of 9970 and 12,950 m−1 cm−1 for the CTD and NCD fusion proteins, respectively. The mass of each purified Pgp3 truncation variant was verified using electrospray ionization mass spectrometry.
Crystallization and X-ray Data Collection
Full-length, CTD, and NCD fusion Pgp3 crystals were grown at 20 °C using the hanging drop vapor diffusion method (24). Selenomethionine-substituted full-length Pgp3 was mixed with an equal volume of reservoir solution containing 30% PEG 550 MME and 0.1 m Tris buffer (pH 8). Irregular prisms appeared within 2 days and were flash cooled in liquid nitrogen using the same solution as the cryoprotectant. Selenomethionine-substituted Pgp3 CTD protein was mixed with an equal volume of solution containing 1.2 m sodium/potassium phosphate (pH 8.2). Hexagonal plates appeared in 2 days. Reservoir solution made 20% (v/v) in glycerol was used as the cryoprotectant for flash cooling. The NCD fusion protein was mixed with an equal volume of solution containing 20% PEG 6000 and 0.1 m trisodium citrate. Rod-shaped crystals appeared within 1 week. Reservoir solution made 8% (v/v) in glycerol was used as the cryoprotectant for flash cooling. All diffraction data were taken at the Northeastern Collaborative Access Team Beamlines 24-ID-E or 24-ID-C at the Advanced Photon Source. The data sets were indexed, processed, and scaled using the HKL-2000 program suite (25).
Structure Determination and Refinement
The program SHELXD (26) identified 22 of the 24 expected selenium sites in the full-length Pgp3 protein. Useful multiwavelength anomalous diffraction phases were calculated in autoSHARP (27) to ∼4.5 Å. Maximum likelihood density modification with solvent flattening, 2-fold noncrystallographic symmetry averaging, and phase extension to 3.1 Å in RESOLVE (28) yielded a partially interpretable electron density map into which the Cα trace of each polypeptide chain of the CTD trimer and a small portion of THCC was constructed using the molecular modeling program COOT (29).
This CTD trace of the full-length protein was used to guide the design of a Pgp3 CTD expression construct, the structure of which was determined using SAD phasing in the program autoSHARP. The 2.0 Å electron density map was of quality sufficient to permit ARP/WARP (30) to build the model automatically, which was subsequently refined using the PHENIX suite of programs (31).
The NCD fusion protein structure was determined by molecular replacement with the program PHASER (32) using the CTD structure from above as the search model. noncrystallographic symmetry averaging over the three trimers in the asymmetric unit permitted the tracing of the intertwined NTD portion of the molecule. The NCD fusion protein structure was refined with the PHENIX program suite.
The newly refined structures of the CTD and the NTD were positioned into the original full-length Pgp3 electron density map with PHASER and refined in PHENIX. Phase combination with phase extension to 3.1 Å in SHARP improved the quality of the electron density map, permitting the THCC to be constructed. Alternating cycles of crystallographic refinement in PHENIX and manual model adjustment in COOT yielded the full-length Pgp3 structure.
Figure Preparation
The figures were created using PyMOL (version 1.5.0.4; Schrödinger). Poisson-Boltzmann electrostatic calculations were performed using the programs PDB2QR (33) and APBS (34).
RESULTS
Pgp3 Is an Elongated, Trimeric Multidomain Protein
Data measurement, processing, and phasing statistics are shown in Table 1. Protein structure refinement statistics are presented in Table 2. The experimental electron density map revealed Pgp3 to be an elongated protein with two globular domains connected by weak density for a coiled-coil. Only the CTD portion of the map permitted confident tracing of the polypeptide chain. Phase combination with partial model phases failed to improve the map, leading us to the divide-and-conquer strategy described under “Experimental Procedures.”
TABLE 1.
Full-length and C-terminal domain experimental phasing statistics
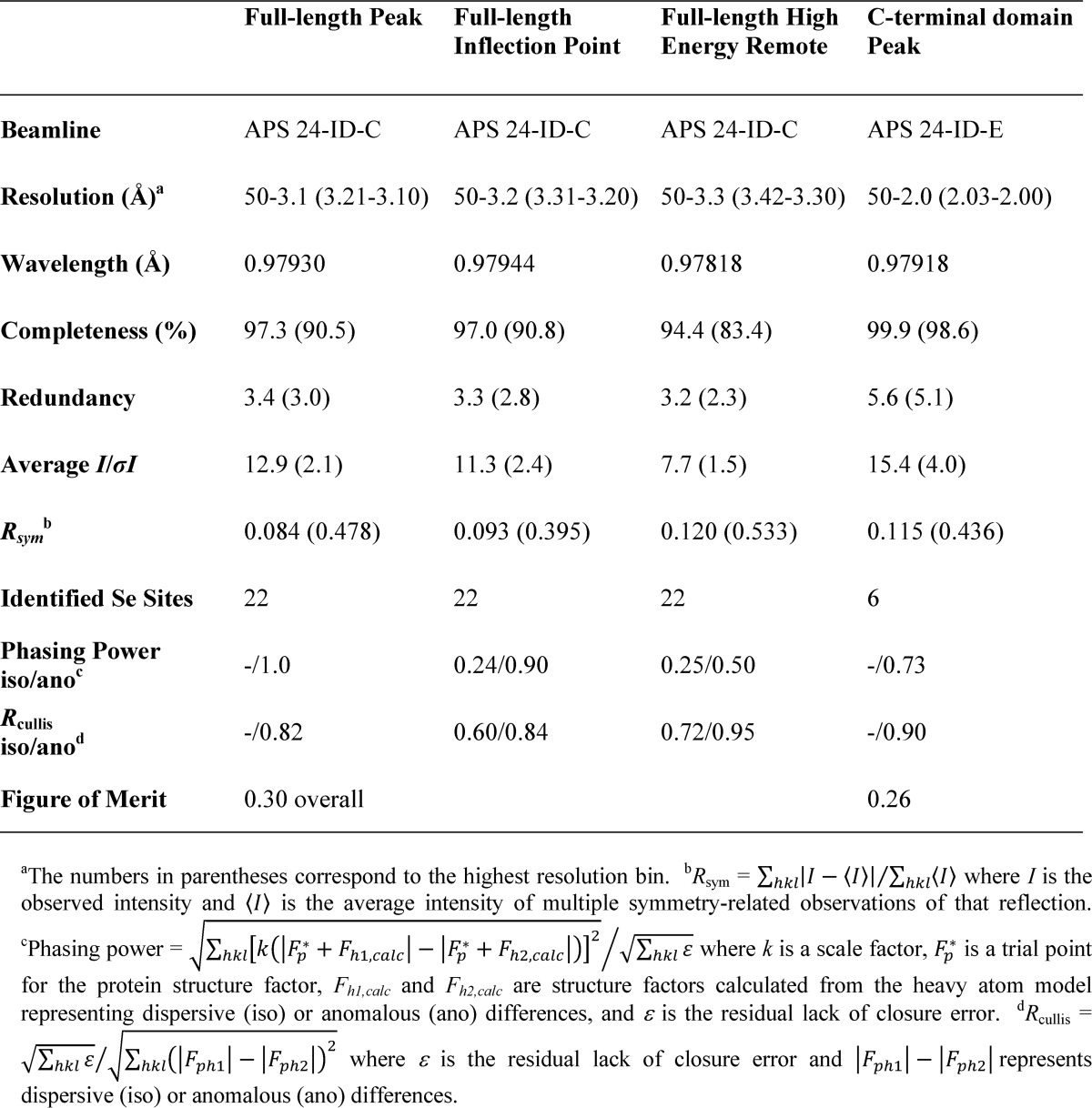
TABLE 2.
Data collection and refinement statistics
| Full-length Pgp3 | C-terminal domain Pgp3 | N-C fused domains Pgp3 | |
|---|---|---|---|
| P21212 | P3221 | P65 | |
| Data collection | |||
| Unit cell | |||
| a (Å) | 188.7 | 78.6 | 146.2 |
| b (Å) | 223.8 | 78.6 | 146.2 |
| c (Å) | 79.2 | 129.1 | 161.0 |
| Resolution range (Å)a | 50–3.1 (3.21–3.1) | 50–2.0 (2.03–2.0) | 30- 2.3 (2.38–2.3) |
| Wavelength (Å) | 0.97930 | 0.97918 | 0.97928 |
| Redundancy | 3.4 (3.0) | 5.6 (5.1) | 7.6 (7.6) |
| Completeness (%) | 97.3 (90.5) | 99.9 (98.6) | 100 (100) |
| I/σI | 12.9 (2.1) | 15.4 (4.0) | 14.6 (3.7) |
| Rsym (%)b | 8.4 (47.8) | 11.5 (43.6) | 13.2 (57.3) |
| Wilson value | 61.0 | 14.3 | 28.0 |
| Refinement | |||
| PDB entry | 4JDM | 4JDN | 4JDO |
| Number of trimers/AU | 2 | 1 | 3 |
| Rcryst (%)c | 23.1 | 15.5 | 18.4 |
| Rfree (%)d | 26.4 | 19.1 | 24.2 |
| RMSD bonds (Å) | 0.012 | 0.008 | 0.009 |
| RMSD angles (°) | 1.433 | 1.156 | 1.268 |
| Ramachandran outliers (%)e | 3.1 | 1.4 | 2.1 |
| No. protein atoms | 11,712 | 3,210 | 14,317 |
| No. solvent atoms | 0 | 305 | 714 |
| No. ligand atoms | 0 | 1 K+, 6 glycerol | 3 Na+ |
| Average protein B-factors (Å2) | 71.7 | 16.6 | 29.4 |
| Average solvent B-factors (Å2) | 24.5 | 31.3 | |
| Average ligand B-factors (Å2) | 21.4 | 33.0 | |
a The number in parentheses is for the highest resolution bin.
b Rsym = Σhkl|I − <I>|/Σhkl<I>, where I is the observed intensity, and <I> is the average intensity of multiple symmetry-related observations of that reflection.
c Rcryst = Σhkl||Fobs − |Fcalc||/Σhkl|Fobs|.
d Rfree = Σhkl||Fobs,t| − |Fcalc||/Σhkl|Fobs,t|, where |Fobs,t| is from a test set not used in the structural refinement.
e Ramachandran plots were calculated using COOT.
The 2.0 Å Structure of the Pgp3 CTD
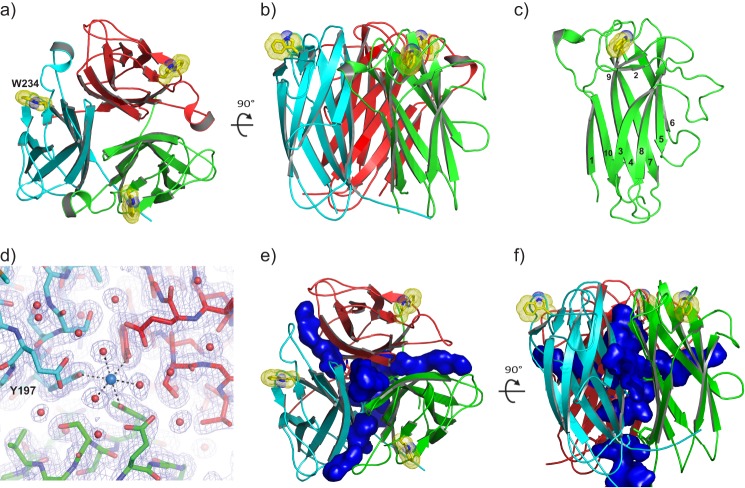
The Pgp3 CTDs form compact, cylindrical, trimeric assemblies similar to those observed in the TNF family of proteins (35) (Figs. 1, a and b). The backbone atoms of the subunits superimpose with RMSDs averaging 0.20 Å. Each subunit possesses a β-barrel jelly roll fold consisting of 10 antiparallel β-strands, with a sheet containing β1, β4, β7, and β10 at the trimer interface and a sheet containing β3, β5, β6, and β8 facing the solvent (Fig. 1c). The short strands β2 and β9 “cap” one end of the β-barrel. β9 harbors Trp-234, the only tryptophan residue in the Pgp3 sequence. Trp-234 sits in a shallow groove formed by β2, β9, and the loop connecting β9 to β10, such that both polar and apolar portions of its indole ring are solvent-exposed. The walls of the depression prevent the Trp-234 side chain from sampling other conformations (see “Discussion”). The loop elements connecting β2 to β3 and β5 to β6 project outward, normal to the barrel's long axis.
FIGURE 1.
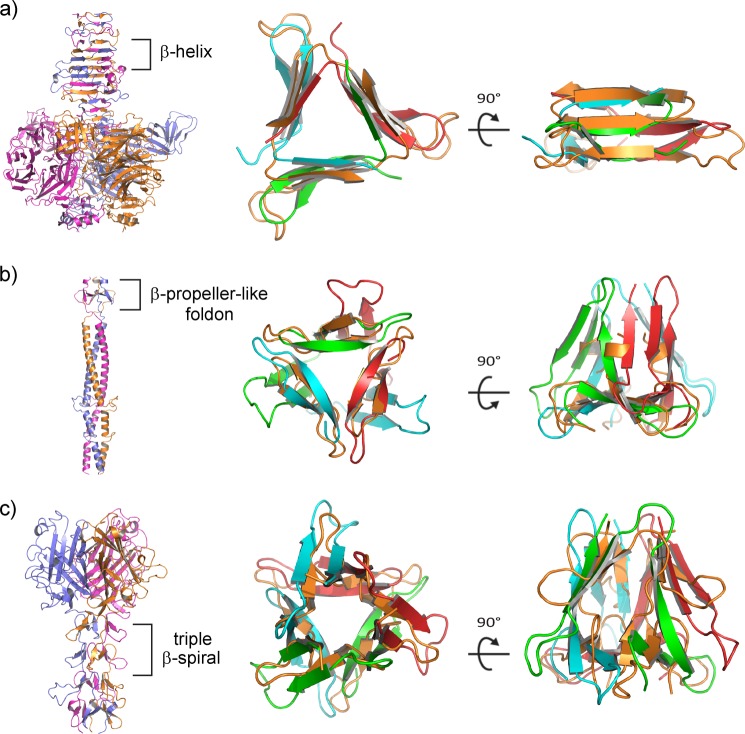
Structure of the Pgp3 CTD trimer (residues 113–264). a, view of the CTD trimer coincident with the molecular 3-fold axis of rotation. Polypeptide chains A, B, and C are colored green, red, and cyan, respectively, here and in all subsequent figures. The solvent-exposed indole rings of Trp-234 residues in the trimer are shown as yellow sticks with the van der Waals radii of the side chain atoms represented as dots. b, view of the CTD trimer normal to the molecular 3-fold axis, rotated as indicated with respect to panel a. c, each CTD monomer possesses a jelly roll β-barrel fold. Individual β-strands are numbered consecutively from the N terminus to the C terminus. d, a σA-weighted 2Fo − Fc electron density map showing a potassium ion bound at the CTD molecular 3-fold symmetry axis in octahedral coordination geometry. The view is the same as in panel a. e, a ribbon diagram of the CTD trimer including a depiction its solvent-accessible cavities shown in blue. Cavities were calculated using a probe radius of 1.4 Å in PyMOL. f, the same as in panel e but rotated 90° about the horizontal axis.
A metal ion coordinated by symmetry-related side chain hydroxyl moieties of Tyr-197 side chains and three water molecules is positioned at the entrance of a solvent-filled channel that penetrates the trimer (Fig. 1d). The octahedral coordination geometry, the presence of 1.2 m sodium/potassium phosphate, and the absence of “difference” features in electron density maps calculated with coefficients Fo − Fc suggest that the metal is a fully occupied potassium ion. Three “portals” at the interfaces between CTD run normal to the central channel, connecting it to the bulk solvent at midpoint of the CTD cylinder (Fig. 1, e and f).
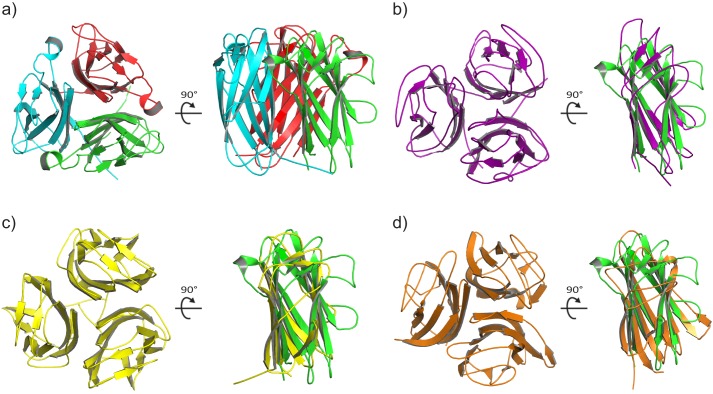
A search of the Protein Data Bank for structural homologs using the program DALI (36) returned well over 100 different proteins with Z-scores greater than 2, despite the lack of significant sequence identity (< 10%). The top 10 scoring proteins are shown in Table 3, and several relevant examples are illustrated in Fig. 2.
TABLE 3.
The top 10 unique hits identified in a DALI structural homology search for Pgp3 CTD residues 118–261
| Molecule | Organism | Z-score | RMSD | Aligned positions | Residues in matched structure | Sequence identity of aligned positions | PDB code |
|---|---|---|---|---|---|---|---|
| Å | % | ||||||
| BC2L-C | B. cenocepacia | 12.1 | 2.1 | 113 | 131 | 10 | 2WQ4 (57) |
| Complement C1Q subcomponent subunit A | Human | 10.3 | 2.7 | 110 | 129 | 11 | 2JG9 (58) |
| Ectodysplasin A | Human | 9.9 | 2.6 | 107 | 145 | 12 | 1RJ7 (69) |
| 30-kDa adipocyte complement-related protein | Mouse | 9.7 | 2.8 | 110 | 137 | 12 | 1C3H (70) |
| Collagen α1 (VIII) chain | Mouse | 9.6 | 3.0 | 110 | 131 | 12 | 1O91 (71) |
| BclA | B. anthracis | 9.6 | 2.7 | 109 | 136 | 12 | 2Z5W (72) |
| TNF-related apoptosis inducing ligand (TRAIL) | Human | 9.5 | 2.5 | 105 | 134 | 10 | 1D2Q (73) |
| Capsid protein VP2 | Infectious pancreatic necrosis virus | 9.2 | 3.0 | 113 | 415 | 14 | 3IDE (74) |
| Minor capsid protein | Enterobacteria phage PRD1 | 9.2 | 3.0 | 113 | 192 | 5 | 1YQ8 (64) |
| Collagen X | Human | 9.1 | 2.9 | 110 | 132 | 15 | 1GR3 (75) |
FIGURE 2.
Structural alignments of the TNF-like CTD with three of the top ten hits coming from a DALI search (36) of the Protein Data Bank. a, orthogonal views of the Pgp3 CTD trimer. b, the Bc2L-C trimer (57) and superposition of a monomer (purple) onto a Pgp3 CTD monomer (green). c, the BclA trimer (72) and superposition of a monomer (yellow) onto a Pgp3 CTD monomer (green). d, The complement component C1q trimer (58) and superposition of a monomer (orange) onto a Pgp3 CTD monomer (green).
The Structure of the Pgp3 NCD Fusion Reveals the Unique NTD Fold
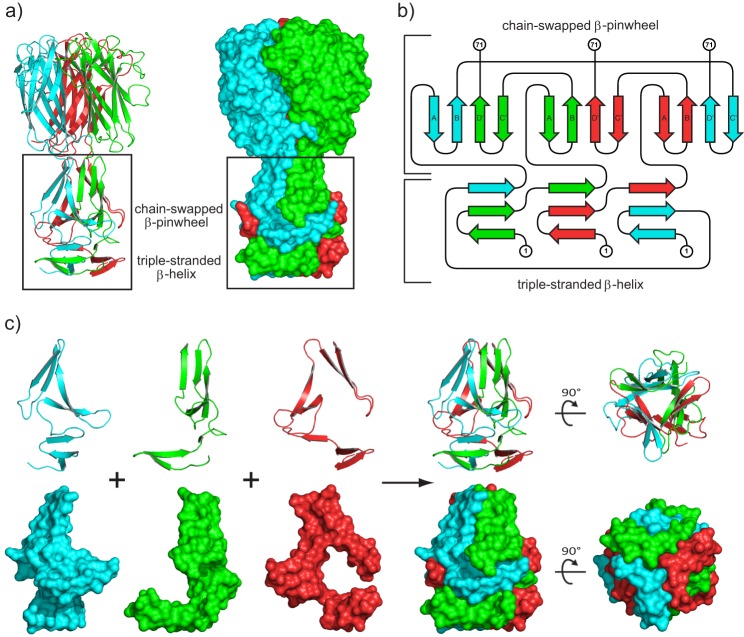
Although the CTD of each subunit is independently folded, residues 1–71 are components of a globular NTD consisting of two modules with internal 3-fold symmetry (Fig. 3a). In the first module, β-hairpins formed by residues 1–23 associate with a strand from an adjacent chain to generate the equivalent of three turns of β-helix. In the second module, residues 32–70 form a closed, three-bladed β-propeller with four-stranded blades. The top portion of Fig. 3b shows that the β-hairpin formed by residues 32–49 swaps in a left-handed fashion to associate with the β-hairpin formed by residues 54–70 of the neighboring chain to complete the blade. The conformation of NTD structural elements coming from each polypeptide is unlikely to exist in isolation given the stabilizing reciprocal interlocking interactions shown in Fig. 3c.
FIGURE 3.
Structure of the Pgp3 NCD fusion protein. a, ribbon diagram and surface representation of the NCD fusion structure (residues 1–71 fused to residues 117–264) with the NTD motifs discussed in the text indicated. b, topology diagram illustrating the unique fold observed in the Pgp3 NTD. N-terminal and C-terminal residues 1 and 71 are labeled for each chain. The canonical β-propeller blade has a fully antiparallel strand pattern A-B-C-D arising from a single polypeptide chain. In contrast, the strand order for the PGP3 blade is A-B-D′-C′ (as shown), where C′ and D′ are strands contributed from a neighboring polypeptide chain, generating a parallel strand interaction at the B-D′ interface. Strand A is the N-terminal strand on the interior of the propeller, but strand C′ is on the exterior rather than D′ at the C terminus characteristic of the canonical arrangement. The topology diagram was created using the program TOPDRAW (76). c, ribbon and surface representations of each NTD monomer showing the interlocking interactions contributed by the individual polypeptide chains.
Full-length Pgp3 Contains a THCC with a Rare Right-handed Twist
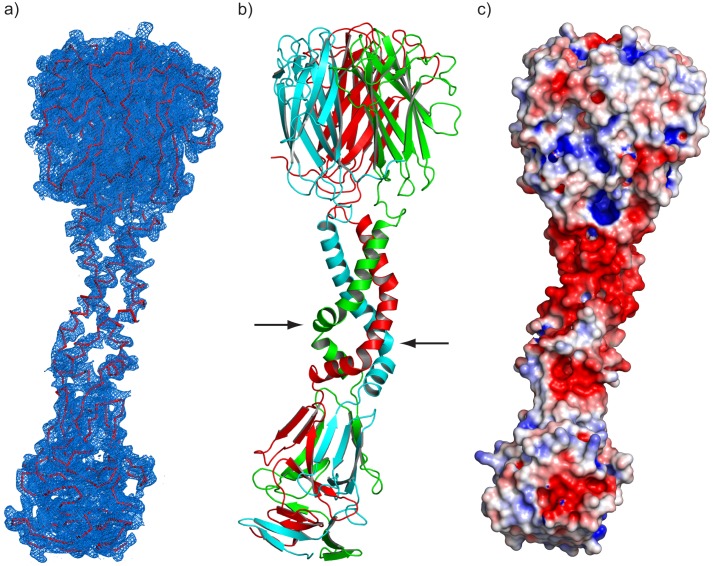
Each 264-residue polypeptide of the Pgp3 trimer is ∼145 Å in length associating to form a parallel THCC that connects the NTD and CTD globular assemblies (Fig. 4a). As shown in Fig. 4b, the presence of a glycine at position 85 acts as a “pivot,” resulting in offset THCCs consisting of residues 73–84 and 86–111. A cluster of 18 aspartic acid residues (90, 94, 98, 103, 106, and 111 from each chain) creates an acidic ring on the surface of the THCC adjacent to the CTD (Fig. 4c).
FIGURE 4.
Structure of full-length Pgp3. a, 3.1 Å σA-weighted 2Fo − Fc electron density map of the full-length Pgp3 homotrimer contoured at 1.25 σ. b, a ribbon diagram of the elongated, bent, full-length Pgp3 structure. The positions of the α-carbon atoms of Gly-85 in two of the three helices are indicated by arrows. c, electrostatic surface potential contoured at ±5 kT. A prominent negatively charged patch (red shading) is observed in the THCC domain adjacent to the beginning of the CTD.
DISCUSSION
The Difficult Structure Determination of Pgp3
The diffuse scattering pathology of the selenomethionine-substituted crystals adversely affected the resolution and quality of the multiwavelength anomalous diffraction-phased experimental electron density map, making the structure determination of the full-length Pgp3 protein a challenge. Its elongated shape and NTD-to-CTD packing interactions (see below) gave rise to a crystal lattice with a solvent content of 75.5% (Vm = 5.0 Å3/Da) (37). This characteristic, combined with the apparent flexibility of the THCC (see below) are the likely source of the crystal pathology. Circumvention of the diffuse scattering problem required the determination of the structures of the globular Pgp3 CTD and NTD components separately to high resolution, positioning the resulting structures back into the 3.1 Å density map of marginal quality, and recalculating the map using combined partial model (184 of 264 residues) and experimental phases. This phase combination exercise substantially improved the 3.1 Å electron density map, permitting the entire full-length Pgp3 protein to be visualized (Fig. 4a).
The Unique Structure of the Pgp3 NTD
The determination and refinement of the NCD fusion structure at a resolution of 2.3 Å permitted the visualization of the unusual Pgp3 NTD. The NTD begins with a β-hairpin in which residues 1–23 of each polypeptide associates with a single strand from an adjacent polypeptide to form what would essentially be the equivalent of three turns of β-helix in a canonical β-helical protein built from a single polypeptide chain (38). Concatenated structural motifs similar to those typically found in viral proteins (Fig. 5) were discovered by eye during perusal of the Structural Classification of Proteins (SCOP) database (39). For example, superposition of the β-helical structural motif formed by residues 1–24 of the Pgp3 NTD trimer and residues 853–877 of the trimeric endosialidase of bacteriophage K1F (Protein Data Bank (PDB) entry 1V0F (40)) reveals the same triple-stranded swapping pattern (compare the lower portions of Fig. 3, a and b, with Fig. 5a). The phage triple β-helix aligns with the Pgp3 β-helix with an root mean square deviation of 2.5 Å over 61 aligned residues of 68 reference residues (Fig. 5a, right panel). Although the above-mentioned region superimposes with a portion of the phage protein that binds sialic acid, the Pgp3 β-helical motif alone does not comprise a complete sugar-binding site found in the K1F phage endosialidase.
FIGURE 5.
Virus-like structural motifs observed in the Pgp3 NTD. a, endosialidase of bacteriophage K1F (PDB entry 1V0F (40)), with the β-helix region indicated (each chain shown with a different color). The bacteriophage β-helix superimposes on the N-terminal β-helix of Pgp3 shown in two orthogonal orientations. The color scheme for Pgp3 polypeptide chains is preserved as above, whereas the virus structure is colored orange for its three chains for clarity in the superpositions (two right panels). b, bacteriophage T4 fibritin with a highlighted foldon motif (PDB entry 1AA0 (46)) superimposed on the lower β-pinwheel region of Pgp3. c, adenovirus fiber with a highlighted portion of its β-spiral region (PDB entry 1Q1U (48)) superimposed on the upper β-pinwheel region of Pgp3.
Immediately C-terminal to the β-helix, residues 32–70 of the Pgp3 NTD form the chain-swapped, three-bladed, β-pinwheel as indicated in the upper portions of in Fig. 3 (a and b). β-Pinwheels differ from canonical β-propellers in that antiparallel strand exchange occurs in the β-propeller between blades within a single polypeptide chain (41). The Pgp3 NTD β-pinwheel differs in that strand exchange between blades comes from adjacent polypeptide chains, resulting in parallel β-strands at the blade center. The four-stranded Pgp3 blade has a twist similar to that observed in canonical β-propeller blades (42), with the inner and outer strands oriented nearly perpendicular to each other. To our knowledge, Pgp3 is the first protein reported to possess a fully closed, three-bladed β-propeller (or chain-swapped β-pinwheel). β-Propellers have been assigned diverse biological roles ranging from enzymatic and signaling functions to DNA-binding/wrapping domains found in prokaryotic type II DNA topoisomerases (43). However, the role(s) for the unique Pgp3 β-pinwheel remain unknown.
The Pgp3 β-pinwheel contains a substructure with similarity to the fibritin foldon domain, the β-propeller-like motif in bacteriophage T4 known to be essential for fibritin trimerization and folding both in vivo and in vitro (44–46). Pgp3 residues 29–48 align with phage T4 foldon residues 463–479 (PDB entry 1AA0) with a root mean square deviation of 2.2 Å over 47 of 57 reference residues (Fig. 5b). The proposed function of the foldon is to promote the rapid trimerization of T4 fibritin (44, 47).
In contrast to the foldon motif, which is structurally similar to the N-terminal portion of the β-pinwheel, the shaft domain of the adenovirus fiber (48) is structurally similar to the C-terminal portion of the Pgp3 β-pinwheel consisting of residues 48–71 (Fig. 5c). Chain swaps of β-strands similar to those in the Pgp3 β-pinwheel are evident in the shaft domain, which contains tandem repeats of β-spiral motifs. Residues 360–392 of the triple β-spiral shaft domain (PDB entry 1QIU) align with residues 29–71 of the Pgp3 β-pinwheel with a root mean square deviation of 3.9 Å over 79 of 99 reference residues (Fig. 5c). The overall size and shape of the adenovirus β-spiral and Pgp3 β-pinwheel are similar, although the viral fold departs from the Pgp3 pattern in that it does not contain a foldon in the same location. Instead, a three-bladed β-propeller-like motif in the spiral occupies this position. A reovirus attachment protein with a triple β-spiral shaft similar to adenovirus was shown to bind sialylated oligosaccharides along the shaft (49). However, the arrangement of the foldon and β-spiral-like strands in Pgp3 are incompatible with binding oligosaccharides at the position equivalent to the sialic acid-binding region of the reovirus protein. The structural similarity of the Pgp3 NTD to motifs found in viral fiber proteins hints at the possibility for a viral origin of the plasmid-encoded Pgp3 gene through the transfer of genetic information from bacteriophage to early chlamydial organisms.
The Flexible Pgp3 THCC Possesses a Rare Right-handed Twist and a Prominent Acidic Ring
The Pgp3 THCC demonstrates an unusual alternating pattern of apolar and polar residue pairs that generates its observed right-handed twist. Although predicted by theoretical calculations, there remains a paucity of examples of right-handed helical coiled-coils coming from natural sources, and those that have been reported are almost invariably four helical coiled-coils (50–53). The N-terminal portion of the Pgp3 THCC consisting of residues 73–84 adopts a larger, hollow shape compared with the C-terminal segment. The short, ∼60 Å pitch calculated using the program TWISTER (54) appears to be a manifestation of the loose internal packing observed in Pgp3 relative to its canonical left-handed THCC counterparts (54, 55). A detailed analysis of the various parameters of the Pgp3 THCC will be provided elsewhere.
The loose packing and apparent flexibility of the Pgp3 THCC is consistent with the diffuse scattering pathology of the Pgp3 crystals, the weak electron density shown in Fig. 4a, the high thermal parameters of its constituent atoms, and the curvature of the Pgp3 THCC that is evident in Figs. 4b and 6a. Although it caused difficulties in the structural study, the pliability of the Pgp3 THCC may be related to its function. As an example, the flexibility of certain pili adhesins has been suggested to permit retention of tight binding to host cell surface receptors while under shear force (56).
FIGURE 6.
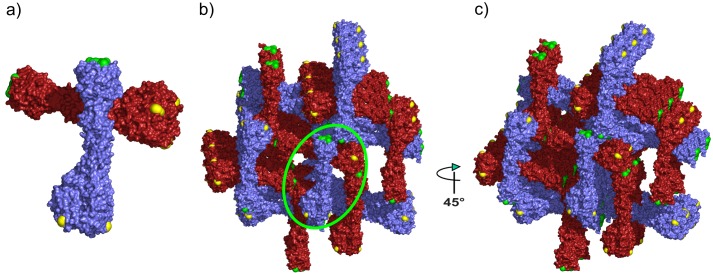
Crystal packing interactions in the full-length Pgp3 crystals. a, the Pgp3 asymmetric unit containing two full-length Pgp3 molecules colored with red and blue surfaces. Trp-234 residues are colored yellow, and Phe-6 residues are colored green. b, crystal packing interactions in the Pgp3 lattice. The orientation is as in panel a. The green circle shows the position of the asymmetric unit in panel a. c, the view is the same as in a but rotated 45° about the vertical axis. The major crystal contact dominating the packing interactions between Phe-6 and Trp-234 is most evident in the head-to-tail interaction between orthogonal Pgp3 trimers in the rightmost portion of this image.
The Pgp3 CTD Resembles the TNF Family of Proteins
Among the 10 closest structural homologs of the Pgp3 CTD as identified by the program DALI (Table 3), the N-terminal domain of Bc2L-C from Burkholderia cenocepacia (57), the C1q component of complement (58), the C-terminal domain of BclA from Bacillus anthracis (59), and collagen C-terminal domains are all known to act in bioadhesion.
Bc2L-C, the most similar structural homolog to the Pgp3 CTD, is a recently identified lectin with specificity for fucosylated human histo-blood group epitopes H-type 1, Lewis b, and Lewis Y (57). The Bc2L-C TNF-α-like domains trigger inerleukin-8 production in cultured airway epithelial cells in a carbohydrate-independent manner and are proposed to play a role in the deregulated proinflammatory response observed in B. cenocepacia lung infections (60), suggesting a given TNF-like trimer can perform more than one function. Full-length and CTD Pgp3 constructs containing an His8 tag were screened against ∼460 distinct mammalian glycans in chip-based glycan array experiments, but both were negative for glycan binding (data not shown). However, the Pgp3 NTD possesses structural motifs similar to sugar-binding modules in viral proteins (e.g., the endosialidase of bacteriophage K1F (40)) and the possibility that the N-terminal His8 tag fused to the full-length Pgp3 protein might have interfered with glycan binding cannot definitively be fully ruled out.
The C1q component of complement demonstrates a high degree of structural similarity to the Pgp3 CTD and plays a key role in innate immunity through recognition of immune complexes and the initiation of the classical complement pathway (61). C1q can directly trigger cellular defense responses, such as chemotaxis, cytokine release, phagocytosis, and cytotoxicity (62). These defense mechanisms are mediated by a variety of receptors present on the host cell surface. Unlike Bc2L-C, C1q fails to induce secretion of IL-8. Additionally, homology to C1q raises the possibility that the secreted Pgp3 could alter complement activation pathways, causing inflammatory responses that may benefit chlamydial spreading.
The structure of the B. anthracis protein BclA (59), the immunodominant protein of its exosporium, draws several parallels to Pgp3 in that its structure reveals a TNF-like C-terminal domain connected to an N-terminal domain (of unknown structure) by a collagen-like triple helix (63). Recent work reveals that the TNF-like domains are positioned distal to the exosporium anchor provided by the BclA NTD, suggesting that it is the TNF-like trimeric assembly that is adhesive and immunogenic (63). The TNF-like domain of Pgp3 is known to be the immunodominant antigen secreted during C. trachomatis infection (18, 19). Thus far, few bacterial proteins have been demonstrated to possess TNF-like topology. Thus, the Pgp3 CTDs, together with those of Bc2L-C and BclA, are the closest bacterial homologs to mammalian TNF superfamily members.
A recent high resolution structure of a bacteriophage (PRD1) spike protein was determined, revealing a striking similarity to TNF superfamily members (64). PRD1 consists of an N-terminal “shaft” and a C-terminal head domain. The structure is clearly similar to that of adenovirus and rheovirus spike proteins, with a conserved assembly of the shaft domains and a similar assembly of the head domains. However, only the PRD1 head domain structure superimposes well on the TNF-like family fold. Perhaps tellingly, the spike proteins of all three viruses are essential for attachment to host cells.
The Pro-inflammatory Pgp3 Trimer May Also Play a Role in Host Cell Adhesion
The trimeric architecture of Pgp3 is reminiscent of viral fiber proteins that function in the attachment of viruses to host cells. The fiber proteins tend to be elongated trimers with an N-terminal globular domain associated with the main body of the viral particle, an extended connector domain, and a globular C-terminal receptor-binding domain/assembly (65, 66). The connectors or shaft domains can be coiled-coils or β-structures such as triple β-spirals and triple β-helices (66). Native trimeric viral fibers are dissociated into monomers by SDS-PAGE buffer only after heating (67, 68). The same is true for all three Pgp3 constructs examined in this study. The interlocking nature of the chain swapping interactions in the NTD (Fig. 3c), as well as the unusual stability known for TNF-like proteins (63), likely confers the SDS-resistant property to the Pgp3 protein despite its loosely packed THCC. Structural similarity of the Pgp3 NTD to motifs found in viral fiber proteins, the similarity of the CTD to the TNF-like proteins that play roles in receptor binding, and the presence of a supercoil connecting domain perhaps hint at its biological role.
Pgp3 Crystal Packing Suggests Two “Hotspots” for Protein-Protein Interactions
As shown in Figs. 1 and 6a, CTD residue Trp-234 is conspicuously displayed in a shallow groove that essentially forces the apolar portion of its indole ring to protrude into solvent. The groove does not permit the Trp-234 indole ring to adopt alternate rotameric conformations. Equally conspicuous is the flat, solvent-exposed surface formed by the meeting of the aromatic rings coming from symmetrical Phe-6 residues of the β-helical motif of the Pgp3 NTD (Fig. 6a). The edges of the six-membered rings meet precisely at the local 3-fold axis of rotation, with the ring planes normal to the long axis of the molecule. This arrangement completely seals the end of the trimer while simultaneously creating a large hydrophobic patch.
The apolar edge of the solvent-exposed indole ring of Trp-234 from one Pgp3 trimer interacts with the flat apolar surface formed by Phe-6 residues in the immediately adjacent Pgp3 trimer. These contacts dominate the critical crystal packing interactions, giving rise to the NTD-to-CTD arrangement that produces large solvent channels in the crystal lattice (Fig. 6). A second packing contact involves Trp-234 at an interface between neighboring CTDs. The solvent-exposed nature of the Trp-234 indole ring and the planar arrangement of the NTD residues Phe-6 appear to be “hotspots” for protein-protein interactions and could conceivably be responsible for the immunogenic nature of the molecule. It is possible that the stalk-like Pgp3 molecule plays a role in adhesion between the chlamydial organism and its host cell by analogy to structural features observed in TNF-like and viral fiber proteins. Under this scenario, the CTD TNF-like domains would likely recognize and adhere to the mammalian host cell, whereas the NTD would adhere to the chlamydial cell after secretion to facilitate cell invasion. In conclusion, the Pgp3 structure determined here will act as a guide in the design of experiments to test the roles of its structural elements in chlamydial infection, host recognition, and receptor binding.
Acknowledgment
We thank Dr. Sergei Strelkov (Katholieke Universiteit Leuven, Belgium) for helpful discussions. Support for the X-ray Crystallography Core Laboratory by the Office of the Vice President for Research at the University of Texas Health Science Center at San Antonio is gratefully acknowledged.
This work was supported, in whole or in part, by National Institutes of Health Grants AI47997 and AI64537 (to G. Z.). This work was also supported by Robert A. Welch Foundation Grant AQ-1399 (to P. J. H.). This work is partially the result of research conducted at the Northeastern Collaborative Access Team beamlines of the Advanced Photon Source, supported by Award RR-15301 from the National Center for Research Resources at the National Institute of Health. Use of the Advanced Photon Source is supported by the U.S. Department of Energy, Office of Basic Energy Sciences under Contract W-31-109-ENG-38. This work was also supported in part by Cancer Therapy & Research Center Cancer Center Support Grant NCI P30CA054174 and Contract Number W911NF-11-1-0136 from the United States Department of Defense to the UTSA/UTHSCSA Center for Excellence in Genomics Research.
The atomic coordinates and structure factors (codes 4DJM, 4DJN, and 4DJO) have been deposited in the Protein Data Bank (http://wwpdb.org/).
- CTD
- C-terminal domain
- NTD
- N-terminal domain
- THCC
- triple-helical coiled-coil
- PDB
- Protein Data Bank
- RMSD
- root mean square deviation.
REFERENCES
- 1. Da Ros C. T., Schmitt Cda S. (2008) Global epidemiology of sexually transmitted diseases. Asian J. Androl. 10, 110–114 [DOI] [PubMed] [Google Scholar]
- 2. den Hartog J. E., Morré S. A., Land J. A. (2006) Chlamydia trachomatis-associated tubal factor subfertility. Immunogenetic aspects and serological screening. Hum. Reprod. Update 12, 719–730 [DOI] [PubMed] [Google Scholar]
- 3. Lu C., Holland M. J., Gong S., Peng B., Bailey R. L., Mabey D. W., Wu Y., Zhong G. (2012) Genome-wide identification of Chlamydia trachomatis antigens associated with trachomatous trichiasis. Invest. Ophthalmol. Vis. Sci. 53, 2551–2559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Stephens R. S. (2003) The cellular paradigm of chlamydial pathogenesis. Trends Microbiol. 11, 44–51 [DOI] [PubMed] [Google Scholar]
- 5. Clifton D. R., Fields K. A., Grieshaber S. S., Dooley C. A., Fischer E. R., Mead D. J., Carabeo R. A., Hackstadt T. (2004) A chlamydial type III translocated protein is tyrosine-phosphorylated at the site of entry and associated with recruitment of actin. Proc. Natl. Acad. Sci. U.S.A. 101, 10166–10171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Engel J. (2004) Tarp and Arp. How Chlamydia induces its own entry. Proc. Natl. Acad. Sci. U.S.A. 101, 9947–9948 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Hybiske K., Stephens R. S. (2007) Mechanisms of host cell exit by the intracellular bacterium Chlamydia. Proc. Natl. Acad. Sci. U.S.A. 104, 11430–11435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Li Z., Chen D., Zhong Y., Wang S., Zhong G. (2008) The chlamydial plasmid-encoded protein pgp3 is secreted into the cytosol of Chlamydia-infected cells. Infect. Immun. 76, 3415–3428 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Seth-Smith H. M., Harris S. R., Persson K., Marsh P., Barron A., Bignell A., Bjartling C., Clark L., Cutcliffe L. T., Lambden P. R., Lennard N., Lockey S. J., Quail M. A., Salim O., Skilton R. J., Wang Y., Holland M. J., Parkhill J., Thomson N. R., Clarke I. N. (2009) Co-evolution of genomes and plasmids within Chlamydia trachomatis and the emergence in Sweden of a new variant strain. BMC Genomics 10, 239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thomas N. S., Lusher M., Storey C. C., Clarke I. N. (1997) Plasmid diversity in Chlamydia. Microbiology 143, 1847–1854 [DOI] [PubMed] [Google Scholar]
- 11. Carlson J. H., Whitmire W. M., Crane D. D., Wicke L., Virtaneva K., Sturdevant D. E., Kupko J. J., 3rd, Porcella S. F., Martinez-Orengo N., Heinzen R. A., Kari L., Caldwell H. D. (2008) The Chlamydia trachomatis plasmid is a transcriptional regulator of chromosomal genes and a virulence factor. Infect. Immun. 76, 2273–2283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Kari L., Whitmire W. M., Olivares-Zavaleta N., Goheen M. M., Taylor L. D., Carlson J. H., Sturdevant G. L., Lu C., Bakios L. E., Randall L. B., Parnell M. J., Zhong G., Caldwell H. D. (2011) A live-attenuated chlamydial vaccine protects against trachoma in nonhuman primates. J. Exp. Med. 208, 2217–2223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. O'Connell C. M., Ingalls R. R., Andrews C. W., Jr., Scurlock A. M., Darville T. (2007) Plasmid-deficient Chlamydia muridarum fail to induce immune pathology and protect against oviduct disease. J. Immunol. 179, 4027–4034 [DOI] [PubMed] [Google Scholar]
- 14. Wang Y., Kahane S., Cutcliffe L. T., Skilton R. J., Lambden P. R., Clarke I. N. (2011) Development of a transformation system for Chlamydia trachomatis. Restoration of glycogen biosynthesis by acquisition of a plasmid shuttle vector. PLoS Pathog. 7, e1002258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Song L., Carlson J. H., Whitmire W. M., Kari L., Virtaneva K., Sturdevant D. E., Watkins H., Zhou B., Sturdevant G. L., Porcella S. F., McClarty G., Caldwell H. D. (2013) Chlamydia trachomatis plasmid-encoded Pgp4 is a transcriptional regulator of virulence-associated genes. Infect. Immun. 81, 636–644 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chen D., Lei L., Lu C., Galaleldeen A., Hart P. J., Zhong G. (2010) Characterization of Pgp3, a Chlamydia trachomatis plasmid-encoded immunodominant antigen. J. Bacteriol. 192, 6017–6024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Comanducci M., Cevenini R., Moroni A., Giuliani M. M., Ricci S., Scarlato V., Ratti G. (1993) Expression of a plasmid gene of Chlamydia trachomatis encoding a novel 28 kDa antigen. J. Gen. Microbiol. 139, 1083–1092 [DOI] [PubMed] [Google Scholar]
- 18. Budrys N. M., Gong S., Rodgers A. K., Wang J., Louden C., Shain R., Schenken R. S., Zhong G. (2012) Chlamydia trachomatis antigens recognized in women with tubal factor infertility, normal fertility, and acute infection. Obstet. Gynecol. 119, 1009–1016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Wang J., Zhang Y., Lu C., Lei L., Yu P., Zhong G. (2010) A genome-wide profiling of the humoral immune response to Chlamydia trachomatis infection reveals vaccine candidate antigens expressed in humans. J. Immunol. 185, 1670–1680 [DOI] [PubMed] [Google Scholar]
- 20. Donati M., Sambri V., Comanducci M., Di Leo K., Storni E., Giacani L., Ratti G., Cevenini R. (2003) DNA immunization with pgp3 gene of Chlamydia trachomatis inhibits the spread of chlamydial infection from the lower to the upper genital tract in C3H/HeN mice. Vaccine 21, 1089–1093 [DOI] [PubMed] [Google Scholar]
- 21. Caspar D. L., Clarage J., Salunke D. M., Clarage M. (1988) Liquid-like movements in crystalline insulin. Nature 332, 659–662 [DOI] [PubMed] [Google Scholar]
- 22. Hendrickson W. A., Horton J. R., LeMaster D. M. (1990) Selenomethionyl proteins produced for analysis by multiwavelength anomalous diffraction (MAD). A vehicle for direct determination of three-dimensional structure. EMBO J. 9, 1665–1672 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Seetharaman S. V., Taylor A. B., Holloway S., Hart P. J. (2010) Structures of mouse SOD1 and human/mouse SOD1 chimeras. Arch. Biochem. Biophys. 503, 183–190 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. McPherson A. (1999) Crystallization of Biological Macromolecules, Cold Spring Harbor Laboratory, Cold Spring Harbor, New York [Google Scholar]
- 25. Otwinowski Z., Minor W. (1997) Processing of x-ray diffraction data collected in oscillation mode. Methods Enzymol. 276, 307–326 [DOI] [PubMed] [Google Scholar]
- 26. Sheldrick G. M. (2008) A short history of SHELX. Acta Crystallogr. A 64, 112–122 [DOI] [PubMed] [Google Scholar]
- 27. Vonrhein C., Blanc E., Roversi P., Bricogne G. (2007) Automated structure solution with autoSHARP. Methods Mol. Biol. 364, 215–230 [DOI] [PubMed] [Google Scholar]
- 28. Terwilliger T. C. (2000) Maximum-likelihood density modification. Acta Crystallogr. D Biol. Crystallogr. 56, 965–972 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Emsley P., Cowtan K. (2004) Coot. Model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 60, 2126–2132 [DOI] [PubMed] [Google Scholar]
- 30. Lamzin V. S., Perrakis A., Wilson K. S. (2001) The ARP/WARP suite for automated construction and refinement of protein models, in International Tables for Crystallography (Rossmann M. G., Arnold E., eds) pp. 720–722, Kluwer Academic Publishers, Dordrecht, The Netherlands [Google Scholar]
- 31. Adams P. D., Afonine P. V., Bunkóczi G., Chen V. B., Davis I. W., Echols N., Headd J. J., Hung L. W., Kapral G. J., Grosse-Kunstleve R. W., McCoy A. J., Moriarty N. W., Oeffner R., Read R. J., Richardson D. C., Richardson J. S., Terwilliger T. C., Zwart P. H. (2010) PHENIX. A comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 66, 213–221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. McCoy A. J., Grosse-Kunstleve R. W., Adams P. D., Winn M. D., Storoni L. C., Read R. J. (2007) Phaser crystallographic software. J. Appl. Crystallogr. 40, 658–674 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Dolinsky T. J., Czodrowski P., Li H., Nielsen J. E., Jensen J. H., Klebe G., Baker N. A. (2007) PDB2PQR. Expanding and upgrading automated preparation of biomolecular structures for molecular simulations. Nucleic Acids Res. 35, W522–W525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Baker N. A., Sept D., Joseph S., Holst M. J., McCammon J. A. (2001) Electrostatics of nanosystems. Application to microtubules and the ribosome. Proc. Natl. Acad. Sci. U.S.A. 98, 10037–10041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Eck M. J., Sprang S. R. (1989) The structure of tumor necrosis factor-α at 2.6 A resolution. Implications for receptor binding. J. Biol. Chem. 264, 17595–17605 [DOI] [PubMed] [Google Scholar]
- 36. Holm L., Rosenström P. (2010) Dali server. Conservation mapping in 3D. Nucleic Acids Res. 38, W545–W549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Matthews B. W. (1968) Solvent content of protein crystals. J. Mol. Biol. 33, 491–497 [DOI] [PubMed] [Google Scholar]
- 38. Yoder M. D., Keen N. T., Jurnak F. (1993) New domain motif. The structure of pectate lyase C, a secreted plant virulence factor. Science 260, 1503–1507 [DOI] [PubMed] [Google Scholar]
- 39. Lo Conte L., Ailey B., Hubbard T. J., Brenner S. E., Murzin A. G., Chothia C. (2000) SCOP. A structural classification of proteins database. Nucleic Acids Res. 28, 257–259 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Stummeyer K., Dickmanns A., Mühlenhoff M., Gerardy-Schahn R., Ficner R. (2005) Crystal structure of the polysialic acid-degrading endosialidase of bacteriophage K1F. Nat. Struct. Mol. Biol. 12, 90–96 [DOI] [PubMed] [Google Scholar]
- 41. Corbett K. D., Shultzaberger R. K., Berger J. M. (2004) The C-terminal domain of DNA gyrase A adopts a DNA-bending β-pinwheel fold. Proc. Natl. Acad. Sci. U.S.A. 101, 7293–7298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Murzin A. G. (1992) Structural principles for the propeller assembly of β-sheets. the preference for seven-fold symmetry. Proteins 14, 191–201 [DOI] [PubMed] [Google Scholar]
- 43. Chen C. K., Chan N.-L., Wang A. H. (2011) The many blades of the β-propeller proteins. Conserved but versatile. Trends Biochem. Sci. 36, 553–561 [DOI] [PubMed] [Google Scholar]
- 44. Boudko S. P., Londer Y. Y., Letarov A. V., Sernova N. V., Engel J., Mesyanzhinov V. V. (2002) Domain organization, folding and stability of bacteriophage T4 fibritin, a segmented coiled-coil protein. Eur. J. Biochem. 269, 833–841 [DOI] [PubMed] [Google Scholar]
- 45. Letarov A. V., Londer Y. Y., Boudko S. P., Mesyanzhinov V. V. (1999) The carboxy-terminal domain initiates trimerization of bacteriophage T4 fibritin. Biochemistry 64, 817–823 [PubMed] [Google Scholar]
- 46. Tao Y., Strelkov S. V., Mesyanzhinov V. V., Rossmann M. G. (1997) Structure of bacteriophage T4 fibritin. A segmented coiled coil and the role of the C-terminal domain. Structure 5, 789–798 [DOI] [PubMed] [Google Scholar]
- 47. Güthe S., Kapinos L., Möglich A., Meier S., Grzesiek S., Kiefhaber T. (2004) Very fast folding and association of a trimerization domain from bacteriophage T4 fibritin. J. Mol. Biol. 337, 905–915 [DOI] [PubMed] [Google Scholar]
- 48. van Raaij M. J., Mitraki A., Lavigne G., Cusack S. (1999) A triple β-spiral in the adenovirus fibre shaft reveals a new structural motif for a fibrous protein. Nature 401, 935–938 [DOI] [PubMed] [Google Scholar]
- 49. Reiter D. M., Frierson J. M., Halvorson E. E., Kobayashi T., Dermody T. S., Stehle T. (2011) Crystal structure of reovirus attachment protein sigma1 in complex with sialylated oligosaccharides. PLoS Pathog. 7, e1002166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Harbury P. B., Plecs J. J., Tidor B., Alber T., Kim P. S. (1998) High-resolution protein design with backbone freedom. Science 282, 1462–1467 [DOI] [PubMed] [Google Scholar]
- 51. Lupas A. (1996) Coiled coils. New structures and new functions. Trends Biochem. Sci 21, 375–382 [PubMed] [Google Scholar]
- 52. Peters J., Baumeister W., Lupas A. (1996) Hyperthermostable surface layer protein tetrabrachion from the archaebacterium Staphylothermus marinus. Evidence for the presence of a right-handed coiled coil derived from the primary structure. J. Mol. Biol. 257, 1031–1041 [DOI] [PubMed] [Google Scholar]
- 53. Stetefeld J., Jenny M., Schulthess T., Landwehr R., Engel J., Kammerer R. A. (2000) Crystal structure of a naturally occurring parallel right-handed coiled coil tetramer. Nat. Struct. Biol. 7, 772–776 [DOI] [PubMed] [Google Scholar]
- 54. Strelkov S. V., Burkhard P. (2002) Analysis of α-helical coiled coils with the program TWISTER reveals a structural mechanism for stutter compensation. J. Struct. Biol. 137, 54–64 [DOI] [PubMed] [Google Scholar]
- 55. Lupas A. N., Gruber M. (2005) The structure of α-helical coiled coils. Adv. Protein Chem. 70, 37–78 [DOI] [PubMed] [Google Scholar]
- 56. Thomas W. E., Trintchina E., Forero M., Vogel V., Sokurenko E. V. (2002) Bacterial adhesion to target cells enhanced by shear force. Cell 109, 913–923 [DOI] [PubMed] [Google Scholar]
- 57. Sulák O., Cioci G., Delia M., Lahmann M., Varrot A., Imberty A., Wimmerová M. (2010) A TNF-like trimeric lectin domain from Burkholderia cenocepacia with specificity for fucosylated human histo-blood group antigens. Structure 18, 59–72 [DOI] [PubMed] [Google Scholar]
- 58. Païdassi H., Tacnet-Delorme P., Garlatti V., Darnault C., Ghebrehiwet B., Gaboriaud C., Arlaud G. J., Frachet P. (2008) C1q binds phosphatidylserine and likely acts as a multiligand-bridging molecule in apoptotic cell recognition. J. Immunol. 180, 2329–2338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Réty S., Salamitou S., Garcia-Verdugo I., Hulmes D. J., Le Hégarat F., Chaby R., Lewit-Bentley A. (2005) The crystal structure of the Bacillus anthracis spore surface protein BclA shows remarkable similarity to mammalian proteins. J. Biol. Chem. 280, 43073–43078 [DOI] [PubMed] [Google Scholar]
- 60. Sulák O., Cioci G., Lameignère E., Balloy V., Round A., Gutsche I., Malinovská L., Chignard M., Kosma P., Aubert D. F., Marolda C. L., Valvano M. A., Wimmerová M., Imberty A. (2011) Burkholderia cenocepacia BC2L-C is a super lectin with dual specificity and proinflammatory activity. PLoS Pathog. 7, e1002238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Ghebrehiwet B., Hosszu K. K., Valentino A., Peerschke E. I. (2012) The C1q family of proteins. Insights into the emerging non-traditional functions. Front. Immunol. 3, 52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Ghebrehiwet B., Peerschke E. I. (2004) cC1q-R (calreticulin) and gC1q-R/p33. Ubiquitously expressed multi-ligand binding cellular proteins involved in inflammation and infection. Mol. Immunol. 41, 173–183 [DOI] [PubMed] [Google Scholar]
- 63. Boydston J. A., Chen P., Steichen C. T., Turnbough C. L., Jr. (2005) Orientation within the exosporium and structural stability of the collagen-like glycoprotein BclA of Bacillus anthracis. J. Bacteriol. 187, 5310–5317 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Merckel M. C., Huiskonen J. T., Bamford D. H., Goldman A., Tuma R. (2005) The structure of the bacteriophage PRD1 spike sheds light on the evolution of viral capsid architecture. Mol. Cell 18, 161–170 [DOI] [PubMed] [Google Scholar]
- 65. van Raaij M. J., Schoehn G., Burda M. R., Miller S. (2001) Crystal structure of a heat and protease-stable part of the bacteriophage T4 short tail fibre. J. Mol. Biol. 314, 1137–1146 [DOI] [PubMed] [Google Scholar]
- 66. Mitraki A., Miller S., van Raaij M. J. (2002) Conformation and folding of novel β-structural elements in viral fiber proteins. The triple β-spiral and triple β-helix. J. Struct. Biol. 137, 236–247 [DOI] [PubMed] [Google Scholar]
- 67. King J., Laemmli U. K. (1971) Polypeptides of the tail fibres of bacteriophage T4. J. Mol. Biol. 62, 465–477 [DOI] [PubMed] [Google Scholar]
- 68. Mitraki A., Barge A., Chroboczek J., Andrieu J. P., Gagnon J., Ruigrok R. W. (1999) Unfolding studies of human adenovirus type 2 fibre trimers. Evidence for a stable domain. Eur. J. Biochem. 264, 599–606 [DOI] [PubMed] [Google Scholar]
- 69. Hymowitz S. G., Compaan D. M., Yan M., Wallweber H. J., Dixit V. M., Starovasnik M. A., de Vos A. M. (2003) The crystal structures of EDA-A1 and EDA-A2. Splice variants with distinct receptor specificity. Structure 11, 1513–1520 [DOI] [PubMed] [Google Scholar]
- 70. Shapiro L., Scherer P. E. (1998) The crystal structure of a complement-1q family protein suggests an evolutionary link to tumor necrosis factor. Curr. Biol. 8, 335–338 [DOI] [PubMed] [Google Scholar]
- 71. Kvansakul M., Bogin O., Hohenester E., Yayon A. (2003) Crystal structure of the collagen α1(VIII) NC1 trimer. Matrix Biol. 22, 145–152 [DOI] [PubMed] [Google Scholar]
- 72. Liu C. Q., Nuttall S. D., Tran H., Wilkins M., Streltsov V. A., Alderton M. R. (2008) Construction, crystal structure and application of a recombinant protein that lacks the collagen-like region of BclA from Bacillus anthracis spores. Biotechnol. Bioeng. 99, 774–782 [DOI] [PubMed] [Google Scholar]
- 73. Cha S. S., Kim M. S., Choi Y. H., Sung B. J., Shin N. K., Shin H. C., Sung Y. C., Oh B. H. (1999) 2.8 A resolution crystal structure of human TRAIL, a cytokine with selective antitumor activity. Immunity 11, 253–261 [DOI] [PubMed] [Google Scholar]
- 74. Coulibaly F., Chevalier C., Delmas B., Rey F. A. (2010) Crystal structure of an Aquabirnavirus particle. Insights into antigenic diversity and virulence determinism. J. Virol. 84, 1792–1799 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Bogin O., Kvansakul M., Rom E., Singer J., Yayon A., Hohenester E. (2002) Insight into Schmid metaphyseal chondrodysplasia from the crystal structure of the collagen X NC1 domain trimer. Structure 10, 165–173 [DOI] [PubMed] [Google Scholar]
- 76. Bond C. S. (2003) TopDraw. A sketchpad for protein structure topology cartoons. Bioinformatics 19, 311–312 [DOI] [PubMed] [Google Scholar]