Abstract
Werner syndrome (WS) is a premature aging disorder where the affected individuals appear much older than their chronological age. The single gene that is defective in WS encodes a protein (WRN) that has ATPase, helicase and 3′→5′ exonuclease activities. Our laboratory has recently uncovered a physical and functional interaction between WRN and the Ku heterodimer complex that functions in double-strand break repair and V(D)J recombination. Importantly, Ku specifically stimulates the exonuclease activity of WRN. We now report that Ku enables the Werner exonuclease to digest through regions of DNA containing 8-oxoadenine and 8-oxoguanine modifications, lesions that have previously been shown to block the exonuclease activity of WRN alone. These results indicate that Ku significantly alters the exonuclease function of WRN and suggest that the two proteins function concomitantly in a DNA damage processing pathway. In support of this notion we also observed co-localization of WRN and Ku, particularly after DNA damaging treatments.
INTRODUCTION
Werner syndrome (WS) is a human autosomal recessive disorder characterized by early onset of premature aging characteristics, including graying and loss of hair, wrinkling and ulceration of skin, atherosclerosis, osteoporosis and cataracts. In addition, WS patients also exhibit an increased incidence of diabetes mellitus type II, hypertension and malignancies (1,2). The gene WRN, defects in which are responsible for WS, was recently cloned (3) and shown to encode a 1432 amino acid protein (WRN) that has both 3′→5′ helicase and 3′→5′ exonuclease activities (4–8). Although WRN appears to play an important role in DNA metabolism, the precise cellular roles of both the helicase and exonuclease activities of WRN remain to be determined.
Cells from patients with WS show premature replicative senesence as compared to cells derived from normal individuals (9). The WS cellular phenotype suggests correlations between faulty DNA metabolism, genomic instability and senescence. WS cells show hypersensitivity to selected DNA-damaging agents, including 4-nitroquinoline-1-oxide (4NQO) (10) and topoisomerase inhibitors (11). As compared to normal cells, WS cells also exhibit increased genomic instability, including higher levels of DNA deletions, translocations and chromosomal breaks (12,13). These studies suggest that WRN plays an important role in DNA metabolism by possibly participating in DNA repair, replication and/or recombination pathways. Like WS, other genetic diseases such as Cockayne syndrome, xeroderma pigmentosum and Bloom syndrome are also characterized by genomic instability, signs of premature aging and increased frequency of cancer. The phenotypes of some of these syndromes appear to be at least partially attributable to defects in specific DNA repair pathways (14).
We have previously identified a strong physical and functional interaction between WRN and Ku, a heterodimer of 68 and 83 kDa subunits (15). This finding has recently been confirmed by an independent laboratory (16). Together with the catalytic subunit of the DNA-dependent protein kinase (DNA-PKcs), Ku is required for repair of double-strand breaks generated during V(D)J recombination and by reactive oxygen species resulting from endogenous metabolism or treatment with ionizing radiation and certain mutagens (17). We have also shown that Ku significantly stimulates the 3′→5′ exonuclease activity of WRN (15), suggesting that WRN and Ku could act in a common pathway of DNA metabolism involving the exonuclease function of WRN. This hypothesis is supported by the observation that mice lacking the Ku83 subunit show a premature aging phenotype similar to WS patients (18). Furthermore, cells deficient in WRN, Ku68 or Ku83 all show genomic instability and undergo premature replicative senescence (19,20).
The main goal of this study was to further examine the interaction between WRN and the Ku complex, particularly with respect to how Ku modifies the enzymatic characteristics of WRN exonuclease. A recent study from our laboratory has shown that the 3′→5′ exonuclease activity of WRN is selectively blocked by certain kinds of modified nucleotides, including apurinic sites, 8-oxoguanine (8-oxoG), 8-oxoadenine (8-oxoA) and cholesterol adducts (21). Here we report that the addition of Ku helped the exonuclease activity of WRN overcome the blockage imposed by 8-oxoA and 8-oxoG adducts but was much less effective in promoting exonucleolytic digestion past an apurinic site or cholesterol moiety. We also show co-immunoprecipitation of purified WRN and Ku and co-localization of WRN and Ku in normal human cells. These results confirm the physical interaction between WRN and Ku and indicate that modification of the exonucleolytic function of WRN by Ku could be important in the processing of specific types of DNA damage.
MATERIALS AND METHODS
Purification of WRN protein
Recombinant human WRN protein was overproduced in baculovirus-infected insect cells and purified to >90% homogeneity essentially as previously described (8). Briefly, the wild-type human WRN cDNA sequence was inserted into a baculoviral DNA construct (Invitrogen, Carlsbad, CA) such that the WRN protein produced contained an N-terminal hexahistidine tag. Sf9 cells were harvested 72–96 h after infection with amplified baculovirus (m.o.i. = 10) and then tested for overproduction of WRN by SDS–PAGE. After cell lysis in mild non-ionic detergent, WRN purification was accomplished by sequential liquid chromatography using DEAE–Sepharose (Pharmacia, Piscatway, NJ), Q-Sepharose (Pharmacia) and Ni–NTA (Qiagen, Valencia, CA) resins. Upon elution from the Ni–NTA affinity resin, 100 µg/ml bovine serum albumin (BSA) was added to peak fractions to stabilize WRN catalytic activities and the protein was stored at –80°C.
Enzymes
Exonuclease III (exo III) and the Klenow fragment of Escherichia coli were purchased from Boehringer Mannheim and T4 polynucleotide kinase was from New England Biolabs (Beverly, MA). Escherichia coli formamidopyrimidine glycosylase (Fpg) and human apurinic/apyrimidinic endonuclease (APE) were gifts from A.Grollman (State University of New York, Stony Brook, NY) and S.Wilson (NIEHS, Research Triangle Park, NC), respectively. Human replication protein A (hRPA) was obtained from M.Kenny (The Albert Einstein Cancer Center, Bronx, NY) and human Ku heterodimer was provided by D.Ramsden (University of North Carolina, Chapel Hill, NC). hRPA and recombinant Ku were purified to near homogeneity as previously reported (22,23).
Immunoprecipitation
Purified WRN and Ku (1 µg each) were incubated together for 1 h at 4°C in IP buffer (50 mM HEPES pH 7.4, 0.1 M NaCl, 0.05% Triton X-100, 1 mM EDTA, 1 mM NaVO4, 1 mM NaF, 1 mM dithiothreitol, 1 mM phenylmethylsulfonyl fluoride and 10 µg/ml each leupeptin, pepstatin A and aprotinin) and pre-cleared with mouse immunoglobulin as described previously (15). The samples were incubated for 1 h at 4°C with either anti-Ku antibody, anti-WRN antibody or mouse immunoglobulin (as a control) and then pre-swollen protein A/G beads (Gibco BRL, Rockville, MD) were added. The immunoprecipitates were pelleted and washed four times with IP buffer, then subjected to SDS–PAGE and western blotting, probing sequentially with anti-WRN, anti-Ku68 and anti-Ku83 antibodies.
DNA substrates
Single-stranded DNA oligomers (32mer, 43mer and 72mer) containing only normal nucleotides were obtained from Gibco BRL. 32mers and 53mers containing modified (8-oxoA, 8-oxoG, an apurinic site or cholesterol) nucleotides were purchased from Midland (Midland, TX). The sequences of the various oligomers and the positions of damaged bases are shown in Table 1. Individual 32mers or 53mers (7 pmol) were 5′-labeled with [γ-32P]ATP (60 µCi, 3000 Ci/mmol) and T4 polynucleotide kinase (10 U) using standard conditions. For construction of a double-stranded DNA substrate with one blunt end and one 3′-recessed (5′-overhang) end, labeled oligomers (undamaged or modified) were mixed with a 2-fold excess of unlabeled 43mer or 72mer, heated together at 90°C for 5 min, then cooled slowly to 25°C. Annealed double-stranded substrates were then separated from unannealed and excess single-stranded oligomers by non-denaturing 12% PAGE. Intact double-stranded DNA substrates were recovered using a Qiaex II gel extraction kit (Qiagen) and stored at 4°C. The presence and location of 8-oxoG or apurinic sites in double-stranded substrates was confirmed using Fpg or APE as described previously (21).
Table 1. DNA oligomers.
The nucleotide modifications are underlined and depicted as follows: G°, 8-oxoG; A°, 8-oxoA; ^, apurinic site; X, cholesterol. The position of the 32P-label at the 5′-end of each 32mer and 53mer is denoted by an asterisk.
The unmodified, unlabeled 43mer and 72mer (to which 32mers and 53mers, respectively, were annealed) are listed in 3′→5′ orientation, aligned with the oligomers above with regard to base pairing structure.
Exonuclease assays
Assays to measure the 3′→5′ exonuclease activity of WRN, exo III and Klenow were carried out as described previously (21). Unmodified or damaged DNA substrates (3 fmol) were incubated with Klenow (2 U), exo III (1 U) or WRN (180–1440 fmol) and with Ku (64–1280 fmol) or hRPA (50–2000 fmol) as indicated for 1 h at 37°C. The reactions were stopped and the DNA digestion products were separated on denaturing 15% polyacrylamide gels and visualized using a PhosphorImager (Molecular Dynamics, Sunnyvale, CA).
Immunofluorescence
SV40-transformed normal human fibroblasts (GM00637) were grown in minimum essential medium (MEM) supplemented with 10% fetal bovine serum, l-glutamine (2 mM), vitamins (1%), amino acids (1%), penicillin (100 U/ml) and streptomycin (100 µg/ml). HeLa cells were grown in DMEM with 10% fetal bovine serum. For both cell lines at least three independent experiments were done. The cells were either untreated or treated with 4NQO at 0.1 µg/ml for 12 h, grown on coverslips for 24 (HeLa) or 48 h (GM00637), then washed with phosphate-buffered saline (PBS) and fixed with freshly prepared 2% paraformaldehyde for 10 min at 20°C. After another wash, the cells were lysed with 0.15% Triton X-100 for 10 min at 4°C. The coverslips were then washed with cold PBS and blocked with 2% BSA in PBS at 20°C. The coverslips were incubated with rabbit polyclonal anti-WRN (1:1000; Novus, Littleton, CO) and goat polyclonal anti-Ku (1:200; Santa Cruz Biotechnology, Santa Cruz, CA) antibodies for 16 h at 4°C, then incubated simultaneously with donkey anti-rabbit Texas red and donkey anti-goat Alexa 588 (Molecular Probes) for 1 h at 20°C. After three washes for 10 min each the coverslips were mounted on Vectashield (Vector Laboratories) and viewed under a Zeiss model 410 laser scan confocal microscope using the green (488 nm) and red (568 nm) channels. The images were analyzed and overlaid using the Metamorph imaging system 4.1 (Universal Imaging). Coincidence of the immunofluorescent signals was determined by combining data from more than 15 cells. SV-40 transformed WS cells (AG11395) were used as controls to check the specificity of the WRN antibody.
RESULTS
Interaction of purified WRN and Ku
We have previously shown that WRN and the Ku heterodimer co-immunoprecipitate from HeLa nuclear extracts using either a WRN or a Ku antibody (15). We wanted to determine whether WRN and the Ku heterodimer could directly interact in vitro. Using an antibody raised against the Ku heterodimer and highly purified recombinant WRN and Ku preparations we were able to immunoprecipitate both the Ku68 and Ku83 subunits as expected. Moreover, this antibody was also effective in pulling down WRN only when Ku was present, while mouse immunoglobulin was not able to precipitate Ku68, Ku83 or WRN (Fig. 1). Similarly, using an antibody to WRN, immunoprecipitation of Ku was accomplished only in the presence of WRN (data not shown). As DNA was undetectable (by Cyber Green staining) in both protein preparations, the interaction between WRN and Ku appears not to be mediated by DNA. This is further supported by the finding that Ku and WRN co-immunoprecipitate from DNase I-treated HeLa nuclear extracts (data not shown; 16). The co-immunoprecipitation of purified recombinant WRN with recombinant Ku confirms the physical interaction shown previously between the native proteins in nuclear extracts. This result also indicates that the interaction between WRN and Ku is not mediated by other components that were present in human nuclear extracts and that possible modifications of either native WRN or Ku that might occur in human cells are not essential for this physical interaction. In general agreement with these results, a recent study demonstrated a physical interaction between the purified recombinant Ku83 subunit and WRN (16).
Figure 1.
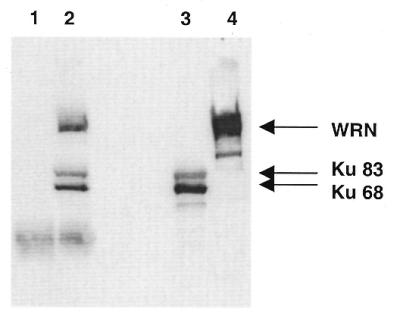
Interaction between purified recombinant Ku and WRN. As described in Materials and Methods, purified recombinant WRN and Ku (1 µg each) were incubated with either mouse immunoglobulin (lane 1) or anti-Ku antibody (lane 2), then precipitated using protein A/G beads. The immunoprecipitates were subjected to SDS–PAGE and western blotting using anti-Ku68, anti-Ku83 and anti-WRN antibodies. Purified Ku heterodimer (lane 3) and WRN (lane 4) were loaded as markers. WRN was not immunoprecipitated by the anti-Ku antibody in the absence of Ku (data not shown).
Effect of Ku on WRN exonuclease digestion of modified DNA substrates
WRN possesses both 3′→5′ helicase and 3′→5′ exonuclease activities (4–6). The 3′→5′ exonuclease activity of WRN requires a single-stranded 5′-overhang (recessed 3′-end) and does not appear to act on single-stranded DNA or double-stranded DNA with blunt ends (6). Notably, WRN helicase activity will not unwind a substrate that does not have a single-stranded 3′-overhang (7). We recently demonstrated that on a DNA substrate (containing only normal nucleotides) with one blunt end and one recessed 3′-end the Ku heterodimer stimulates the exonuclease activity of WRN but has no effect on the 3′→5′ exonuclease activity of either Klenow or exo III (15).
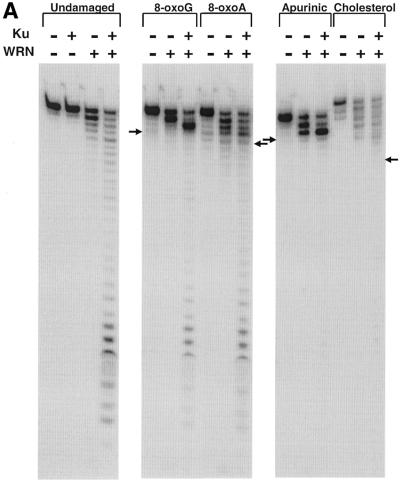
Another earlier study from our laboratory demonstrated that the exonuclease activity of WRN is severely inhibited by the presence of certain modified nucleotides in the strand of the DNA subject to degradation (21). To address a possible role for WRN in conjunction with Ku in processing DNA damage, we wanted to determine whether the stimulatory effect of Ku could help the 3′→5′ exonuclease activity of WRN digest through DNA containing modified nucleotides. Exonuclease digestion of the undamaged substrate as well as substrates containing modified nucleotides (8-oxoA, 8-oxoG, an apurinic site or a cholesterol moiety) by WRN in the presence and absence of Ku is shown in Figure 2A. Ku alone had no exonuclease activity on unmodified DNA (Fig. 2A, left) or DNA containing damaged nucleotides (21; data not shown). As reported previously, Ku significantly stimulated the 3′→5′ exonuclease activity of WRN on an undamaged substrate and, in the absence of Ku, the exonuclease activity of WRN was completely blocked or severely inhibited by substrates containing 8-oxoG, 8-oxoA, an apurinic site or a cholesterol moiety. On the undamaged substrate the extent of digestion in the absence of Ku was proportional to the WRN concentration. However, on the 8-oxoG substrate substantially higher WRN concentrations increased digestion up to but not detectably beyond the modified nucleotide (Fig. 2B). On the 8-oxoG-containing substrate the addition of Ku facilitated a higher extent of digestion by WRN both 3′ to the 8-oxoG nucleotide (resulting in a significant shift in the pause site) and even beyond the 8-oxoG nucleotide. Similarly, the addition of Ku resulted in a significant enhancement of the exonuclease activity of WRN on the substrate containing the 8-oxoA nucleotide. Notably, on the undamaged substrate the pattern of WRN digestion in the presence of Ku differed markedly from that observed when the WRN concentration was increased (compare Fig. 2A and B, left). This suggests that Ku fundamentally changes the interaction of WRN at the 3′-end of a substrate, facilitating digestion of both undamaged and 8-oxoG- and 8-oxoA-containing substrates in a manner that is distinct from simply allowing more molecules of WRN to act at the 3′-end. In contrast, although the addition of Ku increased digestion prior to the apurinic site, only marginal digestion was observed beyond the modified nucleotide (Fig. 2A). Similarly, Ku also did not significantly increase the ability of WRN to digest past a cholesterol moiety. The extent to which Ku can facilitate WRN digestion up to and past each modified nucleotide is quantitated and depicted in Figure 2C. These results clearly show that Ku permits WRN exonuclease to digest through regions containing either 8-oxoG or 8-oxoA, but this effect appears to be specific to the type of damage present in the substrate.
Figure 2.

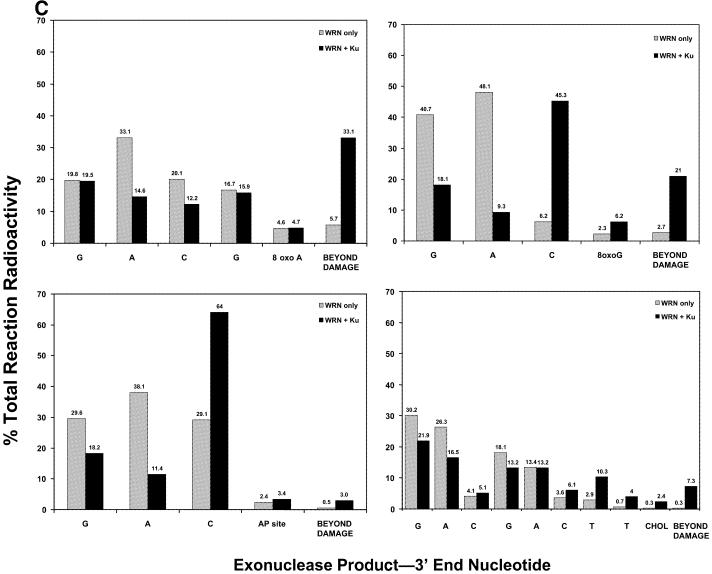
The effect of Ku on the 3′→5′ exonuclease activity of WRN on undamaged and modified DNA substrates. (A) The DNA substrates (32mers annealed to 43mers) and the positions of the modified bases (8-oxo-G, 8-oxoA, an apurinic site or a cholesterol moiety) are indicated in Table 1. The DNA substrates were incubated without enzyme (–WRN) or with WRN (0.18 pmol) in the absence or presence of Ku (64 fmol). Exonuclease reactions and subsequent analysis were carried out as described in Materials and Methods. The position of the degradation product resulting in the modified nucleotide being at the 3′-end of the labeled strand is denoted with an arrow. (B) The undamaged and 8-oxoG substrates were incubated without enzyme or with WRN (0.27–1.1 pmol) and analyzed as described in (A). (Right) The position of the degradation product resulting in the 8-oxoG nucleotide being at the 3′-end is denoted by an arrow. (C) The amounts of radioactivity associated with each labeled DNA product in the 8-oxoA-, 8-oxoG-, apurinic site- and cholesterol-containing substrate reactions from (A) were quantitated using ImageQuant software. The percentage of the total radioactivity associated with each product up to and beyond the modified nucleotide is directly compared for the WRN exonuclease reactions in the presence and absence of Ku. Both the figure and graph demonstrate clearly that Ku permits WRN to digest past certain nucleotides but not others, findings that were reproduced experimentally three or more times for each damaged substrate.
The blockage of WRN exonuclease activity by damaged nucleotides described earlier (21) and above could be related to the proximity of the damage to the 3′-end of the DNA substrate. To determine how damaged nucleotides further from the 3′-end were processed, we designed substrates containing either 8-oxoG or 8-oxoA within the same sequence context but significantly removed from the 3′-end (Table 1). Positioning of either 8-oxoG or 8-oxoA further away from the 3′-end would putatively allow the exonuclease to become maximally active prior to reaching the modification. However, in this context 8-oxoG and 8-oxoA still appeared to be absolute blocks to WRN exonuclease alone and Ku again promoted the ability of WRN to digest past these modifications (Fig. 3). These results indicate that the proximity of the modified nucleotide to the 3′-end is not a factor in either the inhibition of WRN exonuclease activity or in the ability of Ku to stimulate WRN exonuclease activity.
Figure 3.
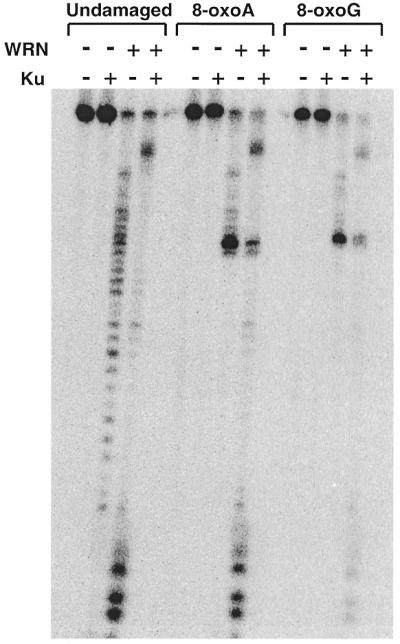
WRN exonuclease activity in the presence and absence of Ku on DNA substrates containing distal 8-oxoA and 8-oxoG modifications. DNA substrates (modified 53mers annealed to unmodified 72mers) and the sites of the 8-oxoA and 8-oxoG nucleotides are shown in Table 1. The substrates were incubated without enzyme or with WRN (1.44 pmol) and/or Ku (1.28 pmol) and reaction products were analyzed as described in Materials and Methods.
Although Klenow can digest through regions of DNA containing an apurinic site, an 8-oxoG lesion or a cholesterol moiety, its exonuclease activity is severely inhibited by the presence of 8-oxoA (21). In contrast to its effect on WRN exonuclease, Ku is unable to assist Klenow in digesting past the 8-oxoA nucleotide (Fig. 4). In addition, exo III is not blocked by the 8-oxoA nucleotide and Ku also has no significant effect on its exonuclease activity (Fig. 4). This strongly suggests that the ability of Ku to assist in digesting past at least 8-oxoA-modified nucleotides is specific to WRN exonuclease.
Figure 4.
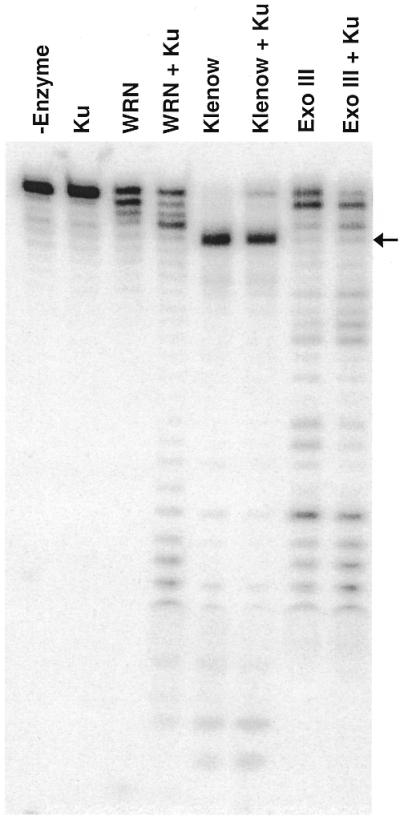
The effect of Ku on the 3′→5′ exonuclease activity of WRN, Klenow and exo III on a substrate containing 8-oxoA. DNA substrate containing 8-oxoA was incubated without enzyme (–enzyme) or with WRN (180 fmol), Klenow (2 U) or exo III (1 U) in the absence or presence of Ku (64 fmol) at 37°C for 1 h. The labeled reaction products were analyzed and visualized as described earlier.
Effect of hRPA on WRN exonuclease digestion
We also examined the effect of hRPA on the exonuclease activity of WRN. hRPA, which binds tightly to single-stranded DNA, has been previously shown to physically and functionally interact with WRN, thereby stimulating its helicase activity on long DNA substrates (24). In contrast to the effect of Ku, hRPA (over a broad concentration range) did not stimulate (but instead may have slightly inhibited) the 3′→5′ exonuclease activity of WRN (Fig. 5). Thus, Ku and hRPA would appear to have distinct roles in stimulating the exonuclease and helicase activities of WRN, respectively.
Figure 5.
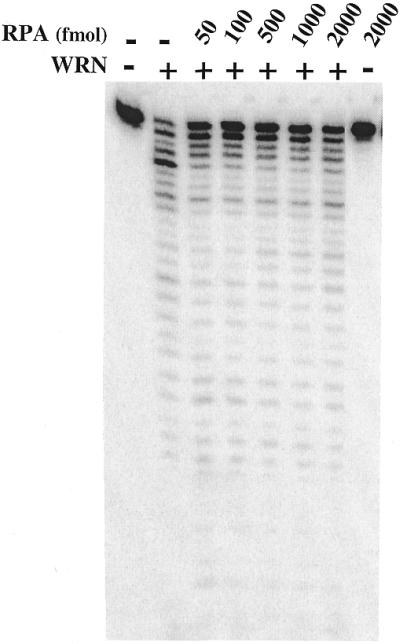
The effect of hRPA on the 3′→5′ exonuclease activity of WRN. Unmodified DNA substrate was incubated without enzyme or with WRN (360 fmol) in the presence of increasing concentrations of hRPA (50–2000 fmol). As a control the DNA substrate was also incubated with hRPA (2000 fmol) in the absence of WRN. The exonuclease reaction products were analyzed and visualized as described earlier.
Co-localization of WRN and Ku
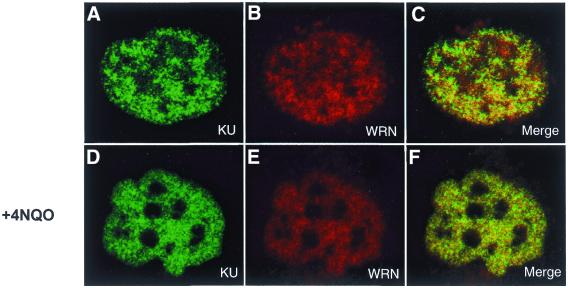
Since we have observed a physical interaction between WRN and Ku in nuclear extracts (15) and now also using purified recombinant proteins, we wanted to determine whether they might be found at the same locations in an intact cell. Wild-type (GM00637 and HeLa) and WRN-deficient (AG11395) cells were either untreated or incubated with 4NQO, a DNA-damaging agent to which WS cells have been shown to be sensitive (10). Immunofluorescence experiments using individual WRN and Ku antibodies and subsequent incubation with different fluorescent tags showed that WRN and Ku are found in multiple abundant foci throughout the nucleus of GM00637 cells. The immunofluorescence intensities from both WRN and Ku antibodies within single representative cells from the untreated and 4NQO-treated cultures are shown in Figure 6. We found no detectable immunofluorescence signal in WS cells after treatment with our WRN antibody, attesting to the specificity of this antibody for native WRN protein (data not shown). In contrast to earlier reports (25), there was no apparent concentration of WRN in nucleolar structures. In fact, WRN (and Ku) appears to be excluded from the nucleolus, partially under non-damaging conditions and completely after 4NQO treatment (Fig. 6D–F). Without 4NQO treatment some of the WRN and Ku foci coincided while others appeared to be independent of one another (Fig. 6A–C). When the cells were treated with 4NQO, the coincidence of WRN and Ku fluorescence increased significantly (Fig. 6F).
Figure 6.
Nuclear localization of Ku and WRN by immunofluorescence. Actively growing SV40-transformed GM00637 cells were untreated (A–C) or incubated with 4NQO (D–F), then fixed and subsequently incubated simultaneously with rabbit anti-WRN and goat anti-Ku as described in Materials and Methods. Anti-rabbit Texas Red-labeled and anti-goat Alexa 588-labeled secondary antibodies were used to visualize WRN and Ku by confocal microscopy at 568 and 488 nm, respectively. The images presented here depict a representative cell from each of the non-treated and 4NQO-treated cultures. In the merged images (C and F) a yellow color appears where WRN (red) and Ku (green) fluorescence signals coincide.
Experiments with HeLa and GM00637 cells were carried out at least three times each. The images were captured in separate channels (green 488 nm, red 568 nm) and overlaid to see the extent of co-localization. Pixels from each image and its corresponding overlaid image were counted using Metamorph imaging system 4.1. For each experiment 15–25 cells were analyzed. Quantitation of co-localized pixels in the merged pictures demonstrated that in undamaged HeLa cells 25 ± 8% of the total WRN fluorescence co-localized with Ku while 20 ± 6% of the total Ku fluorescence co-localized with WRN. In the 4NQO-treated HeLa cells, 38 ± 9% of WRN co-localized with Ku and 32 ± 7% of Ku co-localized with WRN. For undamaged GM00637 cells, 26 ± 6% of WRN co-localized with Ku and 21 ± 4% of Ku co-localized with WRN (Fig. 6C). In the 4NQO-treated GM00637 cells, 41 ± 10% of WRN co-localized with Ku and 35 ± 4.6% of Ku co-localized with WRN (Fig. 6F). Thus, similar results were observed with GM00637 and HeLa cells that both contain wild-type WRN and Ku. These results suggest that WRN and Ku co-localize in a subset of nuclear foci, particularly after treatment with 4NQO, and that the WRN–Ku interaction may be important for processing of DNA damage induced by 4NQO.
DISCUSSION
WRN has both exonuclease and helicase activities, indicating a role in DNA metabolism. As the exonuclease activity is unique to WRN among human RecQ helicase family members, this function is probably crucial to the physiological role of WRN. In support of this notion, the WS phenotype is markedly different than those of Bloom and Rothmund–Thomson syndromes, human genetic diseases that are the result of defects in other RecQ homologs (BLM and RECQ4, respectively). In this study we have demonstrated that a physical interaction with Ku can stimulate the exonucleolytic activity of WRN to remove certain damaged nucleotides that would not be removed by WRN alone. This raises the possibility that WRN and Ku function coordinately in a DNA damage processing pathway.
Immunoprecipitation experiments presented here demonstrate a direct interaction between isolated recombinant WRN and Ku. In addition, there appears to be co-localization of Ku and WRN at defined foci in the nucleus. These results confirm and significantly strengthen previous findings of a physical interaction between WRN and Ku (15,16) in nuclear extracts. Thus, we have extended this observation to in vivo conditions, further supporting the physiological significance of the WRN–Ku interaction. In the absence of DNA-damaging treatments there is partial coincidence of the immunofluorescence of WRN and Ku, indicating that these proteins are not always found together in the cell. However, our observation that the co-localization of WRN and Ku is strongly enhanced after 4NQO treatment supports the notion that these proteins interact in a phase of DNA damage processing in the cell. WS cells are hypersensitive to 4NQO and treatment with this carcinogen results in several types of bulky lesions and increased oxidative damage (including formation of 8-oxoG and 8-oxoA modifications as well as strand breaks) in DNA. Our experiments suggest that WRN and Ku tend to be excluded from nucleoli, particularly after DNA-damaging treatments. Relocalization of WRN away from the nucleolus after DNA-damaging treatments was observed previously (26). However, we must point out that the subcellular compartmentalization of WRN is an area of considerable debate, as previous reports have shown nuclear localization of WRN with either no enrichment or considerable concentrations of WRN associated with nucleoli (25–28). Variations between findings are possibly due to the individual cell types used in various studies and to differences in immunofluorescence protocols. Nevertheless, the direct interaction between WRN and Ku would appear to explain their observed co-localization at a subset of nuclear foci and suggest an in vivo interaction.
Not only do Ku and WRN bind to each other, but this interaction also greatly enhances the exonuclease activity of WRN. Previously we demonstrated that the addition of Ku70/80 greatly stimulates the 3′→5′ exonuclease activity of WRN in vitro on DNA substrates containing normal Watson–Crick base pairs (15), a finding that has recently been reproduced by other investigators (16). In this study we have examined this Ku-mediated stimulation of WRN exonuclease in more detail by analyzing the digestion of substrates containing damaged nucleotides. The presence of 8-oxoA, 8-oxoG, an apurinic site or a cholesterol adduct in the strand subject to degradation severely inhibited the exonuclease activity of WRN (21). Intriguingly, the addition of Ku allows WRN to digest up to and through regions of DNA containing 8-oxoG or 8-oxoA, modifications that block the exonuclease activity of WRN alone. In contrast, Ku is much less effective in permitting WRN exonuclease to digest through DNA containing an apurinic site or a cholesterol adduct. Bulky adducts and apurinic sites are thought to destabilize the double-stranded character of DNA in the proximity of the lesion more so than would minor base modifications such as 8-oxoA or 8-oxoG. Importantly, WRN exonuclease cannot degrade single-stranded DNA or even a duplex substrate containing two mismatched nucleotides at the 3′-end (6). This would suggest that the inhibition of WRN exonuclease by bulky lesions might be due to single-stranded character in the vicinity of the lesion, while the ability of WRN, with the assistance of Ku, to digest through regions containing 8-oxoG- and 8-oxoA-containing nucleotides is due to the maintenance of double-stranded character at the site of these minor base modifications. The 3′→5′ exonuclease activities of Klenow and exo III on an 8-oxoA-containing substrate are not affected by Ku, indicating that this effect of Ku is specific for WRN exonuclease. Our results strongly suggest that the exonuclease function of WRN is altered by its physical interaction with Ku. The effect of Ku could either be to significantly increase the processivity of WRN exonuclease or to direct and possibly tether WRN onto various types of DNA ends, thereby permitting WRN, on occasion, to digest DNA structures that would normally not be suitable substrates. Notably, Ku also permits WRN to digest DNA substrates having blunt ends or single-stranded 3′-tails that are not subject to degradation by WRN exonuclease alone (16). In contrast, Ku appears to have no effect on WRN helicase activity (15) and does not detectably unwind substrates we have tested so far (15). hRPA, a protein that enhances the ability of WRN helicase activity to unwind long duplex DNA substrates (24), does not significantly influence the exonuclease activity of WRN on the (WRN) helicase-resistant substrate used in this study. These observations suggest that the individual functions of WRN can be modulated independently.
The physical and functional interaction of Ku with WRN suggests that these proteins might function in a common DNA metabolic pathway. The idea that WRN and Ku might act together is also supported by similarities in the WS and the Ku80 knockout mouse phenotypes and in the common cellular senescence and genomic instability characteristics of WRN-, Ku70- and Ku80-deficient cells (18–20). Ku is required for V(D)J recombination and double-strand break repair (17). The participation of WRN in V(D)J recombination is unlikely due to the lack of immunodeficiency in WS, but a role in double-strand break repair would be consistent with the increased chromosomal abnormalities observed in WS. The increased frequency of large deletions and translocations (12) and a defect in ligation fidelity (29) observed in WRN-deficient cells could conceivably be due to persistence or perhaps faulty repair of double-strand breaks. Although WRN-deficient cells are hypersensitive to 4NQO and topoisomerase inhibitors that could cause increased strand breakage (10,11), limited investigations have not uncovered any overt sensitivity of WS cells to specific double-strand break-generating agents as is seen in Ku-deficient cells. This could be due to redundancy in WRN function (perhaps by other RecQ members) or to the possibility that loss of WRN function may not contribute significantly to the cell survival end-points measured previously. A role for an unknown exonuclease (and a helicase) in double-strand break repair has been suggested previously (17). Although MRE11 had previously been thought to be this exonuclease, recent experiments suggest that MRE11 and Ku act in different pathways (30). Perhaps WRN is functioning as the exonuclease (and as a helicase as well) in double-strand break repair. In conjunction with the strand break-binding activity of Ku, WRN 3′→5′ exonuclease activity could be involved in processing of the 3′-ends at the sites of breaks. The WRN exonuclease activity could be involved in trimming away single-strand 3′-tails after unwinding of DNA ends and annealing of ends via microhomologies. Alternatively, this processing could be needed to uncover regions of microhomology in the vicinity of the DNA ends that would then anneal, setting the stage for end joining. In these schemes another function of WRN exonuclease might be to leave a suitable 3′-end for gap filling and/or ligation. Our result that WRN exonuclease in the presence of Ku70/80 can act on modified DNA structures implies that part of its function in double-strand break repair might also be to process structures that are not primers for polymerases or joinable by ligases. Notably, DNA strand breaks formed after exposure to reactive oxygen species or other agents often have other structural abnormalities at the exposed ends (31). Such a complementary role for WRN in the Ku-mediated V(D)J recombination pathway would not be required because of the standard nature of the DNA breaks formed during this process. Alternatively, the exonuclease activity of WRN (with or without Ku) could be involved in degradation of heteroduplex DNA during recombinogenic and/or anti-recombinogenic processes. However, the activity of WRN exonuclease on abnormal ends that specifically form during strand-breaking processes has not yet been shown and the requirement for WRN in double-strand break repair or any other DNA metabolic pathway remains to be proven. These types of investigation should provide important information regarding the coordinated functions of WRN with the Ku heterodimer.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Robert Brosh and Patricia Opresko for helpful comments. We appreciate the interaction with the Danish Center for Molecular Gerontology. This work was aided by grant 85-001-13-IRG from the American Cancer Society and grant 7NS-008900 from the Ellison foundation to D.O.
References
- 1.Martin G.M., Austad,S.N. and Johnson,T.E. (1996) Genetic analysis of aging: role of oxidative damage and environmental stresses. Nat. Genet., 13, 25–34. [DOI] [PubMed] [Google Scholar]
- 2.Martin G.M. (1997) The pathobiology of the Werner syndrome. FASEB J., 11, A1449. [Google Scholar]
- 3.Yu C.E., Oshima,J., Fu,Y.H., Wijsman,E.M., Hisama,F., Alisch,R., Matthews,S., Nakura,J., Miki,T., Ouais,S. et al. (1996) Positional cloning of the Werner’s syndrome gene. Science, 272, 258–262. [DOI] [PubMed] [Google Scholar]
- 4.Gray M.D., Shen,J.C., Kamath-Loeb,A.S., Blank,A., Sopher,B.L., Martin,G.M., Oshima,J. and Loeb,L.A. (1997) The Werner syndrome protein is a DNA helicase. Nat. Genet., 17, 100–103. [DOI] [PubMed] [Google Scholar]
- 5.Huang S., Li,B., Gray,M.D., Oshima,J., Mian,I.S. and Campisi,J. (1998) The premature ageing syndrome protein, WRN, is a 3′→5′ exonuclease. Nat. Genet., 20, 114–116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kamath-Loeb A.S., Shen,J.C., Loeb,L.A. and Fry,M. (1998) Werner syndrome protein. II. Characterization of the integral 3′→5′ DNA exonuclease. J. Biol. Chem., 273, 34145–34150. [DOI] [PubMed] [Google Scholar]
- 7.Shen J.C., Gray,M.D., Oshima,J. and Loeb,L.A. (1998) Characterization of Werner syndrome protein DNA helicase activity: directionality, substrate dependence and stimulation by replication protein A. Nucleic Acids Res., 26, 2879–2885. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Orren D.K., Brosh,R.M.,Jr, Nehlin,J.O., Machwe,A., Gray,M.D. and Bohr,V.A. (1999) Enzymatic and DNA binding properties of purified WRN protein: high affinity binding to single-stranded DNA but not to DNA damage induced by 4NQO. Nucleic Acids Res., 27, 3557–3566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Martin G.M., Sprague,C.A. and Epstein,C.J. (1970) Replicative life-span of cultivated human cells. Effect of donor’s age, tissue and genotype. Lab. Invest., 23, 86–92. [PubMed] [Google Scholar]
- 10.Ogburn C.E., Oshima,J., Poot,M., Chen,R., Gollahon,K.A., Rabinovitch,P.S. and Martin,G.M. (1997) An apoptosis-inducing genotoxin differentiates heterozygotic carriers for Werner helicase mutations from wild-type and homozygous mutants. Hum. Genet., 101, 121–125. [DOI] [PubMed] [Google Scholar]
- 11.Lebel M. and Leder,P. (1998) A deletion within the murine Werner syndrome helicase induces sensitivity to inhibitors of topoisomerase and loss of cellular proliferative capacity. Proc. Natl Acad. Sci. USA, 95, 13097–13102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fukuchi K., Martin,G.M. and Monnat,R.J. (1989) Mutator phenotype of Werner syndrome is characterized by extensive deletions. Proc. Natl Acad. Sci. USA, 86, 5893–5897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stefanini M., Scappaticci,S., Lagomarsini,P., Borroni,G., Berardesca,E. and Nuzzo,F. (1989) Chromosome instability in lymphocytes from a patient with Werner’s syndrome is not associated with DNA repair defects. Mutat. Res., 219, 179–185. [DOI] [PubMed] [Google Scholar]
- 14.Bohr V.A., Dianov,G.L., Balajee,A.S., Nehlin,J.O., Gray,M.D., Brosh,R.M.,Jr, Dianova,I., Machwe,A. and Orren,D.K. (1999) DNA repair in human premature aging disorders. In Bohr,V.A., Clark,B.F. and Stevnsner,T. (eds), Molecular Biology of Aging, Alfred Benzon Symposium No. 44. Munksgaard, Copenhagen, Denmark, pp. 262–272.
- 15.Cooper M.P., Machwe,A., Orren,D.K., Brosh,R.M., Ramsden,D. and Bohr,V.A. (2000) Ku complex interacts with and stimulates the Werner protein. Genes Dev., 14, 907–912. [PMC free article] [PubMed] [Google Scholar]
- 16.Li B. and Comai,L. (2000) Functional interaction between Ku and the Werner syndrome protein in DNA end processing. J. Biol. Chem., 275, 28342–28352. [DOI] [PubMed] [Google Scholar]
- 17.Featherstone C. and Jackson,S.P. (1999) Ku, a DNA repair protein with multiple cellular functions? Mutat. Res., 434, 3–15. [DOI] [PubMed] [Google Scholar]
- 18.Vogel H., Lim,D.S., Karsenty,G., Finegold,M. and Hasty,P. (1999) Deletion of Ku86 causes early onset of senescence in mice. Proc. Natl Acad. Sci. USA, 96, 10770–10775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gu Y., Seidl,K.J., Rathbun,G.A., Zhu,C., Manis,J.P., van der Stoep,N., Davidson,L., Cheng,H.L., Sekiguchi,J.M., Frank,K. et al. (1997) Growth retardation and leaky SCID phenotype of Ku70-deficient mice. Immunity, 7, 653–665. [DOI] [PubMed] [Google Scholar]
- 20.Nussenzweig A., Sokol,K., Burgman,P., Li,L. and Li,G.C. (1997) Hypersensitivity of Ku80-deficient cell lines and mice to DNA damage: the effects of ionizing radiation on growth, survival and development. Proc. Natl Acad. Sci. USA, 94, 13588–13593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Machwe A., Ganunis,R., Bohr,V.A. and Orren,D.K. (2000) Selective blockage of the 3′→5′ exonuclease activity of WRN protein by certain oxidative modifications and bulky lesions in DNA. Nucleic Acids Res., 28, 2762–2770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kenny M.K., Schlegel,U., Furneaux,H. and Hurwitz,J. (1990) The role of human single-stranded DNA binding protein and its individual subunits in simian virus 40 DNA replication. J. Biol. Chem., 265, 7693–7700. [PubMed] [Google Scholar]
- 23.Ramsden D.A. and Gellert,M. (1998) Ku protein stimulates DNA end joining by mammalian DNA ligases: a direct role for Ku in repair of DNA double-strand breaks. EMBO J., 17, 609–614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brosh R.M.,Jr, Orren,D.K., Nehlin,J.O., Ravn,P.H., Kenny,M.K., Machwe,A. and Bohr,V.A. (1999) Functional and physical interaction between WRN helicase and human replication protein A. J. Biol. Chem., 274, 18341–18350. [DOI] [PubMed] [Google Scholar]
- 25.Marciniak R.A., Lombard,D.B., Bradley Johnson,F. and Guarente,L. (1998) Nucleolar localization of the Werner syndrome protein in human cells. Proc. Natl Acad. Sci. USA, 95, 6686–6892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gray M.D., Wang,L., Youssoufian,H., Martin,G.M. and Oshima,J. (1998) Werner helicase is localized to transcriptionally active nucleoli of cycling cells. Exp. Cell Res., 242, 487–494. [DOI] [PubMed] [Google Scholar]
- 27.Balajee A.S., Machwe,A., May,A., Gray,M.D., Oshima,J., Martin,G.M., Nehlin,J.O., Brosh,R.M.,Jr, Orren,D.K. and Bohr,V.A. (1999) The Werner syndrome protein is involved in RNA polymerase II transcription. Mol. Biol. Cell, 10, 2655–2668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shiratori M., Sakamoto,S., Suzuki,N., Tokutake,Y., Kawabe,Y., Enomoto,T., Sugimoto,M., Goto,M., Matsumoto,T. and Furuichi,Y. (1999) Detection by epitope-defined monoclonal antibodies of Werner DNA helicases in the nucleoplasm and their upregulation by cell transformation and immortalization. J. Cell Biol., 144, 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Runger T.M., Bauer,C., Dekant,B., Moller,K., Sobotta,P., Czerny,C., Poot,M. and Martin,G.M. (1994) Hypermutable ligation of plasmid DNA ends in cells from patients with Werner syndrome. J. Invest. Dermatol., 102, 45–48. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi-Iwai Y., Sonoda,E., Sasaki,M.S., Morrison,C., Haraguchi,T., Hiraoka,Y., Yamashita,Y.M., Yagi,T., Takata,M., Price,C. et al. (1999) Mre11 is essential for the maintenance of chromosomal DNA in vertebrate cells. EMBO J., 18, 6619–6629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. ASM Press, Washington, DC.