Abstract
Conformational heterogeneity is a defining characteristic of proteins. Intrinsically disordered proteins (IDPs) and denatured state ensembles are extreme manifestations of this heterogeneity. Inferences regarding globule versus coil formation can be drawn from analysis of polymeric properties such as average size, shape, and density fluctuations. Here we introduce a new parameter to quantify the degree of conformational heterogeneity within an ensemble to complement polymeric descriptors. The design of this parameter is guided by the need to distinguish between systems that couple their unfolding-folding transitions with coil-to-globule transitions and those systems that undergo coil-to-globule transitions with no evidence of acquiring a homogeneous ensemble of conformations upon collapse. The approach is as follows: Each conformation in an ensemble is converted into a conformational vector where the elements are inter-residue distances. Similarity between pairs of conformations is quantified using the projection between the corresponding conformational vectors. An ensemble of conformations yields a distribution of pairwise projections, which is converted into a distribution of pairwise conformational dissimilarities. The first moment of this dissimilarity distribution is normalized against the first moment of the distribution obtained by comparing conformations from the ensemble of interest to conformations drawn from a Flory random coil model. The latter sets an upper bound on conformational heterogeneity thus ensuring that the proposed measure for intra-ensemble heterogeneity is properly calibrated and can be used to compare ensembles for different sequences and across different temperatures. The new measure of conformational heterogeneity will be useful in quantitative studies of coupled folding and binding of IDPs and in de novo sequence design efforts that are geared toward controlling the degree of heterogeneity in unbound forms of IDPs.
INTRODUCTION
Proteins undergo disorder-to-order transitions either as units that fold autonomously1 or as intrinsically disordered proteins (IDPs)2 that couple their folding to binding3 or self-assembly.4 The driving forces for and mechanisms of disorder-to-order transitions are governed by the degree of conformational heterogeneity within disordered states and the extent of overlap between conformational ensembles of disordered and ordered states. Therefore, there is growing interest in quantitative studies of disordered states of proteins.5, 6, 7, 8
Studies of disorder in protein folding are focused on characterizing the ensemble of non-native conformations under denaturing as well as native conditions.9, 10, 11 Of interest are questions pertaining to the degree of conformational heterogeneity,12, 13 the balance between intrachain and chain-solvent interactions that define polymeric properties,14, 15, 16 effects of macromolecular crowding,17, 18 intermolecular interactions that lead to protein aggregation,19, 20, 21 and the timescales for conversion between distinct conformations that contribute to internal friction.22 Recent interest has also focused on the topic of IDPs. Their sequences encode preferences for heterogeneous ensembles of conformations as the thermodynamic ground state under standard physiological conditions (aqueous solutions, 150 mM monovalent salt, low concentrations of divalent ions, pH 7.0, and temperature in the 25 ºC–37 ºC range).23, 24 Conformational heterogeneity of IDPs in their unbound forms influences their ability to adopt different folds in the context of binary and multimolecular complexes.25, 26 In IDPs, disorder-to-order transitions are realized by coupling the folding process to either binding or self-assembly providing the heterotypic or homotypic interactions in trans can stabilize the IDP in a specific fold. The stabilities of complexes are thermodynamically linked to the ensemble of conformations that IDPs sample as autonomous units.
Thermodynamic descriptions of disorder-to-order transitions require the use of a suitable order parameter. A bona fide order parameter has to quantify the symmetry that is broken as a result of the disorder-to-order transition. Proteins are polymers and can expand to form low-density conformations that have large interfaces with the surrounding solvent; alternatively, they can collapse to form high-density conformations that minimize the chain-solvent interface. It is well established that is a bona fide order parameter for quantifying density changes that accompany coil-to-globule transitions.27, 28 Here, Rg denotes the radius of gyration and N is the chain length. In protein folding, changes in density are also associated with the acquisition of a homogeneous ensemble of conformations. However, s2 can be used as the sole parameter to monitor folding if and only if proteins follow two-state behavior.29 Theories, simulations, and experiments have established that while s2 is extremely important for understanding the convolution of coil-to-globule transitions with protein folding, it is inadequate for providing a complete description of transitions between unfolded and folded states.30, 31, 32, 33, 34, 35, 36, 37 Recent simulations and experiments have also shown that several IDPs undergo collapse to form globules under standard physiological conditions.38, 39, 40, 41, 42, 43, 44 This preference for globules can be reversed through increases in temperature,45 net charge per residue,46, 47 or concentrations of chemical denaturants.39 Collapse in globule-forming IDPs does not have to imply the acquisition of a homogeneous ensemble of conformations. These results highlight the need for additional parameters that report on overall conformational heterogeneity.
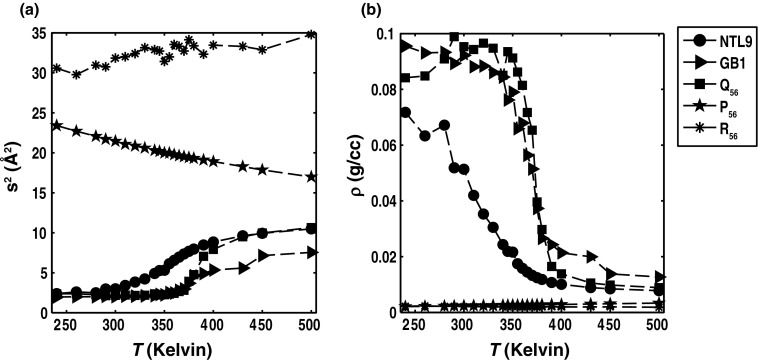
Figure 1 summarizes the temperature dependence of s2 and densities that are obtained from atomistic simulation results for five archetypal systems. Details of the simulations that were used to generate the temperature dependent profiles are discussed in Sec. 2. The N-terminal domain of the ribosomal L9 protein (NTL9) and the B1 domain of protein G (GB1) undergo unfolding transitions as temperature increases. This unfolding is also associated with chain expansion as shown in Figure 1. We compare these profiles to the temperature dependence of s2 and density (ρ) for three homopolymeric systems. These are polyproline (P56), which is intrinsically stiff, polyarginine (R56), which is a highly charged, rod-like polyelectrolyte, and polyglutamine (Q56), which is an intrinsically disordered polar tract. P56 shows weak chain contraction as temperature increases. This feature is consistent with the so-called inverse transition temperature48 that has been observed for poly-L-proline polymers and derives, partially, from the increased fraction of cis peptide bonds at higher temperatures.49 R56 and Q56 show distinct limiting behaviors; the former maintains its rod-like behavior across all temperatures whereas the latter undergoes a globule-to-coil transition as temperature increases. Despite undergoing reversible coil-to-globule transitions, previous simulations and experimental studies demonstrate that collapse does not imply the acquisition of an ensemble of a homogeneous ensemble of conformations, i.e., collapse does not imply folding.50, 51 This in turn implies that while the temperature dependence of s2 provides information regarding coil-to-globule transitions, it fails to discriminate between systems such as NTL9 and GB1 on the one hand and Q56 on the other. Analysis of the temperature dependence of the specific heat capacities (Figure 2), which reports on the temperature dependence of the energy variance, does not provide any additional information that cannot be obtained by analyzing the temperature dependence of density fluctuations.
Figure 1.
Temperature dependence of s2 and density for five archetypal systems. Panel (b) quantifies the temperature dependence of chain density (in units of gm-cm−3), which is calculated as , where MW denotes the molecular weight in gm mol−1.
Figure 2.
Temperature dependence of fluctuations in density and energy for five archetypal systems. Panel (a) shows the temperature dependence of the density fluctuations quantified as the variance of the density distribution for a given temperature, i.e., . Panel (b) shows the temperature dependence of the specific heat capacity. The specific, constant volume heat capacities were calculated as , where MW is the molecular weight and ⟨E⟩ is the ensemble-averaged potential energy for simulated ensembles at a given temperature. Typically, one expects sharp transitions for well-defined order-to-disorder transitions and yet, interestingly, the Q56 system shows the sharpest transition. The relatively broad transitions for NTL9 and GB1 highlight the joint contributions of gradual melting and different degrees of residual local structure in their unfolded states.
In the parlance of energy landscape theory,52, 53, 54 a system such as polyglutamine has a rugged landscape below its collapse transition temperature.55 Indeed, such a scenario has been predicted for IDPs56, 57 and random polypeptide sequences.58, 59 This ruggedness is not registered in measures such as estimates of density or energy fluctuations because distinct conformations of equivalent compactness have negligible energy differences and hence equivalent likelihoods of being accessed. In this scenario, both the energy and density fluctuations will be small and the sharpness of the change in energy and density fluctuations masks the fact that the globule-to-coil transition in a system like polyglutamine might actually be a “disorder-to-disorder” transition where the transition is between distinct classes of heterogeneous conformational ensembles.
In order to detect putative disorder-to-disorder transitions that are masked when analyzing s2 or densities, we need a measure for heterogeneity within a conformational ensemble. For this we introduce a parameter Φ whose design is guided by the need to distinguish between systems that couple their unfolding-folding transitions with coil-to-globule transitions and those systems that undergo coil-to-globule transitions with no evidence of acquiring a homogeneous ensemble of conformations upon collapse. The design of Φ is also intended to accomplish two additional goals for the analysis of results from molecular simulations: (i) To compare the degree of conformational heterogeneity of ensembles obtained for a specific system at different simulation conditions; and (ii) To compare the degree of conformational heterogeneity of ensembles for different polypeptide sequences at equivalent simulation conditions.
The remainder of the narrative is organized as follows: In Sec. 2 we summarize the simulation approach for generating the conformational ensembles that were used to prototype Φ. Section 3 is split into two parts. In part 1, we describe the methodological framework for calculating Φ. In doing so, we discuss the choices made in converging upon the overall approach. In part 2, we use Φ to assess recent simulation results that were reported for the basic regions (bRs) of bZIP transcription factors.60 These results demonstrated the role of sequence contexts in modulating the intrinsic helicities of bZIP-bRs. We show that Φ unmasks the weaknesses inherent to measures of average secondary structure contents as probes for structure and highlight how conformational heterogeneity can prevail in ensembles with high average helicities. We conclude with Sec. 4 that summarizes the uses for Φ in analyzing protein disorder and in de novo sequence design. The discussion also provides a comparison between Φ and other approaches for quantifying conformational heterogeneity.
METHODS
Polypeptide systems included in this work
We simulated homopolymers of glutamine (Q56), proline (P56), and arginine (R56) each 56-residues long. In addition, we included two 56-residue polypeptides, NTL9 and GB1 that adopt well-defined folds at low temperatures. Homopolymers were N-terminally acetylated and C-terminally N′-methylamidated. Atomistic Markov Chain Metropolis Monte Carlo (MC) simulations61 were performed in the canonical ensemble using one polypeptide for each construct. Mobile sodium and chloride ions were included for peptides containing charged residues and the ions were represented explicitly. The salt concentration of ion-containing systems was set to 25 mM. The peptides and ions were enclosed in a spherical droplet of radius 200 Å and the droplet boundary was enforced using a stiff harmonic boundary potential. For the basic region leucine zipper transcription factor (bZIP-bR) peptides we analyzed simulation results from the work of Das et al.60
Details of the metropolis Monte Carlo (MC) simulations
The CAMPARI molecular simulation package (http://campari.sourceforge.net/) in conjunction with the ABSINTH implicit solvation model62 and OPLS-AA/L63 molecular mechanics force field parameters (abs3.2_opls.prm) were used for four out of five sets of simulations. For the poly-arginine system we used the new ion parameters developed by Mao and Pappu64 for the mobile Na+ and Cl− ions. The spatial cutoffs for Lennard–Jones and electrostatic interactions between net-neutral charge groups were set to 10 Å and 14 Å, respectively. No cutoffs were employed for computing the electrostatic interactions for ions and side chain moieties with a net charge. Sodium and chloride ions were modeled explicitly and polypeptides were modeled in atomic detail. The internal degrees of freedom included the backbone ϕ, ψ, ω and side chain χ dihedral angles. Rigid-body moves simultaneously change rotational and translational degrees of freedom of the protein whereas translational moves were applied to alter the positions of mobile ions. Random cluster moves alter the rigid body coordinates of multiple molecules at once. Pucker moves perturb the ring geometry of proline residues.49 The frequencies with which different moves were chosen along with parameters specific to each move type are summarized in the decision tree that is similar to that of Mao et al.47 The starting conformation for homopolymers Q56, P56, and R56 was generated at random from a pre-equilibrated distribution of atomistic self-avoiding random walks. Starting conformations for NTL9 and GB1 for all simulation temperatures were derived from Protein Data Bank (http://www.rcsb.org) IDs 2HBB and 1GB1, respectively. Additional details regarding the setup of the initial folded conformations are as described in Meng et al.14 The bond lengths and bond angles were fixed at values prescribed by Engh and Huber.65
The MC sampling protocol
Simulation results for NTL9, GB1, Q56, P56, and R56 were generated using the following protocol: For each system we performed ten independent MC simulations at each of the following temperatures: T = 240, 260, 280, 290, 300, 310, 320, 330, 340, 345, 350, 355, 360, 365, 370, 375, 380, 390, 400, 430, 450, and 500 K. Each independent simulation used a different random seed to initialize the MC run. A total of 8 × 107 MC steps were used in each independent simulation and of these, the results from the first 2 × 107 steps were discarded as equilibration. Observables were accumulated every 104 MC steps and conformational vectors for the heterogeneity calculation were collected every 105 steps. Thus, an ensemble from a single run that was used to calculate Φ contained 6000 members for each temperature. Reproducibility of the simulation results across multiple independent runs negated the need for using enhanced sampling methods.
The Flory random coil (FRC) model
The FRC reference state was constructed for each polypeptide.66 FRC peptides were represented in all atom detail with the same degrees of freedom as used in the MC simulations described above. FRC conformations were generated by random assignment of sterically allowed combinations of backbone ϕ, ψ, ω and side chain χ dihedral angles while ignoring all inter-residue interactions. Each step of FRC sampling consisted of picking a residue at random then assigning all of the torsional degrees of freedom of the residue to a vector of ϕ, ψ, ω, and χ selected at random from a library of size 104. These libraries were generated for each residue via MC simulations of the corresponding dipeptides in the excluded volume (EV) limit. EV ensembles were generated using atomistic descriptions of the dipeptide while ignoring all non-bonded interactions excepting steric repulsions. A total of 4 × 107 steps were applied in each FRC simulation and resulting polypeptide conformations were accumulated every 105 steps. Ten independent FRC simulations were performed resulting in a total of 4000 reference conformations for each peptide. This pool of conformations is referred to as the FRC ensemble and all members were used in calculating Φ.
RESULTS
Estimating Φ
Our goal is to quantify the degree of conformational heterogeneity given an ensemble of conformations. This requires a method to quantify the degree of similarity between all distinct pairs of conformations within the ensemble. The resultant distribution of pairwise similarity measures is then used to obtain a value for Φ that reports on the degree of conformational heterogeneity within the ensemble. For a chain of N residues, each conformation c is represented as an nd × 1 conformational vector Vc where ,Vc = {d12, d13, …, dN − 1, N}, and each element dij in Vc represents the spatial distance between a unique pair of residues, i and j. For each pair of residues i and j, we calculate . Here, rmi and rnj denote the position vectors of atoms m and n within residues i and j, respectively, and Zij is the number of unique pairwise inter-atomic distances between the two residues. To compare a pair of conformations k and l, we calculate a pairwise dissimilarity measure = 1 – cos(Ωkl) where . An ensemble of nc conformations produces an ensemble of conformational vectors, V1, V2,…etc. These vectors are used to calculate a distribution P(D) of conformational dissimilarity values. Examples of these distributions are shown in Figure 4.
Figure 4.
Temperature dependence of ⟨D⟩ and Φ for the five archetypal systems. Panel (b) includes error bars from a bootstrap analysis whereby 100 distinct bootstrap trials were performed to estimate Φ and the error bars therefore represent standard deviations for the estimate of the mean Φ values.
For a given simulation temperature T, the first moment of the distribution of dissimilarity values, ⟨D⟩, provides an estimate of the most likely value for the degree of conformational heterogeneity within the ensemble. This measure, however, needs calibration in order for it to be used for comparing ensembles across different simulation temperatures or ensembles for different systems. For a given system, the intrinsic conformational properties of amino acids within the sequence place an upper bound on the degree of dissimilarity that is realizable.66 These intrinsic biases need to be accounted for and normalized against if we are to compare the degree of conformational heterogeneity between systems. Furthermore, considerations of chain connectivity might render many of the values for inter-residue distances dij to either be invariant or slowly varying as temperature changes. Accordingly, we use a simulation approximation of the Flory random coil that is based on the rotational isomeric approximation to calibrate the distribution of dissimilarity values obtained for ensembles of a given system. This is accomplished by calculating the pairwise conformational dissimilarity between each conformation k from the ensemble of interest and conformation l drawn from the ensemble of FRC conformations. The latter ensemble varies depending on the amino acid sequence and remains invariant with temperature. Consequently, for each ensemble corresponding to a given simulation temperature T, we obtain two distributions of dissimilarities, viz., the distribution of D-values for pairs of conformations within an ensemble and a distribution of D-values comparing each conformation to an ensemble of FRC conformations (see Figure 3). Averaging over the former yields ⟨D⟩ and averaging over the latter, which is an ensemble of ensembles yields ⟨⟨D⟩⟩FRC. The values of ⟨D⟩ and ⟨⟨D⟩⟩FRC lead to an estimate of the degree of heterogeneity Φ within the ensemble at temperature T. We first compute the ratio H = ⟨D⟩/⟨⟨D⟩⟩FRC and use it to calculate Φ = 1 – H.
Figure 3.

Sample distributions P(D) for two systems at different temperatures. The panel on the left shows P(D) distributions for NTL9 at three different temperatures and the panel on the right shows these distributions for the Q56 system at three different simulation temperatures. In both panels, the solid curves represent intra-ensemble P(D) distributions whereas the dashed curves are for comparisons between conformations within an ensemble at temperature T and conformations drawn from the FRC ensemble.
The Flory random coil model helps us dereference three composition-specific contributions to conformational heterogeneity for a given combination of sequence and simulation conditions. These are (i) trivial differences between residue-specific local conformational preferences, (ii) the effects of chain connectivity, and (iii) differences in heterogeneity that arise due to differences in chain length. The use of the FRC model is akin to the use of an ideal fluid prior in calculations of pair correlation functions in atomic and molecular fluids. The Flory random coil model is an informed prior that accommodates maximal conformational heterogeneity by including and accounting for contributions from local biases. Reference models such as the freely jointed or freely rotating chain models entirely ignore residue-specific local conformational biases. Therefore, their usage would be tantamount to pre-multiplying the value of ⟨D⟩ by a constant pre-factor, the value of which depends on chain length and nothing else. The self-avoiding random walk ensemble or conformations drawn from the so-called excluded volume (EV) limit67 could be used as a reference. However, these conformations are biased in that the ensemble is characterized by correlated fluctuations that afford the unique properties of the EV limit. It is well known that there is a diminution of conformational heterogeneity in the EV limit as compared to the FRC state due to spatial correlations between non-nearest neighbor residues.68 Hence, in choosing a reference state, we select an ensemble that (a) lacks correlations between non-nearest neighbor residues, (b) retains a minimal degree of sequence specificity, and (c) affords the ability to enable quantitative comparisons between different systems and simulation conditions.
In the FRC model, the conformational partition function for the polypeptide is written as a product of partition functions of independent interaction units. All interactions between non-nearest neighbor residues are ignored while the intrinsic conformational preferences of individual residues are captured in terms of weights for each of the possible rotational isomers. The FRC ensemble therefore represents an intuitive upper bound on conformational heterogeneity and helps ensure that Φ is bounded between the values of 0 and 1, 0 ≤ Φ ≤ 1. This property obtains because H ≤ 1 and results from the construction of the reference FRC ensemble, which ensures that ⟨D⟩ ≤ ⟨⟨D⟩⟩FRC. If the degree of intra-ensemble conformational heterogeneity is akin to the upper bound on heterogeneity expected for an FRC ensemble, then the ratio H → 1 and Φ → 0, indicating a maximally heterogeneous ensemble. Conversely, for a homogeneous ensemble of conformations it follows that, D → 0, H → 0, and Φ → 1. Therefore, given two values of ΦA and ΦB for two sequences at identical temperatures such that ΦA > ΦB we infer that when referenced to their respective own Flory random coil states sequence A has higher conformational heterogeneity. It is worth noting that the generation of the FRC reference ensemble adds minimal computational overhead to the overall procedure. Importance sampling methods such as molecular dynamics or Metropolis Monte Carlo sampling are computationally expensive because of the force/energy evaluation that is necessary at each step. In contrast, the FRC reference ensemble is generated using a pre-computed library of rotational isomers for each residue and ignoring all inter-residue interactions. Therefore, the machine time for ensemble generation increases sub-linearly as system size increases. We expect to lower the barrier for generating these reference ensembles by providing a web-based automatic FRC generator at http://pappulab.wustl.edu.
Assessment of conformational ensembles using Φ
Figure 4 shows the variation of D and Φ with temperature for each of the five systems that were introduced in Figure 1. Equivalent inferences can be drawn from the use of either parameter. We focus on the analysis of Φ because of the attributes described above, specifically the ability to use it for comparing different systems at similar sets of conditions. For NTL9 and GB1 the unfolding transition is manifest as a transition between a high value for Φ at low temperatures and a low value for Φ at high temperatures and the transition between these two limits is sharp. The slope of the transition region quantifies the “rate” of change in the degree of conformational heterogeneity with temperature. In contrast to NTL9 and GB1, the temperature dependence of Φ for Q56 is consistent with equivalent degrees of heterogeneity in the high and low temperature regimes. Previous work on polyglutamine led to an estimate of Tθ ≈ 390 K for the theta temperature.45 At T ≈ Tθ, chain-chain and chain-solvent interactions are counterbalanced and statistical properties at the theta temperature resemble that of the FRC model. Accordingly, the temperature dependence of ΦT for Q56 shows a dip and approaches zero near Tθ. Apart from this deviation, the profile of ΦT for Q56 is consistent with the hypothesis of a disorder-to-disorder transition. When combined with the analysis of s2, it becomes clear that the polyglutamine system transitions between two classes of disorder, viz., a heterogeneous ensemble of compact conformations that maximize the density at low temperatures and a heterogeneous ensemble of expanded conformations that minimize the density at high temperatures.
The Φ profiles for NTL9 and GB1 also show dips at intermediate temperatures, and again these temperatures can formally be shown to correspond to the theta temperatures for these systems. This identification is useful in light of the recent results of Hofmann et al.15 They assessed changes in Rg for unfolded molecules as a function of decreasing denaturant concentration for five different systems and found that for unfolded ensembles under native conditions Rg ∼ N0.45±Δ where Δ ≈ 0.05. This implies that, on average, intra-chain and chain-solvent interactions are mutually screened because of generic amino acid compositional biases seen in protein sequences giving rise to the property that the statistics of unfolded ensembles mimic those of polymers at theta temperatures. If this proposed equivalence holds up to scrutiny, then the analysis of ensembles generated for temperatures where Φ → 0 should lead to insights regarding unfolded states sampled under folding conditions.
The intrinsically stiff P56 system shows a linear transition of Φ from a high value to a smaller value. As shown previously,49, 69 the increase in conformational heterogeneity arises mainly due to the increased frequency of generating bends and kinks within polyproline and this is partly due to the increased frequency of sampling cis peptide bonds at higher temperatures. Overall, however, the energy scales that encode chain stiffness in polyproline cannot be overcome by increasing temperature, and the transitions of s2 and Φ reflect this feature.
The results for poly-arginine are particularly relevant for the study of IDPs. Often one uses web-based assessments of the degree of disorder and as shown previously,47 these bioinformatics-based servers70, 71 predict that poly-arginine should be highly disordered. This prediction is the result of conflating high charge content and the resultant high s2 (low density) values to implicitly imply a high degree of conformational heterogeneity. However, we find that Φ > 0.9 across the entire temperature range and this lack of change in Φ with temperature for R56 is consistent with the maintenance of a homogeneous ensemble of rod-like conformations. The degree of chain expansion exceeds that of self-avoiding random walks.47 This expansion results from a combination of long-range electrostatic repulsions and favorable solvation of charged side chains that together give rise to correlated fluctuations and overall rod-like behavior of the chain.72 Chain compaction and increased conformational heterogeneity can be realized by screening the electrostatic repulsions in the presence of high concentrations of salt as was shown previously.47
The preceding analysis shows that it is necessary to use Φ and s2 jointly to characterize the degree and nature of conformational heterogeneity. In systems such as NTL9 the joint use of s2 and Φ highlights the positive coupling between increased conformational heterogeneity and chain expansion (see panel (a) in Figure 5). This is consistent with energy landscape theories that predict strongly funneled landscapes for sequences that fold into well-defined ensembles of self-similar conformations.54 Conversely, polyglutamine and polyarginine show limiting behaviors (panels (b) and (d) of Figure 5). For polyglutamine, joint use of s2 and Φ shows that the globule-to-coil transition observed as temperature increases is consistent with ensembles switching from one class of heterogeneity to another. In the parlance of energy landscape theory, temperature modulates the ruggedness of free energy landscapes. For polyarginine, analysis of the temperature dependence of Φ alone might seem confounding in light of bioinformatics-based prediction that this system should be highly disordered.47, 70 Electrostatic repulsions and the favorable solvation of charges side chains give rise to increased electrostatic persistence lengths, long-range correlated fluctuations, and homogeneous ensembles of rod-like conformations for highly charged systems for all temperatures – an observation that is consistent with experimental data46, 47 and polyelectrolyte theories.73
Figure 5.
(a)–(d) Plots to quantify the assessments of conformational properties that derive from the joint analysis s2 (ordinates) and Φ (abscissae). In each panel, the symbol colors progress from cool to hot as temperature increases.
Application of Φ to assess conformational heterogeneity in IDPs with different secondary structure propensities
In Sec. 3B we showed that the joint use of s2 and Φ provide a more complete picture of conformational heterogeneity. Systems with different degrees of chain compaction can display similar degrees of conformational heterogeneity. Consequently, chain compaction/expansion does not directly imply homogeneous/heterogeneous ensembles. It is also common practice to characterize ensembles in terms of secondary structure propensities because these are accessible to direct inquiry using nuclear magnetic resonance,74 circular dichroism,60 and molecular simulations. Such inquiries often show sequence-specific variations in secondary structure propensities and the question is if higher secondary structure content translates to diminished conformational heterogeneity and vice versa? We answer this question by analyzing recent simulation results60 for the basic regions of bZIP transcription factors.
Basic region leucine zippers (bZIPs) are modular transcription factors that play key roles in eukaryotic gene regulation.75 The basic regions of bZIPs (bZIP-bRs) adopt regular α-helical conformations when bound to DNA.76 Bioinformatics predictions and spectroscopic studies suggest that unbound, monomeric bZIP-bRs are uniformly disordered as autonomous units.77, 78 This assumption was recently tested through quantitative characterization of the conformational preferences of fifteen different bZIP-bRs.60 These were found to have quantifiable preferences for α-helical conformations in their unbound, monomeric forms. This helicity varies from one bZIP-bR to another despite significant sequence similarity of the DNA binding motifs (DBMs). Analysis of the determinants of helicity revealed that intramolecular interactions between DBMs and 8-residue segments directly N-terminal to DBMs are the primary modulators of bZIP-bR helicities. The accuracy of this inference was tested in designed chimeras of bZIP-bRs that have either increased or decreased overall helicities. For a given sequence, the helical propensity at temperature T was calculated using the formula in Eq. 1:
where
| (1) |
and
In Eq. 1, N denotes the number of residues in a bZIP-bR sequence, is the ensemble-averaged probability of finding residue i as part of a helical segment, denotes the number of conformations used for calculating ensemble averages at temperature T, and is a discrete Heaviside function that determines if residue i is part of an α-helical segment in conformation k. A α-helical segment was identified as a stretch that has at least seven consecutive residues with a DSSP (Define Secondary Structure of Proteins)79 designation of “H”, which implies that these residues are part of a regular, hydrogen-bonded α-helix. Panel (a) in Figure 6 shows a plot of Φ against the calculated helicity for seventeen bZIP-bRs that includes 13 naturally occurring bZIP-bRs and four designed chimeric sequences. The results are shown for T = 298 K.
Figure 6.

Assessments conformational heterogeneity in ensembles with different degrees of helical structure. Panel (a) plots Φ against for T = 298 K. The results are shown for 17 naturally occurring and designed sequences. Panel (b) plots Φ against σ2(Rg) and panel (c) plots Φ against σ2(D) for each of the 17 bZIP-bRs.
Naively one might expect a strong positive correlation between an increase in Φ and an increase in helical propensity. We quantified the linear correlation between Φ and helical propensity using the Pearson product moment correlation coefficient. We find a value of r = 5 × 10−4 when we use the Φ and values for all of the sequences listed in panel (a) of Figure 7. We reasoned that the quantification of helicity, which reports on local structural propensities – especially as calculated in Eq. 1 – masks the degree of conformational heterogeneity that is achievable in the ensemble. The seemingly confounding correlation analysis is impacted by the presence of two types bZIP-bRs that either show high average helicity and low Φ as in the bZIP-bR of gcn4 or those that have low overall helicities on average and higher values of Φ (>0.4). We analyzed the correlation between Φ and the variances of the Rg and D distributions, i.e., σ2(Rg) and σ2(D) for each of the 17 sequences. Here, we expect a negative correlation between Φ and the σ2 values because increased conformational heterogeneity should lead to larger fluctuations in chain size and D-values. Panels (b) and (c) in Figure 6 demonstrate these negative correlations. Clearly, locally averaged measures of structure can be misleading because they mask the degree of conformational heterogeneity that can be accommodated within an ensemble despite quantifiable secondary structure content.
Figure 7.
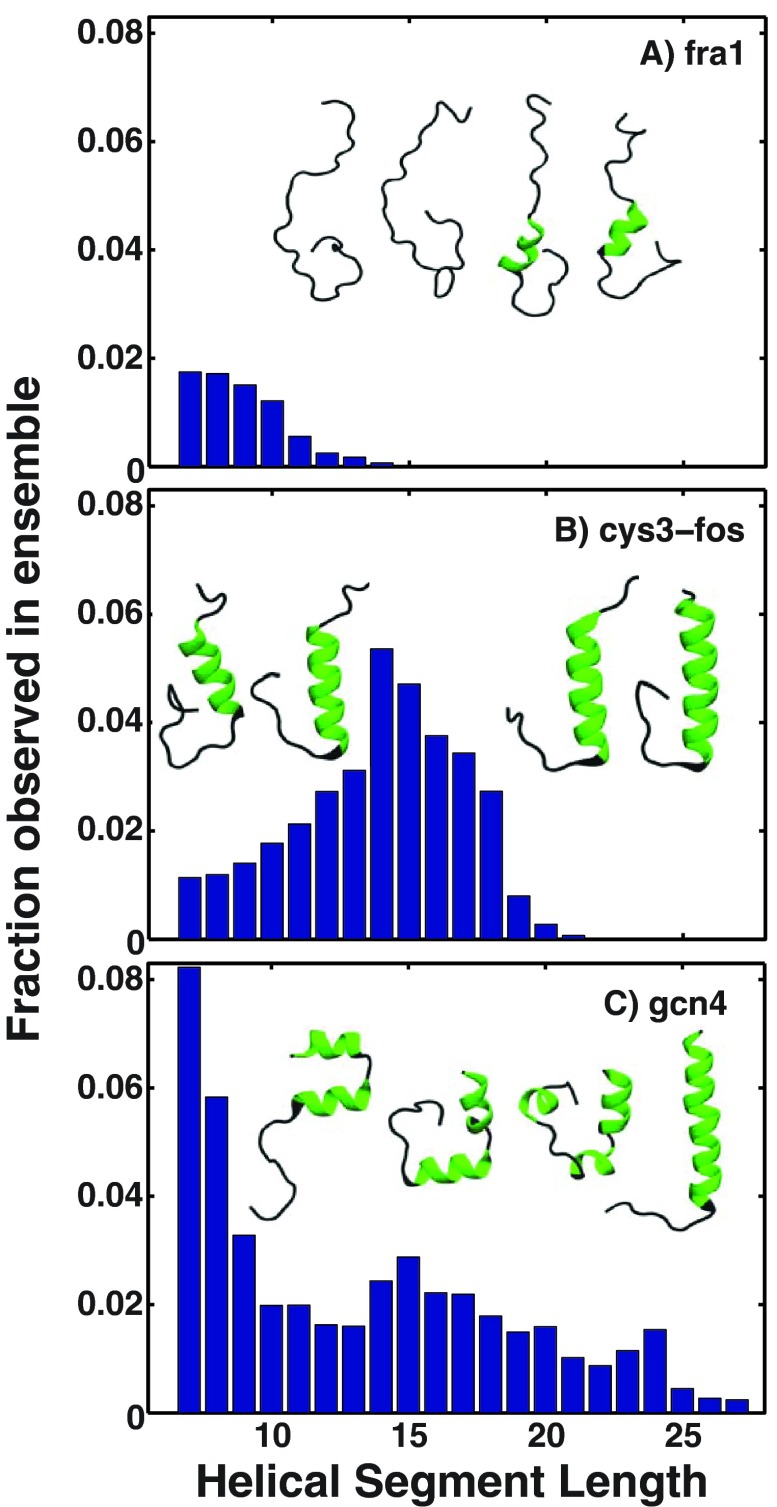
Analysis of conformational heterogeneity in terms of the distribution of helical segment lengths for three of the bZIP-bRs. The figure shows three panels one each for the bZIP-bR of fra1, the chimeric cys3-fos, and gcn4. Each panel shows a histogram of helical segment lengths within the simulated ensembles. A helical segment corresponds to a consecutive stretch of residues in a conformation with a DSSP “H” designation. The value of Φ is dictated by the width of a segment length distribution as opposed to the ensemble-averaged helicity.
Figure 7 shows additional analysis to illustrate the source of the weak correlation between ensemble-averaged helicities and Φ. Panel (a) shows results for the bR of fra1, which has low overall helicity (less than 0.2) and Φ greater than 0.5. The distribution of helical segment lengths, which is narrow, provides an explanation for the higher value of Φ. A similar stretch of 7–10 residues forms helices in roughly 20% of the conformations. Panel (b) shows results for the chimeric bR, cys3-fos, which has a high average helicity and high Φ value (both ≈ 0.6). In this case, fluctuations cause the helical stretch to expand and contract around the central region of the sequence that always spans the DNA binding motif. Panel (c) shows results for the bR of gcn4. Although the ensemble-averaged helicity is high (≈0.6) the ensemble is characterized by a broad distribution of helical segment lengths whereby different sequence stretches fluctuate into and out of helical conformations thus leading to high heterogeneity and a low value (≈0.25) for Φ.
The preceding analysis is important given the previous work Das et al.60 who used de novo sequence design to modulate intrinsic helicities of bZIP-bRs. Inasmuch as this effort was geared toward modulating the bias toward or away from α-helical conformations adopted by bZIP-bRs in their bound states, the current analysis highlights the fact that proper modulation of the degree of disorder in the ensemble requires a joint calculation of helical propensities and Φ. As an illustration of this need for using ensemble heterogeneity as a constraint in de novo sequence design we compare the results for the chimeric fos-gcn4 bR to the wild type gcn4-bR. The former was designed to have lower helicity than gcn4, which is indeed the case. Despite this, the heterogeneity is such that the Φ value is higher in the chimera, which has lower helicity than the wild type gcn4-bR. Such results can confound the objectives of de novo sequence design especially in a setting where helicities are being modulated to impact the driving forces for and mechanisms of coupled folding to binding reactions.
DISCUSSION
Measures such as s2 and fα provide information regarding the global and local conformational preferences, i.e., they help classify the types of local conformational and overall density features of an ensemble of conformations. Conversely, Φ measures the degree of conformational heterogeneity. Hence, combining quantitative analysis of Φ with measures such as s2 and quantification of local secondary structure preferences provides a complete quantitative summary of the degree and nature of conformational heterogeneity for the ensemble in question. This approach also facilitates comparisons between ensembles for different sequences and conditions. Diminished conformational heterogeneity will lead to an increase in Φ whereas increased conformational heterogeneity will decrease the value of Φ. Since this parameter is bounded, the result of normalization using the Flory random coil reference state, quantitative changes to Φ imply quantitative changes to the degree of conformational heterogeneity. This parameter should prove useful in comparative assessments of conformational heterogeneity of conformational ensembles generated for a single system at different temperatures and solution conditions as well as for different systems under similar conditions.
To calculate Φ, we used the first moments ⟨D⟩ and ⟨⟨D⟩⟩FRC. The first moments provide an assessment of the most likely values for the corresponding distribution of conformational dissimilarity values, i.e., P(D). We have also calculated second moments of the underlying distributions. Figure 8 shows the temperature dependence of the variances, i.e., σ2(D) = ⟨(D – ⟨D⟩)2⟩ for each of the five archetypal systems. The variance is high when ⟨D⟩ is high and is low when ⟨D⟩ is low implying that the variance and Φ are negatively correlated. Other than this feature, there is no additional insight to be obtained through analysis of the variance and other higher order moments of P(D) distributions, suggesting that Φ proves to be sufficient for a comparative and quantitative assessment of conformational heterogeneity.
Figure 8.
Temperature dependence of the variance of D calculated from the distributions of D values for each of the five archetypal systems. All inferences regarding conformational heterogeneity that are drawn from analysis of the variance are consistent with those drawn from analysis of Φ.
Our approach to calculate Φ relied on three distinct choices namely, (i) the use of conformational vectors where the elements are inter-residue distances extracted from a specific conformation; (ii) the use of the distribution of pairwise projections of these vectors to calculate the degree of intra-ensemble dissimilarity; and (iii) the use of the FRC model to calibrate the degree of heterogeneity. We discussed the advantages of choice (iii) in the main text. Choices (i) and (ii) lead to an assessment of intra-ensemble conformational dissimilarity. Although these choices are distinct, they are not inherently superior to other methods proposed in the literature. For example, we could have calculated all unique pairwise superpositions of conformations based on least squares optimization and used the resultant distribution of root mean squared deviations (RMSDs)80, 81 as measures of dissimilarities, although the computational expense of these calculations increases substantially with increased number of conformations in an ensemble. One could also use the method of projections to compare conformational vectors comprised of backbone dihedral angles as elements.82 We do not find any intrinsic advantages with using dihedral angle based conformational vectors and this could be used interchangeably with inter-residue distance based conformational vectors. Recent efforts have focused on the use of the number of inter-residue contacts q.83 Each conformation within the ensemble is annotated by its q-number and the distributions of q-numbers, viz., P(q) are analyzed to compare different ensembles to each other. This method, which is analogous to methods used in spin glass theories,84 can be used in conjunction with Φ. It should be noted that the annotation of conformations by q-numbers requires the imposition of an ad hoc criterion for defining contacts, which causes an inherent loss of information. This is in contrast to the conformational vectors Vc used in this work.
Fisher and Stultz85 introduced an order parameter based on information theory to quantify the degree of conformational heterogeneity. In direct analogy with Φ their order parameter O is bounded, i.e., 0 ≤ O ≤ 1; O → 1 for a homogeneous ensemble whereas O → 0 for a maximally heterogeneous ensemble. Their procedure for calculating O uses the weights for individual conformations and pairwise conformational similarities that are based on mean square deviations (MSDs). Molecular simulations such as molecular dynamics and Metropolis Monte Carlo methods are classified as importance sampling methods. Accordingly, they yield a set of conformations sampled from the equilibrium distribution but the weights of individual conformations are generally unknown. Assessment of these weights will require a priori conformational clustering and the assignment of weights is based on cluster sizes / populations. Alternatively one can use a suitable weighted histogram analysis method such as T-WHAM86 to assign weights to individual conformations. The procedure for conformational clustering is based on the calculation of MSDs and there is a nonlinear increase in computational expense with sample size. Further, the intrinsic property of MSDs is such that there are more ways to generate higher values of MSDs than lower ones and this bias also shows a nonlinear dependence on the MSD value and must be taken into account. Finally, the assessment of O uses as reference the average pairwise MSD that result from typical thermal fluctuations around a protein structure and hence the assessment of heterogeneity provided by O is intrinsically different from that Φ. According to the former, the degree of conformational heterogeneity in an ensemble is a normalized effective number of conformations in the ensemble given that thermal fluctuation around a protein structure will yield an average pairwise MSD of ≈2.5Å or higher depending on the temperature. The calculation of Φ makes no assumptions regarding the spatial size of thermal fluctuations around specific conformations because this most certainly depends on the density, i.e., the value of s2 or ρ. Instead the value of Φ quantifies the degree of heterogeneity as the normalized effective number of distinct conformations referenced to a generic, maximally heterogeneous Flory random coil state. It is likely that Φ and O can provide complementary assessments of the degree of disorder in an ensemble, especially in conjunction with assessments of s2 and other measures of local structure that yield insights regarding the type of conformations sampled. This will require improvements to make the calculation of O more efficient so the ensemble size can be expanded beyond the current limitation of ∼300 conformations.85
Practical uses for Φ
The calculation of Φ is designed with two practical purposes in mind. As noted in the Introduction, it is important to have measures of conformational heterogeneity that complement the assessments of ensembles that are obtained by quantification of densities, their fluctuations, and variances in energies. In order to understand the mechanisms of coupled folding and binding of IDPs it would be useful to be able to modulate the degree of disorder in the unbound ensemble using de novo sequence design. The parameter Φ helps in this regard because it provides a direct measure of conformational heterogeneity and can be used to guide sequence design in a way that heterogeneity is either decreased (increased Φ) or increased (decreased Φ).
ACKNOWLEDGMENTS
This work was supported by grants from the National Institutes of Health (5RO1NS056114) and the National Science Foundation (MCB-1121867). We thank Professor Anders Carlsson, Professor Gary Stormo, and two anonymous reviewers for helpful comments and suggestions.
References
- Wolynes P. G., Eaton W. A., and Fersht A. R., Proc. Natl. Acad. Sci. U.S.A. 109, 17770 (2012). 10.1073/pnas.1215733109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker A. K., Lawson J. D., Brown C. J., Williams R. M., Romero P., Oh J. S., Oldfield C. J., Campen A. M., Ratliff C. M., Hipps K. W., Ausio J., Nissen M. S., Reeves R., Kang C., Kissinger C. R., Bailey R. W., Griswold M. D., Chiu W., Garner E. C., and Obradovic Z., J. Mol. Graphics Modell. 19, 26 (2001). 10.1016/S1093-3263(00)00138-8 [DOI] [PubMed] [Google Scholar]
- Dyson H. J. and Wright P. E., Curr. Opin. Struct. Biol. 12, 54 (2002). 10.1016/S0959-440X(02)00289-0 [DOI] [PubMed] [Google Scholar]
- Halfmann R., Alberti S., Krishnan R., Lyle N., O’Donnell C. W., King O. D., Berger B., Pappu R. V., and Lindquist S., Mol. Cell 43, 72 (2011). 10.1016/j.molcel.2011.05.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eliezer D., Curr. Opin. Struct. Biol. 19, 23 (2009). 10.1016/j.sbi.2008.12.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sosnick T. R. and Barrick D., Curr. Opin. Struct. Biol. 21, 12 (2011). 10.1016/j.sbi.2010.11.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vendruscolo M., Curr. Opin. Struct. Biol. 17, 15 (2007). 10.1016/j.sbi.2007.01.002 [DOI] [PubMed] [Google Scholar]
- Mao A. H., Lyle N., and Pappu R. V., Biochem. J. 449, 307 (2013). 10.1042/BJ20121346 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anil B., Li Y., Cho J. H., and Raleigh D. P., Biochemistry 45, 10110 (2006). 10.1021/bi060636o [DOI] [PubMed] [Google Scholar]
- Meng W., Luan B., Lyle N., Pappu R. V., and Raleigh D. P., Biochemistry 52, 2662 (2013). 10.1021/bi301667u [DOI] [PubMed] [Google Scholar]
- Voelz V. A., Jaeger M., Yao S., Chen Y., Zhu L., Waldauer S. A., Bowman G. R., Friedrichs M., Bakajin O., Lapidus L. J., Weiss S., and Pande V. S., J. Am. Chem. Soc. 134, 12565 (2012). 10.1021/ja302528z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoffmann A., Nettels D., Clark J., Borgia A., Radford S. E., Clarke J., and Schuler B., Phys. Chem. Chem. Phys. 13, 1857 (2011). 10.1039/c0cp01911a [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus L. J., Curr. Opin. Struct. Biol. 23, 30 (2013). 10.1016/j.sbi.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Meng W., Lyle N., Luan B., Raleigh D. P., and Pappu R. V., Proc. Natl. Acad. Sci. U.S.A. 110, 2123 (2013). 10.1073/pnas.1216979110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann H., Soranno A., Borgia A., Gast K., Nettels D., and Schuler B., Proc. Natl. Acad. Sci. U.S.A. 109, 16155 (2012). 10.1073/pnas.1207719109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohn J. E., Millett I. S., Jacob J., Zagrovic B., Dillon T. M., Cingel N., Dothager R. S., Seifert S., Thiyagarajan P., Sosnick T. R., Hasan M. Z., Pande V. S., Ruzcinski I., Doniach S., and Plaxco K. W., Proc. Natl. Acad. Sci. U.S.A. 101, 12491 (2004). 10.1073/pnas.0403643101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H.-X., Rivas G., and Minton A. P., Annu. Rev. Biophys. Biomol. Struct. 37, 375 (2008). 10.1146/annurev.biophys.37.032807.125817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elcock A. H., Curr. Opin. Struct. Biol. 20, 196 (2010). 10.1016/j.sbi.2010.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn T. R. and Radford S. E., Arch. Biochem. Biophys. 469, 100 (2008). 10.1016/j.abb.2007.05.015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapidus L. J., Mol. Biosyst. 9, 29 (2013). 10.1039/c2mb25334h [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dedmon M. M., Lindorff-Larsen K., Christodoulou J., Vendruscolo M., and Dobson C. M., J. Am. Chem. Soc. 127, 476 (2005). 10.1021/ja044834j [DOI] [PubMed] [Google Scholar]
- Soranno A., Buchli B., Nettels D., Cheng R. R., Mueller-Spaeth S., Pfeil S. H., Hoffmann A., Lipman E. A., Makarov D. E., and Schuler B., Proc. Natl. Acad. Sci. U.S.A. 109, 17800 (2012). 10.1073/pnas.1117368109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dyson H. J. and Wright P. E., Nat. Rev. Mol. Cell Biol. 6, 197 (2005). 10.1038/nrm1589 [DOI] [PubMed] [Google Scholar]
- Pancsa R. and Tompa P., PLoS ONE 7, e34687 (2012). 10.1371/journal.pone.0034687 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tompa P. and Fuxreiter M., Trends Biochem. Sci. 33, 2 (2008). 10.1016/j.tibs.2007.10.003 [DOI] [PubMed] [Google Scholar]
- Mittag T., Kay L. E., and Forman-Kay J. D., J. Mol. Recognit. 23, 105 (2010). 10.1002/jmr.961 [DOI] [PubMed] [Google Scholar]
- Grosberg A. Y. and Kuznetsov D. V., Macromolecules 25, 1970 (1992). 10.1021/ma00033a022 [DOI] [Google Scholar]
- Sanchez I. C., Macromolecules 12, 980 (1979). 10.1021/ma60071a040 [DOI] [Google Scholar]
- Gianni S., Guydosh N. R., Khan F., Caldas T. D., Mayor U., White G. W. N., DeMarco M. L., Daggett V., and Fersht A. R., Proc. Natl. Acad. Sci. U.S.A. 100, 13286 (2003). 10.1073/pnas.1835776100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherman E. and Haran G., Proc. Natl. Acad. Sci. U.S.A. 103, 11539 (2006). 10.1073/pnas.0601395103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ziv G., Thirumalai D., and Haran G., Phys. Chem. Chem. Phys. 11, 83 (2009). 10.1039/b813961j [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shea J. E. and Brooks C. L., Annu. Rev. Phys. Chem. 52, 499 (2001). 10.1146/annurev.physchem.52.1.499 [DOI] [PubMed] [Google Scholar]
- O’Brien E. P., Brooks B. R., and Thirumalai D., Biochemistry 48, 3743 (2009). 10.1021/bi8021119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nettels D., Gopich I. V., Hoffmann A., and Schuler B., Proc. Natl. Acad. Sci. U.S.A. 104, 2655 (2007). 10.1073/pnas.0611093104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinha K. K. and Udgaonkar J. B., J. Mol. Biol. 353, 704 (2005). 10.1016/j.jmb.2005.08.056 [DOI] [PubMed] [Google Scholar]
- Chou J. J. and Shakhnovich E. I., J. Phys. Chem. B 103, 2535 (1999). 10.1021/jp9839192 [DOI] [Google Scholar]
- Udgaonkar J. B., Arch. Biochem. Biophys. 531, 24 (2013). 10.1016/j.abb.2012.10.003 [DOI] [PubMed] [Google Scholar]
- Crick S. L., Jayaraman M., Frieden C., Wetzel R., and Pappu R. V., Proc. Natl. Acad. Sci. U.S.A. 103, 16764 (2006). 10.1073/pnas.0608175103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mukhopadhyay S., Krishnan R., Lemke E. A., Lindquist S., and Deniz A. A., Proc. Natl. Acad. Sci. U.S.A. 104, 2649 (2007). 10.1073/pnas.0611503104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teufel D. P., Johnson C. M., Lum J. K., and Neuweiler H., J. Mol. Biol. 409, 250 (2011). 10.1016/j.jmb.2011.03.066 [DOI] [PubMed] [Google Scholar]
- Marsh J. A. and Forman-Kay J. D., Biophys. J. 98, 2383 (2010). 10.1016/j.bpj.2010.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jain N., Bhattacharya M., and Mukhopadhyay S., Biophys. J. 101, 1720 (2011). 10.1016/j.bpj.2011.08.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brocca S., Testa L., Sobott F., Samalikova M., Natalello A., Papaleo E., Lotti M., De Gioia L., Doglia S. M., Alberghina L., and Grandori R., Biophys. J. 100, 2243 (2011). 10.1016/j.bpj.2011.02.055 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vaiana S. M., Best R. B., Yau W.-M., Eaton W. A., and Hofrichter J., Biophys. J. 97, 2948 (2009). 10.1016/j.bpj.2009.08.041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A., Lyle N., and Pappu R. V., Biophys. J. 97, 303 (2009). 10.1016/j.bpj.2009.05.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Muller-Spath S., Soranno A., Hirschfeld V., Hofmann H., Ruegger S., Reymond L., Nettels D., and Schuler B., Proc. Natl. Acad. Sci. U.S.A. 107, 14609 (2010). 10.1073/pnas.1001743107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mao A. H., Crick S. L., Vitalis A., Chicoine C. L., and Pappu R. V., Proc. Natl. Acad. Sci. U.S.A. 107, 8183 (2010). 10.1073/pnas.0911107107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tooke L., Duitch L., Measey T. J., and Schweitzer-Stenner R., Biopolymers 93, 451 (2010). 10.1002/bip.21361 [DOI] [PubMed] [Google Scholar]
- Radhakrishnan A., Vitalis A., Mao A. H., Steffen A. T., and Pappu R. V., J. Phys. Chem. B 116, 6862 (2012). 10.1021/jp212637r [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vitalis A., Wang X., and Pappu R. V., J. Mol. Biol. 384, 279 (2008). 10.1016/j.jmb.2008.09.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen S., Berthelier V., Yang W., and Wetzel R., J. Mol. Biol. 311, 173 (2001). 10.1006/jmbi.2001.4850 [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Onuchic J. N., Socci N. D., and Wolynes P. G., Proteins: Struct., Funct., Genet. 21, 167 (1995). 10.1002/prot.340210302 [DOI] [PubMed] [Google Scholar]
- Onuchic J. N., LutheySchulten Z., and Wolynes P. G., Annu. Rev. Phys. Chem. 48, 545 (1997). 10.1146/annurev.physchem.48.1.545 [DOI] [PubMed] [Google Scholar]
- Onuchic J. N. and Wolynes P. G., Curr. Opin. Struct. Biol. 14, 70 (2004). 10.1016/j.sbi.2004.01.009 [DOI] [PubMed] [Google Scholar]
- Vitalis A., Wang X., and Pappu R. V., Biophys. J. 93, 1923 (2007). 10.1529/biophysj.107.110080 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papoian G. A., Proc. Natl. Acad. Sci. U.S.A. 105, 14237 (2008). 10.1073/pnas.0807977105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Potoyan D. A. and Papoian G. A., J. Am. Chem. Soc. 133, 7405 (2011). 10.1021/ja1111964 [DOI] [PubMed] [Google Scholar]
- Camacho C. J. and Thirumalai D., Phys. Rev. Lett. 71, 2505 (1993). 10.1103/PhysRevLett.71.2505 [DOI] [PubMed] [Google Scholar]
- Chan H. S. and Dill K. A., J. Chem. Phys. 99, 2116 (1993). 10.1063/1.465277 [DOI] [Google Scholar]
- Das R. K., Crick S. L., and Pappu R. V., J. Mol. Biol. 416, 287 (2012). 10.1016/j.jmb.2011.12.043 [DOI] [PubMed] [Google Scholar]
- Metropolis A. R. N., Rosenbluth M., Teller A., and Teller E., J. Chem. Phys. 21, 1087 (1953). 10.1063/1.1699114 [DOI] [Google Scholar]
- Vitalis A. and Pappu R. V., J. Comput. Chem. 30, 673 (2009). 10.1002/jcc.21005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kaminski G. A., Friesner R. A., Tirado-Rives J., and Jorgensen W. L., J. Phys. Chem. B 105, 6474 (2001). 10.1021/jp003919d [DOI] [Google Scholar]
- Mao A. H. and Pappu R. V., J. Chem. Phys. 137, 064104 (2012). 10.1063/1.4742068 [DOI] [PubMed] [Google Scholar]
- Engh R. A. and Huber R., Acta Cryst. A47, 392 (1991). 10.1107/S0108767391001071 [DOI] [Google Scholar]
- Flory P. J., Statistical Mechanics of Chain Molecules (Oxford University Press, New York, 1969). [Google Scholar]
- Tran H. T. and Pappu R. V., Biophys. J. 91, 1868 (2006). 10.1529/biophysj.106.086264 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schäfer L., Excluded Volume Effects in Polymer Solutions as Explained by the Renormalization Group (Springer, Berlin, 1999). [Google Scholar]
- Best R. B., Merchant K. A., Gopich I. V., Schuler B., Bax A., and Eaton W. A., Proc. Natl. Acad. Sci. U.S.A. 104, 18964 (2007). 10.1073/pnas.0709567104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Obradovic Z., Peng K., Vucetic S., Radivojac P., and Dunker A. K., Proteins: Struct., Funct., Bioinf. 61, 176 (2005). 10.1002/prot.20735 [DOI] [PubMed] [Google Scholar]
- Ishida T. and Kinoshita K., Bioinformatics 24, 1344 (2008). 10.1093/bioinformatics/btn195 [DOI] [PubMed] [Google Scholar]
- Ha B. Y. and Thirumalai D., Phys. Rev. A 46, R3012 (1992). 10.1103/PhysRevA.46.R3012 [DOI] [PubMed] [Google Scholar]
- Ha B. Y. and Thirumalai D., Macromolecules 28, 577 (1995). 10.1021/ma00106a023 [DOI] [Google Scholar]
- Mittag T. and Forman-Kay J. D., Curr. Opin. Struct. Biol. 17, 3 (2007). 10.1016/j.sbi.2007.01.009 [DOI] [PubMed] [Google Scholar]
- Amoutzias G. D., Veron A. S., Weiner J., Robinson-Rechavi M., Bornberg-Bauer E., Oliver S. G., and Robertson D. L., Mol. Biol. Evol. 24, 827 (2007). 10.1093/molbev/msl211 [DOI] [PubMed] [Google Scholar]
- Ellenberger T. E., Brandl C. J., Struhl K., and Harrison S. C., Cell 71, 1223 (1992). 10.1016/S0092-8674(05)80070-4 [DOI] [PubMed] [Google Scholar]
- Liu J. G., Perumal N. B., Oldfield C. J., Su E. W., Uversky V. N., and Dunker A. K., Biochemistry 45, 6873 (2006). 10.1021/bi0602718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oneil K. T., Hoess R. H., and Degrado W. F., Science 249, 774 (1990). 10.1126/science.2389143 [DOI] [PubMed] [Google Scholar]
- Kabsch W. and Sander C., Biopolymers 22, 2577 (1983). 10.1002/bip.360221211 [DOI] [PubMed] [Google Scholar]
- Vriend G. and Sander C., Proteins: Struct., Funct., Genet. 11, 52 (1991). 10.1002/prot.340110107 [DOI] [PubMed] [Google Scholar]
- Lyman E. and Zuckerman D. M., Biophys. J. 91, 164 (2006). 10.1529/biophysj.106.082941 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mu Y. G., Nguyen P. H., and Stock G., Proteins: Struct., Funct., Bioinf. 58, 45 (2005). 10.1002/prot.20310 [DOI] [PubMed] [Google Scholar]
- Potoyan D. A. and Papoian G. A., Proc. Natl. Acad. Sci. U.S.A. 109, 17857 (2012). 10.1073/pnas.1201805109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parisi G., Phys. Rev. Lett. 50, 1946 (1983). 10.1103/PhysRevLett.50.1946 [DOI] [Google Scholar]
- Fisher C. K. and Stultz C. M., J. Am. Chem. Soc. 133, 10022 (2011). 10.1021/ja203075p [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chodera J. D., Swope W. C., Pitera J. W., Seok C., and Dill K. A., J. Chem. Theory Comput. 3, 26 (2007). 10.1021/ct0502864 [DOI] [PubMed] [Google Scholar]