Abstract
Huntington’s disease (HD) is caused by an abnormal expansion of CAG trinucleotide repeats encoding polyglutamine (polyQ) in the first exon of the huntingtin (htt) gene. Despite considerable efforts, the pathogenesis of Huntington’s disease (HD) remains largely unclear due to a paucity of models that can reliably reproduce the pathological characteristics of HD. Here, we report a neuronal cell model of HD using the previously established tetracycline regulated rat neuroprogenitor cell line, HC2S2. Stable expression of enhanced green fluorescence protein (EGFP) tagged-htt exon 1 (referred to as 28Q and 74Q, respectively) in the HC2S2 cells did not affect rapid neuronal differentiation. However, compared to the cells expressing wild type htt, the cell line expressing mutant htt showed an increase in time-dependent cell death and neuritic degeneration, and displayed increased vulnerability to oxidative stress. Increased protein aggregation during the process of neuronal aging or when the cells were exposed to oxidative stress reagents was detected in the cell line expressing 74Q but not in its counterpart. These results suggest that the neuroprogenitor cell lines mimic the major neuropathological characteristics of HD and may provide a useful tool for studying the neuropathogenesis of HD and for high-throughput screening of therapeutic compounds.
Keywords: Huntington’s disease, pathogenesis, huntingtin, polyglutamine, neuron, neuroprogenitor
Introduction
Huntington’s disease (HD) is an autosomal dominant inherited neurodegenerative disorder that is clinically characterized by chorea, psychiatric dysfunction and cognitive decline (Young, 2003). The underlying molecular cause of HD is due to an expansion of CAG trinucleotide repeats coding for polyglutamine (polyQ) in the first exon of the huntingtin (htt) gene (Lombardi et al., 2009). People carrying the htt gene containing fewer than 35 CAG repeats are not at risk to develop HD, whereas those who have between 35 to 40 repeats have higher risk to develop the disease. Tragically, individuals who have more than 40 CAG repeats will develop HD symptoms with certainty (Bates, 2003). The greater the number of CAG repeats, the earlier the age of onset and the greater the severity of the disorder (Walker, 2007).
The major pathological characteristics of HD include progressive loss of medium-sized spiny neurons (MSNs) that are characterized by their expression of dopamine- and cAMP-regulated phosphoprotein of 32 kDa (DARPP-32) (Ouimet et al., 1984) in the striatum and formation of protein aggregates (Rubinsztein et al., 1999; Vonsattel and DiFiglia, 1998). However, it is still not completely understood why mutant Htt is toxic and why it cannot be efficiently degraded in the neurons. The full length Htt protein contains several protease cleavage sites and is cleaved in vivo, producing N-terminal fragments containing the polyQ repeats that are responsible for the toxicity of mutant Htt. These fragments are aggregation- prone, although whether the aggregates are toxic or protective to neuronal cells is obscure. Several lines of investigation have suggested that multiple mechanisms may underlie the neurotoxic effect of mutant Htt. These include oxidative stress (Stack et al., 2008), excitotoxicity (Graham et al., 2009), impaired mitochondrial integrity (Wang et al., 2009) and energy metabolism (Beal, 2000), dysregulation of calcium homeostasis (Rosenstock et al., 2010) and aberrant activation of transglutaminase (Jeitner et al., 2009), abnormal protein-protein interactions (Li et al., 2007; McGuire et al., 2006), impaired ubiquitin-proteasome system (UPS) (Seo et al., 2004; Wang et al., 2008), and transcriptional dysregulation and altered gene expression (Benn et al., 2008). Although each of these hypotheses is supported by evidence, often the results observed in one HD model cannot be replicated in another HD model system. One partial reason for this is that most models of HD that are currently being used cannot well reproduce the major neuropathological features of the disorder such as progressive neuronal death and aggregation of mutant Htt.
Cell culture models of HD have provided great insight in our understanding of the pathogenesis of HD since they are convenient for performing experiments at the cellular and molecular levels. A variety of cell lines have also been essential for in vitro assay systems for conducting high throughput screening of therapeutic compounds to treat the disorder. However, most cell models of HD that have been developed are usually based on non- neuronal cell lines such as HEK293 cells (Wang et al., 2004b), HeLa cells (Wang et al., 2006), or neuron-like cells such as PC12 (Morris, 1984; van Roon-Mom et al., 2008) and neuroblastoma cell lines (Wyttenbach et al., 2002). Although these cells exhibit some of the pathological features of HD, including Htt aggregation and cytotoxicity, these cells themselves lack basic neuronal properties such as lacking appropriate expression of neuronal markers, absence of large sodium and calcium currents, being unable to fire action potential, and thus are less relevant HD models compared to neuronal cells expressing the disease gene. To overcome these problems, striatal cell lines derived from a mouse model of HD have been generated (Trettel et al., 2000) but it is tedious and time-consuming to differentiate them into mature neurons. We elected to establish a neuronal model of HD by using the previously generated, regulatable rat neuroprogenitor cell line, HC2S2 (Hoshimaru et al., 1996). Several unique characteristics make it a good candidate for modeling HD. First, this cell line was derived from adult rat neuroprogenitor cells and it has been determined that these cells can differentiate into neurons. Second, the cell line is genetically modified, making it conditionally immortalized in culture medium. In the absence of tetracycline the cells continue to proliferate. Third, in the presence of tetracycline the cells are very easily induced to differentiate into neurons and express a variety of neuronal markers including tau, NeuN, neurofilament 200 kDa, glutamic acid decarboxylase, and NMDA receptors (Asahi et al., 1998; Hoshimaru et al., 1996). Finally, differentiated neurons from this cell line have large sodium and calcium currents and fire regenerative action potentials (Hoshimaru et al., 1996). Therefore, the HC2S2 cell line is a “true” neuronal cell line. Here, we describe the modification of the HC2S2 cell line for studying HD. We demonstrate that the modified cell line reproduces the major neuropathological aspects of HD and should be a valuable tool for studying the pathogenesis of HD and for performing high throughput screening of therapeutic compounds for treating the disorder.
Methods
Cell culture, DNA transfection, and fluorescence microscopy
The HC2S2 regulatable rat neuroprogenitor cells were kindly provided by Dr. Fred Gage at the Salk Institute for Biological Studies, La Jolla, California. Procedures for coating plates and cell cultures were based on previously described methods (Hoshimaru et al., 1996). To generate stable cell lines, the HC2S2 cell line was co-transfected with EGFP-htt fusion protein expression constructs (EGFP-htt28Q or EGFPhtt74Q) (Narain et al., 1999; Wang et al., 2006) along with the puromycin resistant pBABE vector (Gloeckner et al., 2009) using a neural stem cell Nucleofector kit and device (Lonza). Transfected cells were selected in culture medium by the presence of 0.25 µg/ml of puromycin in 10 cm petri dishes coated with matrigel (BD Bioscience). Individual cell clones that survived puromycin selection and expressed the EGFP-Htt fusion protein were identified and marked under a Zeiss Axiovert inverted fluorescence microscope. The positive cell colonies composed of 50–400 cells were picked under a phase contrast microscope using a P-200 pipetman and transferred into the matrigel coated 12- or 24-well plates for enlargement in the N2 medium (Hoshimaru et al., 1996). After more than 10 passages, the cell lines that stably expressed the EGFP-Htt fusion proteins that were identified by fluorescence microscopy were used for our experiments.
Immunocytochemical staining
Immunocytochemical staining was done based on our previously described methods (Wang et al., 2009). Briefly, cells were fixed with 4% paraformaldehyde for 25 min, permeabilized with 0.15% Triton X-100 for 12 min and then blocked with 5% bovine serum albumin for 45 min at room temperature. The cells were then incubated with a rabbit anti-DARPP-32 polyclonal antibody (1:100; Santa Cruz Biotechnology Inc.) or a mouse anti-Htt mEM48 antibody (Zhou et al., 2003) followed by incubation with a Cy3 goat anti-rabbit antibody or an Alexa Fluor 546 rabbit anti-mouse antibody (1:200; Jackson ImmunoResearch), respectively. Immunostained cells were observed with a Zeiss Axiovert fluorescence microscope and images were captured with a digital camera and Axiovision software.
Laser scanning confocal microscopy
Confocal microscopy was performed on an Olympus confocal microscope equipped with an argon laser and two HeNe lasers and FluoView 1000 software. 74Q cells were fixed with 4% paraformaldehyde, permeabilized with Triton-X100, and counterstained with DAPI as described before (Wang et al., 2009; Wang and Monteiro, 2007). Images were obtained by scanning the cells under 60× oil objectives.
Neuronal differentiation and oxidative stress reagent treatments
To induce neuronal differentiation, the cell lines stably expressing EGFP-Htt28Q or EGFP-Htt74Q were first cultured in 6-well or 24-well plates coated with either polyornithine (Sigma)/mouse laminin (BD Biosciences) or polyornithine/Matrigel (StemCell Technologies Inc.). They were cultured in 1.5 ml (for 6-well plates) or 0.5 ml (for 24-well plates) of differentiation medium containing F12/DMEM, tetracycline (final concentration 1 µg/ml), and N2 supplement for 3 days and then switched to F12/DMEM supplemented only with tetracycline (final concentration 1 µg/ml) to expedite neuronal differentiation and aging. Fifty percent of the medium in each well was changed every other day. For oxidative stress reagent treatments, neurons were exposed to H2O2 or paraquat at increasing doses for 7 h. The cells were then fixed with 4% paraformaldehyde and subjected to cell death analysis as described previously (Wang et al., 2006).
Quantification of neuritic degeneration and cell death
Neurons that stably express the EGFP-Htt fusion proteins containing either 28Q or 74Q were seeded in 35 mm petri dishes coated with polyornithine/Matrigel at the same cell density. After neuronal induction with tetracycline for 2, 4, 6, and 7 days, the neuritic degeneration was analyzed by phase contrast microscopy. Any neurons that had lost some or all of their neurites were categorized as displaying neuritic degeneration. Cell death was assessed by counting neuronal nuclear fragmentation/shrinkage after the cells were stained with Hoechst 33342 (5 µg/ml) using the previously described methods (Wang et al., 2006; Wang et al., 2004a). After staining, images were captured from different positions in each well with a 20× objective lens under a Zeiss Axiovert fluorescence microscope equipped with a digital camera and Axiovision software. If a cell had a broken nucleus containing two or more parts, it was regarded as a cell with a fragmented nucleus. If a cell had one bright unbroken nucleus but its size was significantly smaller than that of the normal neuronal nuclei, the cell was counted as one with a shrunken nucleus. To prevent bias in judging, each culture was photographed at several different locations, and images were displayed on a computer screen. Observers then evaluated neuritic degeneration or nuclear fragmentation without knowing the identity of the culture. In most cases, at least 1000 cells were counted for each type of treated cells. Alternatively, the cell death was detected by using trypan blue exclusion, staining the cells with a final concentration of 0.05%.
SDS-PAGE, Western blotting and aggregate filter trap assay
SDS-PAGE, Western blotting and filter trap assays were performed as described previously (Wang et al., 2006). The rabbit anti-GFP antibody was used before (Wang et al., 2006) and goat anti-actin polyclonal antibodies were purchased from Santa Cruz Biotechnology. Digitalized Western blot bands were scanned and quantified by measuring pixel number using an automated digitizing system (UN-Scan-it gel, version 6.1).
Statistical analysis
One-way analysis of variance was used for statistical analysis of the experimental results. T-tests were used for comparisons between different groups and p < 0.05 was regarded as statistically significant.
Results
Stable expression of EGFP-tagged htt exon 1 in the neuroprogenitor cell line, HC2S2
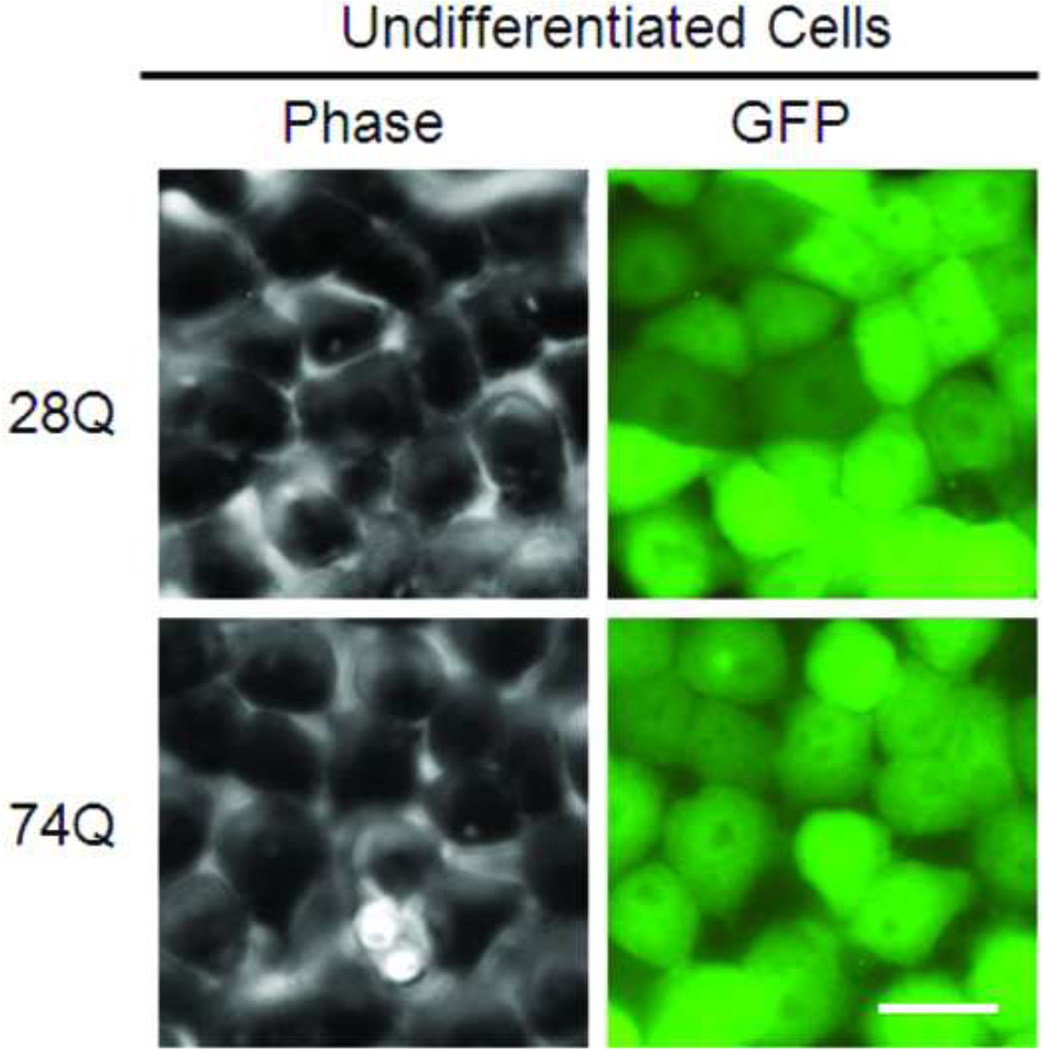
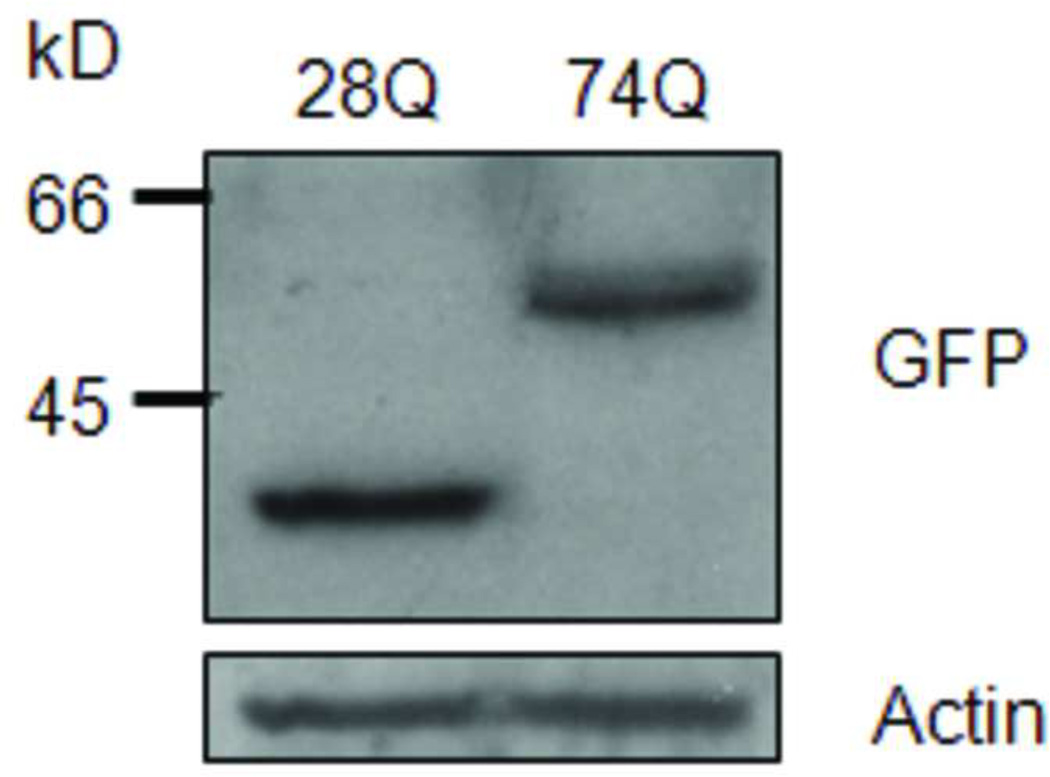
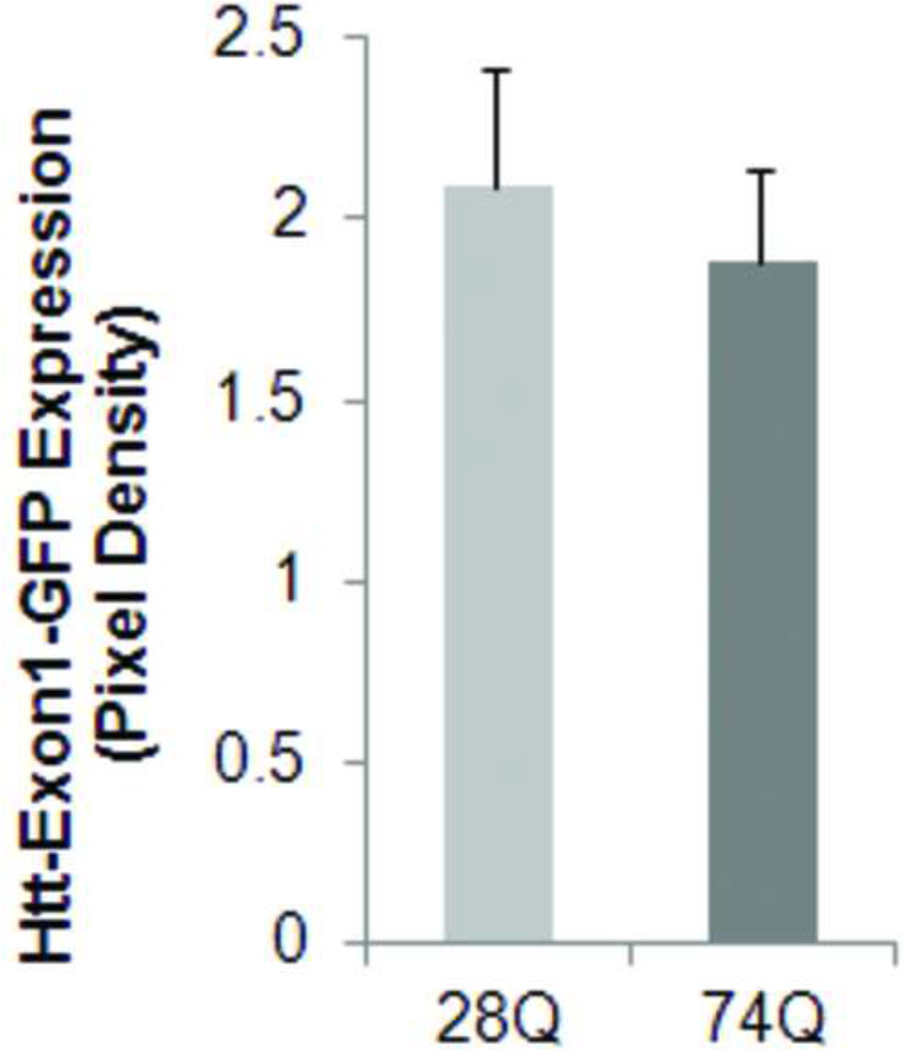
Since a number of reports have revealed that the N-terminal rather than the C-terminal Htt fragment is toxic aggregation-prone in the neurons, we stably transfected the regulatable rat neuroprogenitor cell line, HC2S2, with either an EGFP-htt-exon1- 28Q or EGFP-htt-exon1-74Q plasmid construct (referred to as 28Q and 74Q, respectively) that has previously been described (Narain et al., 1999; Wang et al., 2006). Two cell lines stably expressing either 28Q or 74Q were generated. When grown in the absence of tetracycline, the two cell lines had short processes and did not show any morphological differences (Fig. 1A). Expression of EGFP-htt was evenly distributed and was seen in both the cytoplasm and nucleus (Fig. 1A). At this stage, there were almost no microscopically detectable aggregates present in either cell line. Western blotting showed that the expression level of both EGFP constructs was equivalent in the two stable neuronal cell lines (Fig. 1B, 1C).
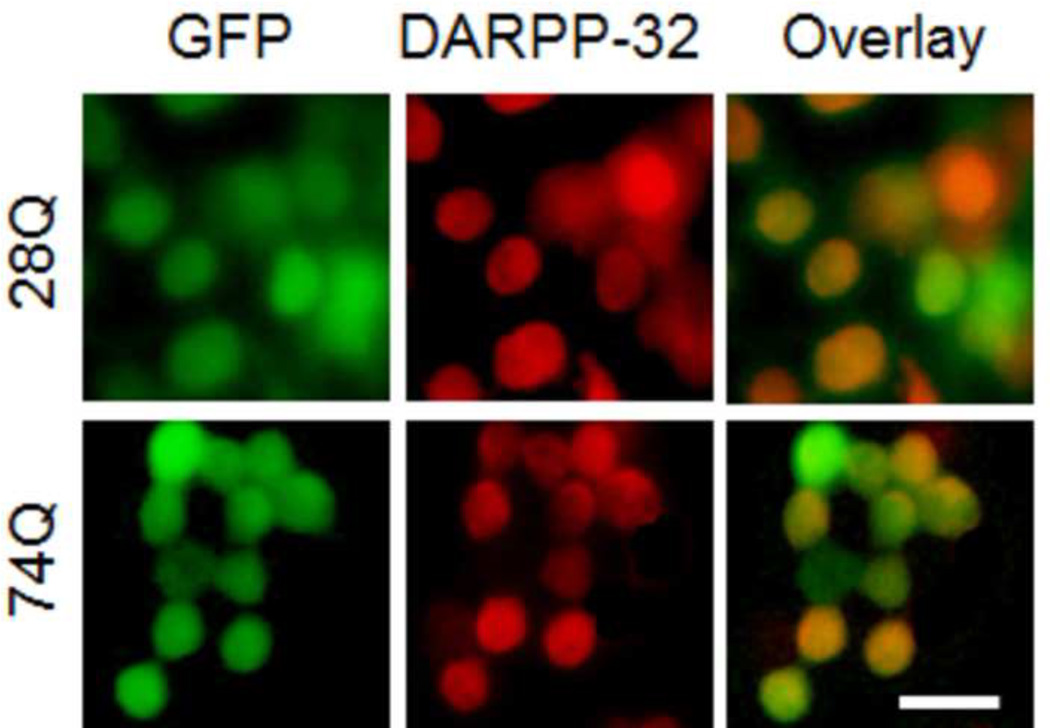
Fig 1. Characterization of neuroprogenitor cell lines stably expressing 28Q or 74Q.
A. Representative images of two stable cell lines, 28Q (top) and 74Q (bottom). Scale bar, 20 µm.
B. Western blot showing the expression of the stably transfected EGFP-htt constructs in the two cell lines. Equal amount of protein from each cell line was loaded on the gel. Actin is shown as a loading control.
C. Quantified expression of EGFP-tagged htt relative to actin showing that there is no significant difference between the 28Q and 74Q htt expression. Data are shown as mean ± SD; n = 3 p > 0.05.
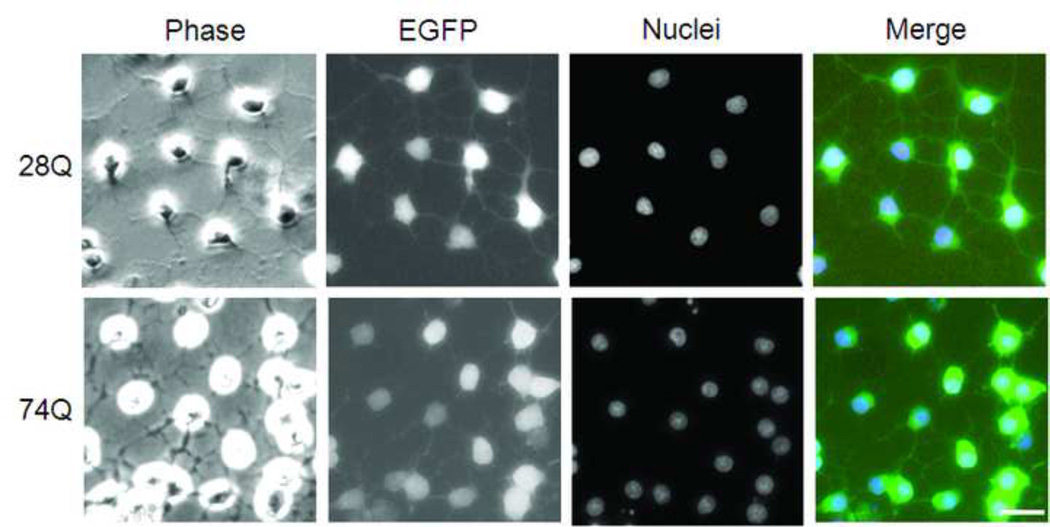
D. Representative images of two stable cell lines after 4 days of induction to differentiate into neurons. Nuclei were labeled with Hoechst 33342 (blue). The merged picture show both Hoechst staining and EGFP (green) expression. Scale bar, 20 µm.
E. Fluorescence microscopy of immunocytochemical staining of DARPP-32 (in red color) in the two cell lines after 2 days of neuronal induction. Scale bar, 20 µm.
When neuronal differentiation was induced by culturing the two cell lines in the presence of tetracycline, both underwent rapid neuronal differentiation and remarkable neurite outgrowth after 4 days (Fig. 1D). There was no evident nuclear fragmentation or shrinkage observed, suggesting an absence of significant neuronal death at this stage (Fig. 1D). Many neurites of the differentiated neurons were also green in color, indicating that, like endogenous Htt, the EGFP-Htt fusion protein was able to be transported to axons and dendrites (Fig. 1D). The EGFP-Htt fusion protein was still distributed evenly inside both types of cells and was found in both the nucleus and cytoplasm (Fig. 1D). EGFP-Htt aggregates were not readily seen under a fluorescence microscope in either cell line at this stage (Fig. 1D). Since the striatal MSNs are the most vulnerable cell population to the toxic mutant Htt, we wondered whether the two types of HD neuronal cell lines express DARPP-32, a neuronal marker for MSNs (Ouimet et al., 1984). After the cells were cultured in differentiation medium containing tetracycline for two days, the expression of DARPP-32 was detectible in majority of the cells by immunocytochemical staining (Fig. 1E, red color). These results suggest that stable transfection of the exogenous EGFP-Htt in the neuroprogenitor cells does not affect their differentiation and that at least some of the neurons express markers of MSNs.
The cell line expressing 74Q is more vulnerable to aging than the cell line expressing 28Q
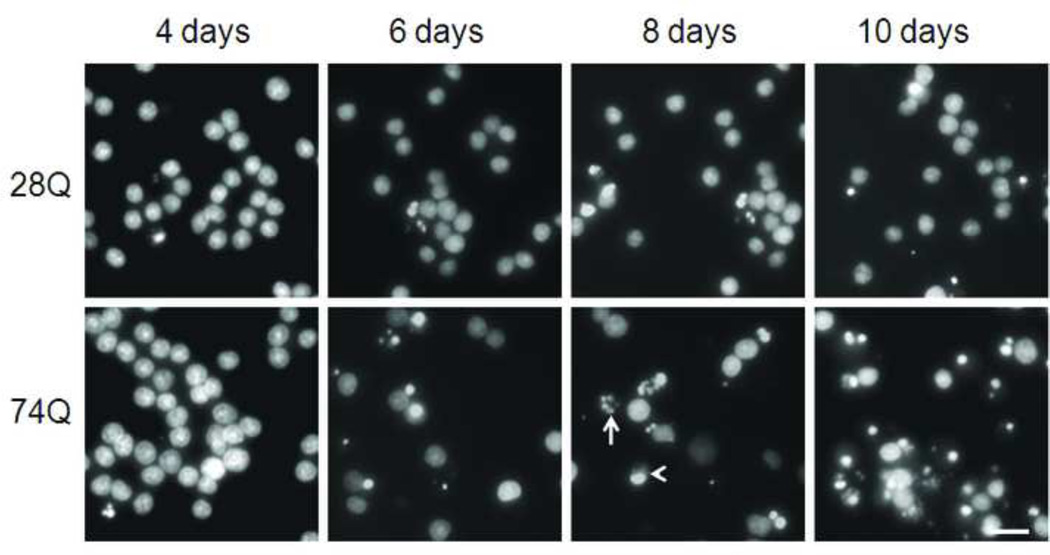
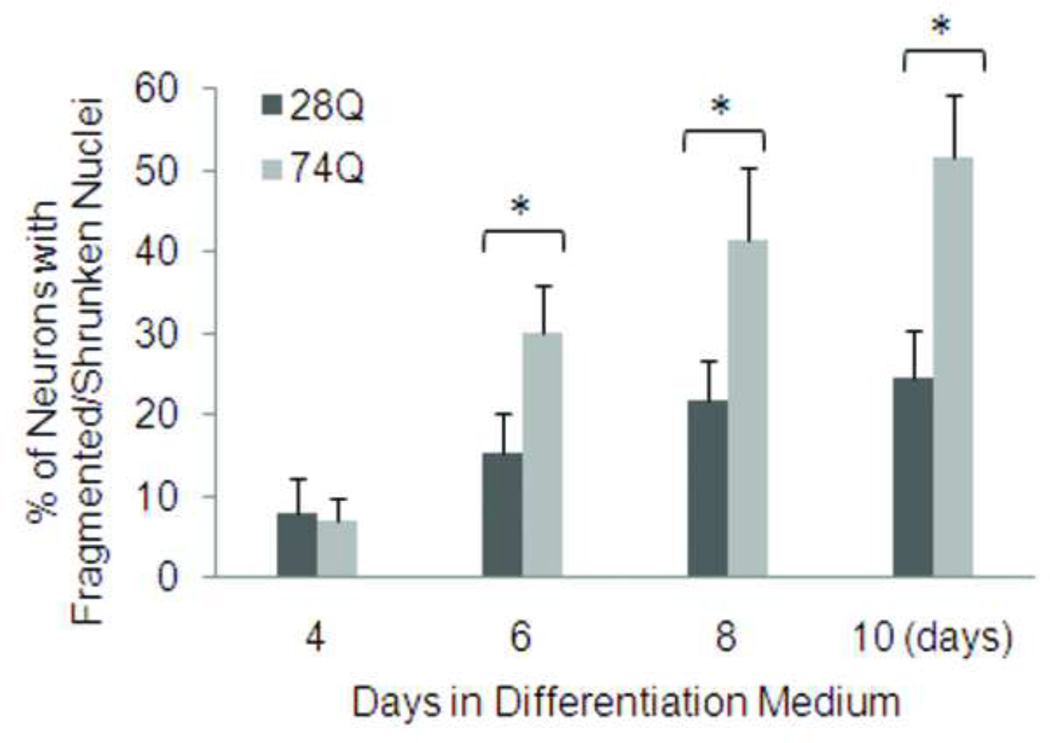
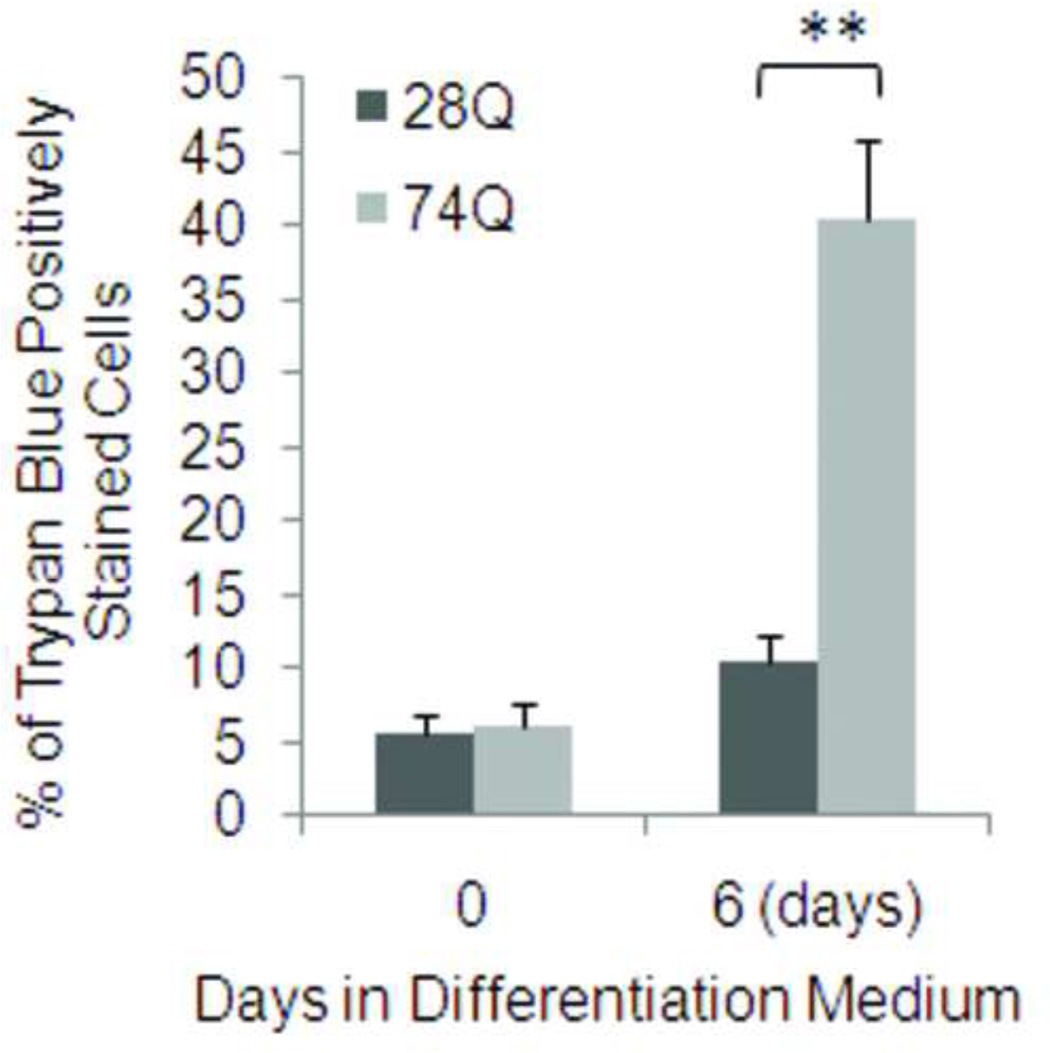
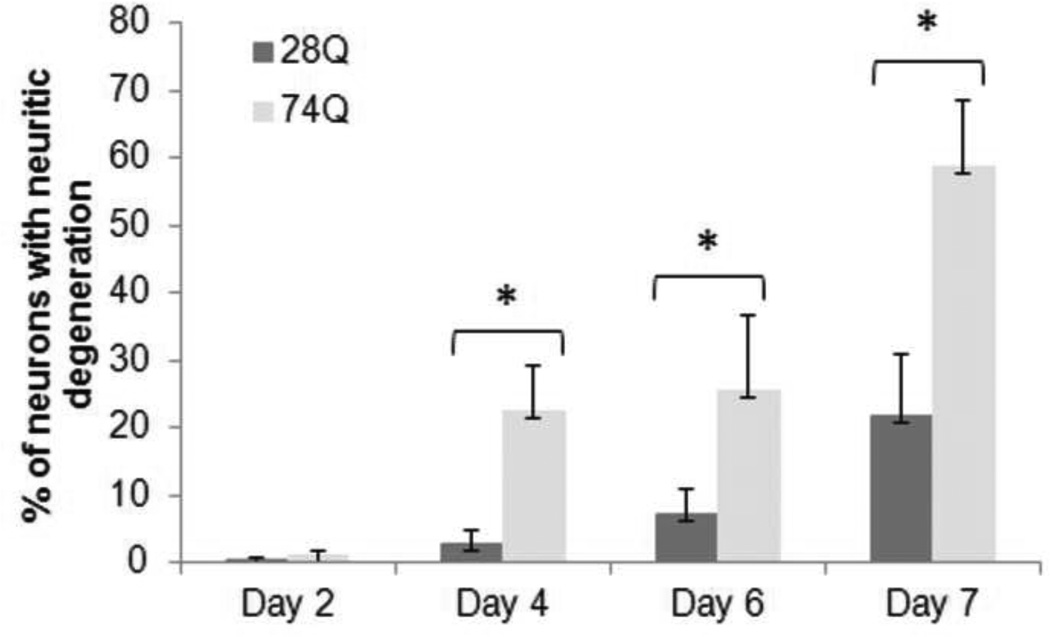
One major characteristic of HD is time-dependent progressive loss of neurons (Bates, 2002). To determine whether the cell line stably expressing EGFP-tagged mutant Htt mimics this characteristic of HD, we examined nuclear fragmentation and/or shrinkage at different time points following neuronal differentiation and aging. As shown in Figs. 2A and 2B, compared to the cell line expressing 28Q, the neuronal cell line expressing 74Q showed remarkable nuclear fragmentation/shrinkage after 6 days of neuronal differentiation, which was confirmed by the trypan blue exclusion test (Fig. 2C). Increasing neuronal death was seen after this stage when neurons became older in the culture (Fig. 2A, 2B). At 6 days, the cell line expressing 74Q only had 17% more nuclear fragmentation/shrinkage than the cell line expressing 28Q. However, the difference increased to 28% by day 10, indicating that the neurotoxicity of the mutant Htt is dependent upon neuronal age.
Fig 2. Vulnerability of the 74Q cell line during neuronal aging.
A. Fluorescence microscopy shows nuclear morphology at different time points following neuronal differentiation. Neuronal nuclei were stained with a DNA-binding dye, Hoechst 33342, before capturing the images. Note that the number of cells with nuclear fragmentation/shrinkage in the 74Q cell line is more than in the 28Q cell line. Arrow, an example of fragmented nucleus. Arrow head, an example of shrunken nucleus. Scale bar, 20 µm.
B. Quantification of cell death as determined by the ratio of neurons with fragmented/shrunken nuclei to the total number of neurons (see Methods). In each group of cells, at least 1000 cells were counted. Data are shown as mean ± SD; n = 3. Asterisk, p < 0.05.
C. Quantification of cell death as determined by the trypan blue exclusion test. In each group of cells, at least 1000 cells were counted. Data are shown as mean ± SD; n = 10. Double asterisk, p < 0.0001.
The cell line expressing 74Q showed more frequent neuritic degeneration than the cell line expressing 28Q
A previous report has shown that neuritic degeneration is found in HD mice, which is associated with neuronal soma degeneration (Li et al., 2001). To detect whether the neuronal cell line carrying the disease gene displays a similar phenotype, we also examined changes in neuritic morphology following neuronal differentiation. As shown in Fig. 3B, the cell line expressing 74Q showed more frequent neuritic degeneration than the cell line expressing 28Q after 4 days of neuronal differentiation. Approximately 60% neurons expressing 74Q showed remarkable neuritic degeneration after 7 days following the neuronal differentiation, whereas most neurites seen in the cell line expressing 28Q remained intact at this stage (Fig. 3A, 3B). Loss of neurites in 74Q neurons appeared to be associated with neuronal death. After 7 days of neuronal differentiation, most of the 74Q neurons lost their neurites, which were associated with evident cell death in the majority of the neurons as indicated by their abnormal nuclear morphology (Fig. 3A, bottom panels). Thus, these results indicate that progressive neuritic degeneration also occurs in the cell line expressing 74Q mutant Htt.
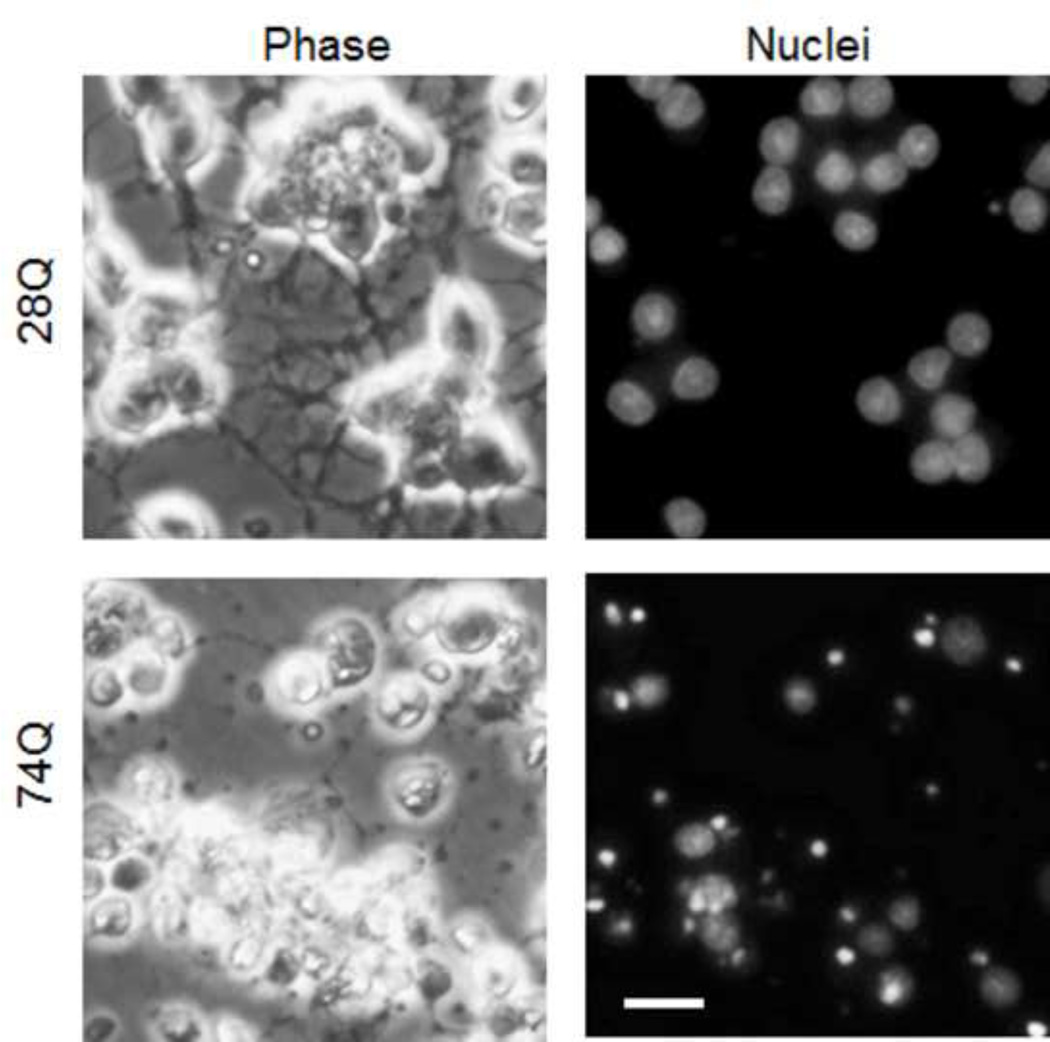
Fig 3. Neuritic degeneration occurs in the 74Q cell line more frequently than in the 28Q cell line.
A. Phase contrast (left panels) and Hoechst 33342 stained nuclei (right panels) images taken at 7 days following neuronal differentiation show that significant neuritic degeneration occurred in the 74Q cell line. Scale bar, 20 µm.
B. Quantification of neuritic degeneration in the neurons differentiated for indicated days. Data are shown as mean ± SD; n = 5; asterisk, p < 0.001.
Expression of mutant Htt makes the cell line more susceptible to exposed oxidative stress
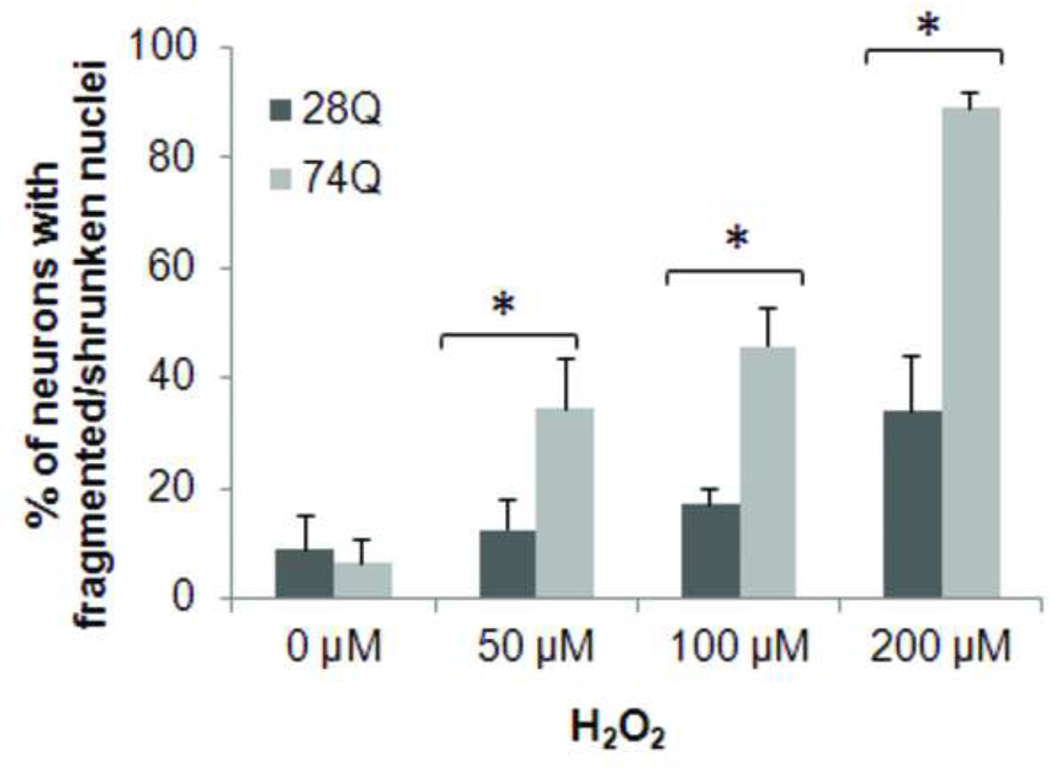
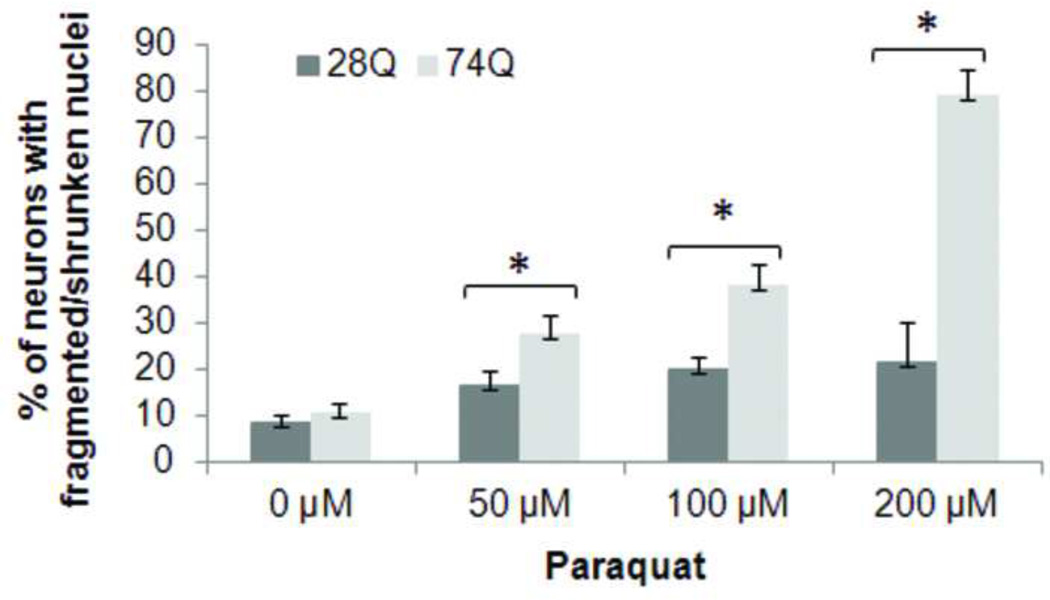
Since a number of observations suggest that oxidative stress is a potential mechanism of neuronal death in HD (Dai et al., 2009; van Roon-Mom et al., 2008), we tested the relative susceptibility of the two cell lines to exposed oxidative stress. Neurons were treated with different concentrations of H2O2 or paraquat, an herbicide that enhances the formation of reactive oxygen species by causing mitochondrial dysfunction (Wernig et al., 2002), and cell death was assessed after 8 hr of treatment. As shown in Figs. 4A and 4B, the cell line expressing 74Q is more vulnerable to both types of oxidative stressors than the cell line expressing 28Q, indicating that oxidative stress may facilitate the neurotoxicity of mutant Htt.
Fig 4. Facilitation of effects of oxidative stress by expression of mutant Htt.
A. Quantification of neuronal death in cells treated with H2O2. Neurons were treated with the indicated concentrations of H2O2 for 8 hr following 4 days of differentiation. Control cells were exposed to PBS. The percentage of cells showing nuclear fragmentation/shrinkage was calculated by determining the ratio of neurons with fragmented/shrunken nuclei to the total cell number. In each group of cells, at least 1000 cells were counted. Data are shown as mean ± SD; n= 5; Asterisk, p < 0.05 .
B. Quantification of neuronal death in cells treated with paraquat. Neurons were treated with the indicated concentrations of paraquat for 8 hr following 4 days of differentiation. Control cells were exposed to PBS. The percentage of cells showing nuclear fragmentation/shrinkage was calculated by determining the ratio of neurons with fragmented/shrunken nuclei to the total cell number. At least 700 cells were counted in each group. Data are shown as mean ± SD; n = 3 Asterisk, p < 0.05.
Protein aggregates are found selectively in the cell line expressing 74Q
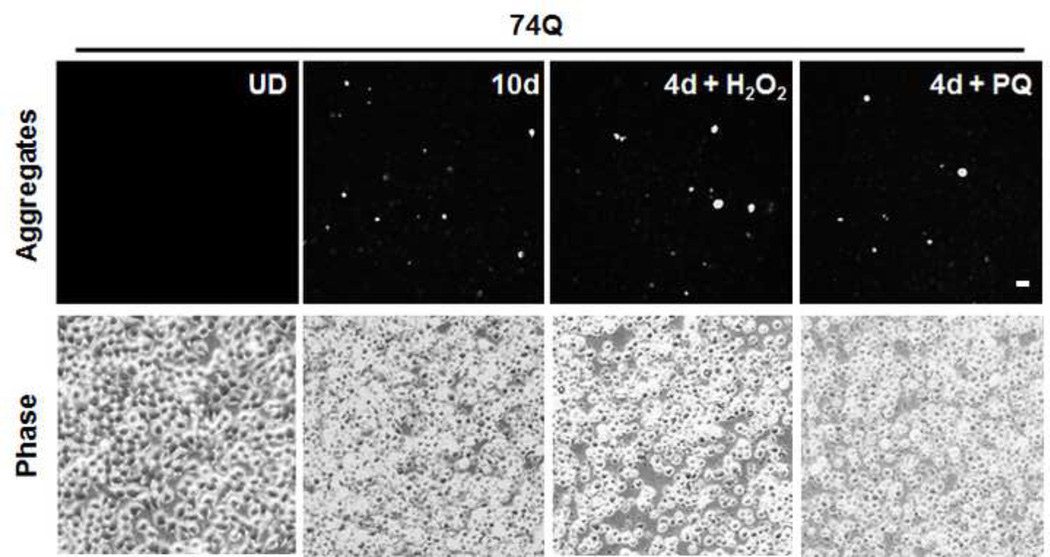
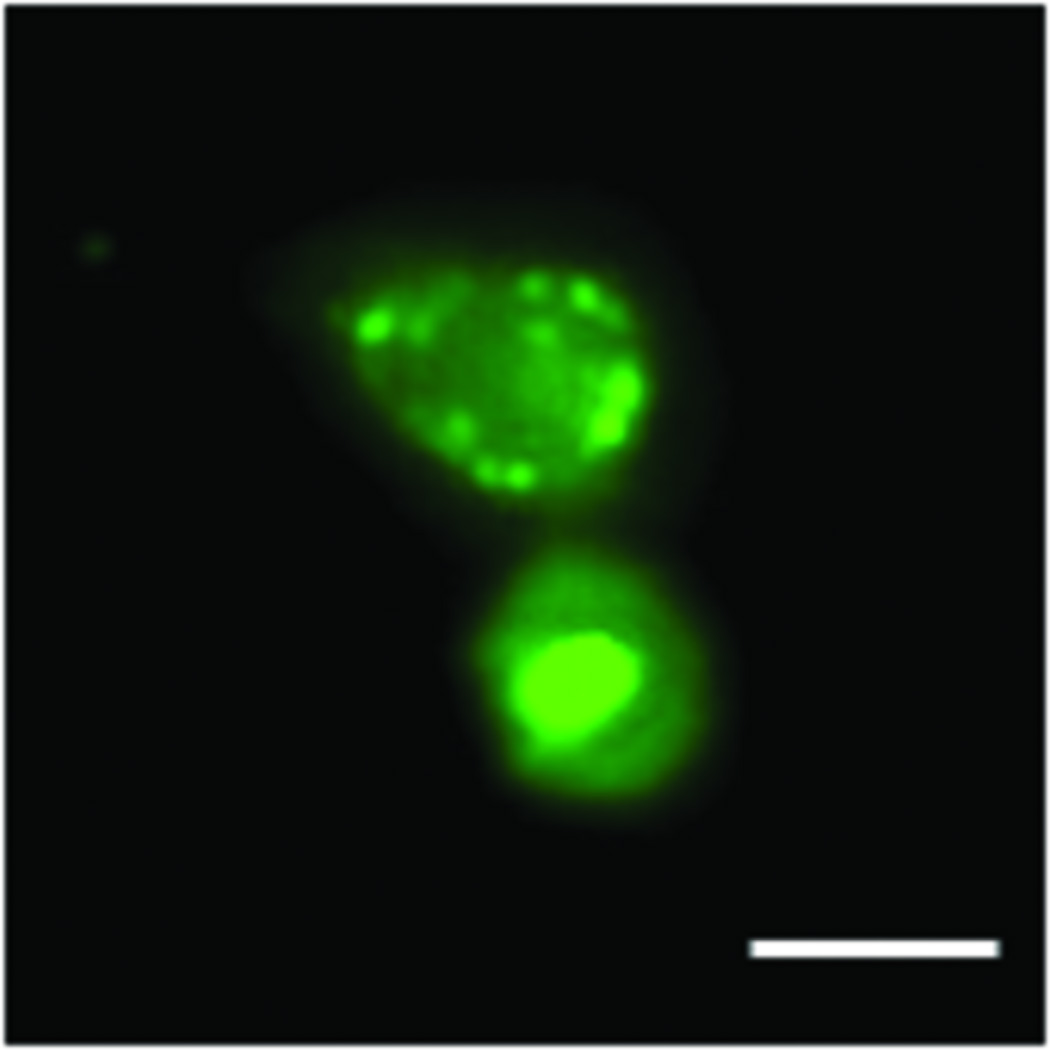
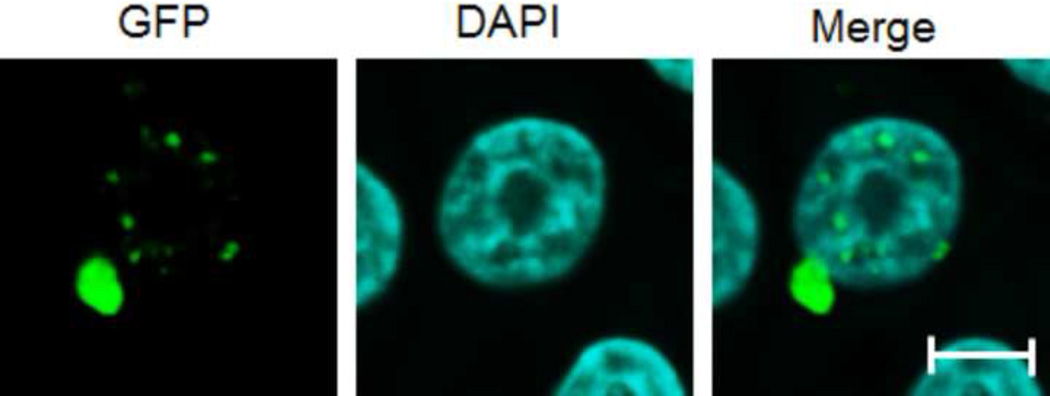
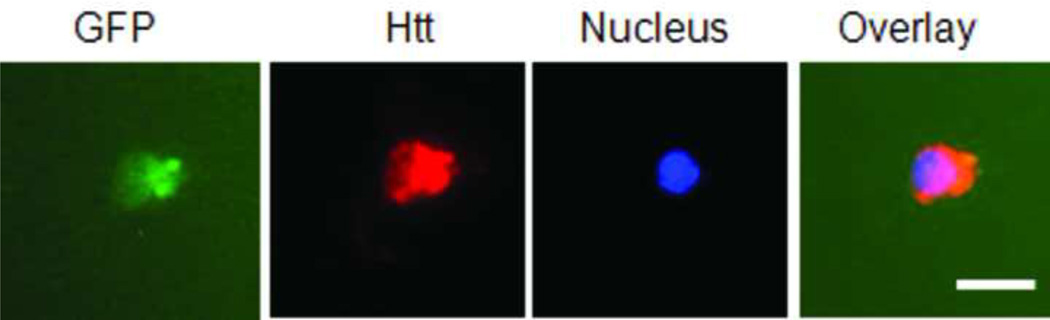
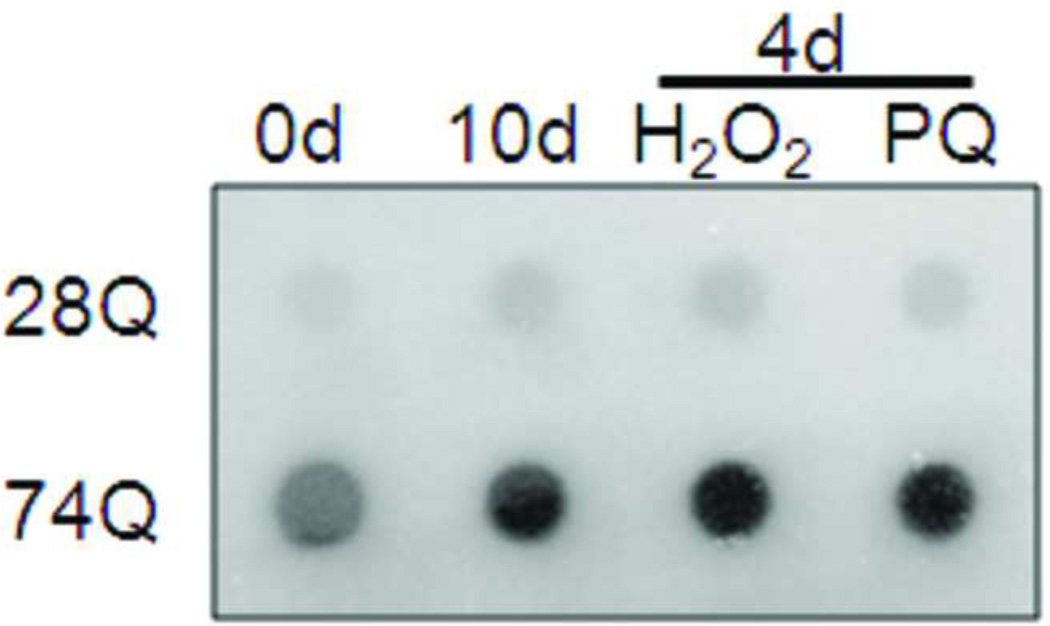
Since mutant protein aggregation is also a major neuropathological hallmark of HD (Rubinsztein et al., 1999), we assessed EGFP-Htt aggregates in the two cell lines. We did not find obvious aggregates in the cell line expressing 28Q in the undifferentiated or differentiated cells (data not shown). As mentioned above, no obvious protein aggregates were seen in the undifferentiated cells expressing 74Q (Fig. 5A) or in the cell line before 6 days of neuronal differentiation (data not shown). Since proteasomal malfunction caused by oxidative stress increases mutant Htt aggregation (Goswami et al., 2006), we accordingly examined mutant Htt aggregation in different unfavorable culture conditions. When the culture medium of the undifferentiated neurons became acidic (pH 6.5) as indicated by loss of the normal culture medium color, large protein aggregates could occasionally be seen in some cells expressing 74Q but not 28Q under a fluorescence microscope (Fig. 5B). The aggregates were seen in both the cytoplasm and in the nucleus in the neurons expressing 74Q (Fig. 5B), which were confirmed by laser scanning confocal microscopy (Fig. 5C) and by immunostaining with the mEM48 antibody (Zhou et al., 2003) (Fig. 5D). Protein aggregates were also microscopically detectible in the neurons that were allowed to undergo 10 days of neuronal differentiation or that received oxidative stress stimuli such as H2O2 or paraquat (Fig. 5A). In contrast, aggregates were not seen in the cell line expressing 28Q under these conditions (data not shown). The aggregates in the cell line expressing 74Q were also detected using a filter trap assay (Fig. 5E). These results indicate that, similar to observations made in in vivo studies, mutant Htt protein in the neuronal cell line is also aggregation-prone, and that aging and oxidative stress can facilitate its aggregation.
Fig 5. Selective protein aggregation in the 74Q cell line.
A. Fluorescence microscopy shows large, bright EGFP-Htt protein aggregates in aged neurons (10 days) or young neurons (4 days) treated with oxidative stress reagents, H2O2 or paraquat (PQ) (100 µM, 8 hr). UD, undifferentiated cells. Scale bar, 20 µm.
B. Fluorescence microscopy showing protein aggregates in the undifferentiated neurons after their culture medium became acidic (pH 6.5). Scale bar, 10 µm.
C. Laser scanning confocal microscopy showing protein aggregates existing in both the nucleus and cytoplasm in the undifferentiated neurons after their culture medium became acidic (pH 6.5). Scale bar, 5 µm.
D. Fluorescence microscopy of neurons that were treated with 100 µM H2O2 for 8 hr after 4 days of neuronal differentiation. The Htt (red color) was immunocytochemically stained with the mEM48 antibody. Scale bar, 10 µm.
E. Filter trap assay with samples from A. For each sample, 30 µg of total protein from the cell lysates was loaded onto the filter. Immunoblotting was performed with an anti-GFP antibody.
Discussion
Here, we established neuroprogenitor cell lines expressing EGFP-Htt exon 1 containing either a normal polyQ tract (28Q) or an expanded polyQ tract (74Q). Without neuronal induction, the undifferentiated neuroprogenitor cells did not show cell death, and the microscopically detectible protein aggregation is relatively minimal. When the cell lines were differentiated into neurons, however, progressive neuronal death and protein aggregates were detected during neuronal aging, both of which were exacerbated by additional oxidative stress stimuli. Intriguingly, neuritic degeneration was also observed and this apparently associated with somal degeneration. These results indicate that the cell lines mimic the major aspects of neuropathological characteristics of HD, progressive neuronal death and formation of protein aggregates.
A previous report indicated that the rat hippocampal neuroprogenitor cell line used here, HC2S2, rapidly exhibits neuronal properties in the presence of tetracycline, expressing a variety of neuronal markers and firing action potential (Hoshimaru et al., 1996). Stable expression of either EGFP-Htt did not disrupt neuronal differentiation as is evidenced by both cell lines expressing DARPP-32 (Fig.1E) and exhibiting typical neuronal morphology upon induction (Fig. 1D). Previous reports have indicated that striatum is not the only brain region containing DARPP-32-positive neurons. It remains to be determined whether DARPP-32-positive neurons outside the striatum are more vulnerable than other types of neurons to the toxicity of mutant Htt during the development of HD. In addition, similar to their parent HC2S2 cell line,
Our results showed that both neuronal death, as indicated by nuclear fragmentation, and neuritic degeneration were time-dependent in the cell culture. It is noticed that the 28Q neurons also appeared a slight increase in cell death when cultured in the differentiation medium over time (Figure 2B), which might reflect the cell culture condition gradually became unfavorable for neuronal survival over time or increasing production of free radicals in the differentiated neurons occurred. However, 74Q expressing neurons showed significantly more cell death than the 28Q neurons after 6 days under the given culture condition, indicating the toxic effects of the mutant Htt. It is possible that the increased production of free radicals may account for this time-dependent neurodegeneration in the cultured neurons due to mitochondrial dysfunction (Browne and Beal, 2004; Browne et al., 1999). When exposed to oxidative stressors, the 74Q neurons were more susceptible than the 28Q neurons, indicating that some effect of mutant Htt on cells makes cell more susceptible to oxidative stress.
Similar to the results obtained from in vivo studies, mutant Htt aggregates were also found in differentiated neurons from the cell line expressing the mutant htt exon 1, although it remains uncertain whether the protein aggregation itself is protective (Arrasate et al., 2004) or deleterious (Davies et al., 1997). Previous data have indicated that after cleavage, it is the N-terminal fragment rather than C-terminal fragment of the mutant protein that shows aggregation-prone property and accumulates in the cells (Li et al., 2000). We did not determine whether the 74Q Htt impaired the ubiquitinproteasome system (UPS) in the cell line expressing 74Q. It will be interesting to determine the UPS activity in the two cell lines and examine whether protein aggregation is associated with an alteration of the UPS activity.
In summary, this report describes an inducible neuroprogenitor cell model of HD, which may provide a useful tool for studying the pathogenesis of HD (for instance, using the cell lines to identify and study the proteins that interact with the mutant Htt) and for high throughput screening of therapeutic compounds.
Acknowledgements
We would like to thank Dr. Robin Miskimins for critically reading the manuscript, Dr. Fred H. Gage for providing the HC2S2 neuronal cell line, Dr. David C. Rubinsztein for providing EGFP-htt constructs, Drs. Joyce Keifer and Fran Day at the South Dakota Imaging Core Facility (supported by NIH P20 RR 015567, which is designated as a Center of Biomedical Research Excellence (COBRE) to Dr. Joyce Keifer) for help in confocal and fluorescence microscopy, and Drs. Huabo Su, and Ms. Lili Guo for providing assistance in fluorescence microscopy. We are also grateful to Dr. Kathleen Eyster and Ms. Sandy Bradley at the South Dakota Genomics Core Facility (supported by NIH INBRE 2 P20 RR016479) for their assistance in using the Nucleofector device. This work was supported by Start-up Funds from the University of South Dakota, a Competitive Research Grant Award of the South Dakota Board of Regents, and a New Faculty Development Award of the University of South Dakota. MJM was supported by an NIH grant GM066287.
References
- Arrasate M, Mitra S, Schweitzer ES, Segal MR, Finkbeiner S. Inclusion body formation reduces levels of mutant huntingtin and the risk of neuronal death. Nature. 2004;431:805–810. doi: 10.1038/nature02998. [DOI] [PubMed] [Google Scholar]
- Asahi M, Hoshimaru M, Hojo M, Matsuura N, Kikuchi H, Hashimoto N. Induction of the N-methyl-D-aspartate receptor subunit 1 in the immortalized neuronal progenitor cell line HC2S2 during differentiation into neurons. J Neurosci Res. 1998;52:699–708. doi: 10.1002/(SICI)1097-4547(19980615)52:6<699::AID-JNR9>3.0.CO;2-3. [DOI] [PubMed] [Google Scholar]
- Bates G. Huntingtin aggregation and toxicity in Huntington's disease. Lancet. 2003;361:1642–1644. doi: 10.1016/S0140-6736(03)13304-1. [DOI] [PubMed] [Google Scholar]
- Bates G, Harper P, Jones L. Huntington's Disease. 3rd edn. Oxford: Oxford University Press; 2002. [Google Scholar]
- Beal MF. Energetics in the pathogenesis of neurodegenerative diseases. Trends Neurosci. 2000;23:298–304. doi: 10.1016/s0166-2236(00)01584-8. [DOI] [PubMed] [Google Scholar]
- Benn CL, Sun T, Sadri-Vakili G, McFarland KN, DiRocco DP, Yohrling GJ, Clark TW, Bouzou B, Cha JH. Huntingtin modulates transcription, occupies gene promoters in vivo, and binds directly to DNA in a polyglutamine-dependent manner. J Neurosci. 2008;28:10720–10733. doi: 10.1523/JNEUROSCI.2126-08.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Browne SE, Beal MF. The energetics of Huntington's disease. Neurochem Res. 2004;29:531–546. doi: 10.1023/b:nere.0000014824.04728.dd. [DOI] [PubMed] [Google Scholar]
- Browne SE, Ferrante RJ, Beal MF. Oxidative stress in Huntington's disease. Brain Pathol. 1999;9:147–163. doi: 10.1111/j.1750-3639.1999.tb00216.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dai Y, Dudek NL, Li Q, Fowler SC, Muma NA. Striatal expression of a calmodulin fragment improved motor function, weight loss, and neuropathology in the R6/2 mouse model of Huntington's disease. J Neurosci. 2009;29:11550–11559. doi: 10.1523/JNEUROSCI.3307-09.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies SW, Turmaine M, Cozens BA, DiFiglia M, Sharp AH, Ross CA, Scherzinger E, Wanker EE, Mangiarini L, Bates GP. Formation of neuronal intranuclear inclusions underlies the neurological dysfunction in mice transgenic for the HD mutation. Cell. 1997;90:537–548. doi: 10.1016/s0092-8674(00)80513-9. [DOI] [PubMed] [Google Scholar]
- Gloeckner CJ, Boldt K, Schumacher A, Ueffing M. Tandem affinity purification of protein complexes from mammalian cells by the Strep/FLAG (SF)-TAP tag. Methods Mol Biol. 2009;564:359–372. doi: 10.1007/978-1-60761-157-8_21. [DOI] [PubMed] [Google Scholar]
- Goswami A, Dikshit P, Mishra A, Mulherkar S, Nukina N, Jana NR. Oxidative stress promotes mutant huntingtin aggregation and mutant huntingtin-dependent cell death by mimicking proteasomal malfunction. Biochem Biophys Res Commun. 2006;342:184–190. doi: 10.1016/j.bbrc.2006.01.136. [DOI] [PubMed] [Google Scholar]
- Graham RK, Pouladi MA, Joshi P, Lu G, Deng Y, Wu NP, Figueroa BE, Metzler M, Andre VM, Slow EJ, et al. Differential susceptibility to excitotoxic stress in YAC128 mouse models of Huntington disease between initiation and progression of disease. J Neurosci. 2009;29:2193–2204. doi: 10.1523/JNEUROSCI.5473-08.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshimaru M, Ray J, Sah DW, Gage FH. Differentiation of the immortalized adult neuronal progenitor cell line HC2S2 into neurons by regulatable suppression of the v-myc oncogene. Proc Natl Acad Sci U S A. 1996;93:1518–1523. doi: 10.1073/pnas.93.4.1518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeitner TM, Pinto JT, Krasnikov BF, Horswill M, Cooper AJ. Transglutaminases and neurodegeneration. J Neurochem. 2009;109(Suppl 1):160–166. doi: 10.1111/j.1471-4159.2009.05843.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Li SH, Johnston H, Shelbourne PF, Li XJ. Amino-terminal fragments of mutant huntingtin show selective accumulation in striatal neurons and synaptic toxicity. Nat Genet. 2000;25:385–389. doi: 10.1038/78054. [DOI] [PubMed] [Google Scholar]
- Li H, Li SH, Yu ZX, Shelbourne P, Li XJ. Huntingtin aggregate-associated axonal degeneration is an early pathological event in Huntington's disease mice. J Neurosci. 2001;21:8473–8481. doi: 10.1523/JNEUROSCI.21-21-08473.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XJ, Friedman M, Li S. Interacting proteins as genetic modifiers of Huntington disease. Trends Genet. 2007;23:531–533. doi: 10.1016/j.tig.2007.07.007. [DOI] [PubMed] [Google Scholar]
- Lombardi MS, Jaspers L, Spronkmans C, Gellera C, Taroni F, Di Maria E, Donato SD, Kaemmerer WF. A majority of Huntington's disease patients may be treatable by individualized allele-specific RNA interference. Exp Neurol. 2009;217:312–319. doi: 10.1016/j.expneurol.2009.03.004. [DOI] [PubMed] [Google Scholar]
- McGuire JR, Rong J, Li SH, Li XJ. Interaction of Huntingtin-associated protein-1 with kinesin light chain: implications in intracellular trafficking in neurons. J Biol Chem. 2006;281:3552–3559. doi: 10.1074/jbc.M509806200. [DOI] [PubMed] [Google Scholar]
- Morris R. Developments of a water-maze procedure for studying spatial learning in the rat. J Neurosci Methods. 1984;11:47–60. doi: 10.1016/0165-0270(84)90007-4. [DOI] [PubMed] [Google Scholar]
- Narain Y, Wyttenbach A, Rankin J, Furlong RA, Rubinsztein DC. A molecular investigation of true dominance in Huntington's disease. J Med Genet. 1999;36:739–746. doi: 10.1136/jmg.36.10.739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ouimet CC, Miller PE, Hemmings HC, Jr, Walaas SI, Greengard P. DARPP-32, a dopamine- and adenosine 3':5'-monophosphate-regulated phosphoprotein enriched in dopamine-innervated brain regions. III. Immunocytochemical localization. J Neurosci. 1984;4:111–124. doi: 10.1523/JNEUROSCI.04-01-00111.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenstock TR, Bertoncini CR, Teles AV, Hirata H, Fernandes MJ, Smaili SS. Glutamate-induced alterations in Ca2+ signaling are modulated by mitochondrial Ca2+ handling capacity in brain slices of R6/1 transgenic mice. Eur J Neurosci. 2010;32:60–70. doi: 10.1111/j.1460-9568.2010.07268.x. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC, Wyttenbach A, Rankin J. Intracellular inclusions, pathological markers in diseases caused by expanded polyglutamine tracts? J Med Genet. 1999;36:265–270. [PMC free article] [PubMed] [Google Scholar]
- Seo H, Sonntag KC, Isacson O. Generalized brain and skin proteasome inhibition in Huntington's disease. Ann Neurol. 2004;56:319–328. doi: 10.1002/ana.20207. [DOI] [PubMed] [Google Scholar]
- Stack EC, Matson WR, Ferrante RJ. Evidence of oxidant damage in Huntington's disease: translational strategies using antioxidants. Ann N Y Acad Sci. 2008;1147:79–92. doi: 10.1196/annals.1427.008. [DOI] [PubMed] [Google Scholar]
- Trettel F, Rigamonti D, Hilditch-Maguire P, Wheeler VC, Sharp AH, Persichetti F, Cattaneo E, MacDonald ME. Dominant phenotypes produced by the HD mutation in STHdh(Q111) striatal cells. Hum Mol Genet. 2000;9:2799–2809. doi: 10.1093/hmg/9.19.2799. [DOI] [PubMed] [Google Scholar]
- van Roon-Mom WM, Pepers BA, t Hoen PA, Verwijmeren CA, den Dunnen JT, Dorsman JC, van Ommen GB. Mutant huntingtin activates Nrf2-responsive genes and impairs dopamine synthesis in a PC12 model of Huntington's disease. BMC Mol Biol. 2008;9:84. doi: 10.1186/1471-2199-9-84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vonsattel JP, DiFiglia M. Huntington disease. J Neuropathol Exp Neurol. 1998;57:369–384. doi: 10.1097/00005072-199805000-00001. [DOI] [PubMed] [Google Scholar]
- Walker FO. Huntington's Disease. Semin Neurol. 2007;27:143–150. doi: 10.1055/s-2007-971176. [DOI] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Karbowski M, Monteiro MJ. Effects of overexpression of huntingtin proteins on mitochondrial integrity. Hum Mol Genet. 2009;18:737–752. doi: 10.1093/hmg/ddn404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Lim PJ, Yin C, Rieckher M, Vogel BE, Monteiro MJ. Suppression of polyglutamine-induced toxicity in cell and animal models of Huntington's disease by ubiquilin. Hum Mol Genet. 2006;15:1025–1041. doi: 10.1093/hmg/ddl017. [DOI] [PubMed] [Google Scholar]
- Wang H, Monteiro MJ. Ubiquilin interacts and enhances the degradation of expanded-polyglutamine proteins. Biochem Biophys Res Commun. 2007;360:423–427. doi: 10.1016/j.bbrc.2007.06.097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Yu SW, Koh DW, Lew J, Coombs C, Bowers W, Federoff HJ, Poirier GG, Dawson TM, Dawson VL. Apoptosis-inducing factor substitutes for caspase executioners in NMDA-triggered excitotoxic neuronal death. J Neurosci. 2004a;24:10963–10973. doi: 10.1523/JNEUROSCI.3461-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Wang CE, Orr A, Tydlacka S, Li SH, Li XJ. Impaired ubiquitin-proteasome system activity in the synapses of Huntington's disease mice. J Cell Biol. 2008;180:1177–1189. doi: 10.1083/jcb.200709080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang WW, Cao R, Rao ZR, Chen LW. Differential expression of NMDA and AMPA receptor subunits in DARPP-32-containing neurons of the cerebral cortex, hippocampus and neostriatum of rats. Brain Res. 2004b;998:174–183. doi: 10.1016/j.brainres.2003.11.034. [DOI] [PubMed] [Google Scholar]
- Wernig M, Tucker KL, Gornik V, Schneiders A, Buschwald R, Wiestler OD, Barde YA, Brustle O. Tau EGFP embryonic stem cells: an efficient tool for neuronal lineage selection and transplantation. J Neurosci Res. 2002;69:918–924. doi: 10.1002/jnr.10395. [DOI] [PubMed] [Google Scholar]
- Wyttenbach A, Sauvageot O, Carmichael J, Diaz-Latoud C, Arrigo AP, Rubinsztein DC. Heat shock protein 27 prevents cellular polyglutamine toxicity and suppresses the increase of reactive oxygen species caused by huntingtin. Hum Mol Genet. 2002;11:1137–1151. doi: 10.1093/hmg/11.9.1137. [DOI] [PubMed] [Google Scholar]
- Young AB. Huntingtin in health and disease. J Clin Invest. 2003;111:299–302. doi: 10.1172/JCI17742. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou H, Cao F, Wang Z, Yu ZX, Nguyen HP, Evans J, Li SH, Li XJ. Huntingtin forms toxic NH2-terminal fragment complexes that are promoted by the age-dependent decrease in proteasome activity. J Cell Biol. 2003;163:109–118. doi: 10.1083/jcb.200306038. [DOI] [PMC free article] [PubMed] [Google Scholar]