Abstract
The selective manipulation of mitochondrial DNA (mtDNA) replication and expression within mammalian cells has proven difficult. One promising approach is to use peptide nucleic acid (PNA) oligomers, nucleic acid analogues that bind selectively to complementary DNA or RNA sequences inhibiting replication and translation. However, the potential of PNAs is restricted by the difficulties of delivering them to mitochondria within cells. To overcome this problem we conjugated a PNA 11mer to a lipophilic phosphonium cation. Such cations are taken up by mitochondria through the lipid bilayer driven by the membrane potential across the inner membrane. As anticipated, phosphonium–PNA (ph–PNA) conjugates of 3.4–4 kDa were imported into both isolated mitochondria and mitochondria within human cells in culture. This was confirmed by using an ion-selective electrode to measure uptake of the ph–PNA conjugates; by cell fractionation in conjunction with immunoblotting; by confocal microscopy; by immunogold-electron microscopy; and by crosslinking ph–PNA conjugates to mitochondrial matrix proteins. In all cases dissipating the mitochondrial membrane potential with an uncoupler prevented ph–PNA uptake. The ph–PNA conjugate selectively inhibited the in vitro replication of DNA containing the A8344G point mutation that causes the human mtDNA disease ‘myoclonic epilepsy and ragged red fibres’ (MERRF) but not the wild-type sequence that differs at a single nucleotide position. Therefore these modified PNA oligomers retain their selective binding to DNA and the lipophilic cation delivers them to mitochondria within cells. When MERRF cells were incubated with the ph–PNA conjugate the ratio of MERRF to wild-type mtDNA was unaffected, even though the ph–PNA content of the mitochondria was sufficient to inhibit MERRF mtDNA replication in a cell-free system. This unexpected finding suggests that nucleic acid derivatives cannot bind their complementary sequences during mtDNA replication. In summary, we have developed a new strategy for targeting PNA oligomers to mitochondria and used it to determine the effects of PNA on mutated mtDNA replication in cells. This work presents new approaches for the manipulation of mtDNA replication and expression, and will assist in the development of therapies for mtDNA diseases.
INTRODUCTION
Most mammalian cells contain hundreds to thousands of mitochondrial DNA (mtDNA) molecules, encoding 13 polypeptides, two ribosomal RNAs and 22 transfer RNAs (1). There are considerable uncertainties about the regulation of mtDNA replication and expression, and progress is hampered because manipulating mtDNA expression in mammalian cells is difficult (2,3). The delivery of appropriate nucleotide oligomers to mitochondria within cells would facilitate the study of mtDNA, just as complementary nucleotide sequences are used as antisense agents to modulate nuclear gene expression (3–8). Up to now no such strategies have been developed for mammalian mitochondria because of the difficulties of delivering oligonucleotides to mitochondria in cells (9).
Peptide nucleic acid (PNA) oligomers are appealing candidates for delivery to mitochondria to manipulate DNA expression (10). These are synthetic DNA analogues with the deoxyribose phosphate backbone replaced by aminoethyl glycine units connected to conventional bases by a methyl carbonyl linker (11). As the PNA base spacing is the same as nucleic acids, they hybridise with complementary DNA and RNA sequences while the neutral PNA backbone increases both binding affinity relative to DNA and resistance to nucleases and proteases (12,13). These attributes make PNA oligomers particularly effective antisense reagents (14–17). However, the unassisted permeation of PNAs through lipid bilayers is negligible (18), although PNAs are taken into cells through vesicle transport or pinocytosis (10,19). Therefore, PNAs are generally delivered to cells using cationic detergents (20), or by conjugation to peptides which catalyse membrane permeation (21–23). While these approaches do deliver PNAs to the cytoplasm (24–26), their subsequent uptake by mitochondria remains a challenge (10).
The selective delivery of PNA oligomers to mitochondria may lead to therapies for mtDNA diseases (2), which are caused by point mutations, deletions or insertions to mtDNA that disrupt oxidative phosphorylation giving rise to neuromuscular defects (27). There is no treatment for these progressive disorders (2), however, most patients harbour both mutated and wild-type mtDNA, therefore selective binding of PNAs to mutated mtDNA may inhibit replication, facilitating propagation of the wild-type mtDNA and reversal of the associated defect (2,3). In a cell-free system PNA oligomers selectively inhibit replication of mutated mtDNA, but not the wild-type sequence that differs by a single base pair (28). Therefore, PNA oligomers have therapeutic potential, but the problem of their delivery to mitochondria within cells remains an obstacle.
To deliver PNAs to mitochondria they must first be transported through the plasma membrane and then taken up across the mitochondrial inner membrane (10). Although PNAs linked to mitochondrial protein targeting sequences are imported through the protein import machinery, these conjugates are difficult to synthesise and their slow uptake into the cytoplasm and vulnerability to proteolysis limits their efficacy (10). A better approach may be to deliver PNAs to mitochondria by conjugation to the triphenylphosphonium cation (6,29). These lipophilic cations easily permeate lipid bilayers and are taken up into the cytoplasm, driven by the plasma membrane potential (∼30–60 mV negative inside) (6,29). From the cytoplasm they are then concentrated several hundred-fold into mitochondria by the large membrane potential across the inner membrane (∼150–180 mV negative inside) (6). Linking neutral molecules as large as 500 Da to triphenylphosphonium cations facilitates their lipid bilayer transport and delivers them to mitochondria within cells (30,31). Therefore, conjugating neutral PNA oligomers (∼3–4 kDa) to lipophilic cations should also drive their uptake into mitochondria within cells. Here we report the synthesis and characterisation of triphenylphosphonium–PNA conjugates, demonstrate their mitochondrial localisation in both isolated organelles and cells, and their selective inhibition of DNA replication in a cell-free system.
MATERIALS AND METHODS
Synthesis of phosphonium–PNA (ph–PNA) conjugates
PNA oligomers with a sequence complementary to the human mtDNA L-chain (np 8339–8349) containing the ′myoclonic epilepsy and ragged red fibres′ (MERRF) A8344G point mutation in the tRNALys gene were synthesised by PerSeptive Biosystems (Framingham, MA) (28). The PNA oligomers used were: H2N-Cys-GTTGGCTCTCT-CO2H and Biotin-OO-GTTGGCTCTCT-O-Cys-CO2H, where the spacer O was 8-amino-3,6-dioxanoic acid. To conjugate PNA to a triphenylphosphonium cation, PNA oligomers (50 nmol) in 50 µl 10 mM HEPES, 1 mM EDTA pH 8.0 were incubated with 2-mercaptoethanol (250 nmol) at 40°C for 1 h, then iodobutyltriphenylphosphonium (IBTP, 500 nmol) in 200 µl HEPES/EDTA:ethanol (4:1) was added and incubated at 40°C for a further 4 h. After quenching with 2-mercaptoethanol (250 nmol) the reaction products were separated by reverse phase HPLC on a C4 analytical column (Vydac, 300 Å, 4.6 × 250 mm), using a Waters 450 HPLC system. A linear gradient starting with 0.1% trifluoroacetic acid (TFA) in water and finishing with 90% acetonitrile and 0.1% TFA was run over 30 min to resolve the reaction products. Ph–PNA conjugate peaks were detected by absorbance at 260 nm, collected, lyophilised and dissolved in water for further analysis. The concentrations of the unmodified PNA and ph–PNA conjugates were determined at 55°C using extinction coefficients at 260 nm of 97 900 M–1 cm–1 and 100 400 M–1 cm–1, respectively.
Matrix-assisted laser desorption/ionisation time of flight mass spectrometry (MALDI-TOF MS)
PNA conjugates (∼0.5 pmol) in water were mixed with ∼0.5 µl 10 mg ml–1 3,5-dimethoxy-4-hydroxycinnamic acid and after crystalisation were analysed by MALDI-TOF MS using a Finnigan MAT Lasermat 2000 instrument by the Protein Microchemistry Facility (Department of Biochemistry, Otago University, Dunedin, New Zealand). Spectra were acquired in positive ion mode using melittin (MW 2846 Da) as an external mass calibrant.
Gel electrophoresis and immunoblotting of PNA conjugates
Ph–PNA conjugates (∼5 nmol) in 20 µl loading buffer (50 mM Tris, 4% SDS, 12% glycerol, 2% 2-mercaptoethanol, 0.01% Coomassie brilliant blue) were separated on a 18.5% Tris–tricine gel (32) using a Bio-Rad Mini Protean system. Gels were then either fixed and stained with Coomassie brilliant blue, or electrotransferred onto 0.2 µm nitrocellulose using a Bio-Rad Mini Trans-Blot system (100 V, 1 h) in transfer buffer (25 mM Tris–HCl pH 8.3, 192 mM glycine, 20% methanol) and then blocked with 2% (w/v) fat-free milk powder in TBS (5 mM Tris–HCl pH 7.4, 20 mM NaCl), 0.1% Tween-20. IgG fractions from rabbit antiserum raised against triphenylphosphonium conjugated to keyhole limpet hemocyanin and from preimmune serum were prepared using a protein A–Sepharose column (Bio-Rad) and diluted 1:500 for immunodetection. Horse radish peroxidase conjugated goat antirabbit IgG (1:20 000, Bio-Rad) was used as a secondary antibody. To detect biotin, horse radish peroxidase conjugated extravidin (1:3000, Sigma) was used. In both cases antibody binding was visualised by chemiluminescence using a Pierce Super Signal R chemiluminescence substrate with Kodak X-OMAT™ AR imaging film.
Mitochondrial incubations
Rat liver mitochondria were prepared by homogenisation in 250 mM sucrose, 5 mM Tris–HCl pH 7.4, and 1 mM EGTA followed by differential centrifugation (33) and the protein concentration determined by the biuret assay using BSA as a standard (34). An electrode selective for the triphenylphosphonium cation was constructed and the voltage relative to a Ag/AgCl reference electrode measured using a Phlips PW9420 voltmeter and recorded on a Maclab data acquisition system (35,36). Mitochondria (2 mg protein) in a 25°C thermostatted chamber were stirred in 2 ml KCl medium (120 mM KCl, 10 mM HEPES pH 7.2, 1 mM EGTA) supplemented with rotenone (2 µg ml–1) and the extramitochondrial concentration of the ph–PNA conjugate measured using the ion-selective electrode. To quantitate ph–PNA conjugate uptake by immunodetection, mitochondria (1 mg protein) were incubated with 1 µM ph–PNA conjugate at 37°C in 250 µl KCl medium supplemented with 10 mM succinate and 50 µM rotenone in the presence or absence of 10 µM carbonyl cyanide p-(trifluoromethoxy) phenyl hydrazone (FCCP). Following incubation the mitochondria were isolated by centrifugation (10 000 g for 1 min) through 300 µl oil (66% silicone oil/34% dioctyl phthalate) into 100 µl 0.25 M sucrose/0.1% Triton X-100 and fractions were analysed by immunoblotting. For crosslinking studies mitochondria (1 mg protein) were suspended in 500 µl 15% sucrose, 10 mM NaCl, 2 mM Tricine–KOH, 40 mM Tris–HCl pH 8.0 and 2.5 mM MgCl2, then treated with DNase I (200 U) for 1 h on ice and the reaction terminated with 5 mM EDTA pH 8.0. The mitochondria were then washed and diluted to 2 mg protein ml–1 in 0.25 M sucrose, 70 mM HEPES pH 7.6, 2 mM EDTA, 50 mM NaCl, 1 mM EGTA, 7 mM 2-mercaptoethanol, 1 mM spermidine and 1 mM PMSF, supplemented with protease inhibitor cocktail mixture (Boehringer Mannheim). After incubation at 37°C for 1 h with 1 µM phosphonium-biotinylated PNA (ph–bioPNA) conjugate in the presence or absence of 10 µM FCCP, formaldehyde was added to 1% and after 4 h at 4°C the reaction was quenched with 125 mM glycine pH 7.0. The mitochondria were resuspended in crosslinking buffer (CB) (0.25 M sucrose, 20 mM HEPES pH 7.6, 2 mM EDTA, 1 mM EGTA) supplemented with 50 mM glycine and 0.5% Tergitol NP-40, diluted 3-fold in CB–0.5% Tergitol NP-40 and the nucleoid protein-enriched fraction was pelleted (110 000 g for 1 h) through a cushion of 20% sucrose, 0.5% Tergitol NP-40, 50 mM HEPES pH 7.5, 50 mM NaCl. The pellet was resuspended in 200 µl CB–1% sarkosyl using a dounce homogeniser and treated with RNase A (50 µg ml–1) for 1 h at room temperature (37). Aliquots (10 µl) were separated by electrophoresis on Tris–tricine gels and biotin was detected by immunoblotting as described previously. That this procedure enriched for the mitochondrial nucleoid was confirmed by PCR of a parallel preparation without formaldehyde treatment (formaldehyde interfered with the PCR) using primers specific for rat mtDNA which generated a 722 bp PCR product (F201–220, AGACGCCTTGCCTAGCCACA; R920–900, GAGGGTGACGGGCGGTGTGT).
Cell culture
143B human osteosarcoma cells and human fibroblasts were grown in Dulbecco’s Modified Eagle’s Medium (DMEM) supplemented with 10% foetal calf serum (FCS) at 37°C in a humidified atmosphere of 95% air/5% CO2. Protein was quantitated by the bicinchoninic acid assay using BSA as a standard (38). For incubations in suspension, 143B cells were harvested using trypsin and 106 cells were suspended in 1 ml DMEM, 10 mM HEPES and 10% FCS. For cell subfractionation, 143B cells were grown to confluence in 24 well tissue culture plates overnight and then incubated with 1 µM ph–PNA in the presence or absence of 10 µM FCCP for 1 h at 37°C and after washing the cells were harvested by scraping in 250 mM sucrose, 20 mM MOPS, 3 mM EDTA pH 6.7, and 1 mg ml–1 digitonin. A mitochondria enriched fraction was prepared from 200 µl crude suspension by centrifugation (10 000 g for 1 min) through 300 µl oil (58% silicone oil/42% dioctyl phthalate) into 100 µl 0.5 M sucrose/0.1% Triton X-100, leaving a cytoplasm enriched upper layer. About 92–96% and 0.3–1% of total citrate synthase and lactate dehydrogenase activities, respectively, were found in the mitochondria enriched fraction (39,40). Both fractions were immunoblotted to detect ph–PNA conjugate localisation.
For immunocytochemistry human fibroblasts were plated onto 13 mm diameter glass coverslips overnight. Following incubation for 4 h at 37°C with 1 µM ph–PNA conjugates in the presence or absence of 10 µM FCCP, cells were fixed with 4% paraformaldehyde in TBS for 30 min, washed with TBS and incubated with 10% FCS/0.1% Triton X-100/TBS (TBST) for 10 min. The IgG fraction of anti-triphenylphosphonium serum (1:500) and anti-cytochrome oxidase (1:100, subunit Iα, mouse monoclonal, Molecular Probes) diluted in TBST were then added and incubated overnight at 4°C. The IgG fraction of preimmune serum was used as a control. After washing with TBS (3 × 5 min) the cells were incubated with appropriate fluorophore-conjugated secondary antibodies diluted in TBS [anti-rabbit IgG Oregon green (1:100), or anti-mouse IgG Texas red (1:400); Molecular Probes] for 15 min in the dark. Streptavidin-CY3 (1:200, Molecular Probes) was used to detect biotin. Cells were then washed in TBS, mounted in DABCO/PVA medium and mounted onto coverslips. Images were acquired using a Bio-Rad MRC 600 laser-scanning confocal microscope using a Nikon Diaphot TMD inverted microscope and Nikon ×60 NA 1.4 oil immersion Plan-Apochromat objective. The 568 and 488 nm lines of a Krypton–Argon laser and K1/K2 filter blocks were used at identical gain, black settings and time frame.
Immunogold electron microscopy
Human fibroblasts in 24 well tissue culture plates were incubated with 200 µl DMEM/FCS with 1 µM ph–PNA in the presence or absence of 10 µM FCCP for 4 h at 37°C, then harvested with trypsin, pelleted and washed in PBS. The pellet was warmed to 37°C and 200 µl fixative (4% formaldehyde, 0.5% glutaraldehyde in 0.2 M cacodylate pH 7.2, 6 mM CaCl2) added and replaced with fresh fixative after 30 s. After incubation at 37°C for 1 h the cell pellet was washed with 200 µl 0.15 M cacodylate pH 7.2, 3 mM CaCl2 (3 × 10 min at room temperature), warmed to 45°C, mixed with 2% agar in 0.1 M cacodylate pH 7.2, and pelleted. After rinsing in PBS (3 × 10 min) the pellets were dehydrated in ethanol, embedded in Lowicryl white and polymerised under UV at 0°C. Ultrathin sections were mounted on nickel grids which were floated on drops of 0.1 M glycine in PBS for 10 min, then 1% BSA in PBS for 15 min, followed by anti-triphenylphosphonium serum overnight at 4°C (1:10 in 1% BSA/1% Tween-20 in PBS). After washes in PBS (3 × 5 min) and 1% BSA/PBS, sections were incubated with colloidal gold particles (10 nm) linked to either goat anti-rabbit IgG (1:5 in 1% Tween-20/PBS, Sigma) or extravidin (1:10 in 1% Tween-20/PBS, Sigma) for 1 h, followed by washing and fixation (2% glutaraldehyde for 15 min). The sections were then stained with osmium tetroxide/uranyl acetate/lead citrate and examined in a CM100 transmission electron microscope.
DNA replication run-off assays
Human mtDNA templates containing the A8344G MERRF point mutation (341 bp), or the corresponding region in wild-type mtDNA (350 bp), were prepared by PCR (28). A 9 bp deletion at the 3′ end of the MERRF sequence caused the size difference between the wild-type and MERRF templates, but did not affect the PNA binding assay (28). The templates were incubated in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 14 mM 2-mercaptoethanol, 150 mM KCl, 1 mM ATP with 100 µM each dATP, dGTP and dTTP and 100 nM specific priming oligodeoxynucleotide (28). After incubation at 70°C for 3 min and cooling to 37°C to allow hybridisation ph–PNA was added along with 10 µM dCTP, 0.33 µM [α-32P]dCTP (3000 Ci/mmol) and a mitochondrial DNA polymerase enriched fraction from bovine liver (1.5 µg protein) (28). After incubation at 37°C for 60 min, replication was terminated with phenol and DNA was ethanol precipitated and analysed on 6% denaturing polyacrylamide gels. Dried gels were exposed to a PhosphorImager cassette (Molecular Dynamics).
PCR–RFLP analysis of mtDNA from MERRF cells
A skin fibroblast culture from a MERRF patient which contained the A8344G mutation in ∼60% of its mtDNA was used (41). Fibroblasts were grown in 96 well plates in DMEM with 10% FCS supplemented with uridine (50 mg l–1) and pyruvate (110 mg l –1) to ∼50% confluence. The medium was supplemented with up to 500 µM ph–PNA or PNA and this was replaced with fresh medium containing PNA or ph–PNA every 3 days for up to 30 days. At the end of the incubation cells were collected by trypsin treatment, lysed in 10 mM Tris–HCl pH 8.3, 50 mM KCl, 1 mM MgCl2 and 0.5% Tween-20, containing proteinase K (100 µg ml–1) for 2 h at 50°C and the enzyme was inactivated by heating to 95°C for 10 min. Myoblasts from a MERRF patient which contained the A8344G mutation in 85% of its mtDNA were grown in 24 well plates in Hams F10 supplemented with 20% FCS, 1% chick embryo extract, 1% penicillin–streptomycin, uridine (50 mg l–1) and pyruvate (110 mg l–1). Cells were seeded at ∼30% confluency and passaged three times in the presence or absence of 10 µM ph–PNA. The medium was replaced with fresh medium containing PNA or ph–PNA every 2 days for a period of 3 weeks. At the end of the incubation the cells were collected by trypsin treatment and DNA isolated by standard procedures. Multiplex PCR was performed on the cell lysates or isolated DNA using two pairs of primers. One pair amplified a region around the MERRF A8344G mutation in human mtDNA. For fibroblasts this was done using primers F8150–8166, CCGGGGGTATACTACC; R8372–8345, GGGGCATTTCACTGTAAAGAGGTGCCGG and for myoblasts the same reverse primer was used but the forward primer was F8191–8210, TGTAAAACGACGGCCAGTAAACCACAGTTTCATGCCCA. PCR generated 223 and 200 bp products for fibroblasts and myoblasts, respectively. The two mismatches in the reverse primer (underlined) enabled the mtDNA around the A8344G point mutation and the corresponding region in wild-type mtDNA to be amplified to the same extent. There is a NaeI restriction site on the mutant mtDNA but not on the wild-type mtDNA. The other primer pair (F516–534, CACACACACCGCTGCTAAC; R1190–1172, GATATGAAGCACCGCCAGG) amplified a 711 bp region, containing a NaeI restriction site in both wild-type and MERRF mtDNA which served as an internal control for complete digestion. Following PCR (30 cycles of 95°C for 5 min, 95°C for 1 min, 56°C for 1 min, 72°C for 1 min; 1 cycle of 72°C for 10 min) an additional PCR cycle was carried out with 30 pmol primers, 5 µCi [α-32P]dCTP (3000 Ci/mmol) and 1 U Taq polymerase (Roche). Labelled products were precipitated with 0.5 vol 7.5 M ammonium acetate, 2 vol 99% ethanol and 0.3 mg.ml–1 glycogen at –80°C for 1 h. The pellet was resuspended and amounts containing 10 000 c.p.m. were digested with 10 U NaeI at 37°C for 12 h. Restriction fragments were separated on a 10% non-denaturing polyacrylamide gel, which was then dried and exposed to Kodak X-OMATTM AR imaging film.
RESULTS
Synthesis and characterisation of ph–PNA conjugates
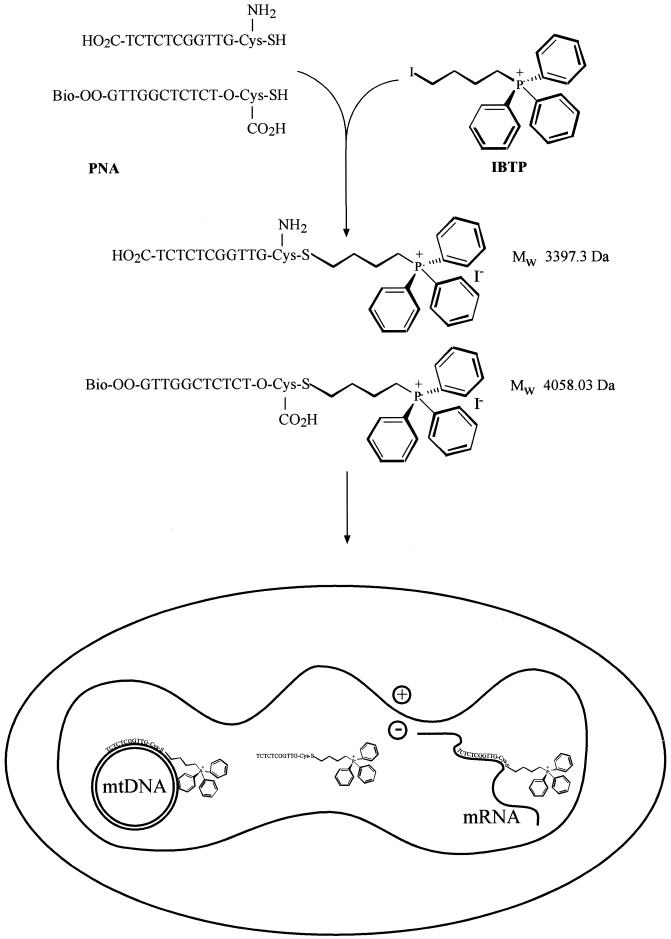
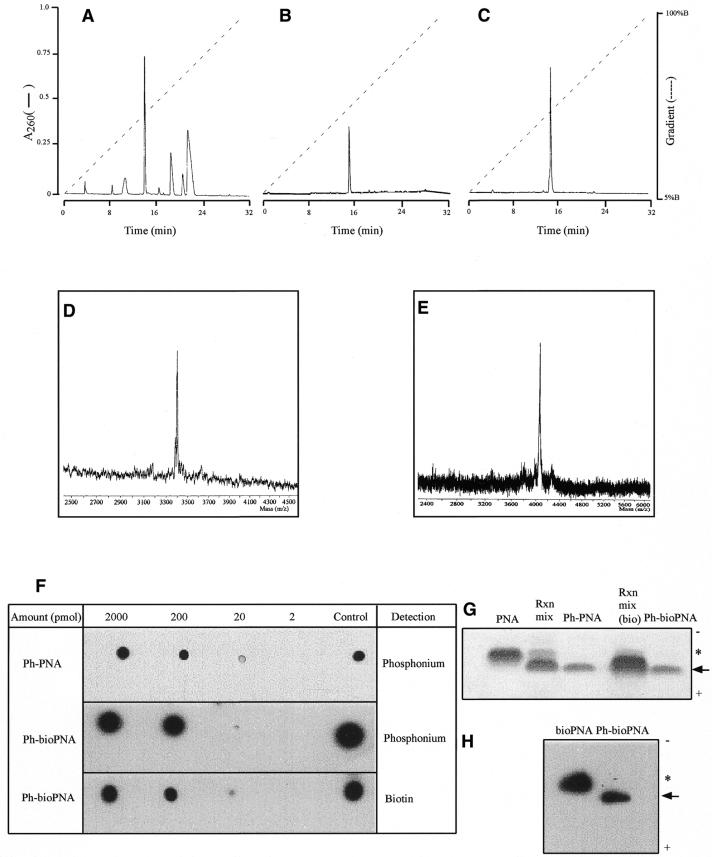
Our strategy of conjugating PNA oligomers to triphenylphosphonium to deliver them to mitochondria is illustrated in Figure 1. Cysteine residues incorporated into the PNA oligomers reacted with IBTP, displacing iodide to form a stable thioether between the PNA and the triphenylphosphonium cation (Fig. 1). The ph–PNA conjugate was purified by reverse phase HPLC (RP-HPLC) of the reaction products (Fig. 2A and B). The same procedure linked a biotinylated PNA (bioPNA) to triphenylphosphonium and the ph–bioPNA was then purified by RP-HPLC (Fig. 2C). The identities of the ph–PNA and ph–bioPNA conjugates were confirmed by MALDI-TOF MS (Fig. 2D and E). To further confirm their composition, and to establish detection protocols, PNA conjugates were adsorbed on nitrocellulose and the triphenylphosphonium moiety was detected using cognate antiserum (Fig. 2F, upper two panels). The biotin moiety of ph–bioPNA was detected by streptavidin binding (Fig. 2F, lower panel). Gel electrophoresis of the PNA conjugates followed by immunoblotting or Coomassie staining showed that the original bioPNA and PNA oligomers (asterisks) were predominantly converted to more rapidly migrating triphenylphosphonium conjugates (arrows) (Fig. 2G and H). Purified ph–PNA or ph–bioPNA gave single bands by this procedure, confirming their purity (Fig. 2G), and after transfer to nitrocellulose the biotin moieties of bioPNA and ph–bioPNA were detected by streptavidin binding (Fig. 2H). The ph–PNA oligomers migrated more rapidly to the anode than unmodified PNA, presumably due to increased SDS binding, despite the incorporation of the positive charge and slight (∼300 Da) increase in molecular weight. In summary, we have synthesised and purified two ph–PNA conjugates and can detect them with antiserum against the triphenylphosphonium moiety and by streptavidin binding to biotin.
Figure 1.
Synthesis of ph–PNA conjugates. The reactions of the cysteine residues of the biotinylated PNA oligomer and the PNA oligomer with IBTP are shown. The sequences and predicted MW values of the products, their uptake into mitochondria and subsequent interactions with mtDNA and mRNA are indicated.
Figure 2.
Purification and characterisation of ph–PNA conjugates. (A) Purification of the ph–PNA reaction products by RP-HPLC. The major peak at ∼15 min, due to the ph–PNA conjugate, was collected, lyophilised, dissolved in water and a sample analysed by RP-HPLC (B). The ph–bioPNA was prepared by a similar procedure (data not shown) and purified by RP-HPLC (C). (D and E) MALDI-TOF analyses of purified ph–PNA and ph–bioPNA, respectively. The observed masses for the ph–PNA (3400 ± 4 Da) and the ph–bioPNA (4054 ± 3 Da) conjugates were within 0.1% of the calculated masses, as expected for external mass calibration. (F) Serial dilutions of the ph–PNA and ph–bioPNA conjugates were adsorbed on nitrocellulose and the triphenylphosphonium moiety detected using antitriphenylphosphonium serum. BSA conjugated to IBTP (∼1 µg protein) was used as a positive control and PNAs not conjugated to IBTP were not detected by this antibody (data not shown). Horse radish peroxidase conjugated to extravidin was used to detect biotin and the bioPNA oligomer (∼5 nmol) was used as a positive control. Unconjugated PNA oligomers were not detected by either procedure (data not shown). (G) Samples (∼5 nmol) of PNA, reaction products prior to purification, and the purified ph–PNA and ph–bioPNA conjugates were separated by Tris–tricine PAGE and stained with Coomassie blue. The precursor PNAs are marked with an asterisk, the ph–PNA conjugates with an arrow and the polarity of electrophoresis is shown. (H) bioPNA and the purified ph–bioPNA conjugate (∼5 nmol of each) were separated by Tris–tricine PAGE, transferred to nitrocellulose and probed for biotin using streptavidin.
Ph–PNA conjugate uptake by isolated mitochondria
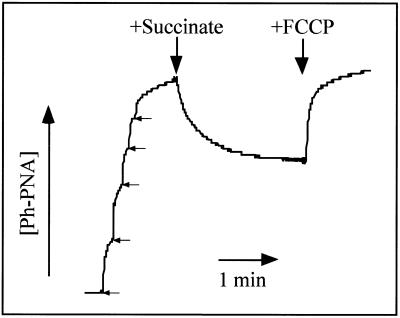
To determine whether the ph–PNA conjugate was taken up by energised mitochondria, we constructed an electrode selective for the triphenylphosphonium moiety. This enabled continuous measurement of the ph–PNA conjugate concentration, as shown by the electrode response to sequential 1 µM additions of ph–PNA (Fig. 3). When succinate was added to establish a membrane potential the ph–PNA conjugate was accumulated into the mitochondrial matrix (Fig. 3). This was confirmed by adding the uncoupler FCCP which abolished the membrane potential leading to efflux of the ph–PNA conjugate from mitochondria back into the medium (Fig. 3). The amount of ph–PNA taken up by mitochondria was ∼2 nmol mg protein–1 (Fig. 3) and the matrix volume is ∼ 0.5–1 µl mg protein–1 (42,43), therefore incubation with micromolar ph–PNA leads to millimolar conjugate concentrations in the mitochondrial matrix. Experiments with the ph–bioPNA conjugate gave similar results to those shown for ph–PNA (data not shown).
Figure 3.
Ph–PNA conjugate uptake measured using an ion-selective electrode. An ion-selective electrode was used to measure the concentration of a ph–PNA conjugate in a mitochondrial suspension. The arrows indicate five sequential additions of 1 µM ph–PNA to calibrate the electrode response. The mitochondria were then energised by addition of succinate (5 mM) and uncoupled with FCCP (10 µM) as indicated.
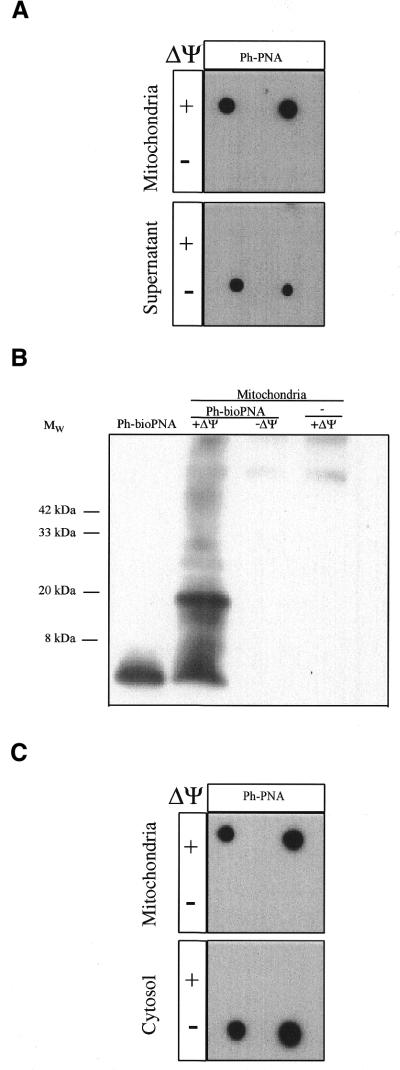
The uptake of the ph–PNA conjugate was also measured by incubation with mitochondria followed by their separation from the incubation medium by centrifugation through oil (Fig. 4). The ph–PNA taken up by mitochondria and that remaining in the supernatant were then detected by immunoblotting of the mitochondrial and supernatant fractions with anti-triphenylphosphonium serum (Fig. 4A). Most of the ph–PNA was accumulated into the matrix of energised mitochondria with little remaining in the supernatant (Fig. 4A). When this was repeated in the presence of FCCP to abolish the membrane potential the ph–PNA was no longer taken up by mitochondria and remained in the supernatant (Fig. 4A).
Figure 4.
Uptake of ph–PNA conjugates by mitochondria. (A) Rat liver mitochondria were incubated with 1 µM ph–PNA in the presence or absence of ΔΨm, (±10 µM FCCP). After incubation the mitochondria were pelleted through oil and the supernatant and mitochondrial fractions were probed with antitriphenylphosphonium serum. (B) A mitochondrial matrix fraction enriched for the nucleoid was isolated from mitochondria which had been incubated ±1 µM bioPNA in the presence or absence of ΔΨm, and then crosslinked with 1% formaldehyde. Samples were separated by Tris–tricine PAGE, transferred to nitrocellulose and probed for biotin. A ph–bioPNA (∼5 nmol) control was also analysed. (C) 143B cells were incubated with 1 µM ph–PNA in the presence or absence of ΔΨm, fractionated with digitionin and separated into mitochondria- and cytoplasm-enriched fractions by centrifugation through oil. Samples from both fractions were then assayed for triphenylphosphonium as above. There was no immuno-reactivity with preimmune serum in these experiments.
We next determined whether the ph–bioPNA conjugate accumulated by energised mitochondria was present in the matrix. To do this we incubated ph–bioPNA with mitochondria, added formaldehyde to crosslink the PNA conjugate to closely associated (≤2 Å) proteins and then isolated a fraction enriched for the mitochondrial nucleoid (Fig. 4B) (37). As the mitochondrial nucleoid is a matrix protein–mtDNA complex, binding of the ph–bioPNA conjugate to nucleoid proteins indicates that the conjugate was present in the matrix. Formaldehyde did crosslink ph–bioPNA to nucleoid proteins in energised mitochondria, leading to several bands containing ph–bioPNA at a higher molecular weight than the conjugate itself (Fig. 4B). None of the high molecular weight bands were found when ph–bioPNA uptake into mitochondria was blocked with FCCP, or when energised mitochondria were treated with formaldehyde in the absence of ph–bioPNA (Fig. 4B). We conclude that ph–PNA conjugates are accumulated several hundred-fold into the mitochondrial matrix driven by the membrane potential.
Uptake of ph–PNA conjugates by mitochondria within cells
To determine whether ph–PNA conjugates were taken up by mitochondria within human cells, we incubated 143B osteosarcoma cells with ph–PNA (Fig. 4C). The cells were then separated into mitochondria- and cytosol-enriched fractions by treatment with digitonin followed by centrifugation through oil (Fig. 4C). This showed that ph–PNA was only found in the mitochondria-enriched fraction (Fig. 4C, upper panel). When the mitochondrial membrane potential (ΔΨm) was dissipated with the uncoupler FCCP the ph–PNA conjugate no longer accumulated in the mitochondria but remained in the cytosol (Fig. 4C, lower panel, –ΔΨ).
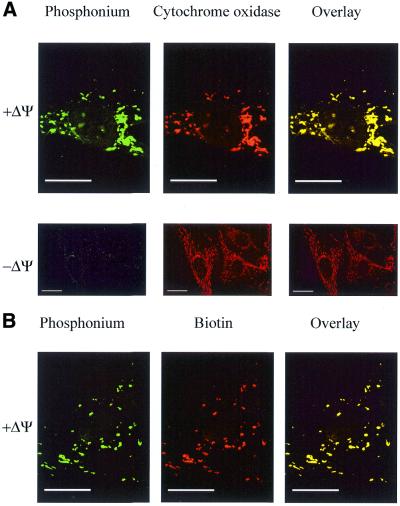
The location of ph–PNA conjugates within human fibroblasts was also determined by fixing cells and visualising the ph–PNA conjugates by confocal immunofluorescence microscopy (Fig. 5). Cells incubated with ph–PNA were probed with an antibody against the mitochondrial enzyme cytochrome oxidase (red) and with antiserum against triphenylphosphonium (green) (Fig. 5A). Overlaying these two micrographs showed that cytochrome oxidase and the ph–PNA conjugate colocalised (yellow), confirming that the ph–PNA conjugate was in mitochondria within cells. The ph–PNA conjugate was not taken up by mitochondria in the presence of FCCP (Fig. 5A, –ΔΨ). The ph–bioPNA conjugate was also accumulated by mitochondria within cells, and when these cells were probed for biotin (red) and the triphenylphosphonium moiety (green), the overlaid micrographs (yellow) showed that both moieties colocalised (Fig. 5B). Therefore, the ph–PNA oligomers are stable within cells and the mitochondrial uptake of the triphenylphosphonium cation is not due to intracellular degradation of the conjugates.
Figure 5.
Mitochondrial localisation of ph–PNA conjugates within fibroblasts by confocal immunofluorescent microscopy. Fibroblasts were incubated with 1 µM ph–PNA (A) or 1 µM ph–bioPNA (B) for 4 h at 37°C, in the presence or absence of ΔΨm. (A) Cells were fixed, incubated with antiserum against triphenylphosphonium (green) and a monoclonal antibody against cytochrome oxidase (red). In the overlaid images yellow indicates co-localisation of triphenylphosphonium and cytochrome oxidase. (B) Cells were fixed and labelled for triphenylphosphonium (green) and biotin (red). In the overlaid images yellow indicates colocalisation of the triphenylphosphonium and biotin. There was no immuno reactivity with preimmune serum in these experiments. Magnification, 1400× (B, A + ΔΨ), 600× (A – ΔΨ); scale bar, 20 µm.
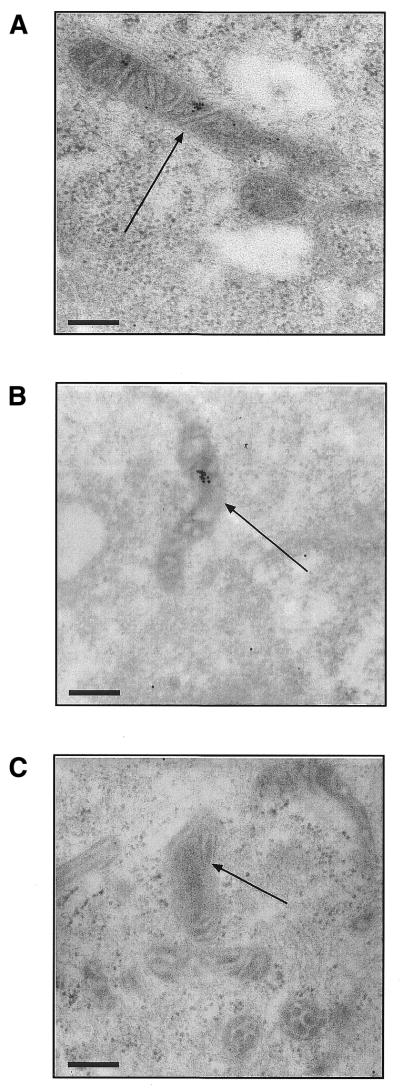
The intramitochondrial location of ph–PNA conjugates within fibroblasts was determined by immunogold electron microscopy (Fig. 6). Cells incubated with ph–PNA conjugates were fixed and incubated with gold-labelled antibodies and visualised by transmission electron microscopy. This showed that both the ph–PNA (Fig. 6A) and the ph–bioPNA conjugates (Fig. 6B) were in the mitochondrial matrix. Mitochondrial accumulation of the ph–PNA conjugates was eliminated with FCCP, although some labelling was seen in the cytoplasm (data not shown). We conclude that the ph–PNA conjugates are taken up by cells and then localise to the mitochondrial matrix driven by the membrane potential.
Figure 6.
Immunogold labelling of ph–PNA conjugates within the mitochondrial matrix. (A) Human fibroblasts were incubated with 1 µM ph–PNA, fixed and the intracellular localisation of triphenylphosphonium detected by immunogold electron microscopy (black dots). (B) Human fibroblasts were incubated with 1 µM ph–bioPNA and the location of the biotin determined by immunogold electron microscopy. (C) Cells were incubated with ph–PNA but the incubation with triphenylphosphonium serum was omitted, while that with the gold-linked antibody was not. Identical electron micrographs were obtained by omitting the ph–PNA conjugates from the incubation. Magnification, 52 000×; scale bar, 200 nm; arrows indicate mitochondria.
Sequence-specific binding and inhibition of DNA replication by ph–PNA conjugates
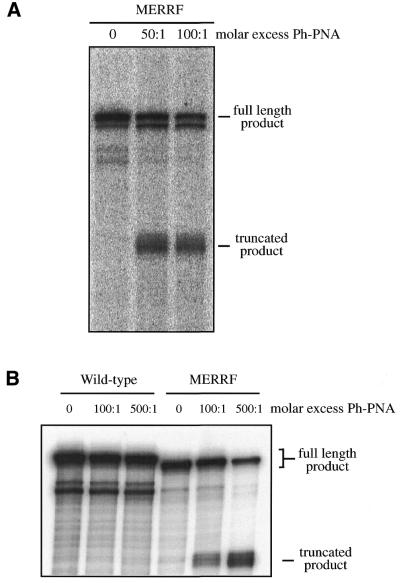
To determine whether conjugation to a phosphonium cation disrupted the selective DNA binding of PNA oligomers, we investigated the effect of ph–PNA on DNA replication in a cell-free system (Fig. 7). The PNA oligomer sequence used is complementary to that around the A8344G tRNALys MERRF mutation in human mtDNA. In a cell-free system a DNA template derived from the mtDNA of a MERRF patient gave the full-length product of 341 bp in a replication run-off assay (Fig. 7A). When this was repeated in the presence of ph–PNA a truncated replication product of 245 bp was formed, due to specific inhibition of DNA replication by binding of ph–PNA to the MERRF A8344G site (Fig. 7A). To investigate the selectivity of binding of ph–PNA to DNA, we prepared a wild-type template that differed from the MERRF template at one nucleotide in the PNA binding region. Incubation with ph–PNA did not inhibit replication of the wild-type DNA and only the full-length product of 350 bp was found (Fig. 7B). The binding affinities of the PNA oligomers and the ph–PNA conjugates to the MERRF DNA template were similar. We conclude that the selective binding of PNA oligomers to particular DNA sequences is not disrupted by conjugation to triphenylphosphonium.
Figure 7.
Selective inhibition of DNA replication by ph–PNA conjugates. (A) An in vitro replication run-off assay from a mtDNA template containing the MERRF mutation was performed. In the absence of ph–PNA this gave a product of 341 nt. In the presence of 50 or 100-fold molar excess of ph–PNA a truncated product of 245 nt was formed. (B) An in vitro replication run-off assay from a wild-type mtDNA template was performed. This template differed from the MERRF template in (A) at a single nucleotide position in the PNA binding region. In the absence of ph–PNA this gave a full-length product of 350 nt (the MERRF DNA template is shorter due to a 9 bp deletion at the 3′ end, downstream of the replication inhibition site). Even in the presence of a 500-fold molar excess of ph–PNA none of the truncated product of 245 nt was found, in contrast to the positive control using the MERRF template. The binding affinities of PNA and ph–PNA to the MERRF PNA templates were 11.3 ± 2.2 and 37 ± 6 nM, respectively (Paul Smith, University of Newcastle upon Tyne, personal communication).
Effect of ph–PNA conjugates on heteroplasmy in MERRF fibroblasts and myoblasts
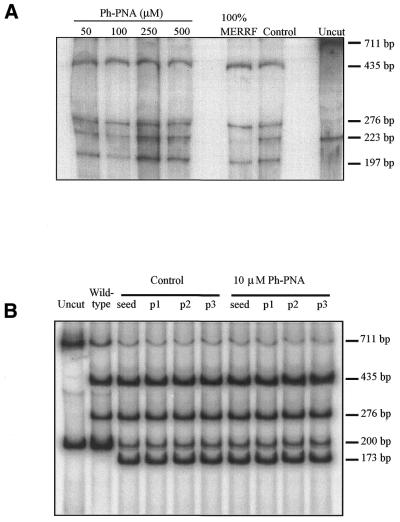
To see if the ph–PNA conjugate could selectively inhibit MERRF, but not wild-type, mtDNA replication in cells we investigated human fibroblasts and myoblasts containing the A8344G MERRF mutation in 60–85% of their mtDNA, respectively (41). The relative proportion of mutated and wild-type mtDNA was measured by a PCR–RFLP assay using the fact that the MERRF mutation introduces a NaeI site that is not present in wild-type mtDNA. As the cells were growing and dividing while their mtDNA content remained stable, each mtDNA molecule replicated several times during these incubations. Even so, in fibroblasts the proportion of mutated and wild-type mtDNA was unchanged after incubation with various concentrations of the ph–PNA conjugate for up to 30 days, as inferred from the ratio of the 197 bp restriction fragment (MERRF) to the uncut 223 bp PCR product (wild-type) (Fig. 8A). Similarly, in myoblasts the ratio of the 173 bp restriction fragment (MERRF) to the uncut 200 bp PCR product (wild-type) was unchanged after incubation with ph–PNA for up to 21 days (Fig. 8B). We conclude that although the ph–PNA conjugates were accumulated several hundred-fold into mitochondria they did not selectively inhibit replication of MERRF mtDNA in fibroblasts or myoblasts.
Figure 8.
PCR–RFLP analysis of mtDNA from MERRF fibroblasts and myoblasts incubated with ph–PNA. For all samples, after incubation the mtDNA was isolated from the cells and the region around the MERRF point mutation amplified by PCR giving a 223 bp product for fibroblasts and a 200 bp product for myoblasts. The MERRF point mutation introduces a NaeI site, leading to 197 and 26 bp fragments for fibroblasts and 173 and 27 bp fragments for myoblasts, following digestion while the wild-type PCR products are uncut. Another mtDNA region of 711 bp containing an NaeI site in both wild-type and MERRF mtDNA was amplified by PCR and digested to 435 and 276 bp fragments as an internal control for complete digestion. (A) Heteroplasmic MERRF fibroblasts were incubated with 50, 100, 250 and 500 µM ph–PNA for 30 days. In addition, control samples from untreated fibroblasts after 30 days (control), samples from homoplasmic MERRF cells (100% MERRF) and undigested samples from untreated cells (uncut) were analysed. (B) Heteroplasmic MERRF myoblasts were seeded and incubated with 10 µM ph–PNA for three passages over 3 weeks. Samples were taken after seeding or after each passage and mtDNA assayed (p1–3). In addition, control samples from untreated myoblasts (control), digested samples from wild-type myoblasts (wild-type) and undigested samples from myoblasts (uncut) were analysed. Data shown are typical results of experiments repeated several times and under a range of ph–PNA concentrations for both fibroblasts and myoblasts.
DISCUSSION
We have shown that our strategy of targeting PNAs to mitochondria within cells by conjugation to a triphenylphosphonium cation is effective. This targeting system overcomes three barriers to delivering PNAs to mitochondria in cells: (i) the lipophilic cation facilitates PNA transport through the lipid bilayers of the plasma membrane and the mitochondrial inner membrane; (ii) the plasma membrane potential drives accumulation of ph–PNA conjugates into the cytoplasm; (iii) the ph–PNA conjugates are then selectively accumulated by energised mitochondria. Furthermore, micromolar ph–PNAs are not cytotoxic, are not degraded within the cell and their nuclear localisation is negligible compared to unmodified or peptide-conjugated PNAs (10). Therefore, lipophilic cations direct the uptake into mitochondria of surprisingly large molecules, a finding that opens the way for the delivery of PNAs and other large bioactive molecules to mitochondria for a range of biomedical uses (30).
The efficacy of PNA oligomers is attributable to their selective nucleic acid binding and this is unaffected by attachment to a triphenylphosphonium cation. In a cell-free system a 50–100-fold molar excess of ph–PNA over MERRF DNA was sufficient to inhibit replication. As cells underwent several cycles of mtDNA synthesis during these experiments, MERRF mtDNA replication should be inhibited if enough of the ph–PNA is accumulated by mitochondria. Micromolar concentrations of ph–PNA were accumulated several hundred-fold by isolated mitochondria, corresponding to ∼2 nmol ph–PNA mg protein–1. As there are ∼7.2 × 109 mitochondria mg protein–1 (44), each containing ∼2.6 mtDNA molecules (45), the molar excess of ph–PNA over mtDNA is ∼104–105-fold, more than 1000× greater than that required to inhibit DNA replication in vitro. Estimating the ph–PNA content of mitochondria in cells is difficult, but the accumulation of ph–PNA into the cytoplasm followed by its concentration by mitochondria should also give millimolar intramitochondrial concentrations, and this has been shown to occur for other phosphonium cations (6). Furthermore, if we assume ∼1000 mtDNA molecules per cell and use the known cell and mitochondrial volumes of MERRF fibroblasts (41), the effective concentration of mtDNA in mitochondria is 1–10 nM. Therefore, only a 10–100-fold accumulation relative to the incubation medium gives a 104–105 molar excess of ph–PNA over mtDNA. Consequently, it was surprising that the ph–PNA conjugate did not inhibit MERRF mtDNA replication in fibroblasts or myoblasts. The reasons for this discrepancy are unclear but there are at least three possible explanations: (i) the ph–PNA is not accumulated by mitochondria in MERRF cells; (ii) PNAs bound to single-stranded mtDNA are displaced by the DNA replication machinery; or (iii) PNAs do not bind to single-stranded mtDNA within cells. The first possibility is unlikely as confocal immunofluorescence microscopy showed that ph–PNA also localised to mitochondria within MERRF fibroblasts (data not shown). While the membrane potential is ∼60 mV lower in MERRF cells (41) the Nernst equation predicts that this would only decrease ph–PNA uptake ∼10-fold, still giving a 103–104-fold molar excess over mtDNA. The second possible explanation, that the mtDNA replication machinery displaces bound PNAs, is plausible, even though in a cell-free system ph–PNA prevented DNA replication. However, for this a partially purified DNA polymerase γ was used and factors lost during preparation may remove PNAs from the path of the advancing replication machinery in mitochondria. The third possiblity, that PNAs do not bind single-stranded DNA during mtDNA replication, seems to us to be the most likely explanation. The classical model for mtDNA replication is that H-strand synthesis starts and is then followed by L-strand synthesis after two-thirds of the old H-strand has been displaced, exposing the L-strand origin of replication (46). The PNA oligomer used here was designed to bind to the L-strand, blocking H-strand replication. The lack of inhibition suggests that any single-stranded mtDNA exposed on the L-strand by the advancing H-strand replication fork is not available to bind PNAs. According to the classical model the displaced H-strand should remain in a single-stranded form available to PNAs (46), however, ph–PNA oligomers with sequences complementary to the H-strand also had no effect on mtDNA replication (unpublished data). Interestingly, the unavailability of the H-strand to complementary PNAs is consistent with a new mtDNA replication model proposed recently in which both mtDNA strands are copied simultaneously (47). We conclude that any single-stranded mtDNA present during mtDNA replication is unavailable for binding to complementary PNAs. This could occur because binding is prevented by proteins associated with the DNA, or because the lifetime of the single-stranded DNA is insufficient. However, PNA oligomers can also be designed to bind specific double-stranded DNA sequences to form a stable triple helix (48). Our newly developed strategy for delivering PNAs to mitochondria gives us a powerful tool to test whether PNA oligomers that bind double-stranded mtDNA can deplete mutated but not wild-type mtDNA. In summary, conjugation of PNA oligomers to lipophilic cations delivers them to mitochondria within cells. We used this approach to show that single-stranded mtDNA is unavailable for binding to complementary PNAs during mtDNA replication. This new strategy for targeting PNAs to mitochondria will lead to a range of novel approaches to manipulate mitochondrial gene expression in cells and facilitate the development of therapies for mtDNA diseases.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Richard Lander for his skilled assistance with the electron microscopy, Paul Smith for measuring the PNA binding affinities and Diana Carne for the MALDI-TOF analyses. This work was supported by grants to M.P.M. and R.A.J.S. from the Marsden Fund administered by the Royal Society of New Zealand and from the Australian Research Council to J.A.W. A.M. is an HRC postgraduate scholar. R.N.L. and D.M.T. thank the Wellcome Trust, the Muscular Dystrophy Campaign and the R.V.I. Hospitals Special Trustees for continued funding.
References
- 1.Larsson N.G. and Clayton,D.A. (1995) Molecular genetic aspects of human mitochondrial disorders. Annu. Rev. Genet., 29, 151–178. [DOI] [PubMed] [Google Scholar]
- 2.Taylor R.W., Chinnery,P.F., Clark,K.M., Lightowlers,R.N. and Turnbull,D.M. (1997) Treatment of mitochondrial disease. J. Bioenerg. Biomembr., 29, 195–205. [DOI] [PubMed] [Google Scholar]
- 3.Chrzanowska-Lightowlers L.Z., Lightowlers,R.N. and Turnbull,D.M. (1995) Gene therapy for mitochondrial DNA defects: is it possible? Gene Ther., 2, 311–316. [PubMed] [Google Scholar]
- 4.Clayton D.A. (1991) Nuclear gadgets in mitochondrial DNA replication and transcription. Trends Biochem. Sci., 16, 107–111. [DOI] [PubMed] [Google Scholar]
- 5.Agrawal S. (1996) Antisense therapeutics. In Walker,J.M., (ed.), Methods in Molecular Medicine. Humana Press, NJ.
- 6.Murphy M.P. (1997) Selective targeting of bioactive compounds to mitochondria. Trends Biotechnol., 15, 326–330. [DOI] [PubMed] [Google Scholar]
- 7.Nakamura K., Hongo,A., Kodama,J., Miyagi,Y., Yoshinouchi,M. and Kudo,T. (2000) Down-regulation of the insulin-like growth factor i receptor by antisense RNA can reverse the transformed phenotype of human cervical cancer cell lines. Cancer Res., 60, 760–765. [PubMed] [Google Scholar]
- 8.Gray G.D., Basu,S. and Wickstrom,E. (1997) Transformed and immortalized cellular uptake of oligodeoxynucleoside phosphorothioates, 3′-alkylamino oligodeoxynucleotides, 2′-o-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochem. Pharmacol., 53, 1465–1476. [DOI] [PubMed] [Google Scholar]
- 9.Ho P.T.C. and Parkinson,D.R. (1997) Antisense oligonucleotides as therapeutics for malignant diseases. Semin. Oncol., 24, 187–202. [PubMed] [Google Scholar]
- 10.Chinnery P.F., Taylor,R.W., Diekert,K., Lill,R., Turnbull,D.M. and Lightowlers,R.N. (1999) Peptide nucleic acid delivery to human mitochondria. Gene Ther., 6, 1919–1928. [DOI] [PubMed] [Google Scholar]
- 11.Hyrup B. and Nielsen,P.E. (1996) Peptide nucleic acids (PNA): synthesis, properties and potential applications. Bioorg. Med. Chem., 4, 5–23. [DOI] [PubMed] [Google Scholar]
- 12.Egholm M., Buchardt,O., Christensen,L., Behrens,C., Freier,S.M., Driver,D.A., Berg,R.H., Kim,S.K., Norden,B. and Nielsen,P.E. (1993) PNA hybridizes to complementary oligonucleotides obeying the Watson–Crick hydrogen-bonding rules. Nature, 365, 566–568. [DOI] [PubMed] [Google Scholar]
- 13.Demidov V.V., Potaman,V.N., Frank,K.M., Egholm,M., Buchard,O., Sonnichsen,S.H. and Nielsen,P.E. (1994) Stability of peptide nucleic acids in human serum and cellular extracts. Biochem. Pharmacol., 48, 1310–1313. [DOI] [PubMed] [Google Scholar]
- 14.Corey D.R. (1997) Peptide nucleic acids: expanding the scope of nucleic acid recognition. Trends Biotechnol., 15, 224–229. [DOI] [PubMed] [Google Scholar]
- 15.Boffa L.C., Morris,P.L., Carpaneto,E.M., Louissaint,M. and Allfrey,V.G. (1996) Invasion of the CAG triplet repeats by a complementary peptide nucleic acid inhibits transcription of the androgen receptor and TATA-binding protein genes and correlates with refolding of an active nucleosome containing a unique AR gene sequence. J. Biol. Chem., 271, 13228–13233. [DOI] [PubMed] [Google Scholar]
- 16.Carlsson C., Jonsson,M., Norden,B., Dulay,M.T., Zare,R.N., Noolandi,J., Nielsen,P.E., Tsui,L.C. and Zielenski,J. (1996) Screening for genetic mutations. Nature, 380, 207. [DOI] [PubMed] [Google Scholar]
- 17.Perry-O’Keefe H., Yao,X.W., Coull,J.M., Fuchs,M. and Egholm,M. (1996) Peptide nucleic acid pre-gel hybridization: an alternative to Southern hybridization. Proc. Natl Acad. Sci. USA, 93, 14670–14675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Nielsen P.E. (1999) Peptide nucleic acids as therapeutic agents. Curr. Opin. Struct. Biol., 9, 353–357. [DOI] [PubMed] [Google Scholar]
- 19.Gray G.D., Basu,S. and Wickstrom,E. (1997) Transformed and immortalized cellular uptake of oligodeoxynuclceoside phosphothioates, 3′-alkylamino oligodeoxynucleotides, 2′-o-methyl oligoribonucleotides, oligodeoxynucleoside methylphosphonates, and peptide nucleic acids. Biochem. Pharmacol., 53, 1465–1476. [DOI] [PubMed] [Google Scholar]
- 20.Ljungstrom T., Knudsen,H. and Nielsen,P.E. (1999) Cellular uptake of adamantyl conugated peptide nucleic acids. Bioconjug. Chem., 10, 965–972. [DOI] [PubMed] [Google Scholar]
- 21.Joliot A., Maizel,A., Rosenberg,D., Tremleau,A., Dupas,S., Volovitch,M. and Prochiantz,A. (1998) Identification of a signal sequence necessary for the unconventional secretion of engrailed homeoprotein. Curr. Biol., 8, 856–863. [DOI] [PubMed] [Google Scholar]
- 22.Derossi D., Calvet,S., Trembleau,A., Brunissen,A., Chassaing,G. and Prochiantz,A. (1996) Cell internalization of the third helix of the antennapedia homeodomain is receptor-independent. J. Biol. Chem., 271, 18188–18193. [DOI] [PubMed] [Google Scholar]
- 23.Pooga M., Hallbrink,M., Zorko,M. and Langel,U. (1998) Cell penetration by transportan. FASEB J., 12, 67–77. [DOI] [PubMed] [Google Scholar]
- 24.Norton J.C., Piatyszek,M.A., Wright,W.E., Shay,J.W. and Corey,D.R. (1996) Inhibition of human telomerase activity by peptide nucleic acids. Nat. Biotechnol., 14, 615–619. [DOI] [PubMed] [Google Scholar]
- 25.Fraser G.L., Holmgren,J., Clarke,P.B.S. and Wahlestedt,C. (2000) Antisense inhibiton of δ-opioid receptor gene function in vivo by peptide nucleic acids. Mol. Pharmacol., 57, 725–731. [DOI] [PubMed] [Google Scholar]
- 26.Nielsen P.E., Egholm,M. and Buchardt,O. (1994) Sequence-specific transcription arrest by peptide nucleic acid bound to the DNA template strand. Gene, 149, 139–145. [DOI] [PubMed] [Google Scholar]
- 27.Wallace D.C. (1999) Mitochondrial diseases in man and mouse. Science, 283, 1482–1488. [DOI] [PubMed] [Google Scholar]
- 28.Taylor R.W., Chinnery,P.F., Turnbull,D.M. and Lightowlers,R.N. (1997) Selective inhibition of mutant human mitochondrial DNA replication in vitro by peptide nucleic acids. Nat. Genet., 15, 212–215. [DOI] [PubMed] [Google Scholar]
- 29.Liberman E.A., Topali,V.P., Tsofina,L.M., Jasaitis,A.A. and Skulachev,V.P. (1969) Mechanism of coupling of oxidative phosphorylation and the membrane potential of mitochondria. Nature, 222, 1076–1078. [DOI] [PubMed] [Google Scholar]
- 30.Murphy M.P. and Smith,R.A.J. (2000) Drug delivery to mitochondria: the key to mitochondrial medicine. Adv. Drug Deliv. Rev., 41, 235–250. [DOI] [PubMed] [Google Scholar]
- 31.Kelso G.F., Porteous,C.M., Coulter,C.V., Hughes,G., Porteous,W.K., Ledgerwood,E.L., Smith,R.A.J. and Murphy,M.P. (2000) Selective targeting of a redox-active ubiquinone to mitochondria within cells: antioxidant and antiapoptotic properties. J. Biol. Chem., 276, 4588–4596. [DOI] [PubMed] [Google Scholar]
- 32.Schagger H. and von Jagow,G. (1987) Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem., 166, 368–379. [DOI] [PubMed] [Google Scholar]
- 33.Chappell J.B. and Hansford,R.G. (1972) Preparation of mitochondria from animal tissues and yeast. In Birnie,G.D. (ed.), Subcellular Components: Preparation and Fractionation. Butterworths, London.
- 34.Gornall A.G., Bardawill,C.J. and David,M.M. (1949) Determination of serum protein by means of the biuret reaction. J. Biol. Chem., 177, 751–766. [PubMed] [Google Scholar]
- 35.Davey G.P., Tipton,K.F. and Murphy,M.P. (1992) Uptake and accumulation of 1-methyl-4-phenylpyridinium by rat liver mitochondria measured using an ion-selective electrode. Biochem. J., 288, 439–443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kamo N., Muratsuga,M., Hongoh,R. and Kobatake,Y. (1979) Membrane potential of mitochndria measured with an electrode sensitive to tetraphenyl phosphonium and relationship between proton electrochemical potential and phosphorylation potential in steady state. J. Membr. Biol., 49, 105–121. [DOI] [PubMed] [Google Scholar]
- 37.Kaufman B.A., Newman,S.M., Hallberg,R.L., Slaughter,C.A., Perlman,P.S. and Butow,R.A. (2000) In organello formaldehyde crosslinking of proteins to mtDNA: Identification of bifunctional proteins. Proc. Natl Acad. Sci. USA, 97, 7772–7777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Smith P.K., Krohn,R.I., Hermanson,G.T., Malia,A.K., Gartner,F.H., Provenzano,M.D., Fujimoto,E.K., Goeke,N.M., Olson,B.J. and Klenk,D.C. (1985) Measurement of protein using bicinchoninic acid. Anal. Biochem., 150, 76–85. [DOI] [PubMed] [Google Scholar]
- 39.Srere P.A. (1969) Citrate synthase. Methods Enzymol., 13, 3–11. [Google Scholar]
- 40.Berry M.N., Edwards,A.M., Barritt,G.J., Grivell,M.B., Halls,H.J., Gannon,B.J. and Friend,D.S. (1991) Isolated Hepatocytes Preparation, Properties and Applications. Elsevier, Amsterdam, The Netherlands.
- 41.James A.M., Wei,Y.H., Pang,C.Y. and Murphy,M.P. (1996) Altered mitchondrial function in fibroblasts containing MELAS or MERRF mitochondrial DNA mutations. Biochem. J., 318, 401–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Brown G.C. and Brand,M.D. (1985) Thermodynamic control of electron flux through mitochondrial cytochrome bc1 complex. Biochem. J., 225, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Brand M.D. (1995) Measurement of mitochondrial protonmotive force. In Rickwood,D. and Hames,B.D. (eds), Bioenergetics–A practical approach. Oxford University Press, NY.
- 44.Robin E.D. and Wong,R. (1988) Mitochondrial DNA molecules and virtual number of mitochondria per cell in mammalian cells. J. Cell Physiol., 136, 507–513. [DOI] [PubMed] [Google Scholar]
- 45.Estabrook R.W. and Holowinsky,A.J. (1961) Studies on the content and organisation of the respiratory enzymes of mitochondria. J. Biochem. Biophys. Cytol., 9, 19–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Clayton D.A. (1992) Transcription and replication of animal mitochondrial DNAs. Int. Rev. Cytol., 141, 217–232. [DOI] [PubMed] [Google Scholar]
- 47.Holt I.J., Lorimer,H.E. and Jacobs,H.T. (2000) Coupled leading- and lagging-strand synthesis of mammalian mitochondrial DNA. Cell, 100, 515–524. [DOI] [PubMed] [Google Scholar]
- 48.Nielsen P.E., Egholm,M. and Buchardt,O. (1994) Evidence for (PNA)2/DNA triplex structure upon binding of PNA to dsDNA by strand displacement. J. Mol. Recognit., 7, 165–170. [DOI] [PubMed] [Google Scholar]