Abstract
Summary
RHD PCR of blood donors may be used to reveal weak D, partial D, DEL and chimeric D+/D− donors among presumed D-negative blood donors. Units donated by such donors pose a definite yet low risk for anti-D immunization of transfusion recipients. The frequency of DEL donors among D-negative donors is 1:350 to 1:2,000 in Europe and up to 1:5 in Asian countries. Different strategies for RHD PCR of blood donors have been used. Probably, the most cost-efficient implementation is replacement of sensitive D antigen testing with the indirect antiglobulin test by RHD PCR in pools which might even reduce total testing cost.
KeyWords: Antigen D, Rh blood group, Blood donors, Indirect antiglobulin test, DEL, Weak D, RHD PCR
Introduction
Soon after the identification of the RH genes, it became apparent that D-positive individuals have two different RH genes, while D-negatives generally only possess one [1]. The additional RH gene present in D-positives was characterized as RHD gene [2, 3] and its presence used to determine the D antigen status [4].
This clear-cut explanation of the D-positive / D-negative antigen status held until 1997, when reports appeared showing that the dictum ‘RtfD-positive equals D-positive was not always true for people of African [5] or Japanese [6] descent. Furthermore, even in a European population, alternative molecular bases for the D-negative phenotype were described [7].
Triggered by these observations, we aimed to determine the molecular bases of D-negative RHD-positive haplotypes in South-Western Germany on a larger scale [8]. For this sake, donors believed to be D-negative were checked by RHD PCR specific for the RHD promoter, RHD intron 4, RHD exon 7, and RHD exon 10. As part of the serologie work-up, the D-negative status was re-evaluated by the indirect antiglobulin test and adsorption/elution testing. Apart from a large range of different RHD-positive D-negative donors, surprisingly a relevant number of donors were D-positive in serologie re-evaluation: Among 754 C- or E-positive donors, 15 DEL and 4 weak or partial D donors were found, and among 7,688 ceddee donors, one chimeric D-positive/ D-negative donor with about 5% D-positive red blood cells (RBC) was detected. A look-back study of his prior donations indicated that both identified D-negative recipients had developed an anti-D. This observation raised the question whether D-negative donors should be checked by RHD PCR to exclude weak D-positive and DEL donors missed by serology.
A General View on Anti-D Immunization
When considering the assumed low immunogenicity of DEL units or comparing the frequency of anti-D immunization by DEL to the often higher frequency of other alloimmunization events [9], two facts on anti-D immunization should be realized:
- First, a prospective evaluation of anti-D immunization by weak D and DEL is difficult. Even after transfusion of normal D-positive units, only about 21–31% of patients develop an anti-D [10, 11, 12]. Weak D and DEL units are likely considerable tobe less immunogenic. If no event is observed among n observations, the upper limit of the confidence interval for the frequency of the event is about 2,996/n [13]. Therefore, a study to exclude an immunogenicity of 2.5% (one tenth of normal D-positive units) would have to include 120 patients. This study size has never been reached in prospective studies [14] or look-back studies [8, 9,15].
- Second, a comparison with immunizations to other RBC antigens may be misleading. Anti-D is the most important cause of hemolytic disease of the newborn. Before the advent of anti-D prophylaxis, this disease occurred in a frequency of up to 1:23 among Rh-negative women [16]. Rh prophylaxis does not work in pre-immunized women. Therefore, even a low-titer anti-D immunization caused by a DEL unit will prevent anti-D prophylaxis from working and thus trigger the devastating cascade of ever increasing anti-D strength with each pregnancy. This mechanism is unique to antigen D, as no prophylaxis is used for the other RBC antigens, and antibodies to other antigens much rarer cause similar harm as a high-titer anti-D.
Evidence for Anti-D Immunization by Weak D Missed in Donor Testing
The immunogenicity of weak D has been questioned [17, 18] as only a few report of this event have been published: None of 49 D-negative recipients of 68 ‘Du units developed an anti-D [14].
However, there is little doubt that transfusion of units donated by weak D donors can cause anti-D immunization in D-negative recipients: Even before the elucidation of the molecular basis of weak D, three anti-D immunization events by ‘weak D’, including a primary immunization by a unit with 800–1,500 antigens/cell, have been documented [19]. These immunizations occurred within less than 2 years when testing for weak D antigens was dropped in the Netherlands [20]. Whereas the underlying alleles of these donors were not evaluated and it might be argued that they represented exceptions carrying special alleles, reports of anti-D immunizations by units from donors with frequent weak D alleles appeared: In New Zealand, a ccD. Ee weak D type 2 donor with about 450 antigens/ cell caused a primary anti-D immunization [21]. The same was shown for a weak D type 1 donor with Cde in trans further suppressing his antigen density to 357 antigens/cell [22]. In addition, an anti-D immunization event by the rare weak D type 26 was caused by a donor with an antigen density as low as 70 antigens/cell [23]. Considering the low likelihood of a look-back study in an inadvertently anti-D-immunized patient, these case reports are likely only the tip of the iceberg.
Evidence for Anti-D Immunization by DEL Donors
Evidence for anti-D immunization by DEL donors exists, although the risk may be considerably lower than for normal D: A female Austrian patient with an unexplained anti-D immunization was shown to have received a DEL unit harboring the rare DEL allele RHD(IVS5–38 del4) [24]. Shortly afterwards a considerable increase of the anti-D titer in a pre-immunized 67-year-old Japanese woman receiving two units with the DEL RHD(K409K) was observed [25]. This observation was important, since RHD(K409K) is the by far most frequent DEL allele in Asians and worldwide. [26]. Four years later, the same allele was shown to cause an anti-D immunization in a 69-year-old Korean male [27]. Still, anti-D immunizations caused by DEL donors seem to be rare events: Only one possible anti-D immunization event was found in a follow-up of 13 units from DEL donors in Denmark [9], and only 3 of 82 D-negative recipients of RHD(93insT) units had developed an anti-D in Canada [15]. In both studies, transfusion of D-positive platelet units was an alternative explanation for the anti-D immunization events, and the immunogenicity of DEL seemed to be lower than that of many other blood group antigens.
Evidence for Anti-D Immunization by RHD+/-Chimeric Donors
The evidence of anti-D immunization by the RHD+/- chimeric donor in the seminal study [8] was convincing, as both identified recipients who were D-negative and not known to possess an anti-D prior to the transfusion had an anti-D after the transfusion. Furthermore, a quick calculation of the number of D-positive RBC revealed that this observation was far from surprising: An RBC unit contains about 220 ml of packed RBC, 5% of which equals 11 ml D-positive RBC. This volume is much higher than the usual volume of D-positive fetal RBC entering the maternal circulation during delivery, a known cause of anti-D immunization. Although the evidence of immunization is obvious, it should be realized that no second D-positive / D-negative chimeric donor was identified in follow-up studies [28, 29, 30, 31], and this event thus seems to be a rarity.
Frequency and Type of Seemingly D-Negative, RHD-Positive Donors
Currently, in the RhesusBase [32] 30 alleles are listed that have been described to be associated with a DEL phenotype (table 1). As the borderline between weak D and DEL is not defined and may vary between laboratories depending on the sensitivity of the indirect antiglobulin test used, some of these alleles may represent weak D with low antigen density. In addition, weak D with low antigen D expression like weak D type 32 [30] or weak partial D like D category VI type I may appear as DEL if a Cde allele is in trans. On the other hand, false-positive results in adsorption/elution technique may also occur [35]; so some alleles observed only once and described as DEL might rather represent D-negative alleles.
Table 1.
Alleles for which a DEL phenotype has been described [32]
| Allele (Trivial name) | Structure | Haplo-type | Mechanism | Phenotype | Distribution* | References | Comments |
|---|---|---|---|---|---|---|---|
| RHD(M1I) | RHD(3G>A) | Variable | loss of start codon | DEL | R (China, Germany) | [33] | |
| RHD(R10W) (Weak D type 61) | RHD(28C>T) | CDe | MIS | DEL, weak D | R | [33] | DEL phenotype if categorized by tube testing; independently characterized as weak D AM412754 |
| RHD(W16R) | RHD(46T>C) | cDE | MIS | DEL | S (Switzerland) | HE999546 | |
| RHD(W16X) | RHD(48G>A) | CDe | STOP in exon 1 | D-negative, DEL | R (Germany) | [8] | reported as DEL (same donor tested on three donations as DEL) in a later study [35] |
| RHD(L18P) | RHD(53T>C) | MIS | DEL | S (China) | [33] | ||
| RHD(93insT) | RHD(93insT) | CDe | FS in exon 1 | DEL | R (Germany, Denmark, Spain) | [34] | DEL status reported in follow-up study[28] |
| RHD(L38X) | RHD(113T>A) | STOP in exon 1 | DEL? | S (Germany) | [35] | ||
| RHD(147delA) | RHD(147delA, IVSl+6delA) | CDe | FS in exon 1 | DEL | S (Germany) | [28] | |
| RHD(IVS1+1G>A) | RHD(IVS1+1G>A) | SPL | DEL | S (Japan) | [36] | ||
| RHD(IVS1-29G>C) | RHD(IVS1-29G>C) | CDe | SPL | DEL | S (Poland) | HE971139 | this allele was initially reported as RHD(IVS2-29G>C) |
| RHD(L84P) | RHD(251T>C) | MIS | DEL | S (China) | [33] | comparably high antigen density | |
| RHD(IVS2-2A>G) | RHD(IVS2-2A>G) | CDe | SPL | DEL | S (Denmark) | [9] | |
| RHD(S112T) | RHD(IVS1-29G>C, 335G>C) | SPL | DEL | S (Switzerland) | HE999547 | both mutations affect the splice site | |
| RHD(A137E) | RHD(410C>A) | CDe | MIS | DEL | S (China) | [37] | DEL status according to tube IAT |
| RHD(L153P) | RHD(458T>C) | cDE | MIS | DEL | S (Germany) | [28] | |
| RHD(IVS3+1G>A) | RHD(IVS3+1G>A) | CDe | SPL | DEL / D-negative / Partial DEL | R (Germany, Denmark, Poland, Austria) | [8] | this allele is almost D-negative |
| RHD(G212R) | RHD(634G>C) | cDe | missense mutation / splice site affected | DEL | S (Germany) | [28] | |
| RHD(IVS5-38del TCTC) | RHD(IVS5-38del TCTC) | CDe | unknown | DEL | S (Austria) | [24] | the IVS5-38 del TCTC polymorphism is also found in RHD alleles with normal antigen strength and therefore not causative [38] |
| RHD(P291R) | RHD(872C>G) | CDe | MIS | DEL | S (Switzerland) | HE999545 | the same missense mutation occurs in the DEL weak D type 4.3 |
| RHD(T201R, F223V, P291R) (weak D type 4.3) | RHD(602C>G,667T >G,819G >A,872C>G) | cDe | MIS | DEL | R (Austria, Germany) | [39] | the DEL phenotype is probably caused by the P291R substitution |
| RHD(M295I) | RHD(885G>T) | CDe | MIS | DEL / weak D | R (Germany) | [8] | borderline DEL / weak D. The same mutation in a cDe haplotype causes the weak D type 11 phenotype. |
| RHD(IVS8-31 T>C) | RHD(IVS8-31 T>C) | CDe | SPL | D-negative/ DEL/ weak D | R (Germany) | [35] | |
| RHD(Y401X) | RHD(1203T>A) | cDE | STOP near 3’ end | DEL D-negative | R | [23] | initially described as D-negative, reported as DEL in another study [28] |
| RHD(D404H) | RHD(1210G>C) | cDE | MIS | DEL | S (Portugal) | JX114749 | |
| RHD(W408R) | RHD(1222T>C) | CDe | MIS | DEL | S (Korea) | [40] | |
| RHD(K409K) | RHD(1227G>A) | CDe | SPL | DEL | Most frequent DEL in Asia [26] | [8] | this allele has initially been misinterpreted as RHD(delEx9) |
| RHD(X418L) | RHD(1252ins T) | CDe | loss of STOP | DEL | R (Austria, Germany) | [23] | |
| RHD-RHCE(2–5)-RHD | RHD-RHCE(2–5)-RHD | CDe? | hybrid allele | Partial D, DEL | S (China) | [33] | the predicted structure is similar to DVI type IV, a partial D frequent in Spain [41]. The reason for the different phenotype is unknown. |
| RHD-RHCE(4–9)- RHD | RHD-RHCE(4-9)-RHD | large hybrid | D-negative, DEL | R (China) | [37] | this allele was initially characterized as D-negative but appeared as DEL in one study [37] | |
| DBU | RHD-RHCE(5-7)-RHD | cDE | loss of D-specific exofacial loops | DEL, Expected partial D | S (Germany) | [28] | |
| RHD(delEx8) | RHD(delEx8) | CDe | 995 bp deletion including exon 8 | DEL | one pedigree (Libanese) | [42] | |
| RHD(delEx9) (obsolete) | the observations of this allele represent RHD(K409K) [43] | ||||||
| RHD(delEx10) | [44] | ||||||
| RHD-RHCE(10) (obsolete?) | RHD-RHCE(10) | [37] | the observation of this allele likely represent RHD(delEx10) |
MIS = Missense mutation; STOP = stop codon; SPL = splice site mutation; FS = frameshift; S = single observation; R = repeatedly observed.
Two main mechanisms may cause a DEL phenotype:
- Classically, a mutation near the splice site hampers normal splicing of RHD. This mechanism was well studied for RHD(K409K), in which a ‘silent 1227G>A substitution in codon 409 is immediately adjacent to the exon 9 / intron 9 boundary. Most transcripts of this DEL lack RHD exon 9 [45]. It is unknown, whether the faint residual D antigen expression is due to D antigen expression by misspliced transcripts or a very low number of correctly spliced transcripts. A similar mechanism is likely to work in RHD(IVS3+1G>A) that in addition seems to lack some specific D epitopes. Although it might be reasoned that the partial D character might be due to a predominance of misspliced transcripts, the exact mechanism is still unknown.
- Some missense mutations have so much impact on the correct expression of RhD so that a DEL phenotype results. Often, these alleles have a borderline weak D / DEL antigen density. An interesting example is RHD (M295l), which displays the weak D type 11 phenotype when it occurs in a cDe haplotype [46] and a DEL phenotype when it occurs in a CDe haplotype [8].
In addition, there is a flurry of different rare mechanisms:
- Some mutations generally expected to destroy any production of intact RhD protein are associated with a DEL phenotype. The most important example is RHD(93insT) which shows a DEL phenotype despite a frameshift. For this allele, it was argued that transcription slipping might result in a low number of wild-type transcripts. Considering the location of this mutation in exon 1 and the DEL phenotype of RHD(M1I) [33] lacking the normal start codon, another explanation is the use of an alternative start codon.
- In RHD(X418L), the stop codon is destroyed resulting in a predicted structure with 71 additional tailing amino acids [23].
A few DEL are characterized by polymorphisms for which it is unknown whether and why they cause the DEL phenotype. For example, one DEL was shown to be associated with an IVS5–38 del4 polymorphism [24], but in a later observation the same polymorphism was observed in a variety of RHD alleles of normal antigen strength [38].
Which RHD PCR Should Be Used?
The use of RHD PCR to detect weak D and DEL donors sero-logically mistyped as D-negative is a task considerably different from other uses of RHD PCR: A very large number of samples have to be tested, imposing considerable pressure on cost and effort. In contrast, only a few positive samples are found, making accessory testing to fully characterize these samples feasible. Therefore, often a two-stage strategy is used that consists of ‘screening detection’ of any RHD-positive sample followed by a more in-depth characterization of only these samples.
For conventional applications, an RHD PCR should rely on testing at least two different RHD-specific polymorphisms, because otherwise hybrid RHD-RHCE-RHD alleles expressing partial D phenotype may be missed. Since most DEL and weak D differ from normal RHD by splice site or missense mutations, they are not affected by this problem, and testing for almost any RHD-specific polymorphism will reveal the vast majority of DEL samples. Therefore, if sensitive serologie testing is continued to detect partial D caused by hybrid alleles, testing a single RHD polymorphism by PCR may be a possible choice to find the DEL samples [28, 35,47].
Another difference to conventional RHD PCR applications is the possibility to start with pooled testing [8, 28, 30, 35, 39, 48]: As long as the samples have initially be checked at least by direct agglutination with anti-D, in Caucasian populations only a minority of samples will be RHD-positive.
While almost any RHD PCR will be sensitive to detect DEL, obtaining specificity is more difficult. Most RHD PCR testing for DEL will also reveal RHD-positive alleles not expressing any D antigen ('D-negative RHD-positive alleles), like those with stop codons. It is nearly impossible to devise a PCR system that will not give a positive result for these alleles, and further characterization of the samples by extensive molecular or serologie testing may be necessary. However, the choice of the polymorphisms used for initial testing may help to reduce the rate of D-negative RHD-positive samples detected:
- Generally, large RHD-CE-D hybrids in which at least RHD exon 4–7 is substituted by RHCE are D-negative. Therefore, using a polymorphism inside this region will avoid detecting these rather frequent D-negative, RHD-positive hybrids.
- In a population with a relevant admixture of individuals of African descent, a relevant number of D-negatives will be RHDψ positive. In such a population, it is advantageous to devise the initial PCR in a way that it does not detect RHDψ.
If an RHD PCR approach is used that restricts initial testing to a polymorphism within the range of RHD exon 4–7, about one third to one half of the detected donors carry these D-negative RHD-positive alleles [28,31] and could thus be kept in the D-negative donor pool.
D-Positive Alleles Missed by PCR
Despite the simplicity to detect most DEL, it must be realized that some alleles encoding for D epitope expression are prone to be missed, especially if the initial ‘screening’ RHD PCR is reduced to a single or a few RHD-specific polymorphisms:
- In almost any approach not specifically devised to detect these alleles, RHCE alleles expressing D epitopes due to mutations unrelated to RHD will be missed. The most pertinent of these alleles are ceRT [49] and ceSL [50].
- Likewise, RHCE alleles expressing D epitopes due to the presence of only one RHD-specific nucleotide like ceCF [51] are missed, if this D-specific polymorphism is not tested for. To a lesser extent, this problem applies to RHCE alleles with short stretches of RHD included, like DHAR in which only exon 5 displays RHD sequence.
- If a single-polymorphism testing strategy is used, short hybrid alleles, generally associated with a partial D phenotype, are missed. For example, testing exon 7 [35] misses DBU, DBT, and many DIV variants; testing intron 4 [28] will miss DVI variants. The relevance of such misses depends on the serologie characteristics of these variants, for example, most DIV variants are readily detected by direct agglutination while DBU represents a DEL phenotype.
Frequency of Missed D-Positive Samples
A survey of published population studies of serologically D-negative donors is given in table 2. Generally, the frequency of DEL among Europeans is 1:350 to 1:2,000, while it gets as high as 1:5 among Asians. It should be noted that in many studies [8, 23,47, 56] weak D or partial D donors were detected among donors previously believed to be D-negative.
Table 2.
Population studies of DEL
| Population | RHD exons screened | D-negative donors screened | DEL detected | Frequency of DEL among D-negative | Most frequent alleles | Number of different DEL alleles found |
|---|---|---|---|---|---|---|
| Brazil [52] | intron 4, exon 10 | 239 | 0 | <1:80 | ||
| Brazil [48] | intron 4, exon 7, pools of 10 | 2,450 | 19 (including weak D) | 1:129 | ||
| Central Europe [23], | 5’ UTR, 3,10 | 1,700 | 15 DEL + 5 weak D / partial D | 1:113 | RHD(M295I), RHD(IVS3+1G>A) | 3 |
| donors with C or E | ||||||
| China [53] | intron 4, exon 7, (5'UTR, exon 10) | 155 (negative on direct agglutination) | 31 (+3 weak D) | 1:5 | RHD(K409K) | 1 |
| China (Shanghai) [37] | (serologie screening) | 1,585 | 279 | 1:6 | RHD(K409K) | |
| Denmark [9] | 5.7.8 | 5,058 (4,932 results available) | 2* | 1:2,029 | 2 | |
| Denmark [47], | exon 10 | 233 | 3 DEL + 1 weak D | 1:78 | 3 | |
| donors with C or E | ||||||
| Germany(South-West) [8], | 5’ UTR, intron 4, exons 7,10 | 754 | 15 DEL + 4 weak D / partial D | 1:50 | RHD(M295I), RHD(K409K) | 3 |
| donors with C or E | ||||||
| Germany(South-West) [8], | 5'UTR, intron 4, exon 10 | 7,688 | 1 chimeric donor | <1:2,500 | ||
| ccddee donors | ||||||
| Germany (South-West) [28] | intron 4 (pools of 20) | 46,133 | 47 | 1:982 | RHD(IVS3+1G>A), RHD(M295I) | 10 |
| Germany (North) [31, 35] | exon 7 (pools of up to 40) | 46,756 | 76 | 1:615 | RHD(IVS3+1G>A), RHD(M295I) | 15 |
| Italy [54] | commercial PCR systems, pools of 5 | 235 | 1 | 1:235 | RHD(M295I) | 1 |
| Korea [43] | intron 4, exon 7 | 126 | 16 + weak D | 1:8 | RHD(K409K) | 1 |
| Korea [40] | 3.5.7.10 plus intron 4 | 264 | 43 | 1:6 | RHD(K409K) | |
| Upper Austria [39] | 4.7.10 in pools of 20 | 2,427 | 3 | 1:809 | RHD(IVS3+1G>A) | 2 |
| Upper Austria [30] | 4.7.10 in pools of 20 | 23,330 | 66 | 1:353 | weak D type 4.3, RHD(IVS3+1G>A) | 6 |
| Thailand [55] | (serologie screening) | 254 | 50 | 1:5 | RHD(K409K) | |
| Tunisia [56] | 488 | 3 weak D, 1 partial D (missed despite IAT) |
In addition, an RHD(IVS3+1G>A) sample was detected but characterized as D-negative.
Possible Testing Strategies
Different blood services introduced RHD PCR for seemingly D-negative blood donors; however, approaches vary. A few considerations will be detailed below.
First-Time Donors versus Repeat Donors
From a logistic point of view, testing repeat donors is much more complicated than testing first-time donors: i) Generally, the sheer number of repeat donors is much higher than those of first-time donors, ii) In order to prevent the repeat testing of the donors, it is necessary to implement a strategy to mark the donors as tested, iii) Often, pooled platelet units are produced exclusively from repeat donors. RHD PCR of those donors will hamper the delivery of the units or necessitate the delivery before D antigen determination is finished.
Based on these considerations and a perceived low cost-efficiency of the RHD PCR, several blood services started by testing exclusively first-time donors. After many years, such strategy should result in a D-negative donor pool largely tested by RHD PCR. However, many donors continue to donate for decades, and the most pertinent examples of missed weak D donors were ‘old-time’ donors [8, 23], who had been tested for weak D many decades ago when the sera were less potent than today and only tube testing was available. Therefore, testing of all donors is recommended, once the logistic basis to exclude repeated testing and delays in platelet unit delivery is established.
C/E-Positive Donors versus All Donors
In many populations, the frequency of DEL among CDe and cDE haplotypes is much higher than among cDe haplotypes. For example, in 6 years RHD PCR in South-Western Germany [28], the DEL frequency among Ccddee donors was 1:51, among ccddEe donors 1:344, among all C or E positive donors 1:67, and among ccddee donors only 1:43,053, more than 600 times lower than among those with C or E. Since the cost per detected donor depends on the number of donors that need to be tested in order to identify one DEL, weak D or chimeric donor, focusing on donors with C or E is much more cost-efficient than testing all donors. On the other hand, in some countries, like Germany, premenopausal women are exclusively supplied with units compatible for all Rh antigens. A ccDELee unit can possibly be transfused to any D-negative premenopausal patient, while only 4% of all premenopausal women are eligible for Ccddee units. Hence, the probability of a ccDELee unit to be transfused to a premenopausal woman is about 25 times larger than that of a Ccddee unit, partly counteracting the increased probability of a DEL unit among Ccddee. Even considering this fact, the cost-efficiency regarding prevention of DEL transfusion to D-negative premenopausal women is about 37 times higher if only donors with C or E are tested.
In conclusion, testing donors with a C or E is considerably more cost-efficient than testing all D-negative donors. However, if the risk of immunization by DEL, weak D, or chimeric donors shall be abolished, testing all donors is necessary.
Add-on Testing versus Replacement of Indirect Antiglobulin Test
Current D antigen determination for blood donors often includes sensitive testing for the first or first few donations followed by confirmatory testing by direct agglutination only. If the RHD PCR is added to such testing regime, there is no risk that the new strategy misses donors that would have been detected by the old serologie strategy. Hence, such ‘add-on’ testing may be realized with very simple and cheap RHD PCR approaches. However, the total cost of D antigen determination is always higher than that of a serologie testing strategy.
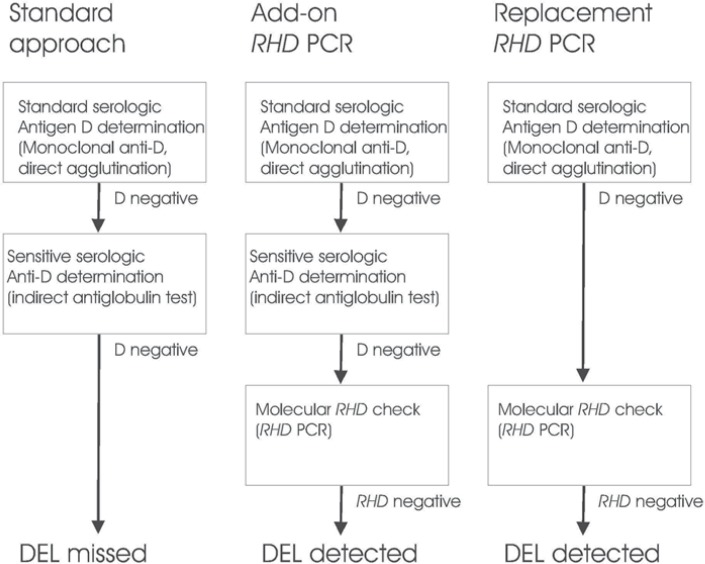
An alternative approach consists of replacement of the sensitive serologie testing by RHD PCR. In this approach, serologie donor testing is reduced to direct agglutination, and all first-time donors negative by direct agglutination are checked by RHD PCR (fig. 1). Obviously, in this setting the RHD PCR must be devised in a form to assure that any relevant D-positive RHD variant that may be missed by direct agglutination is detected. The major advantage of this approach is the elimination of the non-negligible testing cost for D antigen determination in the indirect antiglobulin test.
Fig. 1.
Add-on and replacement RHD PCR. Different approaches for the determination of the antigen D status of first time donors are shown. Left: In standard antigen D determination, donors are first tested with a directly agglutinating monoclonal anti-D, and D-negative donors are then checked by the indirect agglutinin test with an oligoclonal anti-D to detect weak D and partial D antigens. DEL are missed by this approach. Middle: DEL may be detected by adding RHD PCR to the standard D determination. This approach eliminates any risk that D-positive donors detectable by the serologie approach are missed, but of cause the cost must be higher than that of the conventional approach. Right: DEL may be also detected by testing donors D-negative by direct agglutination by RHD PCR. This approach is safe, as long as the RHD PCR is devised in a way that weak D and partial D alle-les are detected, too. The cost of this approach may be higher or lower than that of the standard approach, depending on the relative cost of the indirect antiglobulin test and the RHD PCR used.
Single Donor Testing versus Pooled RHD PCR
As RHD-positive donors are rare among seeming D-negative Caucasian donors, many blood services use pooled RHD PCR. Of course, in populations with a relevant number of individuals of African or Asian descent, RHD-positive D-negative alleles are more frequent, and pooled testing is only advisable if the PCR is devised in a way that the frequent RHD-positive D-negative allele of the population is not detected. A possible pitfall of pooling may be the varying DNA content of different donors units due to different leucocyte concentration. However, most PCR systems are devised in a way that even very small contaminations with D-positive DNA may be detected. It should be remembered that in the initial study on this topic, a donor with only 5% D-positive RBC was found with pooled testing [8].
Further Work-Up of RHD-Positive Units
A drawback of the RHD PCR is the fact that D-negative RHD-positive alleles are detected as well. If these donations shall not be moved to the D-positive donor pool, further work-up is necessary. Serologie analysis of these samples is time-consuming and may be unreliable [31]. From a cost-efficiency perspective, using all RHD-positive units exclusively for D-positive recipients may be a rational choice. Such measure would reduce the D-negative donor pool by about 0.1% but save any additional testing cost.
If the most frequent D-negative alleles of the population are known, donors carrying these alleles can be identified by PCR specifically detecting these alleles; the rare unclassifiable samples might be further analyzed by sequencing. Since this work-up may be very costly, these added costs should not be neglected when switching to RHD PCR is considered. However, these costs will only occur once an RHD PCR-positive donor is detected, reducing their impact on general donor testing. For example, assuming that donors are tested in pools of 20 with a PCR assay at EUR6.00 per PCR, 1 of 1,000 donors is RHD-positive and further work-up is EUR 80.00 per RHD-positive donation, PCR cost per presumed D-negative donation will be EUR 0.30 and work-up cost per presumed D-negative donation EUR 0.08.
Examples of RHD PCR Programs
A survey of current RHD PCR programs is given in table 3. Obviously, RHD PCR is increasingly used, but approaches still vary. The first country in which RHD PCR has become obligatory is Switzerland.
Table 3.
Routine RHD PCR of blood donors
| Blood Service | Testing strategy |
|---|---|
| Red Cross Blood Service Baden Württemberg - Hesse [28] (Germany) | all first time donors, pooled testing for RHD intron 4 |
| Red Cross Transfusion Service of Upper Austria [30] | all first time donors, pooled testing for RHD intron 4.7.10; no IAT |
| NIH (USA) | single donor testing (WA Flegel, Washington, personal communication) |
| Aarhus Hospital (Denmark) [47] | exon 10 for donors with C or E |
| Red Cross Blood Service NSTOB (Germany) | all first time donors, pooled testing for RHD exon 7 |
| Albert Einstein Hospital, São Paulo (Brazil) [48] | |
| Red Cross Blood Service (Bern, Switzerland)* | pooled testing for exons 3.5.10 (H Hustinx, Bern, personal communication) |
| Red Cross Blood Service (Zurich, Switzerland)* | pooled testing intron 4 / exons 5 +7 (until 2011), single donor exon 5, 7 and 3’ untranslated (since 2012) (C Gassner, Zurich, personal communication) |
| Red Cross Blood Service (Innsbruck, Austria) | single donor testing of donors with C or E |
RHD PCR of D-negative donors is mandatory in Switzerland since January 2013, serologie testing by IAT has been dropped.
Cost-Efficiency Considerations
Given the low risk of anti-D immunization by transfusion of DEL units, the cost-efficiency of RHD PCR for blood donors has been disputed. For example, in a recent review, it was concluded that ‘For routine ABO and D determination, DNA testing is more time-consuming, more expensive, prone to misinterpretation, and thus, not an improvement over hemagglutination [57]. Westhoff [58] argued that focusing on D would be inappropriate as long as immunization to K and c would be considered irrelevant in premenopausal women.
The current standard in several European countries, like Germany, includes sensitive D antigen testing of donors and matching for K and c in premenopausal women, and it is unlikely that this standard will be reduced in the near future. Thus, cost-efficiency can be reduced to the question whether using RHD PCR is cost-efficient compared to standard sensitive D antigen testing with indirect antiglobulin test.
As outlined above, the cost-efficiency and imposed workload of RHD PCR for blood donors considerably depends on the approach chosen. The by far most cost-efficient approach is replacement of indirect antiglobulin test testing by RHD PCR and using all RHD-positive units for D-positive recipients. In this approach, workload and testing cost is likely comparable to the ‘old serologie approach and it may be anticipated that with on-going reduction of PCR testing cost, this approach may become - or even may already be - cheaper than a merely serologie testing. Hence, I expect that in the end, indirect antiglobulin testing will be replaced by RHD PCR, not because missing DEL is so risky, but because RHD PCR will become the cheaper way to detect weak D, partial D, and DEL, making any cost-efficiency discussions absurd.
Conclusion
It is obvious that RHD PCR of blood donors may prevent anti-D immunizations and will, albeit rarely, prevent the dismal spiral of ever increasing anti-D in anti-D-immunized pregnant women. Furthermore, RHD PCR may become cheaper than testing donors with the indirect antiglobulin test. For those countries like Germany that decided to go with sensitive testing of donors for the D antigen, the switch from the indirect antiglobulin test to RHD PCR would be an example of a change in testing strategies in which the superior testing strategy ultimately turns out to be the cheaper one.
Disclosure Statement
The author receives royalties as inventor of patents on the molecular genetics of the RH blood group
Acknowledgement
I thank C. Gassner, Zurich, H. Hustinx, Bern, and WA. Flegel, Washington D.C., for sharing data on their recent activities and T.H. Müller, Springe, and A. Seltsam, Springe, for helpful discussion.
References
- 1.Colin Y, Chérif-Zahar B, Le Van Kim C, Raynal V, Van Huffel V, Cartron JP. Genetic basis of the RhD-positive and RhD-negative blood group polymorphism as determined by Southern analysis. Blood. 1991;78:2747–2752. [PubMed] [Google Scholar]
- 2.Le van Kim C, Mouro I, Chérif-Zahar B, Raynal V, Cherrier C, Cartron JP, Colin Y. Molecular cloning and primary structure of the human blood group RhD polypeptìde. Proc Nati Acad Sci USA. 1992;89:10925–10929. doi: 10.1073/pnas.89.22.10925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Arce MA, Thompson ES, Wagner S, Coyne KE, Ferd-man BA, Lublin DM. Molecular cloning of RhD cDNA derived from a gene present in RhD-positive, but not RhD-negative individuals. Blood. 1993;82:651–655. [PubMed] [Google Scholar]
- 4.Bennett PR, Le Van Kim C, Colin Y, Warwick RM, Chérif-Zahar B, Fisk NM, Cartron JP. Prenatal determination of fetal RhD type by DNA amplification. N EnglJ Med. 1993;329:607–610. doi: 10.1056/NEJM199308263290903. [DOI] [PubMed] [Google Scholar]
- 5.Daniels G, Green C, Smart E. Differences between RhD-negative Africans and RhD-negative Europeans. Lancet. 1997;350:862–863. doi: 10.1016/S0140-6736(05)62031-4. [DOI] [PubMed] [Google Scholar]
- 6.Okuda H, Kawano M, Iwamoto S, Tanaka M, Seno T, Okubo Y, Kajii E. The RHD gene is highly detectable in RhD-negative Japanese donors. J Clin Invest. 1997;100:373–379. doi: 10.1172/JCI119543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Avent ND, Martin PG, Armstrong-Fisher SS, Liu W, Finning KM, Maddocks D. Urbaniak SJ. Evidence of genetic diversity underlying Rh D-, weak D (Du), and partial D phenotypes as determined by multiplex polymerase chain reaction analysis of the RHD gene. Blood. 1997;89:2568–2577. [PubMed] [Google Scholar]
- 8.Wagner FF, Frohmajer A, Flegel WA. RHD positive haplotypes in D negative Europeans. BMC Genet. 2001;2:10. doi: 10.1186/1471-2156-2-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krog GR, Clausen FB, Berkowicz A, Jorgensen L, Rieneck K, Nielsen LK, Dziegiel MH. Is current serologie RhD typing of blood donors sufficient for avoiding immunization of recipients? Transfusion. 2011;51:2278–2285. doi: 10.1111/j.1537-2995.2011.03156.x. [DOI] [PubMed] [Google Scholar]
- 10.Frohn C, Dümbgen L, Brand JM, Görg S, Luhm J, Kirchner H. Probability of anti-D development in D-patients receiving D+ RBCs. Transfusion. 2003;43:893–898. doi: 10.1046/j.1537-2995.2003.00394.x. [DOI] [PubMed] [Google Scholar]
- 11.Yazer MH, Triulzi DJ. Detection of anti-D in D− recipients transfused with D+ red blood cells. Transfusion. 2007;47:2197–2201. doi: 10.1111/j.1537-2995.2007.01446.x. [DOI] [PubMed] [Google Scholar]
- 12.Gonzalez-Porras JR, Graciani IF, Perez-Simon JA, Martin-Sanchez J, Encinas C, Conde MP, Nieto MJ, Corral M. Prospective evaluation of a transfusion policy of D+ red blood cells into D− patients. Transfusion. 2008;48:1318–1324. doi: 10.1111/j.1537-2995.2008.01700.x. [DOI] [PubMed] [Google Scholar]
- 13.Sachs L. Statistische Methoden. 5th ed. Berlin: Springer; 1978. [Google Scholar]
- 14.Schmidt PJ, Morrison EG, Shohl J. The antigenicity of the Rho (Du) blood factor. Blood. 1962;20:196–202. [PubMed] [Google Scholar]
- 15.St-Louis R, et al. DEL blood donors alloimmunised patients: the Canadian experience. Vox Sang. 2012;103(suppl 1):14. (abstract 3C-S8–03) [Google Scholar]
- 16.Perry GH, Xue Y, Smith RS, Meyer WK, Caliskan M, Yanez-Cuna O, Lee AS, Gutierrez-Arcelus M, Ober C, Hollox EJ, Tyler-Smith C, Lee C. Evolutionary genetics of the human Rh blood group system. Hum Genet. 2012;131:1205–1216. doi: 10.1007/s00439-012-1147-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kumpel B. Are weak D red blood cells really immu-nogenic? Transfusion. 2006;46:1061–1062. doi: 10.1111/j.1537-2995.2006.00847.x. [DOI] [PubMed] [Google Scholar]
- 18.Schmidt PJ. Are weak D red blood cells really immu-nogenic? Transfusion. 2006;46:2029–2030. doi: 10.1111/j.1537-2995.2006.01012.x. [DOI] [PubMed] [Google Scholar]
- 19.Gorick B, McDougall DCJ, Ouwehand WH, Overbeeke MAM, Tippett P, Hughes-Jones NC, van Rhenen DJ. Quantitation of D sites on selected weak D and partial D’ red cells. Vox Sang. 1993;65:136–140. doi: 10.1111/j.1423-0410.1993.tb02130.x. [DOI] [PubMed] [Google Scholar]
- 20.van Rhenen DJ, Overbeeke MA, Dudok de Wit C, Engelfriet CP. Revised definition of the concept rhesus-negative blood donor; practical experiences (in Dutch) Ned Tijdschr Geneeskd. 1990;134:1005–1007. [PubMed] [Google Scholar]
- 21.Flegel WA, Khull SR, Wagner FF. Primary anti-D immunization by weak D type 2 RBCs. Transfusion. 2000;40:428–434. doi: 10.1046/j.1537-2995.2000.40040428.x. [DOI] [PubMed] [Google Scholar]
- 22.Mota M, Fonseca NL, Rodrigues A, Kutner JM, Cas-tilho L. Anti-D alloimmunization by weak D type 1 red blood cells with a very low antigen density. Vox Sang. 2005;88:130–135. doi: 10.1111/j.1423-0410.2005.00604.x. [DOI] [PubMed] [Google Scholar]
- 23.Gassner C, Doescher A, Drnovsek TD, Rozman P, Eicher NI, Legier TJ, Lukin S, Garritsen H, Kleinrath T, Egger B, Ehling R, Körmöczi GF, Kilga-Nogler S, Schoenitzer D, Petershofen EK. Presence of RHD in serologically D-, C/E+ individuals: a European mul-ticenter study. Transfusion. 2005;45:527–538. doi: 10.1111/j.0041-1132.2004.04211.x. [DOI] [PubMed] [Google Scholar]
- 24.Wagner T, Körmöczi GF, Buchta C, Vadon M, Lanzer G, Mayr WR, Legier TJ. Anti-D immunization by DEL red blood cells. Transfusion. 2005;45:520–526. doi: 10.1111/j.0041-1132.2005.04256.x. [DOI] [PubMed] [Google Scholar]
- 25.Yasuda H, Ohto H, Sakuma S, Ishikawa Y. Secondary anti-D immunization by Del red blood cells. Transfusion. 2005;45:1581–1584. doi: 10.1111/j.1537-2995.2005.00579.x. [DOI] [PubMed] [Google Scholar]
- 26.Shao CP, Maas JH, Su YQ, Köhler M, Legier TJ. Molecular background of Rh D-positive, D-negative, D(el) and weak D phenotypes in Chinese. Vox Sang. 2002;83:156–161. doi: 10.1046/j.1423-0410.2002.00192.x. [DOI] [PubMed] [Google Scholar]
- 27.Kim KH, Kim KE, Woo KS, Han JY, Kim JM, Park KU. Primary anti-D immunization by DEL red blood cells. Korean J Lab Med. 2009;29:361–365. doi: 10.3343/kjlm.2009.29.4.361. [DOI] [PubMed] [Google Scholar]
- 28.Flegel WA, von Zabern I, Wagner FF. Six years experience performing RHD genotyping to confirm D-red blood cell units in Germany for preventing anti-D immunizations. Transfusion. 2009;49:465–471. doi: 10.1111/j.1537-2995.2008.01975.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Körmöczi GF, Förstemann E, Gabriel C, Mayr WR, Schönitzer D, Gassner C. Novel weak D types 31 and 32: adsorption-elution-supported D antigen analysis and comparison to prevalent weak D types. Transfusion. 2005;45:1574–1580. doi: 10.1111/j.1537-2995.2005.00580.x. [DOI] [PubMed] [Google Scholar]
- 30.Polin H, Danzer M, Gaszner W, Broda D, St-Louis M, Proli J, Hofer K, Gabriel C. Identification of RHD alleles with the potential of anti-D immunization among seemingly D− blood donors in Upper Austria. Transfusion. 2009;49:676–681. doi: 10.1111/j.1537-2995.2008.02046.x. [DOI] [PubMed] [Google Scholar]
- 31.Wagner FF, Mardt I, Bittner R, Döscher A. Single adsorption/elution with anti-D may be insufficient to determine the D antigen status of very weak DEL al-leles Transfusion. 2012;52(suppl S3):35A. (abstract S58–030I) [Google Scholar]
- 32.Wagner FF, Flegel WA.The RhesusBase Version 2. www.uni-uIm.de/∼fwagner/RH/RB2 (last accessed April 26,2013).
- 33.Li Q, Ye LY, Guo ZH, Qian M, Zhu ZY. Study on the molecular background of Del phenotype in Chinese population (in Chinese) Zhonghua Yi Xue Yi ChuanXue Za Zhi. 2006;23:486–491. [PubMed] [Google Scholar]
- 34.Müller SP, Bartels I, Stein W, Emons G, Guten-sohn K, Köhler M, Legier TJ. The determination of the fetal D status from maternal plasma for decision making on Rh prophylaxis is feasible. Transfusion. 2008;48:2292–2301. doi: 10.1111/j.1537-2995.2008.01843.x. [DOI] [PubMed] [Google Scholar]
- 35.Wagner FF, Mardt I, Bittner R, Döscher A. RHD PCR of blood donors in Northern Germany: use of adsorption /elution to determine D antigen status. Vox Sang. 2012;103(suppl 1):15. (abstract 3C-S8–04) [Google Scholar]
- 36.Singleton BK, Green CA, Kimura K, Minami A, Okubo Y, Daniels GL. Two new RHD mutations associated with the Del phenotype. Transfus Clin Biol. 2001;8(suppl 1):9s. (abstract) [Google Scholar]
- 37.Li Q, Hou L, Guo ZH, Ye LY, Yue DQ, Zhu ZY. Molecular basis of the RHD gene in blood donors with DEL phenotypes in Shanghai. Vox Sang. 2009;97:139–146. doi: 10.1111/j.1423-0410.2009.01181.x. [DOI] [PubMed] [Google Scholar]
- 38.von Zabern I, Flegel WA. IVS5–38del4 deletion in the RHD gene does not cause a DEL phenotype: relevance for RHD alleles including DFR-3. Transfusion. 2007;47:1552–1555. doi: 10.1111/j.1537-2995.2007.01353.x. [DOI] [PubMed] [Google Scholar]
- 39.Polin H, Danzer M, Hofer K, Gassner W, Gabriel C. Effective molecular RHD typing strategy for blood donations. Transfusion. 2007;47:1350–1355. doi: 10.1111/j.1537-2995.2007.01278.x. [DOI] [PubMed] [Google Scholar]
- 40.Kim JY, Kim SY, Kim CA, Yon GS, Park SS. Molecular characterization of D− Korean persons: development of a diagnostic strategy. Transfusion. 2005;45:345–352. doi: 10.1111/j.1537-2995.2005.04311.x. [DOI] [PubMed] [Google Scholar]
- 41.Esteban R, Monterò R, Flegel WA, Wagner FF, Subirana L, Parrà R, Ribera A,Nogués N. The D category VI type 4 allele is prevalent in the Spanish population. Transfusion. 2006;46:616–623. doi: 10.1111/j.1537-2995.2006.00762.x. [DOI] [PubMed] [Google Scholar]
- 42.Richard M, Perreault J, Constanzo-Yanez J, Kha-lifé S, St-Louis M. A new DEL variant caused by exon 8 deletion. Transfusion. 2007;47:852–857. doi: 10.1111/j.1537-2995.2007.01199.x. [DOI] [PubMed] [Google Scholar]
- 43.Luettringhaus TA, Cho D, Ryang DW, Flegel WA. An easy RHD genotyping strategy for D− East Asian persons applied to Korean blood donors. Transfusion. 2006;46:2128–2137. doi: 10.1111/j.1537-2995.2006.01042.x. [DOI] [PubMed] [Google Scholar]
- 44.Fichou Y, Chen JM, Le Maréchal C, Jamet D, Du-pont I, Chuteau C, Durousseau C, Loirat MJ, Bailly P, Férec C. Weak D caused by a founder deletion in the RHD gene. Transfusion. 2012;52:2348–2355. doi: 10.1111/j.1537-2995.2012.03606.x. [DOI] [PubMed] [Google Scholar]
- 45.Shao CP, Xiong W, Zhou YY. Multiple isoforms excluding normal RhD mRNA detected in Rh blood group Del phenotype with RHD 1227A allele. Transfus Apher Sci. 2006;34:145–152. doi: 10.1016/j.transci.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 46.Wagner FF, Frohmajer A, Ladewig B, Eicher NI, Lon-icer CB, Müller TH, Siegel MH, Flegel WA. Weak D alleles express distinct phenotypes. Blood. 2000;95:2699–2708. [PubMed] [Google Scholar]
- 47.Christiansen M, Sørensen BS, Grunnet N. RHD positive among C/E+ and D− blood donors in Denmark. Transfusion. 2010;50:1460–1464. doi: 10.1111/j.1537-2995.2010.02611.x. [DOI] [PubMed] [Google Scholar]
- 48.Mota M, Dezan M, Valgueiro MC, Sakashita AM, Kutner JM, Castilho L. RHD allelic identification among D-Brazilian blood donors as a routine test using pools of DNA. J Clin Lab Anal. 2012;26:104–108. doi: 10.1002/jcla.21489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wagner FF, Ladewig B, Flegel WA. The RHCE allele ceRT: D epitope 6 expression does not require D-specific amino acids. Transfusion. 2003;43:1248–1254. doi: 10.1046/j.1537-2995.2003.00495.x. [DOI] [PubMed] [Google Scholar]
- 50.Chen Q, Hustinx H, Flegel WA. The RHCE allele ceSL: the second example for D antigen expression without D-specific amino acids. Transfusion. 2006;46:766–772. doi: 10.1111/j.1537-2995.2006.00795.x. [DOI] [PubMed] [Google Scholar]
- 51.Flegel WA, Wagner FF, Chen Q, Schlanser G, Frame T, Westhoff CM, Moulds MK. The RHCE allele ceCF: the molecular basis of Crawford (RH43) Transfusion. 2006;46:1334–1342. doi: 10.1111/j.1537-2995.2006.00901.x. [DOI] [PubMed] [Google Scholar]
- 52.Cruz BR, Chiba AK, Moritz E, Bordin JO. RHD alleles in Brazilian blood donors with weak D or D-negative phenotypes. Transfus Med. 2012;22:84–89. doi: 10.1111/j.1365-3148.2011.01129.x. [DOI] [PubMed] [Google Scholar]
- 53.Chen Q, Li M, Li M, Lu XS, LÜ R, Sun J, Liu Z. Molecular basis of weak D and DEL in Han population in Anhui Province, China. Chin Med J (Engl) 2012;125:3251–3255. [PubMed] [Google Scholar]
- 54.Londero D, Fiorino M, Miotti V, de Angelis V. Molecular RH blood group typing of serologically D-/CE+ donors: the use of a polymerase chain reaction-sequence-specific primer test kit with pooled samples. Immunohematology. 2011;27:25–28. [PubMed] [Google Scholar]
- 55.Srijinda S, Suwanasophon C, Visawapoka U, Pong-savee M. RhC phenotyping, adsorption/elution test, and SSP-PCR: the combined test for D-Elute phenotype screening in Thai RhD-negative blood donors. ISRN Hematol. 2012;2012:358316. doi: 10.5402/2012/358316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Moussa H, Tsochandaridis M, Chakroun T, Jridi S, Abdelneji B, Hmida S, Silvy M, Bailly P, Gabert J, Levy-Mozziconacci A, Jemni-Yacoub S. Molecular background of D-negative phenotype in the Tunisian population. Transfus Med. 2012;22:192–198. doi: 10.1111/j.1365-3148.2012.01142.x. [DOI] [PubMed] [Google Scholar]
- 57.Reid ME. Transfusion in the age of molecular diagnostics. Hematology Am Soc Hematol Educ Program. 2009;171–177 doi: 10.1182/asheducation-2009.1.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Westhoff CM. Molecular genotyping for RHD: what (not) to do? Transfusion. 2007;47:1337–1339. doi: 10.1111/j.1537-2995.2007.01401.x. [DOI] [PubMed] [Google Scholar]