Abstract
The vaccinia virus A43R open reading frame encodes a 168-aminoacid protein with a predicted N-terminal signal sequence and a C-terminal transmembrane domain. Although A43R is conserved in all sequenced members of the orthopoxvirus genus, no non-orthopoxvirus homolog or functional motif was recognized. Biochemical and confocal microscopic studies indicated that A43 is expressed at late times following viral DNA synthesis and is a type-1 membrane protein with two N-linked oligosaccharide chains. A43 was present in Golgi and plasma membranes but only a trace amount was detected in sucrose gradient purified mature virions and none in CsCl gradient purified enveloped virions. Prevention of A43R expression had no effect on plaque size or virus replication in cell culture and little effect on virulence after mouse intranasal infection. Although the A43 mutant produced significantly smaller lesions in skin of mice than the control, the amounts of virus recovered from the lesions were similar.
Keywords: poxvirus, virulence, virus-host interaction, pathogenesis
Introduction
The Poxviridae, a group of large, complex DNA viruses that replicate exclusively in the cytoplasm, are divided into the Chordopoxvirinae and Entomopoxvirinae subfamilies (Moss, 2007). The chordopoxviruses consist of eight genera of which the orthopoxviruses are best known because of two members: variola virus, the causative agent of smallpox, and vaccinia virus (VACV), used to eradicate smallpox by prophylactic immunization. VACV, the prototype poxvirus, encodes ~ 200 genes nearly half of which are present in all chordopoxviruses (Upton et al., 2003). The evolutionarily conserved genes tend to be centrally located within the genome and are mostly involved in essential replication functions, while the genus- and species-specific genes are nearer the ends of the genome and typically involved in host interactions. Proteins that are dispensable for poxvirus replication in cell culture can have a wide variety of roles that directly or indirectly affect virulence by enabling virus spread or resisting immune defenses (Fallon and Alcami, 2006; McFadden, 2005). Studies of such genes can contribute to our understanding of cell biology and immunology and are important for development of vaccines and antivirals.
Since not all VACV genes have been characterized, it seems likely that additional proteins involved in host interactions will be discovered. The A43R open reading frame (ORF) is a candidate as it is embedded within the variable region of the genome. For example, the A41L ORF encodes a chemokine binding protein (Bahar et al., 2008) and the protein encoded by A46R inhibits TLR signaling (Stack et al., 2005). In this study we provided the initial characterization of A43, the product of the A43R ORF, which was expressed after viral DNA replication as a glycosylated protein that associated with Golgi and plasma membranes but was not appreciably incorporated into virus particles. The gene was found to be dispensable for VACV replication in cell culture but caused smaller than normal intradermal lesions in mice.
Results
A43R is conserved among orthopoxviruses
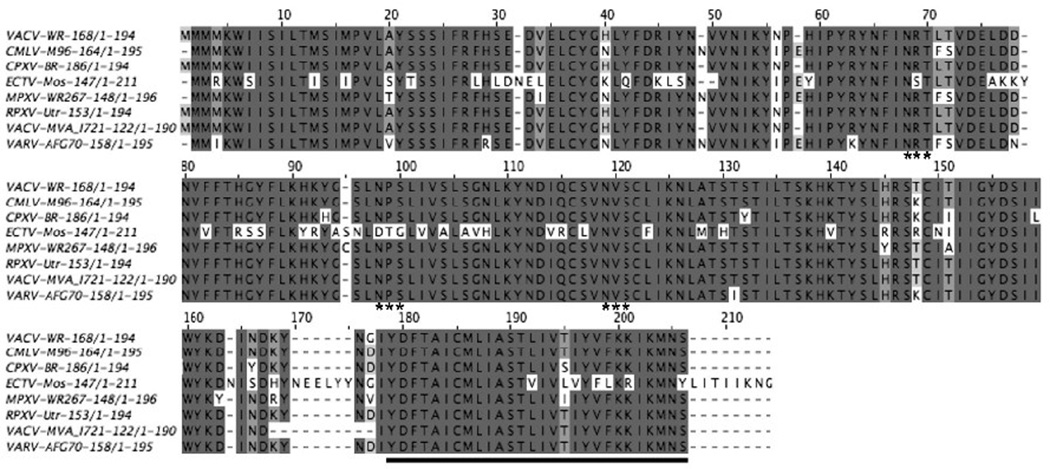
The A43R ORF (VACV WR168) is predicted to encode a 194-amino acid protein with N- and C-terminal hydrophobic domains. The SignalP program (Bendtsen et al., 2004) predicted that the N-terminal hydrophobic region is a signal sequence with cleavage occurring between S22 and S23 (Fig. 1). A43R is highly conserved within all orthopoxviruses; the sequence identity is >94% except for ectromelia virus which has a 78% identity (Fig. 1). However, there are no recognized homologs in any other poxvirus genus, nor are there non-poxvirus homologs or functional motifs to help predict the function of A43.
FIG. 1.
Multiple sequence alignment of A43 orthologs. Jalview (Waterhouse et al., 2009) was used to construct a multiple sequence alignment of A43 orthologs from orthopoxvirus species. The ORFs are indicated after the species and strain names. Abbreviations: VACV-WR, VACV strain WR, CMLV-M96, camelpox virus strain M96; CPXV-BR, cowpox virus strain Brighton; ECTV-Mos, ectromelia virus strain Moscow; MPXV-WR267, monkeypox virus Walter Reed 267; RPXV, rabbitpox virus strain Utrecht; VACV-MVA, VACV strain Modified VACV Ankara; VARV-AFG70, variola virus strain Afghanistan 1970. The asterisks indicate the Asn-X-Ser/Thr potential glycosylation sites. The predicted transmembrane domain is underlined. Note that the numbering includes spaces for alignment so does not correspond precisely to the VACV sequence.
A43 is a glycosylated protein expressed at the late stage of VACV replication
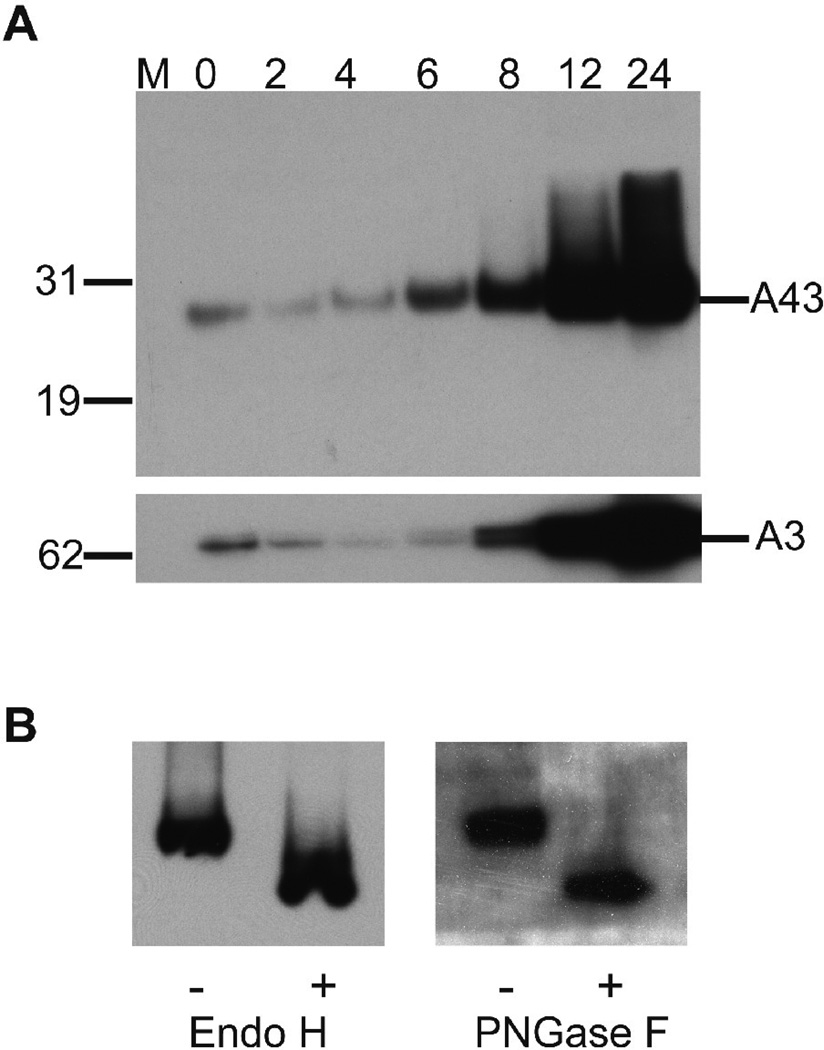
A recombinant virus, in which a V5 epitope tag was added to the C-terminus of A43, was constructed to assist in protein characterization. The region upstream containing the promoter sequence was unaltered so as not to perturb expression. The growth kinetics and plaque phenotype of vA43V5 were similar to that of the parental virus (not shown). The sequence TAAATG, present at the start of the A43R ORF, is a characteristic of late promoters (Davison and Moss, 1989). To experimentally determine the time of synthesis of A43V5, BS-C-1 cells were infected with the recombinant virus and whole cell lysates were prepared at intervals and analyzed by SDS-polyacrylamide gel electrophoresis (PAGE) and Western blotting. A band visualized by binding of the anti-V5 MAb migrated more slowly than the predicted 23-kDa mass of A43V5 (Fig. 2A). The band was detected at 0 time, indicating its presence in the unpurified virus inoculum, but did not increase significantly until 6 h and then continued to increase up to 24 h. This pattern was similar to that of the well-characterized late A3 protein (Fig. 2A). Neither A43 nor A3 were synthesized in the presence of the DNA replication inhibitor AraC (data not shown), which is indicative of VACV proteins expressed at the late stage of infection.
FIG. 2.
A43 synthesis and glycosylation. (A) Western blot analysis of A43 expression kinetics. BS-C-1 cells were infected at a multiplicity of 10 PFU per cell with vA43V5. At 0, 2, 4, 8 12 and 24 h post infection, whole cell lysates were analyzed by SDS-PAGE and Western blotting with an antibody to the V5 epitope tag. The blot was stripped and reprobed with an antibody to the VACV late protein A3. Position and mass in kDA of marker proteins are shown on the left. (B) vA43V5 infected cell lysates were treated with (+) or without (−) Endo H or PNGase F and analyzed by SDS-Page and Western blotting with an antibody to the V5 epitope tag.
Inspection of the predicted amino acid sequence of A43 revealed three potential N-glycosylation sites that could account for the relatively slow electrophoretic migration of A43. To determine the state of glycosylation of A43, a whole cell lysate from vA43V5 infected cells was divided into portions that were untreated or treated with Peptide: N-Glycosidase F (PNGase F) or Endoglycosidase H (Endo H). PNGase F is capable of removing all types of N-linked oligosaccharides, whereas Endo H removes only high mannose and some hybrid types of oligosaccharides. Both glycosidases caused an increase in the mobility of A43V5 as determined by SDS-PAGE and Western blotting (Fig. 2B), consistent with N-glycosylation of the protein.
A43 has two N-linked glycosylation sites
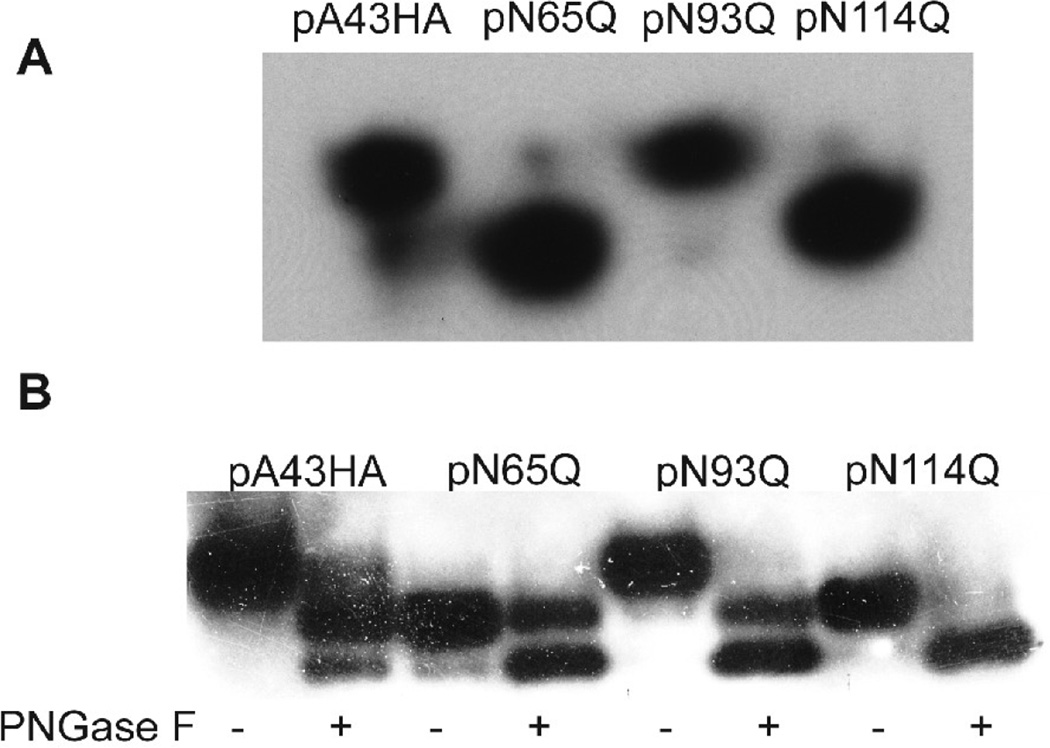
To determine the number and sites of glycosylation, plasmids (pA43HAN65Q, pA43HAN93Q, pA43HAN114Q) were constructed, each with a mutation in one of the three predicted N-linked glycosylation sites of A43 (Fig. 1). Each plasmid expressed A43 under its natural promoter and with a C-terminal HA epitope tag. An additional plasmid (pA43HA) was constructed with none of the glycosylation sites mutated as a control. To analyze the expression of the mutated forms of A43, BSC-1 cells were infected with vΔA43GFP (a recombinant virus with a deletion of A43 to be described below) and then transfected with the plasmids. The cells were lysed and the extracts were analyzed by SDS-PAGE and Western blotting using antibody to the HA tag. If each of the N-X-S/T consensus sequences were glycosylated, then we would expect to see an increase in the mobility of each protein compared to A43HA. However, we found that only two of the three mutated proteins, A43HAN65Q and A43HAN114Q, migrated faster than the unmutated A43HA (Fig. 3A). To confirm and extend this finding, we partially digested the infected/transfected cell lysate with PNGase F and examined the resulting protein migration patterns. Partial digestion of A43HA and A43HAN93Q resulted in two new bands that migrated more rapidly than the uncleaved protein (Fig. 3B), consistent with two N-linked oligosaccharides. Furthermore, the slower of the two new bands migrated in the same position as the undigested A43HAN65Q and A43HAN114Q, indicating removal of a single oligosaccharide. Digestion of A43HAN65Q and A43HAN114Q produced a band that migrated with the faster of the bands produced by digestion of A43HA and A43HAN93Q (Fig. 3B). Taken together, these results proved that A43 was N-glycosylated at positions N65 and N114. The absence of glycosylation at N93 is consistent with the presence of the neighboring proline (Bause, 1983).
FIG. 3.
A43 has two N-linked oligosaccharides. (A) Expression of wild type and mutated A43HA. BS-C-1 cells were infected with vΔA43GFP and transfected with one of four plasmids expressing wild type A43HA or A43HA with mutations N65Q, N93Q, or N114Q. Cell extracts were analyzed by SDS-PAGE and Western blotting with anti-HA antibody. (B) Partial (+) or mock (−) PNGase digestion of cell extracts at 24 h infection with vΔA43GFP and transfection with pA43HA, pN65Q, pN93Q, or pN114Q. The digests were analyzed by Western blotting with antibody to the HA epitope tag.
A43 associates with cellular membranes
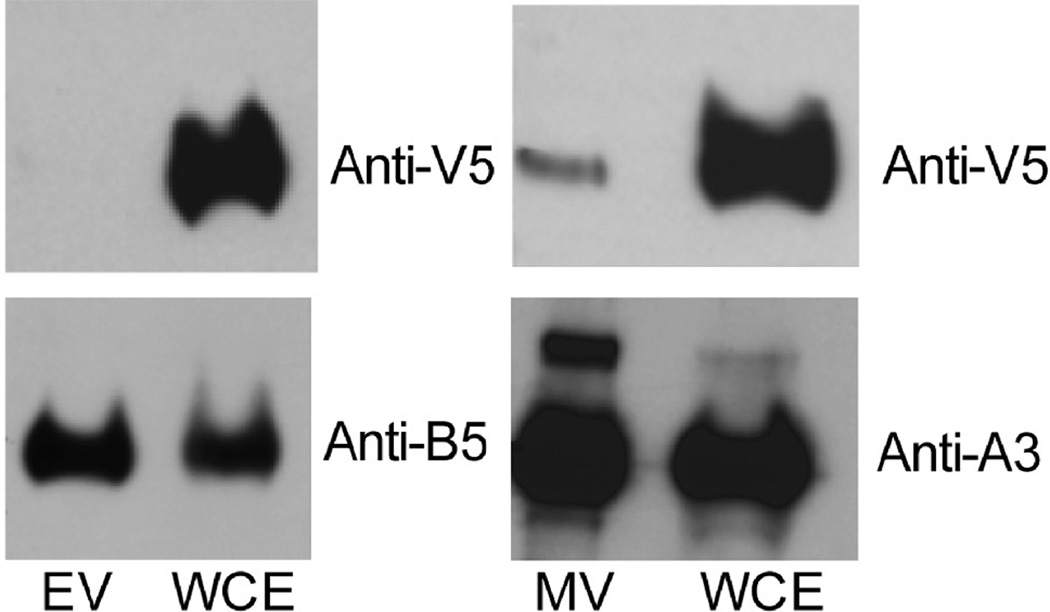
The recombinant virus vA43V5 was used to determine whether A43 is a component of the MV or EV membrane. After RK-13 cells were infected with vA43V5, EVs were purified from the medium on a CsCl density gradient and the MVs were purified from the cell lysate by two sucrose cushions followed by a sucrose density gradient. Purified virions as well as whole cell extracts were analyzed by SDS-PAGE and immunoblotting. A43 was readily detected in whole cell lysate but not in EVs and just barely in MVs (Fig. 4). Both membranes were stripped and re-probed with either the EV protein B5 or the MV core protein A3. A3 and B5 were detected at similar levels in the cell extract and purified MVs and EVs, respectively (Fig. 4). Comparison of the relative amounts of A43, A3 and B5 in extracts and purified virions, suggested that the slight amount of A43 in MVs was adventitious and likely due to some membrane contamination that is invariably present in sucrose gradient purified virions (BM, unpublished). This interpretation is supported by the absence of A43 in CsCl gradient purified EVs. In addition, it would be unprecedented for an MV membrane protein to be N-glycosylated.
FIG. 4.
Analysis of A43 in EV, MV and whole cell extract. BS-C-1 cells were infected with vA43V5; MVs were purified from cell lysates and EVs from the medium. Antibodies to the V5 epitope on A43V5, the EV protein B5, and the MV protein A3 were used to probe Western blots of the whole cell extract (WCE), purified EVs and purified MVs as indicated.
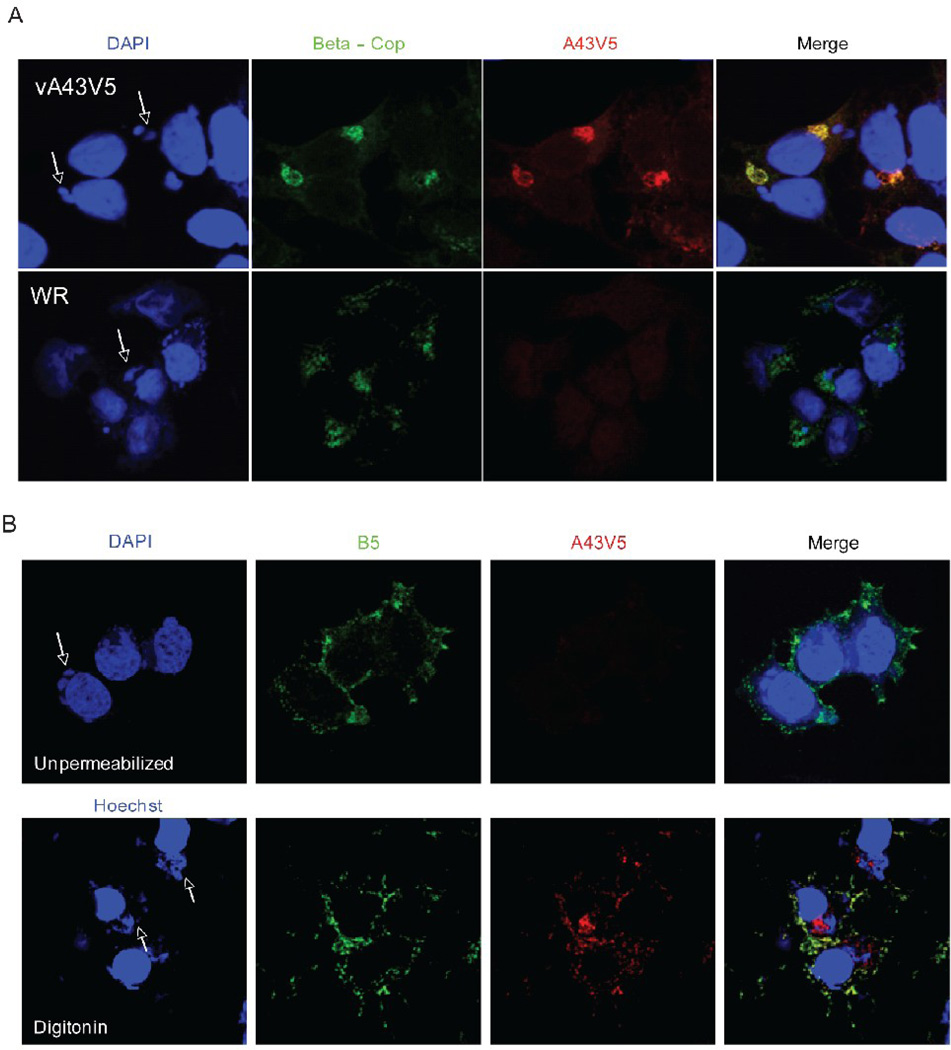
Further experiments were performed to determine whether A43 localized in virus factories or cellular membranes. After infection with vA43V5, the cells were fixed, permeabilized with Triton X-100, and stained with an anti-V5 antibody followed by a fluorescently labeled secondary antibody. 4'–6'-diamidino-2-phenylindole (DAPI) was used to detect DNA in nuclei and cytoplasmic factories, the sites of viral DNA replication and virion assembly. The A43V5 protein predominantly colocalized with β-cop, a specific Golgi complex protein (Fig. 5A). There was negligible background staining of cells infected with VACV WR, which lacks the V5 epitope tag (Fig. 5A). The lack of factory staining was consistent with the absence of A43 in virions. Co-localization with a Golgi membrane marker also occurred when uninfected cells were transfected with a plasmid expressing A43V5 under the cytomegalovirus promoter (data not shown), indicating that the protein has an intrinsic ability to enter the secretory pathway.
FIG. 5.
Intracellular localization and topology of A43. (A) Intracellular localization. HeLa cells were infected with VACV (WR) or vA43V5 at a multiplicity of 0.5 PFU per cell. At 24 h after infection, cells were fixed, permeabilized and stained with antibodies to V5 and β-Cop followed by fluorescently labeled secondary antibodies (red and green respectively). DNA was stained with DAPI (blue). Confocal microscopy images are shown. Arrows point to cytoplasmic virus factories. (B) Topology. Hela cells infected with vA43V5 were fixed and left unpermeabilized (top panels) or permeabilized with digitonin (bottom panels). Cells were then stained with an antibody to the EV protein B5 (green) as well as with an antibody to the V5 epitope tag (red) followed by fluorescently labeled secondary antibodies. DAPI or Hochest were used to stain DNA (blue). Confocal microscopy images are shown. Arrows point to cytoplasmic virus factories.
A43 is a type-1 transmembrane protein
Triton X-100 permeabilizes all cell membranes, allowing antibody binding to both cytoplasmic and luminal domains of proteins. In contrast, digitonin selectively permeabilizes the plasma membrane, making it useful for determining protein topology. After infection with vA43V5, the epitope tag was detected in digitonin-treated cells but not in untreated cells (Fig. 5B). Consequently, A43 must be a type-1 transmembrane protein since the C-terminus containing the V5 tag is cytoplasmic. Therefore, the segment from amino acids 26 to 164 must be luminal, consistent with our finding that A43 is glycosylated at N65 and N104. As a control, the cells infected with vA43V were also stained with an antibody to the luminal domain of B5, a viral type-1 membrane protein that is present at the cell surface and the Golgi complex (Isaacs et al., 1992). As predicted, B5 was detected at the cell surface of intact and digitonin-permeabilized cells (Fig 5B).
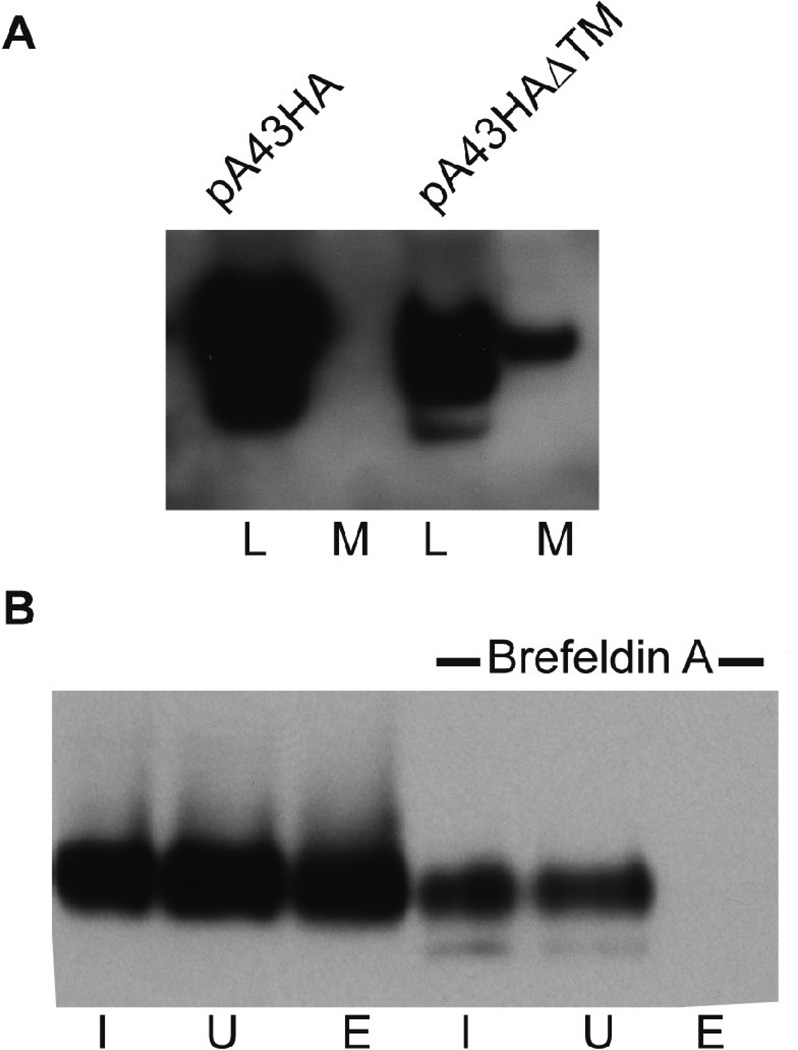
In addition to detecting A43V5 associated with the Golgi apparatus, there was some staining of the plasma membrane, which partially overlapped that of B5 in digitonin-treated cells (Fig. 5B). To confirm the trafficking of A43 through the secretory pathway, we deleted the putative transmembrane domain of A43 in the plasmid pA43HA to form pA43HAΔTM. BSC-1 cells were then infected with vΔA43GFP and transfected with the pA43HA or pA43HAΔTM. After 24 h, the medium was collected and concentrated. The cells were also harvested and lysed. After SDS-PAGE and Western blotting, A43HA was detected only in the cell lysate, whereas A43HAΔTM was detected in the lysate and as a secreted protein in the medium (Fig. 6A).
FIG. 6.
Intracellular trafficking of A43. (A) BS-C-1 cells were infected with vΔA43GFP followed by transfection with plasmids expressing the full length HA-tagged A43 (pA43HA) or a truncated version missing the transmembrane domain (pA43HAΔTM). After 24 h, the proteins in the cell lysates (L) or concentrated medium (M) were analyzed by Western blotting with an antibody to the HA tag. (B) HeLa cells were infected with vA43V5 and brefeldin A was added to one culture after 4 h. At 18 h post infection, the cells were allowed to react with a membrane-impermeable biotin reagent. The biotinylated proteins were affinity purified on NeutrAvidin gel beads. Western blotting with V5 antibody was performed on the input (I), unbound (U) and bound and eluted (E) proteins.
To verify that the intact A43 traffics to the plasma membrane, cells infected with vA43V5 in the presence or absence of brefeldin A were surface labeled with biotin. Brefeldin A was used as a control since it inhibits the transport of proteins from the ER to the Golgi complex. We affinity purified the biotinylated proteins and found that A43V5 was captured exclusively from cells that were not treated with brefeldin A (Fig. 6B). Based on all of the above data, we concluded that A43 transits through the endoplasmic reticulum where N-terminal glycosylation occurs and continues to the Golgi complex where it is concentrated and then to the plasma membrane where the long N-terminal domain is extracellular.
A43 is dispensable for VACV replication and cell-to-cell spread in cultured cells
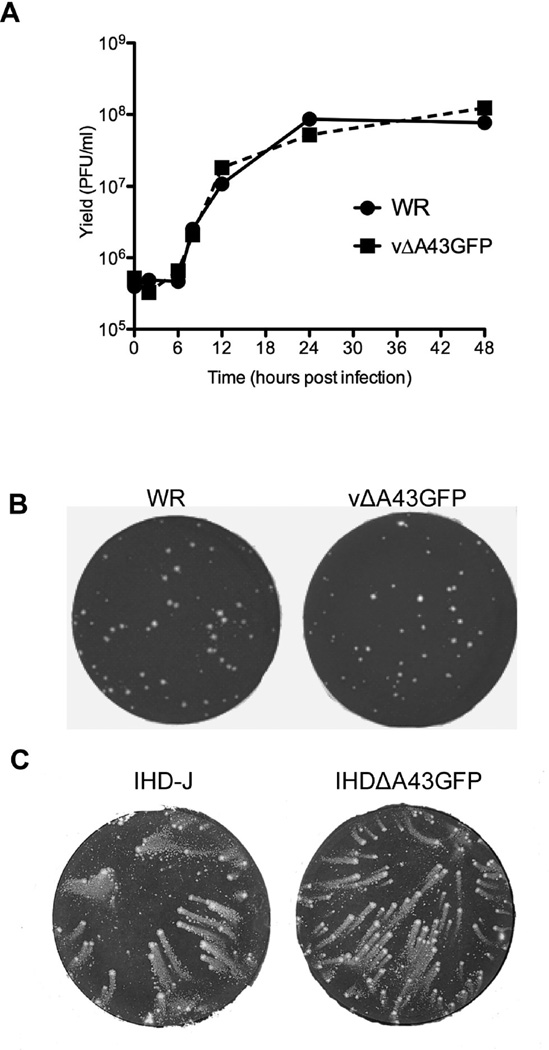
An A43R null mutant was constructed in order to investigate the function of the protein in the VACV life cycle. A43R was deleted from VACV by homologous recombination with a plasmid containing the GFP ORF regulated by the VACV P11 promoter flanked by A44R and A42L sequences. Recombinant virus vΔA43GFP was detected by fluorescent microscopy and clonally purified by repeated plaque isolation, indicating that A43R was non-essential. The deletion of A43R was confirmed by PCR and sequence analysis. BS-C-1 cells were infected with the parental VACV or the vΔA43GFP and the virus yields determined at intervals. The growth curves were virtually superimposable (Fig. 7A) and plaques formed by wild type and vΔA43GFP viruses were similar in BS-C-1 (Fig. 7B) and several other cell lines, namely BHK, CV-1, HeLa, HuTK–, RK13, A549 cells and primary human epidermal keratinocytes (data not shown).
FIG. 7.
Replication of A43R deletion mutant. (A) One-step growth curve. BS-C-1 cells were infected at a multiplicity of 10 PFU/cell with vΔA43GFP and VACV WR. Virus yields were determined at the indicated times post infection by plaque assay. (B) Formation of vΔA43GFP and VACV WR plaques. BS-C-1 cells were infected with vΔA43GFP and VACV WR using a methylcellulose overlay. Cells were fixed and stained with crystal violet at 48 h after infection. (C) Comet formation. BS-C-1 cells were infected with VACV IHD-J or the A43 deletion mutant in the IHD-J background ΔA43GFP using a liquid overlay. After 48 h, the cells were stained as in panel A.
In order to determine whether A43 is important for release of EVs, a second deletion mutant was made in the VACV IHD-J strain background. IHD-J is known for the release of large numbers of EV that form comet-shaped plaques in liquid medium (Payne, 1980) due to a point mutation in the A34R gene (Blasco et al., 1993). As shown in Fig. 7C, deletion of A43R had no effect on comet formation.
Virulence of A43 null mutants in animal models
Since vΔA43GFP showed no apparent replication defect in cell culture, we considered that it might have a role in animal infections. Before initiating such experiments, we made two additional recombinant viruses. A control revertant virus vA43Rev was constructed by replacing the GFP ORF of vΔA43GFP with the wild type A43R ORF to be sure that there were no extraneous mutations that might affect animal studies. The second virus, vA43V5stop, was constructed by replacing the GFP ORF of vΔA43GFP with the A43RV5 ORF containing a point mutation at nucleotide 68 that caused an immediate stop codon. The absence of A43V5 expression was verified by Western blotting (not shown). The purpose of the latter mutant was to rule out any possibility that GFP expression regulated by the P11 promoter has a subtle effect on the virus fitness.
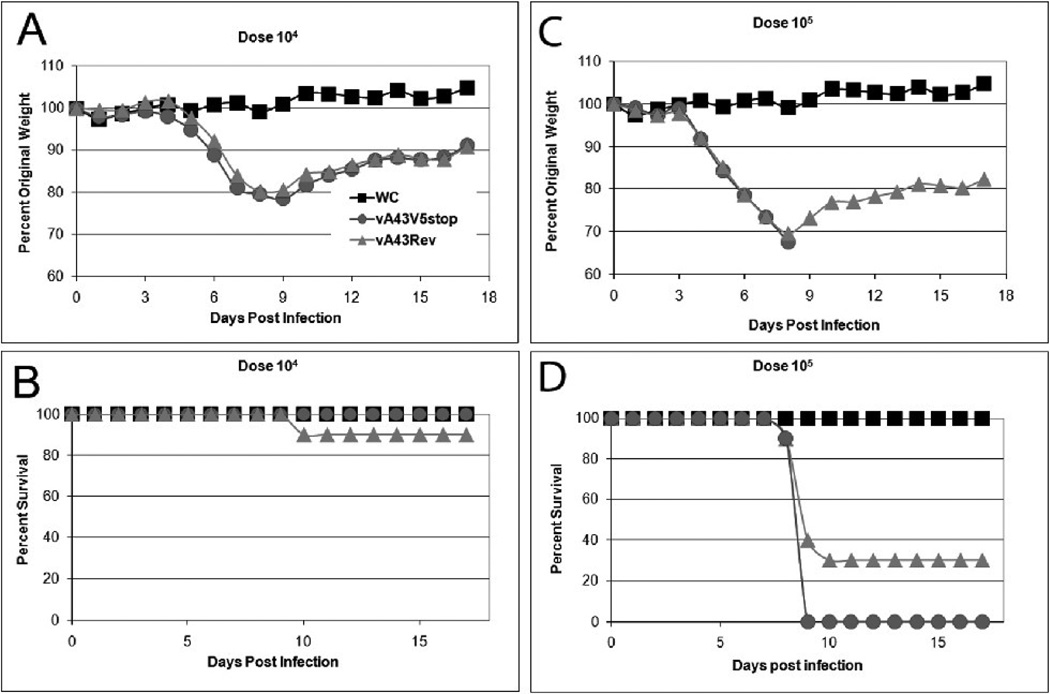
First, we evaluated the virulence of the mutant virus in an intranasal (IN) respiratory model (Turner, 1967). At a dose of 104 PFU, the weight loss curves of groups of ten mice inoculated with vA43Rev and vA43V5stop were superimposable (Fig. 8A). Except for one mouse that had been infected with vA43V5Rev, all of the mice survived (Fig. 8B). At a dose of 105 PFU, three of ten mice receiving vA43Rev survived and none of the mice receiving vA43V5stop survived (Fig. 8C,D). At a dose of 106 PFU, none of the mice in either group survived (not shown). Thus, A43V5 expression had no major effect on VACV virulence in this model.
FIG. 8.
Virulence of A43R null mutant after respiratory infection. Groups (n = 10) of 7-week-old mice remained untreated as weight controls (WC) or were inoculated via the IN route with 104 (A, B) or 105 PFU (C, D) of vA43V5Stop or vA43Rev. Mice were weighed daily for 2 weeks following challenge and were euthanized if they lost 30% of their initial body weight. Percent of original weight was plotted in panels A, C and percent survival in panels B, D.
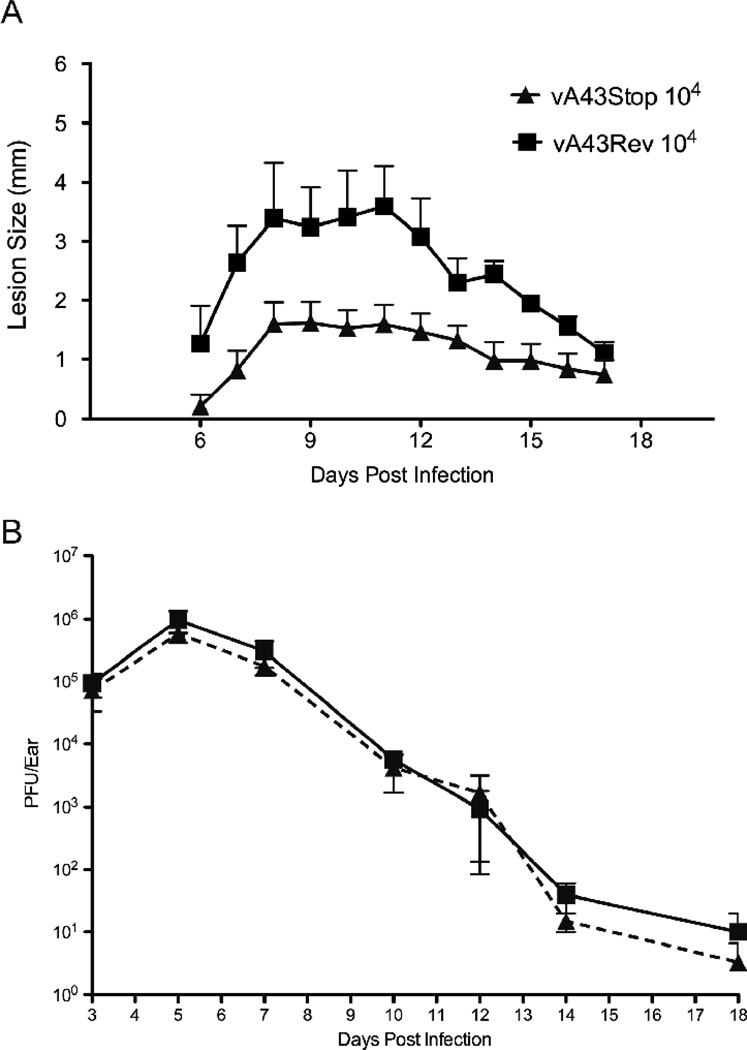
Next, we compared the virulence of vA43V5stop and vA43Rev in a mouse intradermal model (Tscharke and Smith, 1999). Groups of 5 BALB/c mice were inoculated intradermally in the ear pinna with 104 PFU of vA43V5stop or vA43Rev. Once lesions appeared, they were measured daily using digital calipers. The lesions reached a maximal size on day 8 and gradually decreased from day 12 (Fig. 9A). Mice that received vA43stop had significantly smaller lesions over the entire course of the infection (p = 0.001; Mann Whitney test) than the mice that received vA43Rev (Fig. 9A). Examination of hematoxylin and eosin stained sections of lesions from animals infected with the null mutant and revertant revealed proliferation of keratinocytes and infiltration of leukocytes in both cases (data not shown) similar to that previously described (Tscharke and Smith, 1999). The amounts of virus present in the lesions peaked at day 5 and gradually decreased, with no difference between the groups (Fig. 9B). In C57BL/6 mice the lesions produced by vA43V5stop were also smaller than those produced by vA43Rev (p = 0.005), although the lesions of both were larger than in BALB/c mice and the difference was less.
FIG. 9.
Intradermal replication and lesion formation by A43R null mutant. (A) Groups of five Balb/C mice were infected intradermally in the ear pinnae with 104 PFU of vA43Rev or vA43V5Stop. Lesion sizes were determined daily with digital calipers to the nearest 0.5 mm. Standard errors of the mean are shown. (B) Mice were sacrificed and virus titers were determined from three individual ears by plaque assay on BS-C-1 cells. Standard errors are shown.
Discussion
The major conclusions regarding A43 are that the protein (i) is conserved in all orthopoxviruses but not other poxvirus genera or non-poxviruses; (ii) contains no obvious functional motifs; (iii) is a late protein expressed after viral DNA replication; (iii) is not specifically associated with MVs or EVs; (iv) contains N-linked oligosaccharides at two sites; (iv) concentrates in Golgi membranes and traffics to plasma membrane; (v) exhibits a type 1 membrane topology; (vi) is dispensable for replication in tissue culture cells; (vii) null mutant retains virulence in mice by IN route; and (viii) null mutant produces smaller intradermal lesions in mice.
The conservation of A43 homologs in all orthopoxviruses, including the highly passaged MVA strain, led us to think that the protein has a replication function. The predicted late promoter and transmembrane domain, further suggested that A43 might be an EV membrane protein with a role in virus egress. Nevertheless, this was not the case as A43 was not associated with the EV and neither plaque size nor comet formation was altered. Indeed, with regard to tissue culture studies A43R null mutants were indistinguishable in every respect from the parental WR or IHD-J strain of VACV. In view of these results, we considered that A43 is likely involved in host interactions and that the null mutant would be attenuated in animal models. We were surprised therefore to find that the A43 null mutant was as virulent as the revertant in the mouse IN model. However, the null mutant made smaller lesions than the revertant in mouse ear pinnae when injected intradermally. The increased thickness of the lesion is due to proliferation of epidermal cells and leukocyte infiltration (Tscharke and Smith, 1999). We confirmed previous data that replication of VACV in the ear pinna reaches a peak on day 4–5 and the titer starts declining before the lesion reaches a maximal size several days later (Tscharke and Smith, 1999). Despite the difference in lesion size, the titers of mutant and revertant viruses were similar to each other at all time points. The similar titers may be related to the observation that the same amounts of virus are produced when the inocula vary over a 100-fold range (Tscharke and Smith, 1999). Unreduced titers and smaller lesions were also found with the B7R deletion mutant (Price et al., 2000).
Taken together the above characteristics differentiate A43R from other replication dispensable genes. The majority of such genes have early rather than late promoters, which would seem to be generally advantageous if the product interacts with the host. The five previously characterized replication dispensable late genes consist of A14.5L (Betakova et al., 2000), A39R (Comeau et al., 1998; Gardner et al., 2001), A55R (Beard et al., 2006), B7R (Price et al., 2000), I5L (Sood et al., 2008) and F3L (Froggatt et al., 2007). Except for A55R, absence of the gene resulted in attenuation in either the IN (A14.5, I5L) or intradermal (A39, B7R, F3L) mouse model. Loss of A55 actually increased skin lesion size. As pointed out above, the B7R and A43R null mutants exhibited decreased skin lesion size. The retention of virulence in the IN mode by the B7R null mutant was also similar to that of the A43R mutant (Price et al., 2000). There are, however, substantial differences between B7 and A43; B7 localizes in the lumen of the ER whereas A43 is a type 1 membrane protein in Golgi and plasma membranes. It had been suggested that B7 might sequester a cell protein in the ER, though that has not yet been demonstrated. Although A43 might also act by sequestering a cell protein in the Golgi apparatus, its presence on the plasma membrane suggests that it interacts with a cell protein, perhaps a cytokine. Efforts to find an A43 interacting protein would seem worthwhile for future studies.
Material and Methods
Cells and Viruses
BS-C-1 cells were propagated in minimum essential medium with Earle’s salts supplemented with 10% fetal bovine serum, 100 units/ml of penicillin, and 100 µg/ml of streptomycin. HeLa cells were propagated in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and antibiotics as described above. The VACV WR strain and recombinant viruses were prepared as previously described (Earl et al., 1998).
Plasmid and recombinant VACV construction
To construct the recombinant vΔA43GFP, the flanking regions of the A43R ORF were amplified from VACV strain WR genomic DNA template using oligonucleotides ct49 (5'- ATG CTA ATG TCA AGT TTA TTC CAA TAG ATG TCT TAT TAA AAA ACA TAT AT-3'), ct51 (5'-CAT AGA AAA AAA CAA AAT GAA ATT CTT AAA ATT GAC ACT ACA TAT GAA TAT-3'), ct52 (5'- GCA TGG ACG AGC TGT ACA AGT AAT TAT GCT ATG CTA TTA GAA ATG −3'), and ct54 (5'- CTA TTT AGA AAA CCA TCA CTA CTC AAC AAC TAT ACG TTG AAG ATA TCC AAC −3'). The ORF for GFP under the VACV promoter p11 was amplified using primers ct50 (5'- ATA TTC ATA TGT AGT GTC AAT TTT AAG AAT TTC ATT TTG TTT TTT TCT ATG 3') and ct53 (5’- CAT TTC TAA TAG CAT AGC ATA ATT ACT TGT ACA GCT CGT CCA TGC −3'). Primers ct50 and ct51 as well as ct52 and ct53 were designed to complement each other. The above products were used in a second recombinant PCR to yield a GFP ORF flanked by regions up- and downstream of A43R. The resulting PCR product was gel purified and ligated into pCR-BluntII-Topo (Invitrogen, Carlsbad, CA), resulting in pΔA43GFP. The endogenous A43R ORF was replaced with the GFP reporter gene by homologous recombination after transfection (Lipofectamine 2000; Invitrogen) of pΔA43GFP into VACV WR-infected cells. Recombinant viruses expressing GFP were detected with an inverted fluorescence microscope and isolated by three rounds of plaque purification. The correct site of recombination was verified by PCR analysis. A similar procedure was used to delete the A43R ORF from the IHD-J strain of VACV to form IHDΔA43GFP.
Recombinant vA43V5 was derived from vΔA43GFP. The primers ct61 (5'- AAT GCT AAT GTC AAG TTT ATT CCA ATA GAT GTC TTA TTA AAA ACA TAT AT −3'), ct62 (5'- ACC GAG GAG AGG GTT AGG GAT AGG CTT ACC AGA GTT CAT TTT TAT TTT TTT −3'), ct63 (5'- TAT CCC TAA CCC TCT CCT CGG TCT CGA TTC TAC GTA ATT ATG CTA TGC TAT TA −3'), and ct54 were used to amplify A43R from genomic DNA and incorporate a C-terminal V5 tag. Non-fluorescent plaques were picked and clonally purified by repeated isolations.
The A43 revertant virus (vA43Rev) was derived from vΔA43GFP. Primers ct61 and ct54 were used to generate a PCR product containing the A43R gene including 500 bp up- and downstream sequence. The PCR product was gel purified and ligated into pCR-BluntII-Topo (pA43Rev). Homologous recombination was used to replace the GFP gene with the A43R ORF after transfection of pA43Rev into cells infected with vΔA43GFP. Non-GFP-expressing plaques were picked and isolated by three rounds of plaque purification. The correct site of recombination was verified by PCR and sequence analysis. An A43V5− virus (vA43V5Stop) was also derived from vΔA43GFP. A stop codon was generated in the A43R sequence of pA43V5 by using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with PCR oligonucleotides containing the desired mutation. Primers ct79 (5'- CCG GTA TTG GCA TAC AGC TAA TCG ATT TTT AGA TTT CAT TCA GAG −3') and ct80 (5'- CTC TGA ATG AAA TCT AAA AAT CGA TTA GCT GTA TGC CAA TAC CGG −3') were used to change nucleotide 68, resulting in an immediate stop codon. Homologous recombination was used to replace the GFP marker gene with the A43V5Stop sequence after transfection of pA43V5Stop into vΔA43GFP-infected cells. Again, non-GFP-expressing plaques were picked and isolated by three rounds of plaque purification. The correct site of recombination was verified by PCR and sequence analysis.
Primers ct61 (5'- AAT GCT AAT GTC AAG TTT ATT CCA ATA GAT GTC TTA TTA AAA ACA TAT AT −3'), ct88 (5'- TAC CCA TAC GAT GTT CCA GAC TAC GCT TAA TTA TGC TAT GCT ATT AGA AAT −3'), ct89 (5'- TAA TTA AGC GTA GTC TGG AAC ATC GTA TGG GTA AGA GTT CAT TTT TAT TTT TTT −3'), and ct54 were used to amplify A43R from genomic DNA and incorporate a C-terminal HA tag. The PCR product was gel purified and ligated into pCR-BluntII-Topo (pA43HA). pN65Q, pN93Q, and pN114Q were generated in the A43R sequence of pA43HA by using a QuickChange site-directed mutagenesis kit (Stratagene, La Jolla, CA) with PCR oligonucleotides containing the desired mutation. Primers ct101 (5'- CCA TAT AGA TAT AAT TTT ATT CAG CGC ACG TTA ACC GTA GAT GAA C −3') and ct102 (5'- GTT CAT CTA CGG TTA ACG TGC GCT GAA TAA AAT TAT ATC TAT ATG G −3') were used for pN65Q. Primers ct103 (5'- CAC AAA TAT GGT TCA CTT CAG CCT AGT TTG ATT GTC TCA TTA TC −3') and ct104 (5'- GAT AAT GAG ACA ATC AAA CTA GGC TGA AGT GAA CCA TAT TTG TG −3') were used for pN93Q. Primers ct105 (5'- CAA TGC TCA GTA CAG GTA TCG TGT CTC ATT AAA AAT TTG GC −3') and ct106 (5'- GCC AAA TTT TTA ATG AGA CAC GAT ACC TGT ACT GAG CAT TG −3') wereused for pN114Q. pA43HAΔTM was constructed from WR genomic DNA using primers ct64 (5'-TAA TTA AAA TAA AAA GTA ATA TTC ATA TGT AGT GTC AAT TTT AAA TGA TGA −3') and ct90 (5'- TTA AGC GTA GTC TGG AAC ATC GTA TGG GTA ATT ATA CTT GTC ATT TAT ATC TTT AT −3') in a PCR reaction to amplify the A43 coding region under the natural promoter, delete the transmembrane domain and maintain the HA tag sequence. The PCR product was gel purified and ligated into pCR-BluntII-Topo (pA43HAΔTM). All constructed plasmid sequences and mutations were verified by sequence analysis.
Glyscosidase treatment of cell lysate
Cells (106) were washed and lysed in 75 µl of 50 mM Tris pH 7.0, 150 mM NaCl and 1% NP-40 detergent on ice for 20 min. Nuclei were removed by low speed centrifugation and SDS and β-mercaptoethanol were added to achieve final concentrations of 0.5% and 1%, respectively, and heated at 95°C for 10 min. PNGase (2,500 units; New England BioLabs, Ipswich, MA) was added and incubated at 37°C over night or for 1 h to achieve complete or partial digestion, respectively. Digestion with Endo H (New England BioLabs) was carried out similarly overnight. Samples were analyzed by SDS-Page and Western blotting.
EV purification
RK-13 cells were infected with either VACV or vA43V5. At 48 h post-infection, the medium was harvested and clarified by low-speed centrifugation for 20 minutes. EVs were pelleted by centrifugation at 14,000 x g for 1 h at 4°C. The EVs were gently resuspended and further purified by layering over a preformed CsCl gradient as described previously (Domi et al., 2008) and centrifuged at 32,000 rpm in an SW41 rotor for 4 h at 20°C. Fractions collected from the top of the tube were loaded on a 10% polyacrylamide gel and subjected to electrophoresis. The separated proteins were transferred to a polyvinylidene difluoride membrane and analyzed by Western blotting.
MV purification
Infected RK-13 cells were harvested and collected by low-speed centrifugation. Cells were then resuspended and disrupted by Dounce homogenization. Cell suspensions were clarified by low-speed centrifugation and MV purified by centrifugation through two 36% sucrose cushions and one 25–40% sucrose gradient.
SDS-PAGE
Cells were lysed in 0.2% NP-40, 10 mM Tris, pH 7.4, 10 mM CaCl2, 10 mM NaCl containing 8 µg/ml micrococcal nuclease (Worthington Biochemical Corp., Lakewood, NJ) at 4°C for 20 min. After addition of lithium dodecyl sulfate sample buffer and reducing agent (Invitrogen, Carlsbad, CA), cell lysates were heated to 70°C for 10 min. Equal volumes of lysate were analyzed by SDS-PAGE on a 10% bis-Tris-MES [2-(N-morpholino)ethanesulfonic acid]-SDS running buffer (Invitrogen).
Western blot analysis
Proteins separated by SDS-PAGE were electrophoretically transferred to polyvinylidene difluoride membrane (Invitrogen). Membranes were blocked in Tris-buffered saline with 5% nonfat dry milk and 0.05% Tween 20 and then incubated with antibodies for 1 h at room temperature or overnight at 4°C. Protein bands were visualized by chemiluminescence using West-Pico or Dura kits (Pierce Biotechnology Inc., Rockford, IL).
Confocal microscopy
HeLa cells, grown on glass cover slips in 12-well plates were infected at multiplicity of 0.5 PFU per cell. At 24 h post infection, cells were fixed with 4% paraformaldehyde in phosphate-buffered saline (PBS) for 7 min at room temperature, washed three times with PBS, and then permeabilized for 10 min with 0.1% Triton X-100 in PBS at room temperature. Cells were blocked for 1 h with 10% fetal bovine serum in PBS, followed by incubation with primary antibody at room temperature. Cells were washed three times in PBS, followed by incubation with Alexa Fluor-conjugated secondary antibody (Invitrogen) at room temperature. After cells were washed three times with PBS, DNA was stained with DAPI,and coverslips were mounted on slides with Mowiol. Images were collected with a Leica TCS-NT/SP2 inverted confocal microscope system.
Cell surface biotinylation
HeLa cells were infected with vA43V5. At 18 h post infection, cells were washed twice with PBS containing Mg2+ and Ca2+ and then incubated with 0.5 mg/ml of the membrane-impermeable EZ-Link Sulfo-NHS-LC-Biotin (sulfosuccinimidyl-6-[biotinamido] hexanoate) (Pierce) dissolved in PBS on ice for 20 min. Cells were washed with PBS and quenched with 10% fetal bovine serum for 20 min on ice and then washed twice again. Cells were then lysed with NP-40 detergent and the biotinylated proteins were affinity purified on a NeutrAvidin gel (Pierce).
IN infection model
Female BALB/c mice were purchased from Taconic (Germantown,NY) and maintained in a pathogen-free environment in sterile microisolator cages. Groups of 7-week-old mice were anesthetized by inhalation of isoflurane and inoculated via the IN route with a 20-µl suspension of purified VACV into one nostril. Mice were weighed daily for 2 weeks following challenge and were euthanized when they lost 30% of their initial body weight, according to a protocol approved by the National Institute of Allergy and Infectious Diseases Animal Care and Use Committee. The inocula were titered in order to confirm the dose administered.
Ear pinna infection model
Female BALB/c mice were anaesthetized with avertin and inoculated intradermally with 104 PFU of VACV in a 10-µl suspension. Lesions were measured daily with a micrometer. Ears were removed and placed in 2 ml of PBS with 0.05% bovine serum albumin and kept at –80°C until use. Ears were thawed, cut into 1 mm pieces and treated with collagenase for 3–4 h at 37°C, frozen and thawed three times, and sonicated three times for 30 s. Viral titers were determined by plaque assay on BS-C-1 cells.
Acknowledgements
We thank Norman Cooper and Catherine Cotter for providing cells, Heather Hickman for demonstration of intradermal injections, David Tscharke for advice regarding the ear pinnae model and Jon Yewdell for suggesting the brefeldin A control. The work was supported by funds from the Divison of Intramural Research, NIAID, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Bahar MW, Kenyon JC, Putz MM, Abrescia NGA, Pease JE, Wise EL, Stuart DI, Smith GL, Grimes JM. Structure and function of A41, a vaccinia virus chemokine binding protein. Plos Pathogens. 2008:4. doi: 10.1371/journal.ppat.0040005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bause E. Structural requirements of N-glycosylation of proteins. Studies with proline peptides as conformational probes. Biochem. J. 1983;209:331–336. doi: 10.1042/bj2090331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard PM, Froggatt GC, Smith GL. Vaccinia virus keltch protein A55 is a 64 kDa intracellular factor that affects virus-induced cytopathic effect and the outcome of infection in a murine intradermal model. J. Gen. Virol. 2006;87:1521–1529. doi: 10.1099/vir.0.81854-0. [DOI] [PubMed] [Google Scholar]
- Bendtsen JD, Nielsen H, von Heijne G, Brunak S. Improved prediction of signal peptides: SignalP 3.0. J. Mol. Biol. 2004;340:783–795. doi: 10.1016/j.jmb.2004.05.028. [DOI] [PubMed] [Google Scholar]
- Betakova T, Wolffe EJ, Moss B. Vaccinia virus A14.5L gene encodes a hydrophobic 53-amino acid virion membrane protein that enhances virulence in mice and is conserved amongst vertebrate poxviruses. J. Virol. 2000;74:4085–4092. doi: 10.1128/jvi.74.9.4085-4092.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blasco R, Sisler JR, Moss B. Dissociation of progeny vaccinia virus from the cell membrane is regulated by a viral envelope glycoprotein: effect of a point mutation in the lectin homology domain of the A34R gene. J. Virol. 1993;67:3319–3325. doi: 10.1128/jvi.67.6.3319-3325.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comeau MR, Johnson R, DuBose RF, Petersen M, Gearing P, VandenBos T, Park L, Farrah T, Buller RM, Cohen JI, Strockbine LD, Rauch C, Spriggs MK. A poxvirus-encoded semaphorin induces cytokine production from monocytes and binds to a novel cellular semaphorin receptor, VESPR. Immunity. 1998;8:473–482. doi: 10.1016/s1074-7613(00)80552-x. [DOI] [PubMed] [Google Scholar]
- Davison AJ, Moss B. The structure of vaccinia virus late promoters. J. Mol. Biol. 1989;210:771–784. doi: 10.1016/0022-2836(89)90108-3. [DOI] [PubMed] [Google Scholar]
- Domi A, Weisberg AS, Moss B. Vaccinia virus E2L null mutants exhibit a major reduction in extracellular virion formation and virus spread. J Virol. 2008;82:4215–4226. doi: 10.1128/JVI.00037-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Earl PL, Cooper N, Wyatt LS, Moss B, Carroll MW. Preparation of cell cultures and vaccinia virus stocks. In: Ausubel FM, Brent R, Kingston RE, Moore DD, Seidman JG, Smith JA, Struhl K, editors. “Current Protocols in Molecular Biology”. Vol. 2. New York: John Wiley and Sons; 1998. pp. 16.16.11–16.16.13. [Google Scholar]
- Fallon PG, Alcami A. Pathogen-derived immunomodulatory molecules: future immunotherapeutics? Trends Immuno. 2006;27:470–476. doi: 10.1016/j.it.2006.08.002. [DOI] [PubMed] [Google Scholar]
- Froggatt GC, Smith GL, Beard PM. Vaccinia virus gene F3L encodes an intracellular protein that affects the innate immune response. J. Gen. Virol. 2007;88:1917–1921. doi: 10.1099/vir.0.82815-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardner JD, Tscharke DC, Reading PC, Smith GL. Vaccinia virus semaphorin A39R is a 50–55 kDa secreted glycoprotein that affects the outcome of infection in a murine intradermal model. J. Gen. Virol. 2001;82:2083–2093. doi: 10.1099/0022-1317-82-9-2083. [DOI] [PubMed] [Google Scholar]
- Isaacs SN, Wolffe EJ, Payne LG, Moss B. Characterization of a vaccinia virus-encoded 42-kilodalton class I membrane glycoprotein component of the extracellular virus envelope. J. Virol. 1992;66:7217–7224. doi: 10.1128/jvi.66.12.7217-7224.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McFadden G. Poxvirus tropism. Nat. Rev. Microbiol. 2005;3:201–213. doi: 10.1038/nrmicro1099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moss B. Poxviridae: the viruses and their replication. In: Knipe DM, Howley PM, editors. “Fields Virology”. Vol. 2. Philadelphia: Lippincott Williams & Wilkins; 2007. pp. 2905–2946. 2 vols. [Google Scholar]
- Payne LG. Significance of extracellular virus in the in vitro and in vivo dissemination of vaccinia virus. J. Gen. Virol. 1980;50:89–100. doi: 10.1099/0022-1317-50-1-89. [DOI] [PubMed] [Google Scholar]
- Price N, Tscharke DC, Hollinshead M, Smith GL. Vaccinia virus gene B7R encodes an 18-kDa protein that is resident in the endoplasmic reticulum and affects virus virulence. Virology. 2000;267:65–79. doi: 10.1006/viro.1999.0116. [DOI] [PubMed] [Google Scholar]
- Sood CL, Ward JM, Moss B. Vaccinia virus encodes a small hydrophobic virion membrane protein (I5) that enhances replication and virulence in mice. J. Virol. 2008;82:10071–10078. doi: 10.1128/JVI.01355-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stack J, Haga IR, Schroder M, Bartlett NW, Maloney G, Reading PC, Fitzgerald KA, Smith GL, Bowie AG. Vaccinia virus protein Toll-like-interleukin-1 A46R targets multiple receptor adaptors and contributes to virulence. J. Exp. Med. 2005;201:1007–1018. doi: 10.1084/jem.20041442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tscharke DC, Smith GL. A model for vaccinia virus pathogenesis and immunity based on intradermal injection of mouse ear pinnae. J. Gen. Virol. 1999;80:2751–2755. doi: 10.1099/0022-1317-80-10-2751. [DOI] [PubMed] [Google Scholar]
- Turner GS. Respiratory infection of mice with vaccinia virus. J. Gen. Virol. 1967;1:399–402. doi: 10.1099/0022-1317-1-3-399. [DOI] [PubMed] [Google Scholar]
- Upton C, Slack S, Hunter AL, Ehlers A, Roper RL. Poxvirus orthologous clusters: toward defining the minimum essential poxvirus genome. J. Virol. 2003;77:7590–7600. doi: 10.1128/JVI.77.13.7590-7600.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DM, Clamp M, Barton GJ. Jalview Version 2--a multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]