Abstract
TANK-binding kinase 1 (TBK1) has emerged as a novel therapeutic target for unspecified subset of lung cancers. TBK1 reportedly mediates prosurvival signaling by activating NF-κB and AKT. However, we observed that TBK1 knockdown also decreased viability of cells expressing constitutively active NF-κB and interferon regulatory factor 3. Basal phospho-AKT level was not reduced after TBK1 knockdown in TBK1-sensitive lung cancer cells, implicating that TBK1 mediates unknown survival mechanisms. To gain better insight into TBK1 survival signaling, we searched for altered phosphoproteins using mass spectrometry following RNAi-mediated TBK1 knockdown. In total, we identified 2,080 phosphoproteins (4,621 peptides), of which 385 proteins (477 peptides) were affected after TBK1 knockdown. A view of the altered network identified a central role of Polo-like kinase 1 (PLK1) and known PLK1 targets. We found that TBK1 directly phosphorylated PLK1 in vitro. TBK1 phosphorylation was induced at mitosis, and loss of TBK1 impaired mitotic phosphorylation of PLK1 in TBK1-sensitive lung cancer cells. Furthermore, lung cancer cell sensitivity to TBK1 was highly correlated with sensitivity to pharmacological PLK inhibition. We additionally found that TBK1 knockdown decreased metadherin phosphorylation at Ser-568. Metadherin was associated with poor outcome in lung cancer, and loss of metadherin caused growth inhibition and apoptosis in TBK1-sensitive lung cancer cells. These results collectively revealed TBK1 as a mitosis regulator through activation of PLK1 and also suggested metadherin as a putative TBK1 downstream effector involved in lung cancer cell survival.
Keywords: non-canonical IκB kinase, stable isotope labeling by amino acids (SILAC), astrocyte elevated gene-1 (AEG-1)
TANK-binding kinase 1 (TBK1) was originally identified as an NF-κB–activating kinase (1). TBK1 activates the innate immune response through phosphorylation of two transcription factors, NF-κB and interferon regulatory factor 3 (IRF3), in response to proinflammatory cytokines and Toll-like receptor activation (2, 3). Besides its pivotal role in the innate immune response, increasing evidence indicates that aberrant activation of TBK1 and its closest homolog, IκB kinase ε (IKKε), is associated with development of human cancers. IKKε renders cells tumorigenic and is required for breast cancer cell proliferation and survival (4). IKKε has also been found to be amplified or activated in cancers, including lung cancer, ovarian cancer, glioma, and breast cancer (4–7).
Mechanisms that activate TBK1 and the downstream proteins and pathways regulated by TBK1 in cancer remain incompletely understood. Some studies have placed TBK1 downstream of RAS signaling, increasing the potential for TBK1-targeting therapeutics in cancer. The RalB GTPase engages TBK1 with oncogenic RAS to support cancer cell survival, suggesting that TBK1 is an unappreciated RAS downstream protein involved in cancer cell survival (8). A systemic RNAi screening study revealed that TBK1 is specifically required for survival of lung cancer cells harboring oncogenic KRAS mutations and further suggested that BCL-XL is important for TBK1-sensitive lung cancer cell survival (9). However, another study found both KRAS mutant cell lines to be insensitive to TBK1 inhibition and epidermal growth factor receptor (EGFR) mutant lung cancer cell lines to be sensitive to TBK1 inhibition (10). Studies also identified AKT as a novel substrate that is directly phosphorylated by TBK1 and mediates its prosurvival role (10, 11). Collectively, these studies argue that the genetic or phenotypic determinants for TBK1 dependency and survival mechanisms mediated by TBK1 remain to be elucidated. Identifying these determinants, especially in the context of RAS mutations, could have significant implications for targeted therapy of cancer.
To gain global insight into the TBK1 signaling network, especially signaling involved in cancer cell survival, we combined protein knockdown using short hairpin RNA specifically targeting TBK1 (shTBK1) with quantitative phosphoproteomics. Using this approach, we developed a TBK1-regulated phosphoproteomic signature and found that TBK1 knockdown leads to decreased phosphorylation of Polo-like kinase 1 (PLK1) and metadherin (MTDH). Interestingly, PLK1 has been identified as a synthetic lethal interacting protein in cancer cells harboring oncogenic KRAS (12), and MTDH expression is known to be induced by oncogenic HRAS (13). Our phosphoproteomics approach thus revealed novel links between TBK1 and the signaling network maintaining survival of RAS-mutant cancer cells.
Results
NF-κB, IRF3, and AKT Are Not Essential for TBK1-Mediated Survival Signaling.
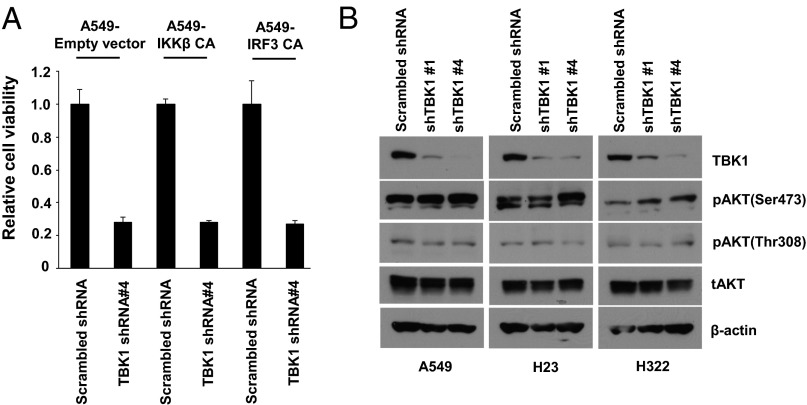
We introduced two different TBK1 shRNAs to H23 and A549 lung adenocarcinoma cell lines, which harbor oncogenic KRAS mutations. We observed that shTBK1 reduced cell viability and induced poly(ADP-ribose) polymerase (PARP) cleavage (SI Appendix, Fig. S1). Of note, both phenotypes were dependent on knockdown efficiency of two different hairpins. To delineate molecular mechanisms by which TBK1 mediates survival signaling, we first tested whether known TBK1 downstream targets, NF-κB and IRF3, are responsible for maintaining cancer cell survival (1–3). We generated A549 cells that stably express constitutively active (CA) IκB kinase β (IKKβ), which is a key kinase for NF-κB activation by proinflammatory cytokines, and CA-IRF3 (14, 15). Ectopic expression of the mutant proteins was confirmed by Western blotting (SI Appendix, Fig. S2). We also confirmed that cells expressing CA-IKKβ and -IRF3 expressed significantly higher levels of NF-κB and interferon (IFN)-β–Luc reporter genes as well as their target genes interleukin-8 (IL-8) and CXCL10, respectively (SI Appendix, Fig. S3). Using these stable cell lines, we tested whether the CA mutant proteins could rescue loss of cell viability induced by TBK1 knockdown. We found that neither CA-IKKβ nor IRF3 could rescue cell death induced by TBK1 loss, indicating that NF-κB and IRF3 are not key molecules in survival signaling driven by TBK1 (Fig. 1A). We next tested whether TBK1 is responsible for maintaining basal activity of NF-κB or AKT in lung cancer cells. AKT has been recently reported as a direct TBK1 substrate that mediates prosurvival signaling (10, 11). However, TBK1 knockdown failed to decrease either basal or tumor necrosis factor (TNF)-α–induced NF-κB–DNA binding or AKT phosphorylation in TBK1-sensitive lung cancer cells (SI Appendix, Fig. S4; Fig. 1B). These data suggest that TBK1 potentially activates unknown signaling pathways to maintain lung cancer cell survival that may be independent of NF-κB, IRF3, and AKT.
Fig. 1.
Known TBK1 targets are not essential for TBK1 survival signal. (A) Loss of TBK1 decreases cell viability of A549 lung adenocarcinoma cells independent of NF-κB and IRF3 activity. A549 cells stably expressing CA-IKKβ (S177/181D) or IRF3 (S396D) were infected with lentivirus encoding control shRNA (scrambled shRNA) or hairpins targeting TBK1 (shTBK1 #4). Cell viability was examined 6 d after lentivirus infection. Shown are triplicate experiments ±1 SE. (B) Loss of TBK1 does not decrease basal AKT phosphorylations in TBK1-sensitive lung cancer cell lines. Whole cell extracts were prepared 3 d after infection.
Quantitative Phosphoproteome Analysis of TBK1 Signaling in KRAS-Mutant Lung Cancer Cells.
To gain better insight into the TBK1 survival signaling network in lung cancer cells, we analyzed global phosphoproteomic changes after knocking down TBK1. We chose the A549 lung adenocarcinoma cell line, because A549 cells are dependent on TBK1 for survival (SI Appendix, Fig. S1). It is possible that caspase activation after TBK1 loss would disrupt the cellular proteome in a nonspecific manner, which may hinder identification of key survival proteins regulated by TBK1 (16). Thus, we examined TBK1 knockdown efficiency and PARP cleavage at 24, 48, 72, and 96 h after lentivirus infection to decide on an appropriate time point for harvesting whole cell extracts for phosphoproteomic analysis. From this experiment, we found that PARP cleavage was induced from 72 h after infection and endogenous TBK1 was significantly depleted starting from 48 h after infection (SI Appendix, Fig. S5). These data indicate that 48 h after infection is the most appropriate time point to prepare samples for phosphoproteomics.
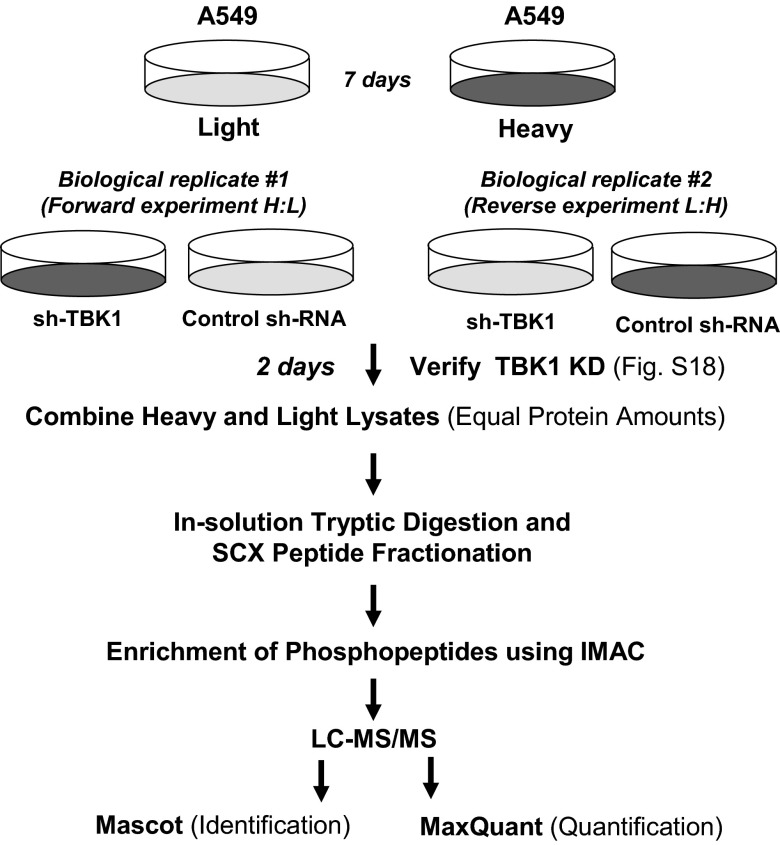
To define TBK1-regulated signaling with quantitative phosphoproteomics, we used Stable Isotope Labeling by Amino Acids in Cell Culture (SILAC) (17). The SILAC methodology eliminates many preanalytical variables that can induce quantification error because the two cell populations are mixed after treatment and treated as a single sample in all subsequent steps. The schematic of the SILAC-based TBK1 phosphoproteomics is described in Fig. 2. From this approach, we identified 4,621 unique phosphopeptides, which corresponded to 2,080 unique phosphoproteins (SI Appendix, Fig. S6; Dataset S1). We focused on phosphopeptides whose quantity is significantly affected by loss of TBK1. We defined a TBK1-regulated phosphopeptide as one that has had quantity changed by greater than an average of ±1.5-fold and was greater than an average of one SD away from the average deviation observed within the data. From this analysis, we observed that loss of TBK1 regulated phosphorylation of 385 proteins (477 peptides). A selected list of TBK1-regulated phosphopeptides is shown in Table 1 (with the full list shown in Dataset S2). Selected MS data were validated by Western blotting (SI Appendix, Fig. S7) and visual inspection of extracted ion chromatograms (SI Appendix, Fig. S8), which showed increased phosphorylation of EGFR, Met, and ERK1/2 and decreased phosphorylation of p70S6K.
Fig. 2.
Schematic of the SILAC-based quantitative phosphoproteomics approach to TBK1 signaling.
Table 1.
Selected TBK1-regulated phosphopeptides
| Protein name | Gene name | Fold change | Phosphopeptide sequence | Site |
| Kinase | ||||
| Ribosomal protein S6 kinase | RPS6KB1 | −3.21 | TPVpSPVKFSPGDFWGR* | S424 |
| Mitogen-activated protein kinase 3 | MAPK3/ERK1 | 4.83 | IADPEHDHTGFLTEpYVATR | Y204 |
| Hepatocyte growth factor receptor | MET | 2.93 | DMYDKEpYYSVHNK | Y1234 |
| Epidermal growth factor receptor | EGFR | 2.73 | GSTAENAEpYLR* | Y1197 |
| Polo-like kinase 1 | PLK1 | −1.63 | KKpTLCGTPNYIAPEVLSK† | T210 |
| Phosphatase | ||||
| Protein tyrosine phosphatase, type 12 | PTPN12 | 1.51 | NLpSFEIK | S435 |
| Protein tyrosine phosphatase, type 14 | PTPN14 | 1.83 | HKYVSGSpSPDLVTR | S594 |
| Transcription regulators | ||||
| Metadherin/astrocyte elevated gene-1 | MTDH/AEG-1 | −1.63 | SETSWEpSPKQIK* | S568 |
| Metadherin/astrocyte elevated gene-1 | MTDH/AEG-1 | 1.83 | LSSQISAGEEKWNpSVSPASAGKR | S306 |
| V-myc myelocytomatosis viral oncogene homolog | MYC | 1.70 | KFELLPTPPLSPpSRR | S64 |
| Jun proto-oncogene | JUN | 2.09 | NSDLLTpSPDVGLLK | S63 |
Amino acids in boldface represent phosphorylated sites.
Changed in both biological replicates. Fold changes indicated are average of two biological replicates.
The phosphosite predicted from MaxQuant was T214, but the actual phosphosite was T210 based on visual inspection of MS/MS spectra.
Global View to TBK1-Regulated Phosphoproteome.
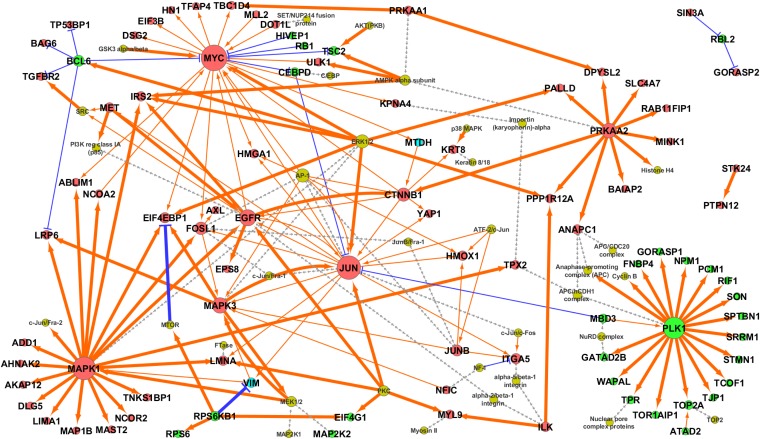
Next, we performed bioinformatics analyses of TBK1-regulated phosphoproteins to gain a global view of signaling networks regulated by TBK1. These analyses included Gene Ontology (GO) pathway enrichment analysis, motif analysis using the motif-x algorithm (18), and GeneGo MetaCore pathway analysis (trace pathways, 1 step). The resulting MetaCore network was further combined with phosphoprotein abundance direction changes obtained from TBK1 phosphoproteomics experiments (using majority voting for proteins with multiple phosphopeptide observations), keeping only those nodes (up/down) and edges [transcriptional regulation, (de)phosphorylation] consistent with experimental observation. This process results in a “self-consistent” network, in which all abundance changes are consistent with all directed interactions.
From GO pathway enrichment analysis, we found that 3 of the top 10 enriched GO hits in the down-regulated phosphoproteins included pathways involved in cell-cycle regulation, especially those involved in G1/S transition and mitosis (SI Appendix, Table S1). Motif analysis revealed that proline-directed motifs (pS/T-P) were enriched in the TBK1-regulated phosphopeptides (SI Appendix, Fig. S9), suggesting that TBK1 might be involved in the regulation of cell-cycle-regulating proline-directed kinases such as cyclin-dependent kinases or mitogen-activated protein kinases. Most importantly, the self-consistent network generated by MetaCore pathway analysis revealed that TBK1 knockdown decreases the PLK1 phosphorylation network (Fig. 3). Table 2 shows the selected list of TBK1–PLK1 subnetwork components, which revealed that TBK1 knockdown leads to decreased activating phosphorylation of PLK1 (Thr-210) and its mitotic target proteins (19–29). These results prompted us to validate our idea that TBK1 is involved in mitotic regulation through activating PLK1.
Fig. 3.
Self-consistent network composed of TBK1-regulated phosphoproteins. Red nodes indicate increased phosphoproteins; green nodes indicate decreased phosphoproteins; bright blue nodes (MTDH, VIM) are uncertain direction due to peptides in the same protein that were differently regulated after TBK1 knockdown; and dim yellow nodes indicate genes/complexes added by GeneGo Metacore to connect significantly changed nodes but were either not observed to be significantly changed (MTOR, MAP2K1, SRC) or are complexes for which no data were available. Orange arrows indicate activation/phosphorylation, and blue lines indicate inhibition/dephosphorylation. Gray dashed lines indicate membership within a protein complex. Phosphorylation is indicated by thick lines, and transcriptional regulation is indicated by narrow lines. Node size corresponds to number of edges connecting to the node.
Table 2.
Selected list of TBK1–PLK1 subnetwork components
| Gene name | Phosphopeptide sequence | Fold change | Mitotic function | Ref(s). |
| NPM1 | CGSGPVHIpSGQHLVAVEEDAESEDEEEEDVK | −1.79 | Centrosome replication | 20 |
| PCM1 | SDGpSENLCTPQQSR | −1.56 | Centrosome assembly and function | 21, 22 |
| STMN1 | RApSGQAFELILSPR | −1.97 | Mitotic spindle regulation | 23 |
| TOP2A | IKNENTEGpSPQEDGVELEGLK | −1.61 | Chromatin condensation | 24, 25 |
| TPR | GVQGPLNVpSLSEEGKSQEQILEILR | −1.51 | Mitotic spindle checkpoint | 26 |
| WAPAL | RPEpSPpSEISPIKGSVR | −1.77 | Regulator of sister chromatid cohesion | 27, 28 |
| GORASP1 | KPPGpTPPPSALPLGAPPPDALPPGPTPEDpSPSLETGSR | −1.58 | Postmitotic assembly of Golgi stacks | 29 |
Amino acids in boldface represent phosphorylated sites.
TBK1 Is Activated at Mitosis and Phosphorylates PLK1.
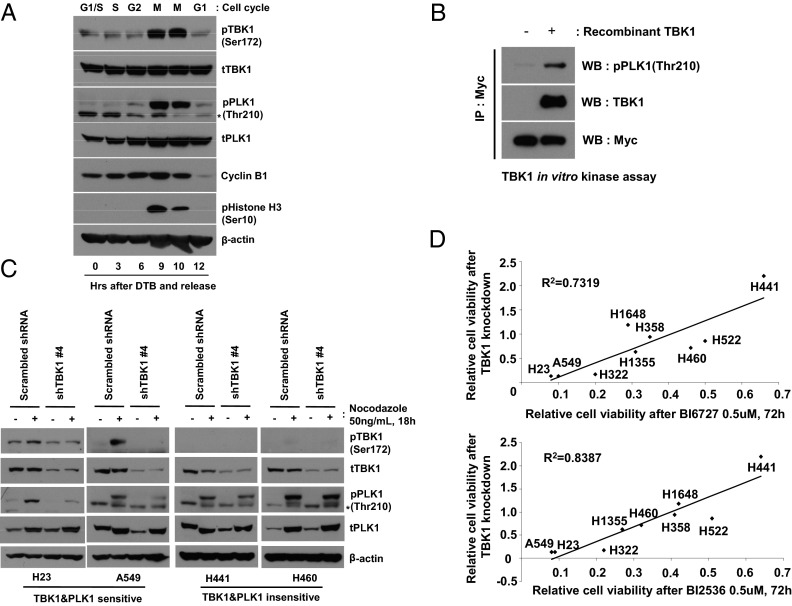
We first tested whether TBK1 activity is induced in a cell-cycle-dependent manner, particularly at mitosis, similar to other mitotic kinases such as cyclin-dependent kinase 1 (CDK1), PLKs, and Aurora kinases. A549 cells were released from double thymidine-induced G1-S boundary arrest, and activation of TBK1 and PLK1 was monitored. TBK1-activated phosphorylation (Ser-172) coincided with phosphorylation of histone H3, which is indicative of mitosis, and the kinetics of TBK1 activation is similar to PLK1 phosphorylation at Thr-210 (Fig. 4A). Of note, we found that TBK1 directly phosphorylated PLK1 in vitro, indicating that PLK1 is a mitotic TBK1 substrate (Fig. 4B). To examine the role of TBK1 in PLK1 activation in more detail, mitotic phosphorylation of PLK1 was examined after TBK1 knockdown. We observed that TBK1 was phosphorylated at mitotic cells, and mitotic phosphorylation of PLK1 was impaired by TBK1 knockdown in H23 and A549 cells, both of which are sensitive to TBK1 loss (Fig. 4C). Interestingly, neither mitotic phosphorylation of TBK1 nor TBK1 regulation of PLK1 phosphorylation was observed in two TBK1-resistant cells, H441 and H460 (Fig. 4C), suggesting that proper activation of PLK1 through TBK1 is important for survival of TBK1-sensitive lung cancer cells. To further validate this finding, we extended our approach to a larger number of lung cancer cell lines. We hypothesized that, if TBK1-mediated PLK1 activation is important for a subset of lung cancer cell survival, lung cancer cells sensitive to TBK1 knockdown would be also sensitive to PLK1 inhibition. To test this hypothesis, we performed shRNA-mediated knockdown of TBK1 in nine lung cancer cell lines, which were also treated with two different PLK1 inhibitors, BI6727 (Volasertib) and BI2536, and then examined cell viability. From this experiment, we observed that lung cancer cells that are sensitive to TBK1 knockdown were highly correlated with sensitivity to PLK1 inhibition (R2 = 0.7319 for BI6727 and 0.8387 for BI2536; Fig. 4D and SI Appendix, Fig. S10). These data strongly support the notion that TBK1-mediated PLK1 phosphorylation plays a pivotal role in maintaining survival of a subset of lung cancer cells.
Fig. 4.
TBK1 is a mitotic kinase involved in PLK1 activation. (A) TBK1 is activated in a cell-cycle-dependent manner. A549 cells were cell-cycle-synchronized by double thyimidine block (DTB) and released into fresh medium, and then whole cell extracts were prepared at the indicated times. Cell-cycle stages were verified by cyclin B1 and phospho-histone H3 (Ser-10) immunoblotting. *Nonspecific. (B) TBK1 directly phosphorylates PLK1. TBK1 in vitro kinase experiments were performed by using recombinant TBK1 (420 ng) and immunopurified Myc-tagged-kinase dead PLK1 (K82R) from 293FT cells as a substrate. (C) Impaired mitotic phosphorylation of PLK1 by TBK1 knockdown. Cells were infected with control or shTBK1 lentiviruses and then exposed to nocodazole (50 ng/mL) the next day for 18 h. (D) Analysis of correlation between TBK1 and PLK1 sensitivity in lung cancer cells. Nine non-small-cell lung cancer (NSCLC) cell lines were exposed to two different PLK1 inhibitors, BI6727 and BI2536 (0.5 µM, 72 h), or infected with control or shTBK1 lentiviruses (6 d), and then cell viability was examined. Each cell line’s remaining cell viability compared with control was plotted, and correlation coefficient (R2) was calculated.
TBK1 Regulates Phosphorylation of MTDH, an Important Survival Protein in Lung Cancer Cells.
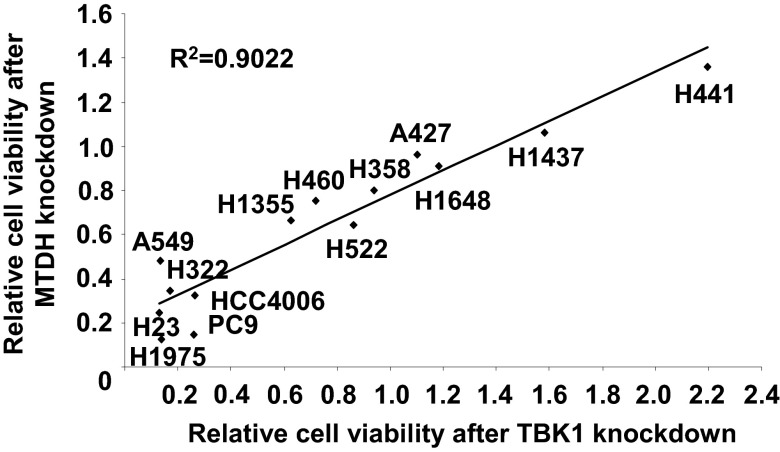
We tried to identify survival proteins regulated by TBK1 in addition to PLK1. To this aim, we searched for hits whose loss would recapitulate shTBK1’s phenotype, especially in the down-regulated phosphoproteins. In particular, phosphorylation of serine 568 on MTDH/Astrocyte Elevated Gene-1 (AEG-1) was reduced after TBK1 knockdown in both replicates (Table 1). MTDH is known as a downstream target of oncogenic HRAS and has been shown to be often dysregulated in human cancers (13, 30, 31). We found that high expression of MTDH is associated with poor prognosis in early-stage lung cancer (SI Appendix, Fig. S11), suggesting the importance of this protein in lung cancer aggressiveness. We next tested whether TBK1 could directly phosphorylate MTDH. We were unable to find evidence that TBK1 can directly phosphorylate MTDH (SI Appendix, Fig. S12). This result suggests that other Ser/Thr kinases possibly mediate this phosphorylation. To assess the functional significance of MTDH in TBK1-mediated survival signaling, we first tested whether loss of MTDH could produce the same results as the TBK1 knockdown phenotype. H23 and A549 cells were infected with lentiviruses encoding two different hairpins targeting MTDH. We found that loss of MTDH induced PARP cleavage and reduced cell viability (SI Appendix, Fig. S13). Consistent with TBK1–PLK1 above, we used the same approach to test our hypothesis that, if MTDH is a key TBK1 downstream target for survival, then lung cancer cells sensitive to TBK1 knockdown are also sensitive to MTDH knockdown. We performed shRNA-mediated knockdown of MTDH, TBK1, and KRAS in 14 lung cancer cell lines with defined EGFR and KRAS mutation status. The knockdown efficiency was validated by Western blotting in two different cell lines, H23 and H1437, which represent sensitive and resistant to loss of those survival proteins, respectively (SI Appendix, Fig. S14). From this experiment, we observed that lung cancer cells that are sensitive to MTDH knockdown were highly correlated with sensitivity to TBK1 knockdown (R2 = 0.9022; Fig. 5 and SI Appendix, Fig. S15). We observed less correlation between TBK1 sensitivity and KRAS sensitivity (R2 = 0.5271; SI Appendix, Fig. S16) and MTDH sensitivity and KRAS sensitivity (R2 = 0.3829; SI Appendix, Fig. S17). These data suggest that MTDH is an important mediator of TBK1 signaling to maintain survival in lung cancer cells.
Fig. 5.
Analysis of correlation between TBK1 and MTDH sensitivity in lung cancer cells. Fourteen NSCLC cells were infected with lentiviruses encoding control shRNA (scrambled shRNA), hairpins targeting MTDH (shMTDH#4), or TBK1 (shTBK1#4). Cell viability was measured 6 d after lentivirus infection. Each cell line’s remaining cell viability compared with control was plotted, and correlation coefficient (R2) was calculated.
Discussion
We describe a system-wide approach to decipher TBK1-mediated survival signaling networks in lung cancer cells. Our TBK1-regulated phosphoproteome revealed unexpected TBK1 function in regulating the mitotic kinase, PLK1. PLK1 contributes to multiple stages of mitosis, including CDK1 activation, spindle assembly, anaphase-promoting complex/cyclosome activation, and cytokinesis (32). Given the mitotic roles of PLK1, we speculate that loss of TBK1 possibly allows abnormality in mitosis (e.g., aberrant chromosome segregation), leading to mitotic cell death. Furthermore, cancer cell vulnerability to mitotic stress or alterations in cell-cycle phase may determine cellular responses to inhibition of TBK1 signaling. Interestingly, our results linking TBK1 to downstream PLK1 signaling integrated two previous RNAi screens identifying TBK1 and PLK1 as a synthetic lethal interaction partners with oncogenic KRAS (9, 12). Our results thus suggest that targeting PLK1 or the mitotic machinery would be an effective strategy against lung cancer cells that harbor oncogenic KRAS or for which survival is dependent on TBK1.
From our phosphoproteomics experiments, we could not identify phosphopeptides corresponding to conventional TBK1 substrates, including IRF3, IRF7, RelA, and IκB. This result may be due to lack of proinflammatory stimuli in our experimental condition. It is possible that TBK1 mediates phosphorylation of cancer-cell-specific survival proteins (e.g., PLK1, MTDH), whereas it phosphorylates proinflammatory substrates (e.g., IRF3, RelA) in normal cells in response to immune stimuli. Molecular mechanisms determining this specificity deserve further attention. One possibility is that TBK1 forms cancer-cell-specific protein–protein interaction (PPI) networks. The comprehensive TBK1–PPI network has been studied in immune cells (33); thus, a similar approach could be used to clarify the context-dependent TBK1–PPI network.
Although not fully addressed in this work, we observed that TBK1 knockdown increased phosphorylation of oncogenic kinases, including EGFR, Met, and ERK1/2 (SI Appendix, Figs. S7 and S8; Table 1). This result suggests that cancer cells activate compensatory growth/survival-promoting mechanisms driven by activation of multiple protein kinases, including EGFR, Met, and ERK, in response to loss of TBK1 expression. One common reason for failure of kinase-targeted cancer therapy is that cancer cells activate alternative routes of the kinase pathway for survival (34, 35). The molecular mechanisms underlying compensatory activation of these receptor tyrosine kinases (RTKs) after TBK1 knockdown are not clear. Duncan et al. reported that pharmacological inhibition of MEK leads to transcriptional up-regulation of RTKs (34), but this characteristic may not be the case for TBK1 because we did not see any change in total EGFR and Met levels after knocking down TBK1. Another possibility is that loss of TBK1 would inhibit function of phosphatases targeting RTKs. We found that loss of TBK1 increases phosphorylation of tyrosine phosphatases (PTPN12, PTPN14) whose loss activates protooncogenic RTKs, including EGFR, HER2, and platelet-derived growth factor receptor (PDGFR) (36). Thus, it is possible that loss of TBK1 induces inhibitory phosphorylation on tyrosine phosphatases, thereby activating compensatory RTKs.
Collectively, our global view regarding the TBK1-regulated phosphoproteome revealed a TBK1 function in mitosis as well as a unique survival protein whose phosphorylation is regulated by TBK1.
Materials and Methods
A549 cells were metabolically labeled with the SILAC protein quantitation kit (Pierce). After 7 d of labeling, cells were infected with lentiviruses encoding control hairpin (scrambled shRNA) or shRNA specific for TBK1 (shTBK1 #4), and then TBK1 knockdown was confirmed before proceeding to proteomics experiments (SI Appendix, Fig. S18). After we checked the level of label incorporation with in-solution digestion and liquid chromatography–tandem mass spectrometry (LC-MS/MS), a total of 1.6 mg of mixed cell lysates (0.8 mg of control and 0.8 mg of shTBK1) was mixed and digested with trypsin. The resulting tryptic peptides were fractionated by strong cation exchange chromatography. Each fraction was subjected to phosphopeptide enrichment by using immobilized metal affinity chromatography (Phos-Select; Sigma-Aldrich). LC-MS/MS was performed on a nanoflow liquid chromatograph (U3000; Dionex) interfaced with an electrospray hybrid ion-trap mass spectrometer (LTQ-Orbitrap; Thermo). Relative quantification was performed by using MaxQuant (Version 1.1.1.25). Additional details can be found in SI Appendix, SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. Paul Fisher (Virginia Commonwealth University) for MTDH expression plasmid and fruitful discussion; and Rasa Hamilton for editorial assistance with the manuscript. The work was supported in part by grants from the National Functional Genomics Center; National Institutes of Health Moffitt Lung Cancer Specialized Program of Research Excellence Grant P50-CA119997; a gift from the DeBartolo family; and the Proteomics, Molecular Genomics, Analytic Microscopy, and Flow Cytometry Core Facilities at the H. Lee Moffitt Cancer Center and Research Institute. The Moffitt Proteomics Core Facility is supported by US Army Medical Research and Material Command Award DAMD17-02-2-0051 continuing as W81XWH-08-2-0101 for a National Functional Genomics Center; National Cancer Institute Award P30-CA076292 as a Cancer Center Support Grant; and the Moffitt Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. J.R.H. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1220674110/-/DCSupplemental.
References
- 1.Tojima Y, et al. NAK is an IkappaB kinase-activating kinase. Nature. 2000;404(6779):778–782. doi: 10.1038/35008109. [DOI] [PubMed] [Google Scholar]
- 2.Shen RR, Hahn WC. Emerging roles for the non-canonical IKKs in cancer. Oncogene. 2011;30(6):631–641. doi: 10.1038/onc.2010.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Chau TL, et al. Are the IKKs and IKK-related kinases TBK1 and IKK-epsilon similarly activated? Trends Biochem Sci. 2008;33(4):171–180. doi: 10.1016/j.tibs.2008.01.002. [DOI] [PubMed] [Google Scholar]
- 4.Boehm JS, et al. Integrative genomic approaches identify IKBKE as a breast cancer oncogene. Cell. 2007;129(6):1065–1079. doi: 10.1016/j.cell.2007.03.052. [DOI] [PubMed] [Google Scholar]
- 5.Guo JP, et al. Deregulation of IKBKE is associated with tumor progression, poor prognosis, and cisplatin resistance in ovarian cancer. Am J Pathol. 2009;175(1):324–333. doi: 10.2353/ajpath.2009.080767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Guan H, et al. IKBKE is over-expressed in glioma and contributes to resistance of glioma cells to apoptosis via activating NF-κB. J Pathol. 2011;223(3):436–445. doi: 10.1002/path.2815. [DOI] [PubMed] [Google Scholar]
- 7.Guo J, et al. IKBKE is induced by STAT3 and tobacco carcinogen and determines chemosensitivity in non-small cell lung cancer. Oncogene. 2013;32(2):151–159. doi: 10.1038/onc.2012.39. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 8.Chien Y, et al. RalB GTPase-mediated activation of the IkappaB family kinase TBK1 couples innate immune signaling to tumor cell survival. Cell. 2006;127(1):157–170. doi: 10.1016/j.cell.2006.08.034. [DOI] [PubMed] [Google Scholar]
- 9.Barbie DA, et al. Systematic RNA interference reveals that oncogenic KRAS-driven cancers require TBK1. Nature. 2009;462(7269):108–112. doi: 10.1038/nature08460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ou YH, et al. TBK1 directly engages Akt/PKB survival signaling to support oncogenic transformation. Mol Cell. 2011;41(4):458–470. doi: 10.1016/j.molcel.2011.01.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Xie X, et al. IkappaB kinase epsilon and TANK-binding kinase 1 activate AKT by direct phosphorylation. Proc Natl Acad Sci USA. 2011;108(16):6474–6479. doi: 10.1073/pnas.1016132108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Luo J, et al. A genome-wide RNAi screen identifies multiple synthetic lethal interactions with the Ras oncogene. Cell. 2009;137(5):835–848. doi: 10.1016/j.cell.2009.05.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee SG, Su ZZ, Emdad L, Sarkar D, Fisher PB. Astrocyte elevated gene-1 (AEG-1) is a target gene of oncogenic Ha-ras requiring phosphatidylinositol 3-kinase and c-Myc. Proc Natl Acad Sci USA. 2006;103(46):17390–17395. doi: 10.1073/pnas.0608386103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang J, et al. NF-kappa B RelA subunit is crucial for early IFN-beta expression and resistance to RNA virus replication. J Immunol. 2010;185(3):1720–1729. doi: 10.4049/jimmunol.1000114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zheng Y, Vig M, Lyons J, Van Parijs L, Beg AA. Combined deficiency of p50 and cRel in CD4+ T cells reveals an essential requirement for nuclear factor kappaB in regulating mature T cell survival and in vivo function. J Exp Med. 2003;197(7):861–874. doi: 10.1084/jem.20021610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dix MM, et al. Functional interplay between caspase cleavage and phosphorylation sculpts the apoptotic proteome. Cell. 2012;150(2):426–440. doi: 10.1016/j.cell.2012.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ong SE, et al. Stable isotope labeling by amino acids in cell culture, SILAC, as a simple and accurate approach to expression proteomics. Mol Cell Proteomics. 2002;1(5):376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- 18.Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23(11):1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- 19.Jang YJ, Ma S, Terada Y, Erikson RL. Phosphorylation of threonine 210 and the role of serine 137 in the regulation of mammalian polo-like kinase. J Biol Chem. 2002;277(46):44115–44120. doi: 10.1074/jbc.M202172200. [DOI] [PubMed] [Google Scholar]
- 20.Krause A, Hoffmann I. Polo-like kinase 2-dependent phosphorylation of NPM/B23 on serine 4 triggers centriole duplication. PLoS ONE. 2010;5(3):e9849. doi: 10.1371/journal.pone.0009849. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Dammermann A, Merdes A. Assembly of centrosomal proteins and microtubule organization depends on PCM-1. J Cell Biol. 2002;159(2):255–266. doi: 10.1083/jcb.200204023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hames RS, et al. Dynamic recruitment of Nek2 kinase to the centrosome involves microtubules, PCM-1, and localized proteasomal degradation. Mol Biol Cell. 2005;16(4):1711–1724. doi: 10.1091/mbc.E04-08-0688. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Budde PP, Kumagai A, Dunphy WG, Heald R. Regulation of Op18 during spindle assembly in Xenopus egg extracts. J Cell Biol. 2001;153(1):149–158. doi: 10.1083/jcb.153.1.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Adachi Y, Luke M, Laemmli UK. Chromosome assembly in vitro: Topoisomerase II is required for condensation. Cell. 1991;64(1):137–148. doi: 10.1016/0092-8674(91)90215-k. [DOI] [PubMed] [Google Scholar]
- 25.Uemura T, et al. DNA topoisomerase II is required for condensation and separation of mitotic chromosomes in S. pombe. Cell. 1987;50(6):917–925. doi: 10.1016/0092-8674(87)90518-6. [DOI] [PubMed] [Google Scholar]
- 26.Lee SH, Sterling H, Burlingame A, McCormick F. Tpr directly binds to Mad1 and Mad2 and is important for the Mad1-Mad2-mediated mitotic spindle checkpoint. Genes Dev. 2008;22(21):2926–2931. doi: 10.1101/gad.1677208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kueng S, et al. Wapl controls the dynamic association of cohesin with chromatin. Cell. 2006;127(5):955–967. doi: 10.1016/j.cell.2006.09.040. [DOI] [PubMed] [Google Scholar]
- 28.Gandhi R, Gillespie PJ, Hirano T. Human Wapl is a cohesin-binding protein that promotes sister-chromatid resolution in mitotic prophase. Curr Biol. 2006;16(24):2406–2417. doi: 10.1016/j.cub.2006.10.061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lin CY, et al. Peripheral Golgi protein GRASP65 is a target of mitotic polo-like kinase (Plk) and Cdc2. Proc Natl Acad Sci USA. 2000;97(23):12589–12594. doi: 10.1073/pnas.220423497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brown DM, Ruoslahti E. Metadherin, a cell surface protein in breast tumors that mediates lung metastasis. Cancer Cell. 2004;5(4):365–374. doi: 10.1016/s1535-6108(04)00079-0. [DOI] [PubMed] [Google Scholar]
- 31.Kikuno N, et al. Knockdown of astrocyte-elevated gene-1 inhibits prostate cancer progression through upregulation of FOXO3a activity. Oncogene. 2007;26(55):7647–7655. doi: 10.1038/sj.onc.1210572. [DOI] [PubMed] [Google Scholar]
- 32.Archambault V, Glover DM. Polo-like kinases: Conservation and divergence in their functions and regulation. Nat Rev Mol Cell Biol. 2009;10(4):265–275. doi: 10.1038/nrm2653. [DOI] [PubMed] [Google Scholar]
- 33.Goncalves A, et al. Functional dissection of the TBK1 molecular network. PLoS ONE. 2011;6(9):e23971. doi: 10.1371/journal.pone.0023971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Duncan JS, et al. Dynamic reprogramming of the kinome in response to targeted MEK inhibition in triple-negative breast cancer. Cell. 2012;149(2):307–321. doi: 10.1016/j.cell.2012.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nazarian R, et al. Melanomas acquire resistance to B-RAF(V600E) inhibition by RTK or N-RAS upregulation. Nature. 2010;468(7326):973–977. doi: 10.1038/nature09626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun T, et al. Activation of multiple proto-oncogenic tyrosine kinases in breast cancer via loss of the PTPN12 phosphatase. Cell. 2011;144(5):703–718. doi: 10.1016/j.cell.2011.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.