Abstract
The Arabidopsis PENETRATION RESISTANCE 3 (PEN3) ATP binding cassette transporter participates in nonhost resistance to fungal and oomycete pathogens and is required for full penetration resistance to the barley powdery mildew Blumeria graminis f. sp. hordei. PEN3 resides in the plasma membrane and is recruited to sites of attempted penetration by invading fungal appressoria, where the transporter shows strong focal accumulation. We report that recruitment of PEN3 to sites of pathogen detection is triggered by perception of pathogen-associated molecular patterns, such as flagellin and chitin. PEN3 recruitment requires the corresponding pattern recognition receptors but does not require the BAK1 coreceptor. Pathogen- and pathogen-associated molecular pattern-induced focal accumulation of PEN3 and the PENETRATION RESISTANCE 1 (PEN1) syntaxin show differential sensitivity to specific pharmacological inhibitors, indicating distinct mechanisms for recruitment of these defense-associated proteins to the host–pathogen interface. Focal accumulation of PEN3 requires actin but is not affected by inhibitors of microtubule polymerization, secretory trafficking, or protein synthesis, and plasmolysis experiments indicate that accumulation of PEN3 occurs outside of the plasma membrane within papillae. Our results implicate pattern recognition receptors in the recruitment of defense-related proteins to sites of pathogen detection. Additionally, the process through which PEN3 is recruited to the host–pathogen interface is independent of new protein synthesis and BFA-sensitive secretory trafficking events, suggesting that existing PEN3 is redirected through an unknown trafficking pathway to sites of pathogen detection for export into papillae.
Keywords: plant, PAMP-triggered immunity, focal protein accumulation, vesicle
Plants defend themselves against pathogens through suites of layered but overlapping defense responses. Perception of nonself elicitor molecules, referred to as pathogen-associated molecular patterns (PAMPs), initiates the first wave of defense responses known collectively as PAMP-triggered immunity (1). Initial defense responses associated with PAMP-triggered immunity seem to be aimed at limiting the ability of the invading pathogen to enter plant tissues and become established. These responses include closure of stomata to limit pathogen entry (2) and local reinforcement of the cell wall at sites of interactions with pathogens through deposition of callose-rich papillae (3). Deposition of papillae has been observed in response to bacterial, fungal, and oomycete pathogens and has been proposed to act as a physical barrier, limiting access of pathogenic microorganisms to the cytosol of plant cells (4, 5).
In addition to providing structural reinforcements, papillae may also be sites of accumulation of secreted antimicrobial compounds. The PENETRATION RESISTANCE 2 myrosinase and the plasma membrane (PM) -localized ATP binding cassette (ABC) transporter PEN3 (synonyms PDR8 and ABCG36) have been proposed to participate in the synthesis and export of antimicrobial metabolites (6–9). Arabidopsis mutants lacking the PEN3 transporter allowed the nonhost powdery mildew Blumeria graminis f. sp. hordei (Bgh), a biotrophic fungal pathogen of barley, to penetrate epidermal cells and form haustorial feeding structures more frequently than WT plants (9). Additionally, pen3 mutants were more susceptible to the cosmopolitan nectrotrophic fungus Plectosphaerella cucumerina and the oomycete potato late blight pathogen Phytophthora infestans. A functional PEN3-GFP fusion protein showed strong focal accumulation at sites of attempted penetration by Bgh appressoria, highlighting the targeted nature of this early defense response (9).
Although deposition of papillae seems to be an important early component of plant antimicrobial defenses, the mechanisms and cellular components involved in the targeting and recruitment of these cell surface defenses to the host–pathogen interface remain unclear. In this study, we evaluated the ability of purified PAMPs to elicit focal accumulation of PEN3-GFP and used pharmacological inhibitors to probe the cellular processes required for recruitment of PEN3 and the PEN1 syntaxin (10) to sites of papilla deposition. Our results suggest that pattern recognition receptors (PRRs) mark the site of pathogen attack and initiate recruitment of PEN3, PEN1, and potentially, other defense-related proteins to sites of pathogen detection. We provide evidence that focal accumulation of PEN3 occurs outside the cell and within papillae and that the process of PEN3 recruitment and export does not occur through polarized secretion, suggesting the participation of an as yet undefined trafficking pathway.
Results and Discussion
PAMP Perception Elicits PEN3 Focal Accumulation in Leaf Mesophyll Cells.
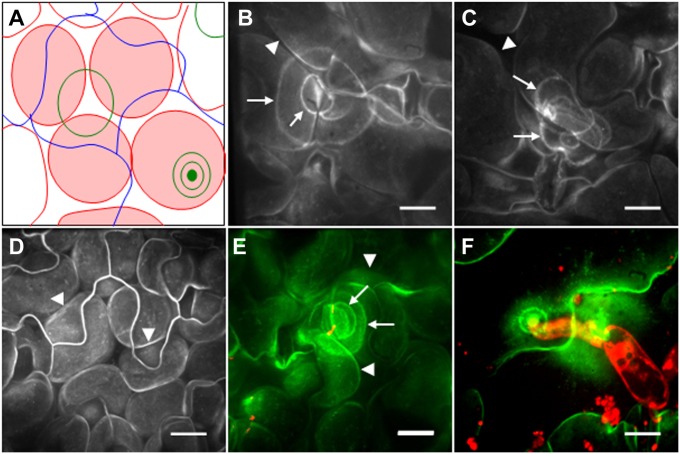
The PEN3 ABC transporter shows strong focal accumulation at sites of attempted penetration by powdery mildew fungi (9). We hypothesized that local perception of PAMPs might initiate focal accumulation of PEN3 and other defense-related proteins to sites of interaction with pathogens. To test this hypothesis, we monitored the localization of a functional PEN3-GFP fusion (9) in Arabidopsis leaf tissue after treatment with flg22 peptide, a PAMP derived from bacterial flagellin, or hydrolyzed chitin, a PAMP associated with cell walls of fungal pathogens and insect pests. In untreated and H2O control-treated leaves, PEN3-GFP was constitutively present and uniformly distributed throughout the PM in both epidermal and mesophyll cells (Fig. 1D), consistent with previous findings (9). Syringe infiltration of either 5 μM flg22 or 100 μg/mL chitin elicited focal accumulation of PEN3-GFP into ring-like structures and bull’s eye patterns in mesophyll cells (Fig. 1 B and C). The patterns observed in mesophyll cells are similar to the focal accumulations of PEN3-GFP previously observed in epidermal cells at sites of attempted penetration by the barley powdery mildew Bgh (9) (Fig. 1F). To guide readers in the interpretation of our confocal images, we have included a cartoon illustration showing features commonly observed in z-projected images of the mesophyll cell layer (Fig. 1A). Similarly, infiltration of leaves with Escherichia coli expressing the fluorophore Discosoma sp. red fluorescent protein (dsRED), a nonpathogenic bacterium carrying numerous PAMPs, elicited focal accumulation of PEN3-GFP into rings around sites of bacterial clusters (Fig. 1E). The observed rings and bull’s eye structures were specifically caused by PEN3-GFP focal accumulation and not autofluorescence, because we did not observe these structures in flg22-treated nontransgenic plants (Fig. S1A). The observation of discrete PEN3-GFP focal accumulation after syringe infiltration of PAMP solutions, a process expected to fill the apoplastic space relatively evenly with PAMP solution, was unexpected. One possibility that may explain the discretely localized response is that, after evaporation of the infiltrated liquid, the delivered solutes are not evenly distributed within the apoplastic space. To determine if PAMPs show uneven distribution after syringe infiltration, we infiltrated a solution of fluorescent-labeled TAMRA-flg22 into Col-0 leaves and imaged leaves at 3 h postinfiltration after all signs of leaf water soaking were gone. We observed uneven distribution of TAMRA-flg22, with regions of high relative concentrations of fluorescent peptide (Fig. S2). We propose that uneven distribution of solute after syringe infiltration results in high local concentrations of PAMP in some regions of the apoplast, causing discrete focal accumulation of PEN3-GFP at these sites.
Fig. 1.
PAMP- and pathogen-triggered focal accumulation of PEN3. (A) Illustration of features typically seen in z-projected confocal images of the mesophyll layer from flg22-treated leaves. Red, mesophyll cell outlines; red shading, fluorescence from mesophyll cell PM; green, PEN3-GFP focal accumulations; blue, outlines of overlying puzzle piece-shaped epidermal cells. (B) Localization of PEN3-GFP in mesophyll cells 24 h after syringe infiltration of 5 μM flg22. (C) Localization of PEN3-GFP in mesophyll cells 24 h after syringe infiltration of 100 μg/mL chitin. (D) Localization of PEN3-GFP in mesophyll cells 24 h after syringe infiltration of H2O. (E) Localization of PEN3-GFP in mesophyll cells 24 h after syringe infiltration of 108 cfu/mL E. coli (dsRED). Clusters of E. coli (dsRED) appear red (or yellow where an overlapping green signal is present), and PEN3-GFP appears green. (F) Localization of PEN3-GFP in epidermal cells 24 h after inoculation with Bgh. Propidium iodide-stained fungal structures appear red, and PEN3-GFP appears green. Arrows in B–E indicate PEN3-GFP focal accumulation. Arrowheads indicate PEN3-GFP–labeled PM outlines of leaf mesophyll cells. Note that rings of PEN3-GFP accumulation are distinct from mesophyll cell outlines and that, in some images (such as D), outlines of overlying, puzzle piece-shaped epidermal cells are visible. B–F are z-projected confocal images. (Scale bars: 20 μm.)
To determine the timeframe during which PEN3-GFP focal accumulation occurs after PAMP treatment, we performed a time course experiment. We monitored PEN3-GFP localization at 3, 6, 9, 12, and 24 h after syringe infiltration of 5 μM flg22 and counted the number of PEN3-GFP focal accumulations observed in 20 randomly selected microscope fields of view. We began to observe focal accumulation of PEN3-GFP at 6 h after flg22 treatment, and the number of focal accumulations increased progressively during the course of the experiment (Fig. S3).
Flg22 treatment also elicited focal accumulation of GFP-PEN1 (11), a defense-associated syntaxin, and we observed similar ring type deposits of the β-1,3 glucan cell wall polymer callose, a common constituent of papillae, after staining flg22-infiltrated leaf tissue with aniline blue (Figs. S1B and S4D). To determine if the observed callose rings induced by flg22 treatment colocalize with rings of PEN3 accumulation, we used the callose binding fluorophore sirofluor to label callose in live leaf tissue after treatment with flg22 peptide (12). Sirofluor labeling of callose in flg22-treated leaves expressing PEN3-GFP revealed patterns of callose deposits that largely colocalized with rings of PEN3-GFP focal accumulation (Fig. S1 C–E). Similarly, callose deposits in papillae showed significant colocalization with focal accumulations of PEN3-GFP at sites of attempted penetration by Bgh (Fig. S1 F–I).
PAMP-Induced Focal Accumulation of PEN3 Is Dependent on Pattern Recognition Receptors but Does Not Require BAK1.
To confirm that PAMP-elicited PEN3 focal accumulation is dependent on the corresponding PRRs, we transformed the flagellin receptor mutant fls2 (SALK_093905) (13) and the chitin receptor mutant cerk1-2 (14, 15) with the PEN3 promoter-PEN3-GFP construct. fls2 plants expressing PEN3-GFP did not exhibit focal accumulation of PEN3-GFP in response to infiltration with 5 μM flg22 (Fig. 2A) but retained the ability to target PEN3-GFP in response to 100 μg/mL chitin (Fig. 2B). Similarly, cerk1 plants expressing PEN3-GFP failed to target PEN3-GFP in response to chitin treatment (Fig. 2C) but retained the ability to target PEN3-GFP in response to flg22 (Fig. 2D). These results show that perception of PAMPs by PRRs is sufficient to elicit focal accumulation of PEN3 and suggest that PRRs can provide spatial information at the PM to initiate recruitment of papilla-associated defenses to the host–pathogen interface.
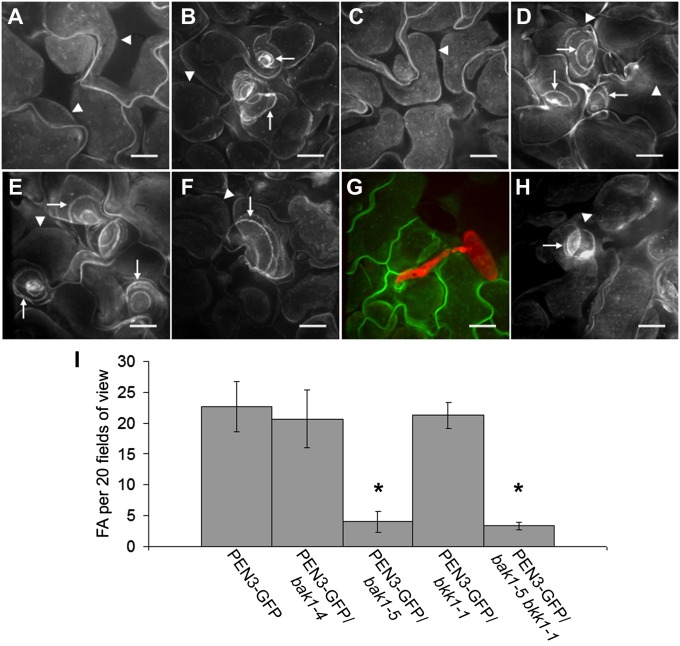
Fig. 2.
Involvement of PRRs FLS2 and CERK1 and coreceptors BAK1 and BKK1 in focal accumulation (FA) of PEN3. (A) Localization of PEN3-GFP in fls2 mutant leaves 24 h after syringe infiltration with 5 μM flg22. (B) Localization of PEN3-GFP in fls2 mutant leaves 24 h after syringe infiltration with 100 μg/mL chitin. (C) Localization of PEN3-GFP in cerk1 mutant leaves 24 h after syringe infiltration with 100 μg/mL chitin. (D) Localization of PEN3-GFP in cerk1 mutant leaves 24 h after syringe infiltration with 5 μM flg22. (E) Localization of PEN3-GFP 24 h after syringe infiltration with 5 μM flg22. (F) Localization of PEN3-GFP in bak1-4 mutant leaves 24 h after syringe infiltration with 5 μM flg22. (G) Localization of PEN3-GFP in bak1-4 mutant leaves 24 h after inoculation with Bgh. (H) Localization of PEN3-GFP in bak1-5 bkk1-1 double mutant leaves 24 h after syringe infiltration with 5 μM flg22. (I) Quantitative display of flg22-induced PEN3-GFP FAs in WT background (PEN3-GFP), bak1-4, bak1-5, and bkk1-1 mutant backgrounds, or bak1-5 bkk1-1 double mutant background. FAs were enumerated for 20 random microscope fields of view per leaf for three leaves per line. Error bars represent SD (n = 3), and asterisks indicate significant differences (P < 0.05; Tukey posthoc test). A–H are z-projected confocal images. Arrowheads in A–F and H indicate PEN3-GFP–labeled PM outlines of leaf mesophyll cells, and arrows indicate PEN3-GFP FA. (Scale bars: 20 μm.)
On flg22 perception, FLS2 forms a complex with the BRI1-associated receptor kinase 1 (BAK1) receptor-like kinase, and bak1 mutants display impaired responsiveness to flg22 (16, 17). To test the involvement of BAK1 in PAMP-induced relocalization of PEN3, we stably transformed our PEN3 promoter-PEN3-GFP fusion construct into the T-DNA null bak1-4 (SALK_116202) (16) mutant and monitored the localization of PEN3-GFP in response to flg22 treatment or Bgh inoculation. Syringe infiltration of bak1-4 (PEN3-GFP) leaves with 5 μM flg22 resulted in focal accumulations of PEN3-GFP (Fig. 2F) indistinguishable from accumulations observed in plants carrying a functional BAK1 (Fig. 2E). We quantitatively assessed the impact of mutation in bak1 on flg22-induced PEN3-GFP focal accumulation and found no significant difference in the frequency of PEN3-GFP focal accumulation in the bak1-4 mutant background, showing that BAK1 is dispensable for flg22-mediated focal accumulation of PEN3 (Fig. 2I). Additionally, BAK1 was also dispensable for recruitment of PEN3 to fungal penetration sites, because PEN3-GFP in bak1-4 showed normal accumulation at sites of attempted penetration by Bgh (Fig. 2G). The observation that BAK1 is completely dispensable for focal accumulation of PEN3-GFP induced by flg22 treatment was unexpected, because bak1 mutant plants are significantly impaired in numerous flg22-induced responses, including generation of reactive oxygen species, activation of mitogen-activated protein kinases such as MPK3 and MPK6, and induction of gene expression changes (14, 15). Recently, a semidominant allele, bak1-5, was discovered that exhibited more severe defects in PAMP-induced responses than the null bak1-4 (18). A bak1-5 bak1-like 1 (bkk1-1) double mutant was found to be almost completely unresponsive to the PAMP peptides flg22 and elf18 (derived from bacterial EF-Tu), indicating that BKK1 may participate in PRR complexes and may be partially redundant with BAK1 in this capacity (19). We assessed flg22-induced focal accumulation of PEN3-GFP in the bak1-5 bkk1-1 double mutant background and the bak1-5 and bkk1-1 single mutant backgrounds to determine if bak1-5 could exert a dominant negative effect on PEN3 focal accumulation and if BKK1 was important for PRR-directed PEN3 recruitment. The bak1-5 bkk1-1 double mutant retained the ability to initiate focal accumulation of PEN3-GFP on flg22 treatment; however, quantitative analysis revealed a significant reduction in the frequency of PEN3-GFP focal accumulations observed (Fig. 2 H and I). Interestingly, the bak1-5 single mutant was impaired in PEN3-GFP focal accumulation to a similar degree as the bak1-5 bkk1-1 double mutant, and the bkk1-1 mutant showed no apparent defect in flg22-induced focal accumulation of PEN3 (Fig. 2I). These results indicate that, (i) although BAK1 is not required for flg22-induced focal accumulation of PEN3-GFP, the semidominant bak1-5 allele can exert a dominant negative effect on flg22-induced PEN3 recruitment and (ii) BKK1 is not likely to play a role in flg22-induced recruitment of PEN3.
bak1-5 bkk1-1 Double Mutant Exhibits Reduced PEN3-GFP Focal Accumulation After Bacterial Inoculation.
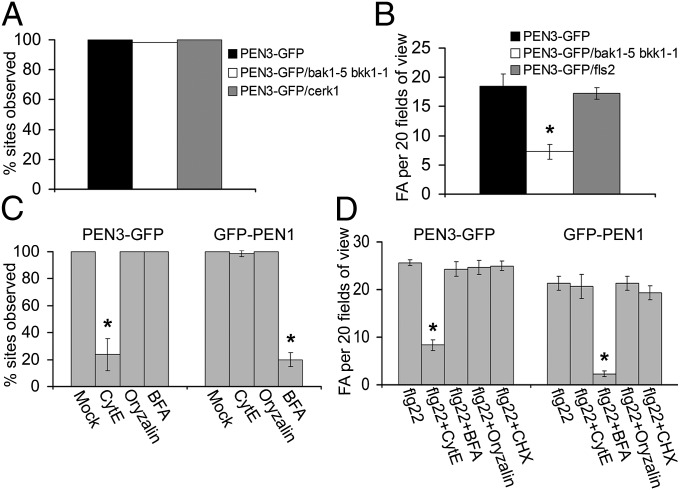
Because the bak1-5 bkk1-1 double mutant plants were significantly impaired in flg22-induced focal accumulation of PEN3-GFP, we sought to determine if this mutant showed a reduction in PEN3-GFP focal accumulation in the context of an interaction with an intact microbial pathogen to gain evidence that PAMP perception is an important signal required for focal accumulation of defenses during plant–microbe interactions. We first assessed focal accumulation of PEN3-GFP at Bgh penetration sites in the bak1-5 bkk1-1 double mutant and the cerk1 chitin receptor mutant. We did not observe a reduction in recruitment of PEN3-GFP to Bgh penetration sites in either mutant background, and neither mutant showed any impairment in penetration defense against the fungus (Fig. 3A and Fig. S5). It is likely that many PAMP and damage-associated molecular pattern signals in addition to chitin are perceived during powdery mildew penetration, and therefore, the loss of the single PRR does not have a significant impact on recruitment of defenses. Some of the PAMP and damage-associated molecular pattern signals generated at fungal penetration sites may be perceived through PRR complexes that are not impacted by the dominant negative activity of bak1-5. Alternatively, we cannot rule out the possibility that other signals besides PAMPS are perceived at powdery mildew penetration sites to initiate recruitment of PEN3-GFP. We next assessed focal accumulation of PEN3-GFP after inoculation of the bak1-5 bkk1-1 double mutant or the fls2 flagellin receptor mutant with type III secretion-deficient Pseudomonas syringae pv. tomato (Pst) DC3000 hrcC or E. coli. We did not observe an apparent reduction in PEN3-GFP focal accumulation in the fls2 mutant; however, the bak1-5 bkk1-1 mutant was significantly impaired in PEN3-GFP focal accumulation induced by Pst DC3000 hrcC and E. coli (Fig. 3B and Fig. S6A). These results indicate that additional bacterial PAMPs other than flagellin are important for initiating PEN3 focal accumulation and show that the reduced PAMP responsiveness of bak1-5 bkk1-1 is correlated with reduced PEN3-GFP focal accumulation after bacterial inoculation, suggesting that PAMP perception is an important signal required for recruitment of PEN3 to plant–bacteria interaction sites. We also observed increased multiplication of both Pst DC3000 hrcC and E. coli in bak1-5 bkk1-1 mutants (Fig. S6 B and C). Additionally, the pen3-1 mutant was found to be more susceptible to Pst DC3000 hrcC and partially compromised in protection conferred by pretreatment with flg22 before inoculation with Pst DC3000, suggesting that PEN3 is important for full PTI against bacteria (20).
Fig. 3.
Quantitative evaluation of mutant and pharmacological inhibitor effects. (A) Frequencies at which PEN3-GFP was observed at Bgh penetration sites in WT background (PEN3-GFP; black), bak1-5 bkk1-1 double mutant (white), or cerk1 mutant (gray). Accumulation of PEN3-GFP was scored at 50 infection sites per leaf for three leaves per plant line. (B) Frequencies of PEN3-GFP FAs in WT background (PEN3-GFP; black), bak1-5 bkk1-1 double mutant (white), or fls2 mutant (gray) leaves at 24 h after inoculation with 108 cfu/mL Pst DC3000 hrcC bacteria. FAs were enumerated for 20 random microscope fields of view per leaf for three leaves per line. (C) Frequencies at which PEN3-GFP or GFP-PEN1 were observed at Bgh penetration sites in leaves pretreated with mock, cytochalasin E (CytE), oryzalin, or BFA. Accumulation of PEN3-GFP or GFP-PEN1 was scored at 50 infection sites per leaf for three leaves per plant line/treatment. (D) Frequencies of flg22-induced PEN3-GFP or GFP-PEN1 FAs in leaves treated with 5 μM flg22, flg22 + cytE, flg22 + BFA, flg22 + oryzalin, or flg22 + CHX. FAs were enumerated for 20 random microscope fields of view per leaf for three leaves per treatment. Error bars in all panels represent SD (n = 3), and asterisks indicate significant differences (P < 0.05; Tukey posthoc test).
Effects of Pharmacological Inhibitors on Focal Accumulation of PEN3-GFP and GFP-PEN1.
To investigate the mechanisms underlying recruitment of PEN3 and PEN1 to sites of attempted penetration by powdery mildew fungi and focal accumulations elicited by PAMP perception, we used pharmacological inhibitors to disrupt specific cellular functions and monitored the localization of PEN3-GFP or GFP-PEN1 in response to inoculation with Bgh or treatment with flg22 peptide.
Bgh conidia typically germinate and begin to form appressoria by about 6 h postinoculation (hpi), and by about 14 hpi, immature haustoria are observable (21). We, therefore, treated leaves with inhibitors 1 h before inoculation with Bgh conidia and performed control experiments to verify the efficacy of the inhibitors within the timeframe during which PEN3 and PEN1 are recruited to penetration sites. To test the requirement for intact actin microfilaments, we used the pharmacological agent cytochalasin E to disrupt the actin cytoskeleton (22). Syringe infiltration of 2 μg/mL cytochalasin E caused significant disruption of actin filaments, which was visualized in Arabidopsis leaves expressing GFP-talin (23) by 3 h postinfiltration, and the disruption persisted for at least 24 h after infiltration (Fig. S7A). To test the effect of actin disruption on recruitment of PEN3-GFP or GFP-PEN1 to sites of attempted penetration by Bgh, we syringe-infiltrated leaves with 2 μg/mL cytochalasin E 1 h before inoculation with Bgh and observed the localization of GFP fusions at Bgh penetration sites 24 hpi. In control-treated leaves, we observed significant focal accumulation of PEN3-GFP and GFP-PEN1 at or near the tips of fungal appressoria, where attempted penetration occurs (Fig. S4 A and C). Disruption of actin filaments by cytochalasin E blocked accumulation of PEN3-GFP at most penetration sites (Fig. 3C and Fig. S4A). In contrast to the results observed for PEN3-GFP, we found that actin disruption by cytochalasin E had little or no effect on the recruitment of GFP-PEN1 to Bgh penetration sites (Fig. 3C and Fig. S4C). These results are consistent with previous findings, in which the barley PEN1 homolog ROR2 was found to localize to penetration sites in barley cells in which actin filaments were disrupted by overexpression of the actin depolymerizing factor HvADF3 (24). We obtained similar results with another actin inhibitor, latrunculin B (Figs. S7A and S8A) (25). These results suggest that PEN3 and PEN1 are recruited to powdery mildew penetration sites through distinct mechanisms and may partially explain the increased penetration success of the wheat powdery mildew on Arabidopsis treated with actin microfilament inhibitors (26).
To further test the involvement of actin filaments in the focal accumulation of PEN3, we used delayed treatment of cytochalasin E to determine if actin is required for retention of PEN3 at penetration sites or only for initial recruitment. As noted, Bgh conidia begin to form immature haustoria by about 14 hpi (21). Therefore, we treated Bgh-inoculated PEN3-GFP leaves with cytochalasin E 16 hpi and subsequently determined the presence of PEN3-GFP at Bgh penetration sites 40 hpi. Treatment with cytochalasin E at 16 hpi did not affect focal accumulation of PEN3-GFP at Bgh penetration sites (Fig. S4A), suggesting that actin filaments are required only for the initial recruitment of PEN3 to penetration sites and not for retention of PEN3 at these sites. Additionally, we tested the requirement for actin filaments in PAMP-mediated focal accumulation of PEN3 or PEN1. Cytochalasin E impaired flg22-induced formation of PEN3-GFP rings when coinfiltrated with flg22 but did not impact focal accumulation of GFP-PEN1 (Fig. 3D and Fig. S4 B and D). We occasionally observed aggregates and punctate localization of PEN3-GFP in some mesophyll cells of leaves coinfiltrated with flg22 and cytochalasin E; however, such aggregates were also observed in leaves infiltrated with cytochalasin E alone (Fig. S4B), suggesting that they were the result of inhibitor treatment rather than induced by PAMP perception. Again, we obtained similar results with the alternative actin inhibitor latrunculin B (Fig. S8D). We conclude that actin filaments are required for both PAMP- and Bgh-induced focal accumulation of PEN3-GFP but not focal accumulation of GFP-PEN1.
To test the role of microtubules in recruitment of PEN3 or PEN1 to Bgh penetration sites, we used the inhibitor oryzalin to disrupt microtubules (27). Syringe infiltration of 1 mM oryzalin disrupted microtubules by 6 hpi, and disruption persisted for at least 24 h as observed in Arabidopsis leaves expressing a GFP–β-tubulin fusion protein (28) (Fig. S7B). Syringe infiltration of oryzalin 1 h before inoculation with Bgh had no effect on the recruitment of either PEN3-GFP or GFP-PEN1 to penetration sites (Fig. 3C and Fig. S4 A and C). Additionally, oryzalin treatment did not impact flg22-induced recruitment of PEN3-GFP or GFP-PEN1 (Fig. 3D and Fig. S4 B and D), suggesting that microtubules are not required for focal accumulation of PEN3 or PEN1. Similar results were obtained with the inhibitor colchicine (Figs. S7B and S8 B and E) (27).
PEN3 is constitutively expressed and present in the PM of Arabidopsis leaves and modestly transcriptionally induced in response to pathogen challenge (9). Therefore, focal accumulation of PEN3 may arise from redistribution or accumulation of already existing protein, from polarized secretion of newly synthesized PEN3 in vesicles, or by endocytosis of PEN3 from the PM and redelivery to sites of pathogen detection. To determine if recruitment of PEN3 requires secretory trafficking, we monitored focal accumulation of PEN3 in the presence of the trafficking inhibitor brefeldin A (BFA) (29). Under our treatment conditions, syringe infiltration of 100 μg/mL BFA caused intracellular accumulation of a secretion-targeted GFP variant (30) by 4 h after infiltration, and intracellular accumulation of secretion-targeted GFP variant persisted for at least 24 h (Fig. S7C), showing the efficacy of the inhibitor. BFA treatment had no impact on PEN3-GFP focal accumulation at Bgh penetration sites or after flg22 treatment (Fig. 3 C and D and Fig. S4 A and B). Consistent with previous findings, BFA treatment impaired recruitment of GFP-PEN1 to Bgh penetration sites and promoted a significant increase in the frequency of haustorium formation by Bgh (Fig. 3C, Fig. S4C, and S9) (31). Additionally, coinfiltration of BFA with flg22 impaired PAMP-induced focal accumulation of GFP-PEN1 (Fig. 3D and Fig. S4D). Similar results were obtained with the vesicle trafficking inhibitor monensin (Figs. S7C, S8 C and F, and S9) (32). These results show that delivery of PEN3 to sites of pathogen detection is not dependent on BFA- or monensin-sensitive secretory trafficking and provide additional evidence that PEN1 and PEN3 are recruited to sites of pathogen detection through distinct trafficking mechanisms.
To test the requirement for de novo synthesis in focal accumulation of PEN3 or PEN1, we used cycloheximide (CHX) (33) to disrupt protein synthesis. Under our treatment conditions, syringe infiltration of 5 μg/mL CHX blocked incorporation of 35S-labeled methionine and cysteine into newly synthesized proteins (Fig. S7D). Syringe infiltration of leaves with 5 μg/mL CHX 1 h before inoculation with Bgh impaired the development of Bgh appressoria on the leaf surface. Therefore, we were not able to assess the effect of CHX on recruitment of PEN3-GFP or GFP-PEN1 to sites of attempted Bgh penetration. However, we frequently observed accumulation of both PEN3-GFP and GFP-PEN1 at the primary germ tubes of germinated Bgh conidiospores, showing that focal accumulation of PEN3 to sites of interaction with Bgh can occur in the presence of CHX (Fig. S4 A and C). Additionally, coinfiltration of 5 μg/mL CHX with 5 μM flg22 did not impair recruitment of either PEN3-GFP or GFP-PEN1 into ring-type focal accumulations (Fig. 3D and Fig. S4 B and D). These data suggest that focal accumulation of PEN3 is accomplished by actin-dependent recruitment of existing PEN3 to sites of pathogen detection and not dependent on polarized secretion of newly synthesized protein.
Fluorescence Recovery After Photobleaching Indicates That PEN3 Focal Accumulation Does Not Occur Through a Continuous Recruitment Process.
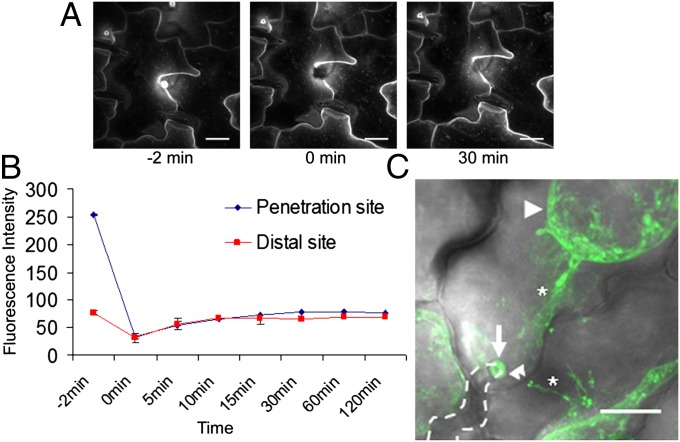
Asymmetric distributions of proteins can be maintained through endocytic cycling with constant redelivery of endocytosed protein to a specific membrane domain to maintain a high concentration of the protein at that membrane site. This mechanism is responsible for the polar distribution of the PIN-FORMED auxin efflux transporters in root cells (34). To determine if focal accumulation of PEN3 is maintained at powdery mildew penetration sites through a continuous recruitment process, we evaluated fluorescence recovery after photobleaching (FRAP) of PEN3-GFP at Bgh penetration sites. After photobleaching at a distal membrane site, PEN3-GFP fluorescence signal recovered within 30 min (Fig. 4B). However, when PEN3-GFP was bleached at Bgh penetration sites, the signal recovered to a level of fluorescence similar to the level observed at distal membrane sites within 30 min, but it did not recover to the level observed at the penetration site before photobleaching after 2 h (Fig. 4 A and B). These results indicate that PEN3 is not continuously delivered to penetration sites but delivered one time and retained at these sites. Additionally, the recovery of fluorescence signal at penetration sites to levels similar to fluorescence levels observed at distal membrane sites indicates that the PM remains dynamic at penetration sites. Our results are consistent with previous findings, in which FRAP of PEN1 at fungal penetration sites indicated a single delivery event rather than continuous delivery of PEN1 to sites of pathogen detection (24). Thus, PEN3 and PEN1 are both delivered to the host–pathogen interface in a single event over a relatively narrow timeframe, albeit by distinct delivery processes.
Fig. 4.
FRAP and plasmolysis analysis of PEN3-GFP FA at powdery mildew penetration sites. (A) Z-projected confocal micrographs collected before, immediately after, and 30 min after photobleaching of the PEN3-GFP signal at a single Bgh penetration site (24 hpi). (Scale bars: 20 μm.) (B) Quantitative analysis of PEN3-GFP fluorescence intensity at a Bgh penetration site and a distal membrane site before (−2 min), immediately after (0 min), and at various time points after photobleaching. Fluorescence intensity at each site was determined as described in Materials and Methods. Error bars indicate SD (n = 10). (C) PEN3-GFP signal at a Bgh penetration site after plasmolysis in 0.85 M NaCl. PEN3-GFP signal (green) from a z-projected confocal micrograph is overlaid onto the corresponding bright-field image. The arrow indicates PEN3-GFP FA at the fungal penetration site, the arrowhead indicates PEN3-GFP fluorescence at the retracted protoplast PM, the double arrowheads indicate the epidermal cell wall boundary, the dashed line indicates the position of the Bgh appressorial germ tube, and asterisks indicate Hechtian strands. (Scale bar: 20 μm.)
Plasmolysis Indicates That Focal Accumulation of PEN3 at Penetration Sites Is Extracellular.
Recent studies have suggested that PEN3 and PEN1 are found outside the cell in papillae, potentially within membrane structures such as exosomes or multivesicular bodies observed in or near papillae (12, 35). To determine if focal accumulation of PEN3 occurs within the PM or outside of the cell within the papilla, we performed plasmolysis and observed PEN3-GFP localization at Bgh penetration sites after plasmolysis-induced retraction of the protoplast PM. After plasmolysis, focal accumulations of PEN3-GFP remained in papillae in the extracellular space, indicating that PEN3 is exported into papillae (Fig. 4C). The persistence of PEN3-GFP fluorescence within papillae suggests that the GFP moiety is protected from the low pH environment of the apoplast, most likely indicating that it is present within the lumen of a membrane-bounded compartment within the papilla as suggested previously for PEN1 (12).
The PEN3 transporter has been proposed to export antimicrobial metabolites into papillae (6, 7, 9). The observed extracellular localization of PEN3 within papillae raises a number of questions about the role of PEN3 in antifungal defense. If PEN3 is indeed present on membrane compartments within papillae, it is possible that these compartments are loaded with antimicrobial compounds, which are subsequently exported from the membrane compartments into the papilla by PEN3. Alternatively, PEN3 may participate in loading such compartments with antimicrobial compounds before their export into papillae. Meyer et al. (12) hypothesized that export of PEN1 into papillae may occur through exosomes generated by fusion of multivesicular bodies with the PM and subsequent release of intralumenal vesicles into the papilla. Additionally, transmission EM studies have revealed the presence of membrane-bound paramural bodies containing intralumenal vesicles outside the cell in or near papillae (35–37). One possibility is that PEN1 and PEN3 are exported into papillae in the same membrane compartment, although such a hypothesis must account for the discrepancies in the requirement for actin filaments and BFA-sensitive secretory trafficking for focal accumulation of these proteins. PEN1 and PEN3 may be recruited to the same endomembrane compartment through distinct mechanisms. Alternatively, PEN1 and PEN3 may be exported into papillae in distinct compartments.
Our observation that flg22-induced focal accumulation of PEN3 occurred independently of BAK1 was unexpected. Whether PRR-directed recruitment of defenses to the cell surface requires a coreceptor and what the identity of such a coreceptor is remain to be determined. Future studies should resolve the specific mechanisms underlying recruitment of defense proteins, such as PEN1 and PEN3, to the host–pathogen interface and shed light on the role of PRRs in providing the spatial information of pathogen detection at the cell surface and initiating defense targeting.
Materials and Methods
PAMP and Pharmacological Inhibitor Treatments.
The flg22 peptide (amino acid sequence QRLSTGSRINSAKDDAAGLQIA) was synthesized by EZBiolab at a purity level of ≥70%. Stocks were prepared by dissolving the peptide in H2O at a concentration of 10 mM, stored at −20 °C, and subsequently diluted in H2O to the specified concentration for experiments. Chitin solutions were prepared by dissolving hydrolyzed chitin purified from crab shells (Sigma Aldrich) in H2O at a concentration of 100 μg/mL followed by brief centrifugation to remove insoluble material. PAMP solutions were infiltrated into Arabidopsis leaves using 1-mL needleless syringes pressed against the abaxial leaf surface. Pharmacological inhibitors were diluted in H2O to the specified concentrations for experiments. Arabidopsis leaves were treated with inhibitor solutions by syringe infiltration as described above. Details of inhibitor preparation and handling are described in SI Materials and Methods.
Powdery Mildew Penetration Assays and Callose Staining.
Bgh conidiospores were inoculated onto Arabidopsis leaves as described (38). At 48 hpi, three leaves were excised and cleared of pigment by incubation in 100% EtOH at 65 °C for 20 min. Cleared leaves were rinsed in H2O, transferred to aniline blue solution [0.01% (wt/vol) aniline blue in 150 mM K2HPO4, pH 9.5], and stained for 4–6 h at room temperature (39). Stained leaves were mounted on microscope slides in 70% (vol/vol) glycerol and observed on a Leica DMI 5000 B epifluorescence microscope with a 20× objective and A4 filter cube (365 ± 25-nm excitation filter, 400-nm dichroic and 450-nm long-pass emission filter). Penetration sites were scored as having either papillae only (small callose spots) or haustoria (large callose encasements) and papillae to determine the frequency of fungal penetration success. Callose staining after flg22 treatment was performed using the methods described above. Sirofluor labeling of callose in live tissue was carried out as described previously (12).
Confocal Microscopy and FRAP.
Plants expressing fluorescent protein fusions were observed on a confocal microscope consisting of a Leica DMI 6000 B inverted microscope (Leica Microsystems) fitted with a Yokogawa CSU-10 spinning disk confocal attachment (Yokogawa Electric Corporation) and a Photometrics QuantEM 512SC EM-CCD camera (Photometrics). Pieces of Arabidopsis leaves expressing GFP fusion proteins were cut and mounted in H2O on microscope slides. Samples were illuminated with a 488-nm diode laser and observed using a 525 ± 25-nm emission filter. For Bgh inoculated tissue, samples were mounted in 0.005% (wt/vol) propidium iodide in H2O to stain fungal structures (40). For plasmolysis experiments, leaf samples were mounted in 0.85% (wt/vol) NaCl and incubated 10 min before imaging. Propidium iodide, TAMRA-flg22, and dsRED were excited with a 561-nm diode laser and observed using a 620 ± 30-nm emission filter. Sirofluor was excited with a 405-nm diode laser and observed using a 480 ± 20-nm emission filter. Microscope control and image acquisition were accomplished using Metamorph software (Molecular Devices). Image processing was performed using ImageJ software (rsbweb.nih.gov/ij/) and Photoshop (Adobe). Z projections were typically created from 50 to 150 optical sections (z distance = 0.3 μm) by maximum projection using ImageJ. Photobleaching for FRAP was carried out on a region of interest of PEN3-GFP focal accumulation or a distal PM site using a MAG Biosystems FRAP-3D system equipped with a 473-nm diode laser. Images were collected 2 min before bleaching, immediately after bleaching, and at 5, 10, 15, 30, 60, and 120 min after bleaching. Fluorescence intensity at each time point for 10 pixels for each bleached site was determined using ImageJ software and averaged. Intensity values were normalized to background PM fluorescence for comparison.
Supplementary Material
Acknowledgments
We thank David Ehrhardt for assistance with confocal microscopy, Christian Voigt for providing E. coli (dsRED), and Cyril Zipfel for providing bak1-5 bkk1-1 mutant seeds. Funding was provided, in part, by the Carnegie Institution for Science, National Institutes of Health Postdoctoral Fellowship F32-GM0834393 (to W.U.), and National Science Foundation Grants 0519898 (to S.C.S.) and 0929226 (to S.C.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1218701110/-/DCSupplemental.
References
- 1.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 2.Melotto M, Underwood W, Koczan J, Nomura K, He SY. Plant stomata function in innate immunity against bacterial invasion. Cell. 2006;126(5):969–980. doi: 10.1016/j.cell.2006.06.054. [DOI] [PubMed] [Google Scholar]
- 3.Hoefle C, Hückelhoven R. Enemy at the gates: Traffic at the plant cell pathogen interface. Cell Microbiol. 2008;10(12):2400–2407. doi: 10.1111/j.1462-5822.2008.01238.x. [DOI] [PubMed] [Google Scholar]
- 4.Brown I, Mansfield J, Bonas U. hrp genes in Xanthomonas campestris pv. vesicatoria determine ability to suppress papilla deposition in pepper mesophyll cells. Mol Plant Microbe Interact. 1995;8(6):825–836. [Google Scholar]
- 5.Schmelzer E. Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 2002;7(9):411–415. doi: 10.1016/s1360-1385(02)02307-5. [DOI] [PubMed] [Google Scholar]
- 6.Bednarek P, et al. A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science. 2009;323(5910):101–106. doi: 10.1126/science.1163732. [DOI] [PubMed] [Google Scholar]
- 7.Clay NK, Adio AM, Denoux C, Jander G, Ausubel FM. Glucosinolate metabolites required for an Arabidopsis innate immune response. Science. 2009;323(5910):95–101. doi: 10.1126/science.1164627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lipka V, et al. Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science. 2005;310(5751):1180–1183. doi: 10.1126/science.1119409. [DOI] [PubMed] [Google Scholar]
- 9.Stein M, et al. Arabidopsis PEN3/PDR8, an ATP binding cassette transporter, contributes to nonhost resistance to inappropriate pathogens that enter by direct penetration. Plant Cell. 2006;18(3):731–746. doi: 10.1105/tpc.105.038372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Collins NC, et al. SNARE-protein-mediated disease resistance at the plant cell wall. Nature. 2003;425(6961):973–977. doi: 10.1038/nature02076. [DOI] [PubMed] [Google Scholar]
- 11.Assaad FF, et al. The PEN1 syntaxin defines a novel cellular compartment upon fungal attack and is required for the timely assembly of papillae. Mol Biol Cell. 2004;15(11):5118–5129. doi: 10.1091/mbc.E04-02-0140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Meyer D, Pajonk S, Micali C, O’Connell R, Schulze-Lefert P. Extracellular transport and integration of plant secretory proteins into pathogen-induced cell wall compartments. Plant J. 2009;57(6):986–999. doi: 10.1111/j.1365-313X.2008.03743.x. [DOI] [PubMed] [Google Scholar]
- 13.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 14.Miya A, et al. CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc Natl Acad Sci USA. 2007;104(49):19613–19618. doi: 10.1073/pnas.0705147104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Petutschnig EK, Jones AME, Serazetdinova L, Lipka U, Lipka V. The lysin motif receptor-like kinase (LysM-RLK) CERK1 is a major chitin-binding protein in Arabidopsis thaliana and subject to chitin-induced phosphorylation. J Biol Chem. 2010;285(37):28902–28911. doi: 10.1074/jbc.M110.116657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 17.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104(29):12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schwessinger B, et al. Phosphorylation-dependent differential regulation of plant growth, cell death, and innate immunity by the regulatory receptor-like kinase BAK1. PLoS Genet. 2011;7(4):e1002046. doi: 10.1371/journal.pgen.1002046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roux M, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23(6):2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Xin X-F, Nomura K, Underwood W, He SY. Induction and suppression of PEN3 focal accumulation during Pseudomonas syringae pv. tomato DC3000 infection of Arabidopsis. Mol Plant Microbe Interact. 2013;26(8):861–867. doi: 10.1094/MPMI-11-12-0262-R. [DOI] [PubMed] [Google Scholar]
- 21.Kerby K, Somerville S. Enhancement of specific intercellular peroxidases following inoculation of barley with Erysiphe graminis f. sp. hordei. Physiol Mol Plant Pathol. 1989;35(4):323–337. [Google Scholar]
- 22.Yahara I, Harada F, Sekita S, Yoshihira K, Natori S. Correlation between effects of 24 different cytochalasins on cellular structures and cellular events and those on actin in vitro. J Cell Biol. 1982;92(1):69–78. doi: 10.1083/jcb.92.1.69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Takemoto D, Jones DA, Hardham AR. GFP-tagging of cell components reveals the dynamics of subcellular re-organization in response to infection of Arabidopsis by oomycete pathogens. Plant J. 2003;33(4):775–792. doi: 10.1046/j.1365-313x.2003.01673.x. [DOI] [PubMed] [Google Scholar]
- 24.Bhat RA, Miklis M, Schmelzer E, Schulze-Lefert P, Panstruga R. Recruitment and interaction dynamics of plant penetration resistance components in a plasma membrane microdomain. Proc Natl Acad Sci USA. 2005;102(8):3135–3140. doi: 10.1073/pnas.0500012102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Spector I, Shochet NR, Kashman Y, Groweiss A. Latrunculins: Novel marine toxins that disrupt microfilament organization in cultured cells. Science. 1983;219(4584):493–495. doi: 10.1126/science.6681676. [DOI] [PubMed] [Google Scholar]
- 26.Yun B-W, et al. Loss of actin cytoskeletal function and EDS1 activity, in combination, severely compromises non-host resistance in Arabidopsis against wheat powdery mildew. Plant J. 2003;34(6):768–777. doi: 10.1046/j.1365-313x.2003.01773.x. [DOI] [PubMed] [Google Scholar]
- 27.Morejohn LC. The molecular pharmacology of plant tubulin and microtubules. In: Loyd CW, editor. The Cytoskeletal Basis of Plant Growth and Form. London: Academic; 1991. pp. 29–43. [Google Scholar]
- 28.Shaw SL, Kamyar R, Ehrhardt DW. Sustained microtubule treadmilling in Arabidopsis cortical arrays. Science. 2003;300(5626):1715–1718. doi: 10.1126/science.1083529. [DOI] [PubMed] [Google Scholar]
- 29.Satiat-Jeunemaitre B, Cole L, Bourett T, Howard R, Hawes C. Brefeldin A effects in plant and fungal cells: Something new about vesicle trafficking? J Microsc. 1996;181(Pt 2):162–177. doi: 10.1046/j.1365-2818.1996.112393.x. [DOI] [PubMed] [Google Scholar]
- 30.Zheng H, Kunst L, Hawes C, Moore I. A GFP-based assay reveals a role for RHD3 in transport between the endoplasmic reticulum and Golgi apparatus. Plant J. 2004;37(3):398–414. doi: 10.1046/j.1365-313x.2003.01969.x. [DOI] [PubMed] [Google Scholar]
- 31.Nielsen ME, Feechan A, Böhlenius H, Ueda T, Thordal-Christensen H. Arabidopsis ARF-GTP exchange factor, GNOM, mediates transport required for innate immunity and focal accumulation of syntaxin PEN1. Proc Natl Acad Sci USA. 2012;109(28):11443–11448. doi: 10.1073/pnas.1117596109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhang GF, Driouich A, Staehelin LA. Effect of monensin on plant Golgi: Re-examination of the monensin-induced changes in cisternal architecture and functional activities of the Golgi apparatus of sycamore suspension-cultured cells. J Cell Sci. 1993;104(Pt 3):819–831. doi: 10.1242/jcs.104.3.819. [DOI] [PubMed] [Google Scholar]
- 33.Kerridge D. The effect of actidione and other antifungal agents on nucleic acid and protein synthesis in Saccharomyces carlsbergensis. J Gen Microbiol. 1958;19(3):497–506. doi: 10.1099/00221287-19-3-497. [DOI] [PubMed] [Google Scholar]
- 34.Steinmann T, et al. Coordinated polar localization of auxin efflux carrier PIN1 by GNOM ARF GEF. Science. 1999;286(5438):316–318. doi: 10.1126/science.286.5438.316. [DOI] [PubMed] [Google Scholar]
- 35.An Q, Hückelhoven R, Kogel K-H, van Bel AJE. Multivesicular bodies participate in a cell wall-associated defence response in barley leaves attacked by the pathogenic powdery mildew fungus. Cell Microbiol. 2006;8(6):1009–1019. doi: 10.1111/j.1462-5822.2006.00683.x. [DOI] [PubMed] [Google Scholar]
- 36.An Q, Ehlers K, Kogel K-H, van Bel AJE, Hückelhoven R. Multivesicular compartments proliferate in susceptible and resistant MLA12-barley leaves in response to infection by the biotrophic powdery mildew fungus. New Phytol. 2006;172(3):563–576. doi: 10.1111/j.1469-8137.2006.01844.x. [DOI] [PubMed] [Google Scholar]
- 37.Micali CO, Neumann U, Grunewald D, Panstruga R, O’Connell R. Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell Microbiol. 2011;13(2):210–226. doi: 10.1111/j.1462-5822.2010.01530.x. [DOI] [PubMed] [Google Scholar]
- 38.Zimmerli L, Stein M, Lipka V, Schulze-Lefert P, Somerville S. Host and non-host pathogens elicit different jasmonate/ethylene responses in Arabidopsis. Plant J. 2004;40(5):633–646. doi: 10.1111/j.1365-313X.2004.02236.x. [DOI] [PubMed] [Google Scholar]
- 39.Vogel J, Somerville S. Isolation and characterization of powdery mildew-resistant Arabidopsis mutants. Proc Natl Acad Sci USA. 2000;97(4):1897–1902. doi: 10.1073/pnas.030531997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ramonell K, et al. Loss-of-function mutations in chitin responsive genes show increased susceptibility to the powdery mildew pathogen Erysiphe cichoracearum. Plant Physiol. 2005;138(2):1027–1036. doi: 10.1104/pp.105.060947. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.