Abstract
Approximately 70% of patients with non–small-cell lung cancer present with late-stage disease and have limited treatment options, so there is a pressing need to develop efficacious targeted therapies for these patients. This remains a major challenge as the underlying genetic causes of ∼50% of non–small-cell lung cancers remain unknown. Here we demonstrate that a targeted genetic dependency screen is an efficient approach to identify somatic cancer alterations that are functionally important. By using this approach, we have identified three kinases with gain-of-function mutations in lung cancer, namely FGFR4, MAP3K9, and PAK5. Mutations in these kinases are activating toward the ERK pathway, and targeted depletion of the mutated kinases inhibits proliferation, suppresses constitutive activation of downstream signaling pathways, and results in specific killing of the lung cancer cells. Genomic profiling of patients with lung cancer is ushering in an era of personalized medicine; however, lack of actionable mutations presents a significant hurdle. Our study indicates that targeted genetic dependency screens will be an effective strategy to elucidate somatic variants that are essential for lung cancer cell viability.
Keywords: driver mutations, MAPK pathway, signal transduction, siRNA screen
Gain-of-function (GOF) mutations in enzymes (such as kinases) confer increased activation of signaling pathways and lead to unregulated cellular proliferation and survival (1, 2). Inhibition of mutationally altered oncogenic kinases, such as EGFR or EML4-ALK (echinoderm microtubule-associated protein-like 4 fused to anaplastic lymphoma kinase), results in significant increases in lung cancer cell death and inhibition of proliferation (3, 4). Further highlighting the importance of somatic GOF mutations, patients with lung cancer with mutationally active EGFR are generally responsive to EGFR inhibitors, whereas patients with increased expression of EGFR by mechanisms such as amplification are generally not responsive to EGFR inhibitors (5, 6). This emphasizes the importance of identifying somatic mutations in enzymes that alter their activity so that they become essential for maintaining lung cancer cell viability. A lack of “actionable” mutations, which we define as mutations in enzymes that are amenable to pharmacological inhibition, is slowing our progress in administration of personalized therapies to patients with lung cancer. This has recently been highlighted by the Biomarker-integrated Approaches for Targeted Therapy for Lung cancer Elimination (BATTLE) trial, in which real-time biopsies were used to assess the mutational status of patients with lung cancer, but the actionable mutations and choice of targeted therapies to administer to patients is extremely limited (7). As personalized diagnostic approaches to lung cancer become increasingly feasible, progress must be made to provide novel targets and biomarkers to treat many of these patients.
Computational approaches to identify functionally important somatic variants dominate the literature, but it is unclear how many of the predicted driver mutations will actually be relevant and required to maintain cancer cell survival (8, 9). Labor-intensive functional studies must be performed to validate many of these potential drivers (8). More recently, several approaches are coming to the forefront to more efficiently identify genetic dependencies from cancer cells for which whole genome or exome sequencing has been performed (10). These include cross-species comparisons, insertional mutagenesis screens, and RNAi screening (10). There are benefits and deficiencies with many of these approaches, as highlighted by Eifert et al. (10). Our targeted approach benefits from a small screening strategy, a custom on-target siRNA library for efficient knockdown of all genes harboring nonsynonymous somatic mutations, and a defined endpoint. For the defined endpoint, somatic variants that are identified as essential are generated in the laboratory, and these variants are tested to assess relative activation of downstream pathways to determine if the somatic variants are GOF mutants. These studies were expanded to determine if other mutations in the target enzymes in primary tumors are similarly GOF mutations. Finally, we assess whether targeting these somatic variants or their downstream targets with small-molecule inhibitors will result in specific killing of the lung cancer cells, suggesting the enzymes could act as biomarkers. The use of this strategy allowed us to identify mutations in three different kinases (FGFR4, MAP3K9, and PAK5) that are activating toward the ERK pathway. We demonstrate that targeted depletion of the kinases with siRNA results in reduced viability of the lung cancer cells and attenuation of signaling in the relevant downstream pathways. Highlighting the importance of these kinases in promoting cell survival, pharmacological inhibition of the target kinases or downstream targets results in robust responses specifically in the lung cancer cell lines harboring the mutations. This suggests that patients with similar mutations may respond similarly to the targeted therapies used in our study.
Results
Targeted Genetic Dependency Screen.
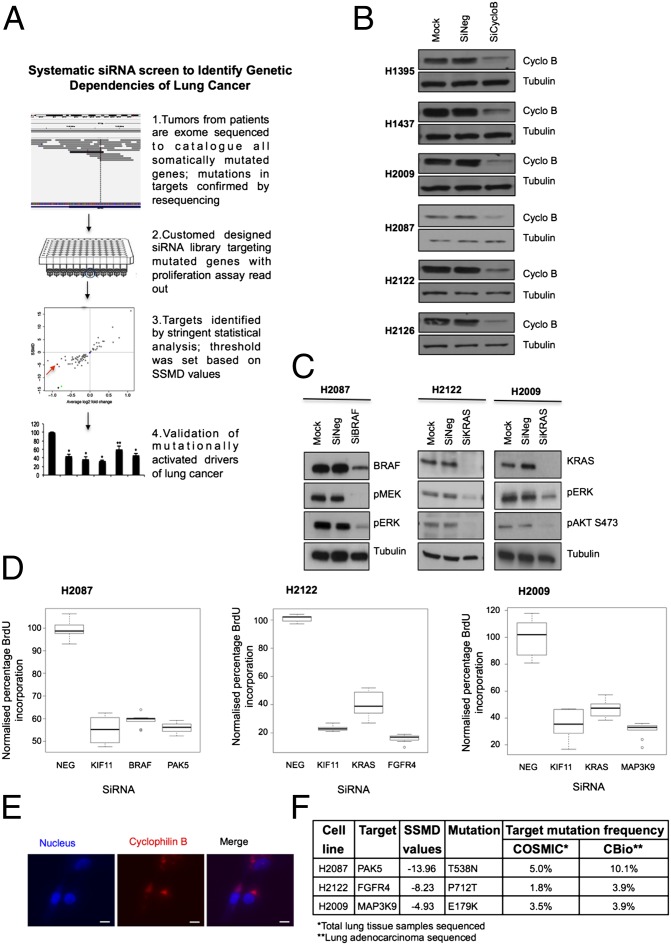
By depleting lung cancer cells of all proteins that carry nonsynonymous somatic mutations and monitoring effects on proliferation, we aimed to identify novel genes that harbor GOF mutations (Fig. 1A; methods and statistical analysis summarized in SI Appendix, Fig. S1 A and B). We evaluated six NSCLC cell lines that were exon-sequenced by the Sanger Institute (11); these cell lines harbored, on average, 65 nonsynonymous somatic mutations, and each cell line had a unique cellular morphology [genes and mutations listed in SI Appendix, Table S1, based on the Catalogue of Somatic Mutations in Cancer (COSMIC); summarized in SI Appendix, Fig. S1C, with cell morphologies displayed in SI Appendix, Fig. S1D] (11, 12). All cell lines were adherent and amenable to transient siRNA knockdown based on depletion of cyclophilin B (Fig. 1B; summarized in SI Appendix, Fig. S1E). As proof of principle for our approach, we depleted mutationally activated KRAS from H2009 (KRASG12A) and H2122 (KRASG12C) cells and monitored inhibition of downstream signaling. Depletion of KRASG12A or KRASG12C resulted in attenuation of signaling in the PI3K and ERK pathways (Fig. 1C). Additionally, we depleted mutationally altered BRAF from H2087 cells; knockdown of BRAFL597V attenuated ERK pathway activation (Fig. 1C) (13). Consistent with these results, depletion of KRAS and BRAF in the respective cell lines led to an inhibition of proliferation (Fig. 1D), and these mutationally activated enzymes served as a positive controls for screens in these cell lines [in addition to KIF11 (kinesin family member 11), an essential protein required for cell division] (14). We verified siRNA was successfully transduced into the cells in each assay by using DY-547-cyclophilin B siRNA (Fig. 1E) (15). The proliferative effects observed from depleting the mutated proteins for all cell lines are highlighted in SI Appendix, Tables S2–S7. Statistical plots for BrdU assays used to establish mean values, z-scores, and strictly standardized mean differences (SSMD) for the H2087, H2009, and H2122 cell lines are displayed in SI Appendix, Fig. S2 A–I (siRNA oligonucleotides are listed in SI Appendix, Table S9). SSMD scores (β) were used to identify proteins for which depletion led to a statistically significant decrease in BrdU incorporation. Based on SSMD’s probability interpretation, cutoff criteria were established at β ≤ −5 for strong regulators (corresponding to an error rate <1%) and β ≤ −3 for intermediate regulators of proliferation (corresponding to an error rate <2.5%; statistical methods included in SI Appendix) (16–19). It is likely that other candidates that fell outside this stringent selection may also prove fruitful; however, more downstream validation would be required. This work highlighted three therapeutically tractable target kinases that scored high in our screen: FGFR4, MAP3K9 (MLK1), and PAK5 (PAK7), from the H2122, H2009, and H2087 cell lines, respectively (Fig. 1D and SI Appendix, Tables S2–S4); the relative mutational frequencies of these genes in lung cancer are listed in Fig. 1F. The other three NSCLC cell lines we evaluated (H1395, H1437, and H2126) displayed less significant fold-change and SSMD values (SI Appendix, Tables S5–S7); therefore, we chose to focus on the targets identified in the cell lines listed earlier.
Fig. 1.
Genetic dependency screen identifies three potential genetic drivers of lung cancer. (A) Schematic of siRNA targeted screen carried out on a panel of six NSCLC cell lines. (B) Transfection efficiency for each cell line was optimized by using 100 nM siRNA against cyclophilin B (SiCycloB), 100 nM of nontargeting siRNA (SiNeg), and transfection reagent alone (Mock). (C) Cell line-specific positive controls BRAF and KRAS were knocked down by using 100 nM siRNA in respective NSCLC cells. (D) Box-and-whisker plots of statistically validated targets identified from BrdU screens, with data normalized to nontargeting siRNA (NEG), plotted with KIF11 as assay positive control, and BRAF (H2087) and KRAS (H2122, H2009) as cell line-specific positive control. (E) DY-547-cyclophilin B siRNA was used as an indicator of efficient transfection. Cell nuclei were stained with Hoechst 33342. Images captured for H2009 cells are illustrated as an example. (Scale bars: 10 µm.) (F) Validated targets identified from targeted screen based on SSMD values in three NSCLC cell lines and the frequency of their mutations following sequencing of lung cancer cells. Data compiled from COSMIC and cBio Cancer Genomics Portal databases.
GOF Mutations in PAK5 Enhance Activation of the ERK Pathway.
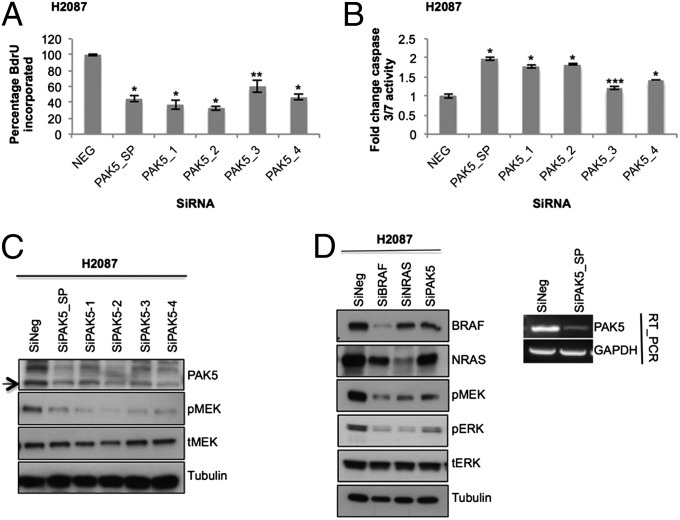
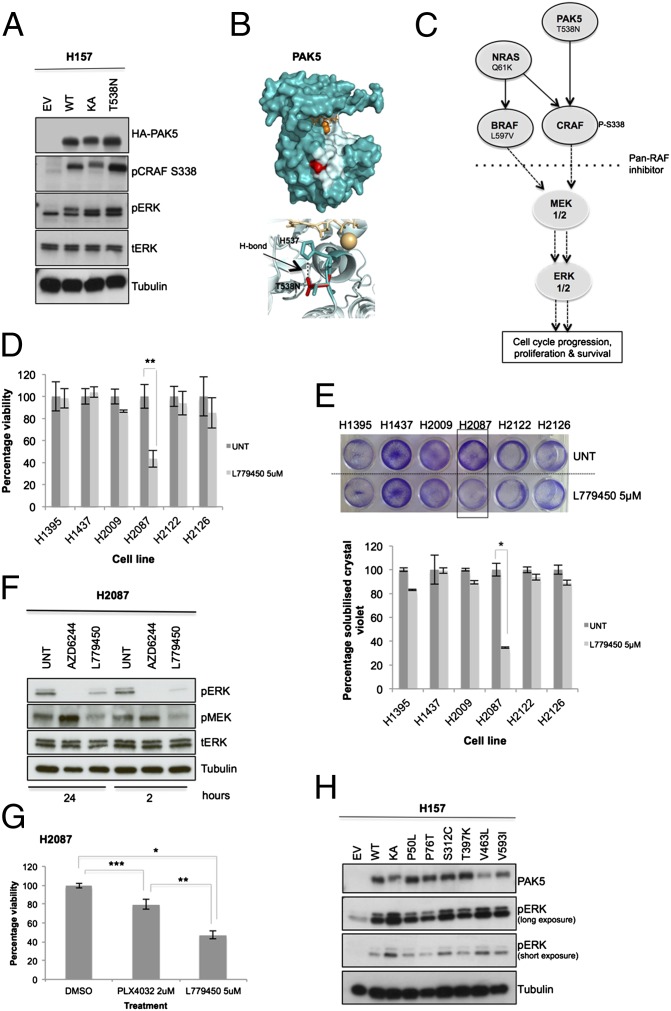
As an example of the methods used in our screen, we will describe our strategy for the H2087 cell line. This cell line harbors 79 nonsynonymous somatic mutations in 79 different genes, among 4,700 genes sequenced (11). A normal cell line established from the blood of the same patient was sequenced and used to establish somatic events (11). We depleted this cell line of transcripts that carried nonsynonymous mutations (corresponding to 79 different proteins) by using on-target siRNA SMARTpools and monitored effects on proliferation. For the H2087 cell line, PAK5 was identified as a mutated protein required for proliferation. Based on our screening criteria, PAK5 had the most significant SSMD value for the H2087 cell line (SI Appendix, Table S2) and was further verified with a caspase assay (SI Appendix, Table S8 and Fig. S2 J–L, show statistical analysis). PAK5 is a member of the class II p21 activated family of kinases (20) and is an upstream regulator of CRAF (RAF-1) (21). Recent cancer genomic sequencing studies indicate that PAK5 is mutated in 5% to 10% of lung cancer cases (Fig. 1F). A deconvolution experiment was performed by using each siRNA singularly from the siRNA pool to verify on-target specificity of individual oligos. The single siRNA oligos significantly suppressed cell proliferation (P < 0.01), induced apoptosis, decreased expression of the PAK5 kinase, and attenuated signaling in the MAPK kinase (MEK)/ERK pathway (Fig. 2 A–C). Importantly, depletion of PAK5 with the siRNA pool dampened MEK/ERK signaling to a level comparable to depleting other mutationally activated upstream regulators of the MEK/ERK pathway present in this cell line, which include BRAFL597V and NRASQ61K (Fig. 2D and SI Appendix, Fig. S1C). To assess if the PAK5T538N mutation (heterozygous mutation verified by resequencing) is a GOF mutation, we expressed this mutant and compared with WT PAK5 and a kinase active (KA) PAK5 mutant (S573N, based on homology with a PAK4KA mutant S445N) (22). The PAK5T538N mutant had increased activity toward CRAF in comparison with PAK5WT and PAK5KA, and this correlated with an increase in ERK pathway activation (Fig. 3A). The mutation in PAK5 at Thr538 is located in a surface accessible region that is likely involved in substrate interactions [based on comparison with protein kinase A (PKA), T538 lies in the substrate binding groove] and may enhance an interaction between CRAF and PAK5 (Fig. 3B) (13). The H2087 lung cancer cell line has accumulated several mutations in the RAF/MEK/ERK pathway (including PAK5T538N, NRASQ61K, and BRAFL597V), resulting in hyperactivation of the ERK pathway, potentially in a manner analogous to a BRAF V600E mutation (Fig. 3C) (13). To determine if pharmacological inhibition of the RAF kinases (ARAF, BRAF, and CRAF) would promote cell death, we treated H2087 cells with a pan-RAF inhibitor (L779450). We observed a significant decrease (P < 0.01) in cell viability specifically in the H2087 cells and not in the five additional NSCLC cell lines included in our screen (Fig. 3 D and E; Fig. 3F demonstrates that L779450 suppresses signaling in the ERK pathway in H2087 cells). Consistent with multiple mutations acting in concert to activate all RAFs, the BRAF-specific inhibitor PLX4032 was less toxic to the H2087 cells than the pan-RAF inhibitor (Fig. 3G). Lack of toxicity could also be attributed to the paradox effect whereby inhibitor-bound BRAF can act as a scaffold to enhance the activation of CRAF by NRASQ61K (23, 24). Finally, we determined if other mutations in PAK5 are activating toward the ERK pathway. We observed that three additional PAK5 somatic mutations (S312C, V463L, and V593I, identified in primary lung tumors by COSMIC) are activating toward the ERK pathway compared with WT PAK5 (Fig. 3H).
Fig. 2.
On-target validation of PAK5 in H2087 cells. (A and B) PAK5 was knocked down in H2087 (SMARTpool 100 nM; oligos 1–4, 50 nM). Plates were subjected to (A) BrdU assay and (B) caspase 3/7 activity apoptosis assay. Data (n = 9) were normalized to nontargeting siRNA (NEG). Error bars indicate SD. P values were calculated by using one-tailed paired Student t test (*P < 0.005, **P < 0.01, and ***P < 0.05 vs. nontargeting siRNA control). (C) On-target effect of PAK5 SMARTpool (100 nM) and individual oligos (50 nM) was also verified by Western blotting. (D) H2087 cells were reverse-transfected with 100 nM siRNA as indicated (BRAF and SMARTpool siNRAS and siPAK5). Knockdown of PAK5 was confirmed by RT-PCR.
Fig. 3.
Characterization of PAK5 kinase as an actionable mutated target. (A) Empty vector (EV), WT PAK5, KA S573N, and T538N mutant were overexpressed in H157 cells, and lysates were analyzed by Western blotting. (B, Upper) Crystal structure of PAK5 kinase domain (Protein Data Bank ID code 2F57) with ATP and magnesium ions shown as orange sticks and spheres, respectively. Likely substrate binding region is shown in pale blue, based on observed binding sites for other protein kinases, including PKA (PDB ID IQ24). The mutation site of T538N, shown in red, is found at the bottom of this binding region. (B, Lower; red sticks) Analysis of the structure shows that this mutation allows more hydrogen-bonding opportunities with the neighboring histidine residue (H537; blue sticks) compared with the WT construct. (C) Mutationally activated PAK5, NRAS, and BRAF hyperactivate the MEK/ERK pathway to drive proproliferative signals and promote lung cancer cell survival. (D) The panel of six NSCLC cell lines was treated with 5 µM of the pan-RAF inhibitor L779450 for 72 h, and cytotoxicity was determined by MTT assay. Data (n = 9) were normalized to untreated cells (UNT; DMSO, <0.01%). (E, Upper) Cytotoxicity was also confirmed by colony formation assay with crystal violet staining and (E, Lower) quantified by solubilization (n = 6). (F) H2087 cells were treated with AZD6244 (3 µM) or L779450 (5 µM) for indicated time points. Inhibition of downstream substrates pMEK and pERK was analyzed. (G) H2087 cells were treated with BRAF-specific inhibitor PLX4032 (2 µM) or pan-RAF inhibitor L779450 (5 µM). Cell viability was assessed by MTT (n = 9). (H) PAK5 mutants in lung cancer identified from the COSMIC database were overexpressed in H157 cells. Three mutants—S312C, V463L, and V593I—were identified to be activating at levels comparable to KA (S573N). Error bars show SD. P values were calculated by using one-tailed paired Student t test (*P < 0.005, **P < 0.01, and ***P < 0.05 vs. DMSO control).
FGFR4 Is Essential for Lung Cancer Cell Survival and Proliferation.
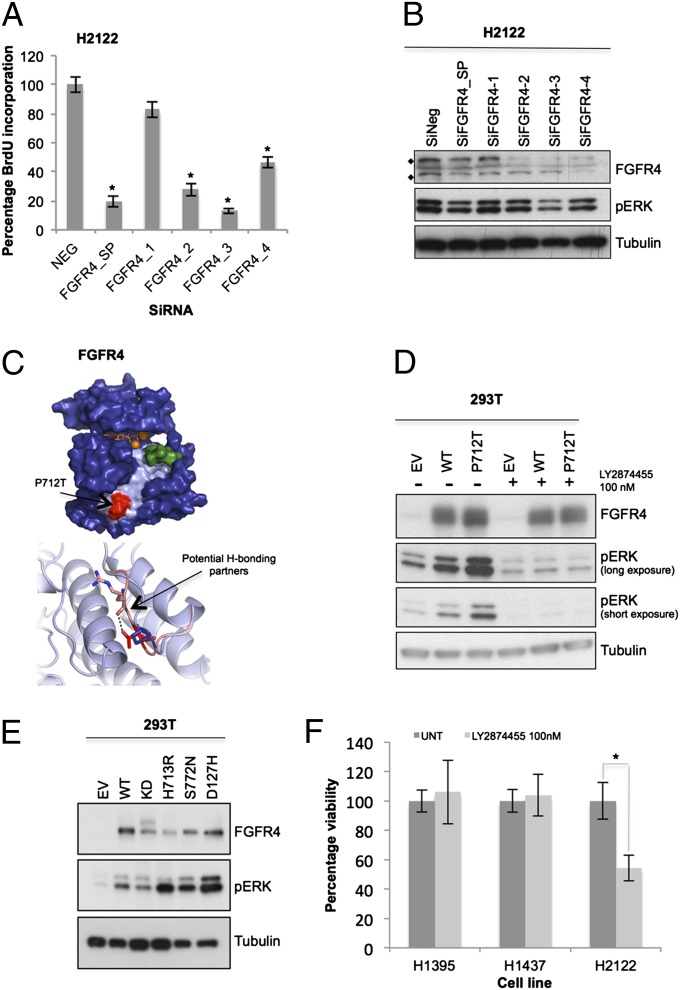
FGFR4 was identified as a significant driver of proliferation in the H2122 NSCLC cell line (SI Appendix, Table S3 and Fig. S2 D–F). This cell line harbors a P712T heterozygous somatic mutation (verified by resequencing) that lies in a surface-accessible PXXP motif in the C-terminal region of the kinase domain (Fig. 4C) (25). The FGFR family of receptor tyrosine kinases regulates myriad cellular fate decisions, including proliferation and cell survival (26). Depletion of FGFR4 from the H2122 cell line by using single siRNAs from the siRNA pool resulted in a significant decrease in BrdU incorporation (P < 0.001) and promoted cell death with three of four of the siRNA oligos (Fig. 4A and SI Appendix, Fig. S3A). Furthermore, depletion of FGFR4 from H2122 cells attenuated signaling in the PI3K/Akt and ERK pathways (Fig. 4B and SI Appendix, Fig. S3 B and C). To test if the P712T mutation is a GOF mutation, we expressed this mutant in HEK293T and H157 cells and observed hyperactivation of the ERK pathway compared with WT FGFR4 (Fig. 4D and SI Appendix, Fig. S3D). Treatment of cells with the FGFR-specific inhibitor LY2874455 abrogated FGFR4-dependent activation of the ERK pathway by WT and mutant FGFR4, indicating the P712T mutation does not alter drug binding (Fig. 4D). Additional FGFR4 mutants are also activating toward the ERK pathway compared with WT FGFR4 [Fig. 4E; note that H713R and S772N were observed in different lung cancer cell lines and the D127H mutation was observed in two distinct ovarian and three distinct hematopoietic cancer cell lines in the Cancer Cell Line Encyclopedia (CCLE) (12)]. We then tested if pharmacological inhibition of FGFR4 would promote cell death. Treatment of H2122 cells with the FGFR inhibitor LY2874455 resulted in a significant loss of cell viability (Fig. 4F) compared with two lung cancer cell lines that lacked mutations in FGFR family members (Fig. 4F; the inhibitor also decreased ERK signaling in the H2122 cells; SI Appendix, Fig. S3E). These data suggest that patients who harbor GOF mutations in FGFR4 may be responsive to FGFR inhibitors.
Fig. 4.
FGFR4 is a mutationally activated receptor tyrosine kinase essential for lung cancer cell proliferation. (A) FGFR4 was knocked down in H2122 cells (SMARTpool 100 nM; oligos 1–4, 25 nM). Plates were subjected to BrdU assay or MTT assay (SI Appendix, Fig. S3A), and data were normalized to nontargeting siRNA (NEG). (B) On-target effect of FGFR4 SMARTpool (100 nM) and individual oligos (25 nM) was also verified by Western blotting. Protein lysates were probed for FGFR4 protein level (♦, FGFR4 unmodified at 88 kDa and modified at 125 KDa) and inhibition of downstream signals of pERK compared with the nontargeting siRNA (SiNeg). (C, Upper) Model of the FGFR4 tyrosine kinase domain based on the crystal structure of FGFR2 (PDB ID 2PVF) bound to ATP and magnesium ions (orange sticks and spheres, respectively). Partial substrate (green) is bound into the potential binding pocket (pale blue). P712T mutation site is shown in red. (C, Lower) Potential additional hydrogen-bonding partners of the mutant structure (R670) as pink sticks compared with WT. (D) Empty vector (EV), FGFR4 WT, and P712T mutant were expressed ectopically in 293T cells, with or without treatment with the FGFR inhibitor LY2874455, 30 min before lysis. (E) FGFR4 cancer mutants were also generated and overexpressed in 293T cells for 48 h, compared with empty vector, WT, and kinase-dead K503M (KD). Lysates were collected, and downstream ERK phosphorylation was analyzed by Western blotting. (F) H2122 cells (FGFR4P712T) and H1395 and H1437 (FGFRWT) were treated with FGFR inhibitor and assayed by MTT. Data (n = 9) were normalized to untreated cells (UNT; DMSO, <0.01%). Error bars show SD. P values were calculated by using one-tailed paired Student t test (*P < 0.005 vs. nontargeting siRNA or untreated).
GOF Mutation in MAP3K9 Leads to Preferential Activation of the MEK/ERK Pathway.
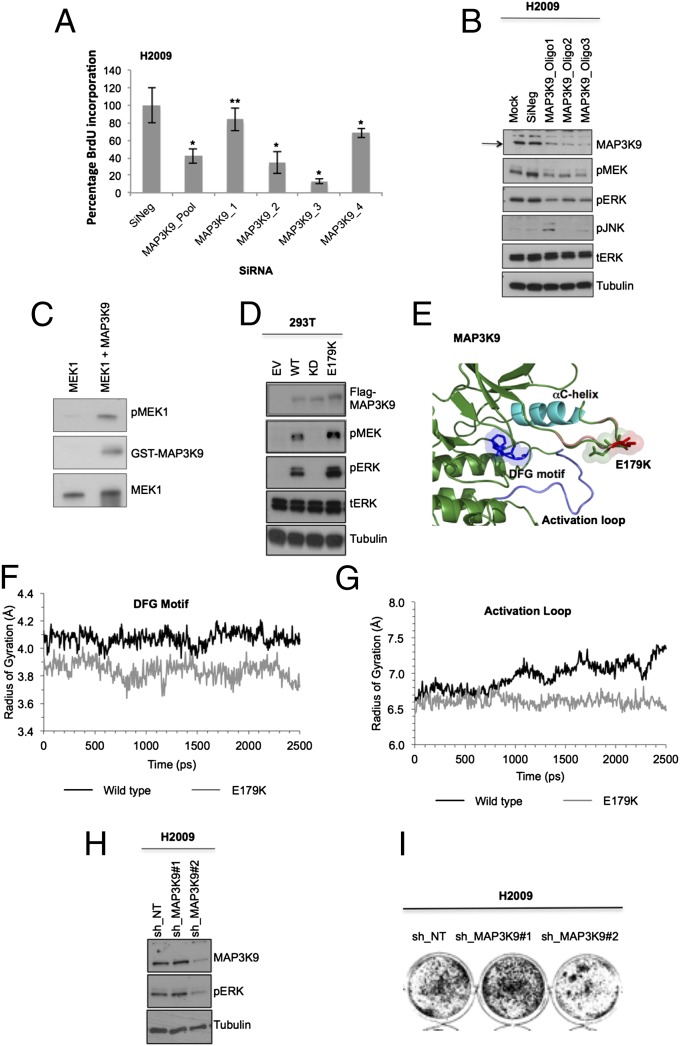
Finally, MAP3K9 was evaluated from the H2009 cells based on its potential as a drug target (27) and the fold change and SSMD values from the siRNA screen (SI Appendix, Table S4 and Fig. S2 G–I). MAP3K9 was identified to have a heterozygous somatic mutation (E179K) based on results from CCLE, and this was verified by resequencing. The CCLE database became available before screening this last cell line, and we therefore included MAP3K9 in our screen (SI Appendix, Table S1). MAP3K9 is a member of the mixed lineage family of kinases and is composed of an SH3 domain, Ser/Thr kinase domain, and a Cdc42/Rac interactive binding (CRIB) domain (28). This kinase is an upstream activator of the JNK and ERK pathways (29, 30). The role of MAP3K9 in cancer is not well defined, but mutations in this kinase were recently found in 15% of patients with metastatic melanoma (29). We performed a deconvolution experiment and observed that three of four siRNA oligos were specific for MAP3K9 and significantly decreased proliferation (P < 0.005; Fig. 5A and accompanying Western blot to demonstrate specificity of siRNAs; SI Appendix, Fig. S3F). Depletion of MAP3K9 from H2009 cells resulted in a significant decrease in MEK/ERK pathway activation, but no significant changes in the JNK pathway (Fig. 5B and SI Appendix, Fig. S3F). To test if MAP3K9 could directly phosphorylate MEK in vitro, we performed a kinase assay and observed that purified MAP3K9 kinase domain can phosphorylate kinase-dead MEK1 (Fig. 5C) (31). To determine if the E179K mutation is a GOF mutation, we expressed MAP3K9E179K in HEK293T cells along with a WT and kinase-dead MAP3K9 (D294A). Compared with WT, the MAP3K9E179K has increased activity toward the MEK/ERK pathway (Fig. 5D). The glutamate (E in sequence below) at position 179 lies in a flexible, unstructured, and highly polar loop (ARHDPDEDISQT) in the N-lobe of the kinase domain. Mutation from an acidic to a basic residue is likely to impact the preferred conformation of this loop (Fig. 5E). Molecular-dynamic simulations highlighted a role for this mutation in stabilizing the activation loop and DFG motif compared with MAP3K9WT (Fig. 5 F and G), leading to stabilization of an active conformation (32). Consistent with our results, induced depletion of MAP3K9 (shRNA2) resulted in attenuated MEK/ERK pathway signaling and a concomitant decrease in cell viability (Fig. 5 H and I). Combined, these data suggest that mutationally altered MAP3K9 is required to promote proliferation in the H2009 cells.
Fig. 5.
MAP3K9 harbors an activating mutation maintaining H2009 cell proliferation. (A) MAP3K9 was knocked down in H2009 cells (SMARTpool 100 nM; oligos 1–4, 50 nM). Plates were subjected to BrdU assay, and data were normalized to nontargeting siRNA (NEG). Error bars show SD. P values were calculated by using one-tailed paired Student t test (*P < 0.005 and **P < 0.05 vs. nontargeting siRNA control). (B) MAP3K9 was transiently knocked down in H2009 cells by different siRNA oligos at 100 nM (SI Appendix, Table S9), and downstream substrate phosphorylation was analyzed by Western blotting. (C) Kinase-inactive MEK1 and purified GST-MLK1 kinase domain (isolated from baculovirus-infected insect cells) were subjected to in vitro kinase assay. (D) Empty vector (EV), WT, kinase-dead (KD) D294A, and E179K mutants were expressed ectopically in 293T cells. Lysates were collected and analyzed by Western blotting to evaluate MEK/ERK activation relative to WT and kinase-dead. (E) Structural image of MAP3K9 highlighting the position of the mutation site (green/red sticks) in comparison with the DFG motif (dark blue sticks), αC-helix (cyan), and activation loop (blue). The region containing the E179K mutation (pink and green bubble) is located close to the activation loop (blue). Mutation site is housed within a highly polar loop. Mutation from the acidic glutamic acid (green sticks) to a basic lysine residue (red sticks) is likely to impact the preferred conformation. This is likely to disrupt potential hydrogen bonding opportunities occurring between E179 (green sticks) and activation loop, thus stabilizing activation loop in a potentially active confirmation. Image created in PyMol version 1.3. (F and G) Radius of gyration of the (F) DFG motif and (G) activation loop within WT MAP3K9 (black line) compared with MAP3K9E179K (gray line). Data show a reduction in the radius of gyration within the E179K mutation, suggesting a greater stability within these regions allowing less movement. Data shown are the average of three repeats generated by using GROMACS version 4.5.3. (H and I) MAP3K9 knockdown was induced with 1 mM IPTG in H2009 cells. Level of MAP3K9 knockdown and downstream ERK phosphorylation was evaluated by (H) Western blotting relative to nontargeting control (shNT), and effect on foci formation was assessed by (I) colony formation assay stained with crystal violet (scanned plate converted to black-and-white image).
FGFR4, MAP3K9, and PAK5 Mutation Positive Cell Lines Are Responsive to MEK Inhibition.
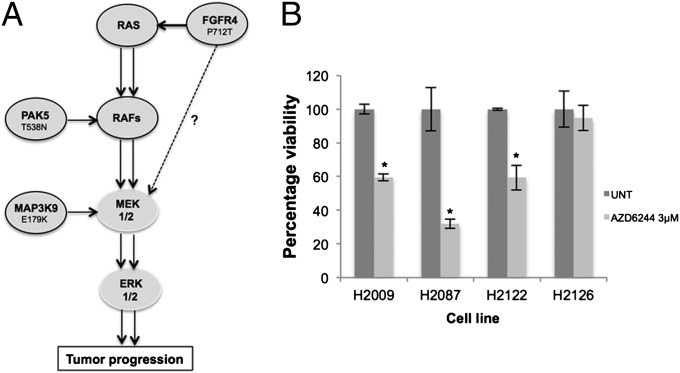
Cumulatively, our data highlight the importance of the MEK/ERK pathway in maintaining lung cancer cell proliferation and viability, and illustrate that there are multiple mechanisms to promote and enhance activation of this pathway (Fig. 6A). To determine if MEK inhibition would promote apoptosis in these three lung cancer cell lines, relative to the H2126 lung cancer cell line, which has no identifiable mutations in the MEK/ERK pathway, we treated the cells with a MEK inhibitor, AZD6244 (Fig. 6B). Consistent with our previous knockdown data, inhibition of the MEK/ERK pathway increased cell death specifically in those cell lines with mutations in FGFR4, MAP3K9, and PAK5, whereas the H2126 cell line was unaffected by MEK inhibition. In summary, our results suggest that patients with lung cancer with FGFR4, MAP3K9, or PAK5 mutations could be stratified for treatment with MEK inhibitors.
Fig. 6.
(A) Proposed mechanism of pathway hyperactivation in lung cancer. Intermediate driver mutants cooperate with previously established driver mutants such as BRAF and KRAS to further activate the MAPK proproliferative pathway. FGFR4P712T mediates pathway activation upstream of KRAS and possibly via additional intermediate substrates other than RAFs, PAK5T538N mediates activation at the RAF level, and MAP3K9E179K acts as a direct MEK kinase. (B) Four NSCLC cell lines (H2009, H2087, H2122 harboring MEK/ERK activating mutants, and H2126 with no identifiable MEK/ERK activating mutant) were treated with the MEK inhibitor AZD6244 at 3 µM, and cytotoxicity was determined by MTT assay. Data (n = 9) were normalized to untreated cells (UNT; DMSO, <0.01%). Error bars show SD. P values were calculated by using one-tailed paired Student t test (*P < 0.05 vs. untreated control).
Discussion
Cancer genomic studies are providing spatiotemporal portraits of the process of clonal evolution that each tumor undergoes en route to a malignant phenotype (33). For this information to become clinically relevant, we must adapt strategies to identify actionable mutations in the branch zones of the tumor as well as truncal regions (33). Our study provides a functional genomics approach that can be personalized to the mutational landscape of an individual’s tumor, thereby allowing the identification of mutationally activated essential genes in real time. Our targeted genetic dependency screen identifies unique actionable mutations, highlighting the potential of this approach to identify mutationally activated low frequency oncogenes, which could serve as biomarkers and possibly future targets for therapeutic intervention.
The three targets identified by our screen use different mechanisms to activate the MEK/ERK pathway (Fig. 6A), and the specific acquisition of these unique GOF mutations is likely dependent on the genetic makeup of the specific tumor. For example, the H2087 lung cancer cell line has accumulated several intermediate activating mutations in the MEK/ERK pathway that result in robust activation of this pathway. Given the importance of the cellular localization of signaling complexes, it may be that different mutationally activated complexes are used to achieve differing cellular outputs. For example, activation of the MEK/ERK pathway by PAK5T538N/CRAF could promote increased cell survival, whereas activation of the MEK/ERK pathway by BRAFL597V may increase cellular proliferation. For the P712T GOF mutation in FGFR4, this mutation occurs in a cell line with an activating KRAS mutation. Combining activated FGFR4 with activated KRAS may specifically direct mutant KRAS to activate the MEK/ERK pathway, suggesting that the presence of other mutations in a tumor may direct the signaling outputs of mutationally activated KRAS and have implications for combination therapies (34). Finally, we observed that the MAP3K9E179K mutation is a GOF mutation toward the MEK/ERK pathway and not the JNK pathway. The exact mechanism of activation is unknown, but our in vitro data suggest that MAP3K9 is directly activating MEK (Fig. 5C).
Although we focused on kinases as a result of their pharmacological tractability, several other interesting candidates were identified from our screen. For example, in the H2087 cell line, GATA2 was identified as an important driver of proliferation. The mutation in GATA2, P125T, is located in the transactivation domain, and it is interesting to speculate whether this mutation may promote activation of GATA2-mediated transcription. Indeed, GATA2 has been identified as an essential protein in KRAS-driven lung cancers, and GOF mutations in GATA2 were identified in 7 of 85 patients with chronic myelogenous leukemia in recent studies (35, 36). In addition, we identified a small GTPase RasL10b, as a potential mutationally activated enzyme in the H2122 cell line. It is unknown if this protein plays a role in tumorigenesis, but if the mutations promote the activation of the GTPase in an analogous manner to KRAS codon 12 and 13 activating mutations, the protein could promote activation of signaling pathways to enhance proliferation (37). Although we were able to successfully identify important functional somatic variants by using our approach, one limitation is that we did not include more complex genetic alterations such as translocations, and there is always a possibility that mutations can be missed from any global exon-based sequencing approach. In addition, false-positive findings as a result of depletion of essential house-keeper genes harboring nonfunctional passenger mutations could prove problematic; therefore, we emphasize the importance of assessing the functional impact of the genetic alterations on the target gene.
There is an urgent need for the identification and characterization of actionable mutations in lung cancer (4, 7). As patients’ exomes are increasingly being sequenced in real time, we will be presented with exciting opportunities to evaluate the genetic alterations that are promoting lung cancer cell viability (7). However, if clinicians are to provide targeted therapies aimed at these genetic alterations, then we must begin to catalogue actionable mutations, which can be used to dictate patient therapy. Here we provide a targeted approach to identify actionable mutations, and, combined, our data indicate that patients with FGFR4, MAP3K9, or PAK5 mutations could benefit from treatment with a MEK inhibitor [such as trametinib (GSK1120212), which is currently being used in clinical trials]. Alternatively, these findings could encourage the development of FGFR4, MAP3K9, and PAK5 inhibitors to be used in the clinic.
Methods
Statistical analysis, structural modeling, and other additional methods are described in SI Appendix.
Cells were reverse transfected with DharmaFECT1 (Thermo Scientific) using optimized conditions (SI Appendix, Fig. S1E) in 96-well plates arrayed with 100 nM siRNA oligos seeded from resuspended custom-designed siRNA library (Thermo Scientific; SI Appendix, Table S9). After 72 h, cells were assayed for BrdU incorporation (BrdU Cell Proliferation Assay; Calbiochem) or apoptosis (Apo-ONE Homogeneous Caspase-3/7 assay; Promega) according the to manufacturers’ protocols (SI Appendix, Fig. S1A). Each experimental setup was seeded in triplicate, including siGLO cyclophilin B transfection indicator control (Thermo Scientific), and at least three independent experiments were carried out (n = 9).
Supplementary Material
Acknowledgments
We thank Dr. Angeliki Malliri, Dr. Tony Hunter, and members of the Signalling Networks in Cancer Group for critical review of the manuscript. This research was fully supported by Cancer Research UK.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. I.K.M. is a guest editor invited by the Editorial Board.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1305207110/-/DCSupplemental.
References
- 1.Hammerman PS, et al. Mutations in the DDR2 kinase gene identify a novel therapeutic target in squamous cell lung cancer. Cancer Discov. 2011;1(1):78–89. doi: 10.1158/2159-8274.CD-11-0005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Yang P, et al. Clinical features of 5,628 primary lung cancer patients: Experience at Mayo Clinic from 1997 to 2003. Chest. 2005;128(1):452–462. doi: 10.1378/chest.128.1.452. [DOI] [PubMed] [Google Scholar]
- 3.Gazdar AF. Epidermal growth factor receptor inhibition in lung cancer: The evolving role of individualized therapy. Cancer Metastasis Rev. 2010;29(1):37–48. doi: 10.1007/s10555-010-9201-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Gerber DE, Minna JD. ALK inhibition for non-small cell lung cancer: From discovery to therapy in record time. Cancer Cell. 2010;18(6):548–551. doi: 10.1016/j.ccr.2010.11.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Dahabreh IJ, et al. Somatic EGFR mutation and gene copy gain as predictive biomarkers for response to tyrosine kinase inhibitors in non-small cell lung cancer. Clin Cancer Res. 2010;16(1):291–303. doi: 10.1158/1078-0432.CCR-09-1660. [DOI] [PubMed] [Google Scholar]
- 6.Lynch TJ, et al. Activating mutations in the epidermal growth factor receptor underlying responsiveness of non-small-cell lung cancer to gefitinib. N Engl J Med. 2004;350(21):2129–2139. doi: 10.1056/NEJMoa040938. [DOI] [PubMed] [Google Scholar]
- 7.Kim ES, et al. The BATTLE trial: Personalizing therapy for lung cancer. Cancer Discov. 2011;1(1):44–53. doi: 10.1158/2159-8274.CD-10-0010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Brognard J, Zhang YW, Puto LA, Hunter T. Cancer-associated loss-of-function mutations implicate DAPK3 as a tumor-suppressing kinase. Cancer Res. 2011;71(8):3152–3161. doi: 10.1158/0008-5472.CAN-10-3543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kaminker JS, et al. Distinguishing cancer-associated missense mutations from common polymorphisms. Cancer Res. 2007;67(2):465–473. doi: 10.1158/0008-5472.CAN-06-1736. [DOI] [PubMed] [Google Scholar]
- 10.Eifert C, Powers RS. From cancer genomes to oncogenic drivers, tumour dependencies and therapeutic targets. Nat Rev Cancer. 2012;12(8):572–578. doi: 10.1038/nrc3299. [DOI] [PubMed] [Google Scholar]
- 11.Greenman C, et al. Patterns of somatic mutation in human cancer genomes. Nature. 2007;446(7132):153–158. doi: 10.1038/nature05610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barretina J, et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature. 2012;483(7391):603–607. doi: 10.1038/nature11003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wan PT, et al. Cancer Genome Project Mechanism of activation of the RAF-ERK signaling pathway by oncogenic mutations of B-RAF. Cell. 2004;116(6):855–867. doi: 10.1016/s0092-8674(04)00215-6. [DOI] [PubMed] [Google Scholar]
- 14.Kittler R, et al. An endoribonuclease-prepared siRNA screen in human cells identifies genes essential for cell division. Nature. 2004;432(7020):1036–1040. doi: 10.1038/nature03159. [DOI] [PubMed] [Google Scholar]
- 15.Berezhna SY, Supekova L, Supek F, Schultz PG, Deniz AA. siRNA in human cells selectively localizes to target RNA sites. Proc Natl Acad Sci USA. 2006;103(20):7682–7687. doi: 10.1073/pnas.0600148103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhang XD. A new method with flexible and balanced control of false negatives and false positives for hit selection in RNA interference high-throughput screening assays. J Biomol Screen. 2007;12(5):645–655. doi: 10.1177/1087057107300645. [DOI] [PubMed] [Google Scholar]
- 17.Birmingham A, et al. Statistical methods for analysis of high-throughput RNA interference screens. Nat Methods. 2009;6(8):569–575. doi: 10.1038/nmeth.1351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang XD. A pair of new statistical parameters for quality control in RNA interference high-throughput screening assays. Genomics. 2007;89(4):552–561. doi: 10.1016/j.ygeno.2006.12.014. [DOI] [PubMed] [Google Scholar]
- 19.Zhang XD. Illustration of SSMD, z score, SSMD*, z* score, and t statistic for hit selection in RNAi high-throughput screens. J Biomol Screen. 2011;16(7):775–785. doi: 10.1177/1087057111405851. [DOI] [PubMed] [Google Scholar]
- 20.Eswaran J, Soundararajan M, Kumar R, Knapp S. UnPAKing the class differences among p21-activated kinases. Trends Biochem Sci. 2008;33(8):394–403. doi: 10.1016/j.tibs.2008.06.002. [DOI] [PubMed] [Google Scholar]
- 21.Wu X, Carr HS, Dan I, Ruvolo PP, Frost JA. p21 activated kinase 5 activates Raf-1 and targets it to mitochondria. J Cell Biochem. 2008;105(1):167–175. doi: 10.1002/jcb.21809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Qu J, et al. Activated PAK4 regulates cell adhesion and anchorage-independent growth. Mol Cell Biol. 2001;21(10):3523–3533. doi: 10.1128/MCB.21.10.3523-3533.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Heidorn SJ, et al. Kinase-dead BRAF and oncogenic RAS cooperate to drive tumor progression through CRAF. Cell. 2010;140(2):209–221. doi: 10.1016/j.cell.2009.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Poulikakos PI, Zhang C, Bollag G, Shokat KM, Rosen N. RAF inhibitors transactivate RAF dimers and ERK signalling in cells with wild-type BRAF. Nature. 2010;464(7287):427–430. doi: 10.1038/nature08902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pawson T, Warner N. Oncogenic re-wiring of cellular signaling pathways. Oncogene. 2007;26(9):1268–1275. doi: 10.1038/sj.onc.1210255. [DOI] [PubMed] [Google Scholar]
- 26.Eswarakumar VP, Lax I, Schlessinger J. Cellular signaling by fibroblast growth factor receptors. Cytokine Growth Factor Rev. 2005;16(2):139–149. doi: 10.1016/j.cytogfr.2005.01.001. [DOI] [PubMed] [Google Scholar]
- 27.Wang LH, Johnson EM., Jr Mixed lineage kinase inhibitor CEP-1347 fails to delay disability in early Parkinson disease. Neurology. 2008;71(6):462. doi: 10.1212/01.wnl.0000324506.93877.5e. [DOI] [PubMed] [Google Scholar]
- 28.Gallo KA, Johnson GL. Mixed-lineage kinase control of JNK and p38 MAPK pathways. Nat Rev Mol Cell Biol. 2002;3(9):663–672. doi: 10.1038/nrm906. [DOI] [PubMed] [Google Scholar]
- 29.Stark MS, et al. Frequent somatic mutations in MAP3K5 and MAP3K9 in metastatic melanoma identified by exome sequencing. Nat Genet. 2012;44(2):165–169. doi: 10.1038/ng.1041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kant S, et al. TNF-stimulated MAP kinase activation mediated by a Rho family GTPase signaling pathway. Genes Dev. 2011;25(19):2069–2078. doi: 10.1101/gad.17224711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Anastassiadis T, Deacon SW, Devarajan K, Ma H, Peterson JR. Comprehensive assay of kinase catalytic activity reveals features of kinase inhibitor selectivity. Nat Biotechnol. 2011;29(11):1039–1045. doi: 10.1038/nbt.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Van Der Spoel D, et al. GROMACS: Fast, flexible, and free. J Comput Chem. 2005;26(16):1701–1718. doi: 10.1002/jcc.20291. [DOI] [PubMed] [Google Scholar]
- 33.Gerlinger M, et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med. 2012;366(10):883–892. doi: 10.1056/NEJMoa1113205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zakrzewska M, et al. ERK-mediated phosphorylation of fibroblast growth factor receptor 1 on Ser777 inhibits signaling. Sci Signal. 2013;6(262):ra11. doi: 10.1126/scisignal.2003087. [DOI] [PubMed] [Google Scholar]
- 35.Kumar MS, et al. The GATA2 transcriptional network is requisite for RAS oncogene-driven non-small cell lung cancer. Cell. 2012;149(3):642–655. doi: 10.1016/j.cell.2012.02.059. [DOI] [PubMed] [Google Scholar]
- 36.Zhang SJ, et al. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci USA. 2008;105(6):2076–2081. doi: 10.1073/pnas.0711824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lin ZY, Chuang WL. Genes responsible for the characteristics of primary cultured invasive phenotype hepatocellular carcinoma cells. Biomed Pharmacother. 2012;66(6):454–458. doi: 10.1016/j.biopha.2012.04.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.