Abstract
Humans are colonized by immense populations of viruses, which metagenomic analysis shows are mostly unique to each individual. To investigate the origin and evolution of the human gut virome, we analyzed the viral community of one adult individual over 2.5 y by extremely deep metagenomic sequencing (56 billion bases of purified viral sequence from 24 longitudinal fecal samples). After assembly, 478 well-determined contigs could be identified, which are inferred to correspond mostly to previously unstudied bacteriophage genomes. Fully 80% of these types persisted throughout the duration of the 2.5-y study, indicating long-term global stability. Mechanisms of base substitution, rates of accumulation, and the amount of variation varied among viral types. Temperate phages showed relatively lower mutation rates, consistent with replication by accurate bacterial DNA polymerases in the integrated prophage state. In contrast, Microviridae, which are lytic bacteriophages with single-stranded circular DNA genomes, showed high substitution rates (>10−5 per nucleotide each day), so that sequence divergence over the 2.5-y period studied approached values sufficient to distinguish new viral species. Longitudinal changes also were associated with diversity-generating retroelements and virus-encoded Clustered Regularly Interspaced Short Palindromic Repeats arrays. We infer that the extreme interpersonal diversity of human gut viruses derives from two sources, persistence of a small portion of the global virome within the gut of each individual and rapid evolution of some long-term virome members.
Keywords: metagenomics, microbiome, diversity generating retroelement, CRISPR
There are an estimated 1031 viral particles on earth, and human feces contain at least 109 virus-like particles per gram (1–3). Many of these are identifiable as viruses that infect bacteria (bacteriophages), but the great majority remains unidentified. Even today, gut virome samples taken from different human individuals still yield mostly novel viruses (4–8), and only a small minority of viral ORFs resembles previously studied genes (7).
Bacteriophages are of biomedical importance because of their ability to transmit genes to their bacterial hosts, thereby conferring increased pathogenicity, antibiotic resistance, and perhaps new metabolic capacity (4, 5, 9, 10). Despite their importance, the forces diversifying bacteriophage genomes in human hosts have not been studied in detail. Humans show considerable individual variation in the bacterial lineages present in their guts (11–13); this variation likely is one reason for the differences in their phage predators (5–8, 14). The large differences in phage populations among individuals also may be influenced by within-individual viral evolution.
To investigate the origin and nature of human viral populations, we carried out a detailed study of a single human gut viral community. Ultra-deep longitudinal analysis of DNA sequences from the viral community, combined with characterization of the host bacteria, revealed rapid change over time and begins to specify some of the mechanisms involved.
Results
Sample Collection, Viral Purification, and DNA Sequencing.
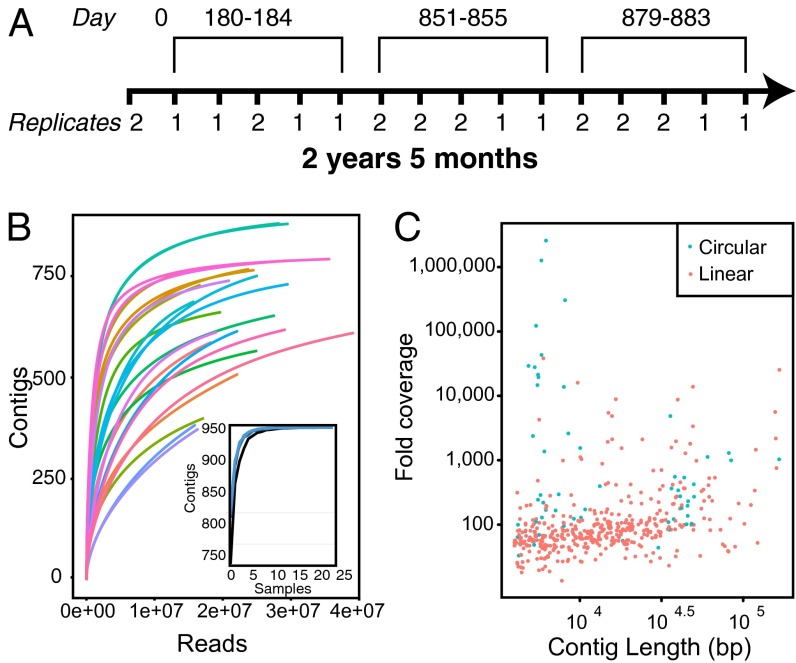
Stool samples (n = 24) were collected from a healthy male at 16 time points spread over 884 days (Fig. 1A). For eight of the time points, two separate samples taken 1 cm apart were purified and sequenced independently to allow estimation of within-time point sample variation. Virus-like particles were extracted by sequential filtration, Centricon ultrafiltration, nuclease treatment, and solvent extraction. Purified viral DNA was subjected to linear amplification using Φ29 DNA polymerase, after which quantitative PCR showed that bacterial 16S sequences were reduced to less than 10 copies per nanogram of DNA, and human sequences were reduced to below 0.1 copies per nanogram, the limit of detection. Paired-end reads then were acquired using Illumina HiSeq sequencing, yielding more than 573 million reads (Q ≥ 35; mean read length, 97.5 bp), with 15–39 million reads per sample (Table S1). No attempt was made to study gut RNA viruses, which also are known to exist, although some samples were dominated by abundant plant RNA viruses ingested with food (15).
Fig. 1.
Longitudinal analysis of the human gut virome from a single individual. (A) Timeline of sample collection. Note that at some time points, two separate portions of the stool sample, taken approximately 1 cm apart, were processed and sequenced independently to assess reproducibility. (B) Rarefaction analysis of sampling depth by number of reads; detection of each contig is scored as positive if 50% of the contig is covered by sequence reads. (Inset) Contig recovery. The x-axis is the number of samples included (black line: 2 million reads; blue line: 15 million reads). (C) Contig spectrum, relating the lengths of the contigs assembled in bas pairs (x-axis) to the depth of coverage (y-axis). Circular contigs are shown as blue and linear contigs as red.
Sequence reads from each sample were first assembled individually using MetaIDBA (16). When reads were aligned back onto contigs generated within each sample, only 71% of reads could be aligned. Improved contigs then were generated using a hybrid assembly method combining all samples, taking advantage of the fact that viruses that are rare at one time point may be abundant at another. After this step, 97.6% of the reads could be aligned to contigs, allowing assessment of within-contig diversity. Rarefaction (collector’s curve) analysis showed that the detection of these contigs was saturated at <107 reads per sample and at 7–10 samples (Fig. 1B), well below our sampling effort. After quality filtering and manual editing, 478 contigs showed >20-fold coverage (median, 82-fold); from the purification results, we infer these contigs to be mostly or entirely DNA viruses (Fig. 1C). Sixty contigs assembled as closed circles (ranging in size from 4–167 kb), an indication of probable completion of these genome sequences, providing an estimate of the viral population size and composition in unprecedented detail. One circular genome was sequenced independently using the Sanger method and was confirmed to have the structure predicted from the Solexa/Illumina data (SI Methods). The abundance of each contig at each time point was measured by the proportion of reads that aligned to it, normalized to the length of each contig. The correlation coefficient between replicate samples from the same time point was at least 0.99, indicating a high degree of reproducibility (Fig. S1).
Viral Groups Detected.
Taxonomic analysis of these contigs indicated recovery of Microviridae, Podoviridae, Myoviridae, and Siphoviridae, but contigs with taxonomic attributions were a minority, only 13%, emphasizing the enormous sequence variation present in bacteriophages. Microviridae (the group including ΦX174) predominated, but this predominance could be a consequence of favored amplification by Φ29 polymerase of the small circular genomes that characterize this group.
The most abundant contigs were mostly retained over the duration of the experiment. Because there are many possible pairwise comparisons between time points, distances between time points analyzed (Fig. 2A, x-axis) were compared with Jaccard index values (Fig. 2A, y-axis), which score shared membership, over all of the possible pairwise comparisons of time points. On average, more than 80% of contigs were found in common between the time points separated by 850 d (points at the right side of the plot), the longest time intervals compared.
Fig. 2.
Stability and change in the gut virome of the individual studied. (A) Conserved membership in the viral community over time intervals analyzed using the Jaccard index. Because many pairwise comparisons are possible between the 24 time points, we plotted shared membership for all pairs of time points as a function of the length of time between each pair. The x-axis shows the time interval between time points, and the y-axis shows shared membership in the two communities compared summarized using the Jaccard index. Perfect identity yields a value of 1, and complete divergence yields a value of 0. (B) Comparison of substitution rates among viral families. Temperate phages are shown in blue, and lytic phages are in red. The viral families studied are shown at the bottom; substitution rates on the y-axis are substitutions per base, per day. Only contigs with clear taxonomic attributions were analyzed; such contigs comprise a minority of all contigs.
No contigs corresponded to known viruses infecting eukaryotic cells. To investigate the possible presence of eukaryotic cell viruses further, we aligned the raw sequence reads to the National Center for Biotechnology Information viral genome database. Thirty-two percent coverage was seen for Gyrovirus in one time point, and pooling reads over all time points yielded 42% coverage. Gyrovirus is a Circovirus genus with very small genome sizes (∼2.3 kb) recently reported to infect humans (17). However, the number of reads aligning was modest (10 total), and in no case did both reads of the paired end reads align. Because of these results, and in addition to the small target size, we believe that the detection of Gyrovirus is uncertain. All other animal cell virus genomes showed <10% coverage, so detection is questionable. The rarity of eukaryotic virus sequences is typical of gut virome samples from healthy individuals (4–6, 18, 19), emphasizing the tremendous size of the bacteriophage populations of the gut.
Host Bacteria.
To allow tracking of the bacterial hosts, for three of the time points we also sequenced a total of 5.2 Gb of DNA purified from unfractionated stool, which yields predominantly bacterial DNA. Attribution of bacterial lineages using MetaPhlAn (20) showed members of the Bacteroides and Firmicutes phyla to be the most abundant community members (Fig. S2). Bacterial community membership and taxonomic proportions showed only modest variation over time.
Longitudinal Base Substitution in Viral Contigs.
The depth of sequence information available and the quality of the viral contigs allowed a detailed assessment of the rates of accumulation of base substitutions. For each viral contig at each time point, the extent of nucleotide polymorphism was determined by aligning reads within each sample. The extent of nucleotide substitution then was compared for each contig between time points, and substitution frequencies were correlated with biological features.
Substitution rates varied with viral family and replication style (Fig. 2B). The Microviridae showed the highest substitution rate (P < 0.004). Microviridae package ssDNA genomes, which have been reported to show higher mutation rates than dsDNA genomes in vitro (21, 22), and this study confirms this result in a human host. The Podo-, Myo-, and Siphoviridae all package dsDNA genomes and showed lower substitution frequencies. The lowest substitution rates were seen for temperate bacteriophage (P = 0.015, Kruskal–Wallace test), which can integrate into the host bacterial genome. Temperate phages were identified as contigs satisfying at least one of three criteria: (i) encoding integrase genes, (ii) homologs present as prophage in sequenced bacterial genomes, or (iii) annotated as resembling previously studied temperate phage (5). When integrated, temperate phage DNA is replicated by high-fidelity bacterially encoded machinery, and temperate phage also may undergo fewer lytic replication cycles; both result in lower substitution rates. Temperate bacteriophage showed significantly lower substitution rates even when Microviridae were excluded from the comparison (P = 0.044). There was no significant difference in rates among the families of large dsDNA viruses.
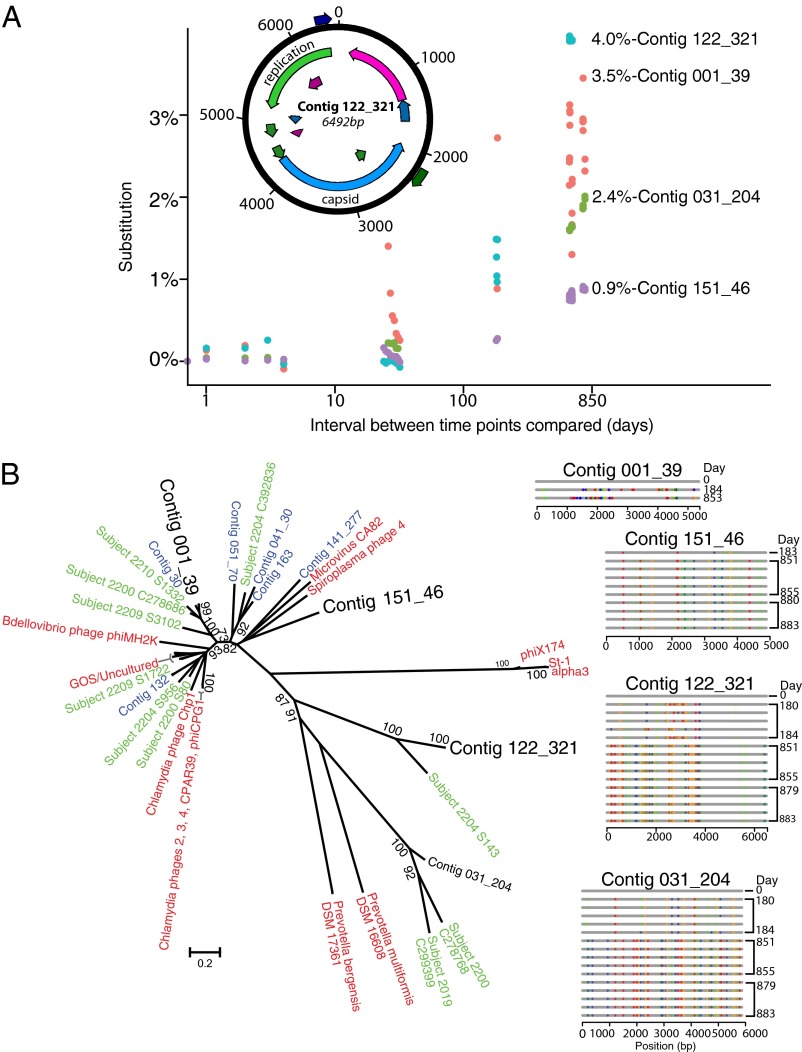
The four contigs with the highest rate of nucleotide substitution were all members of the Microviridae (Fig. 3A). The main variant for each lineage showed 1–4% nucleotide substitutions over the course of the experiment (more than one substitution per 105 nt per day). An alternative explanation for these high substitution rates could be the immigration of new closely related Microviridae into the community. To investigate this possibility, we reconstructed the consensus genome for the four contigs at multiple time points and aligned them against a large collection of Microviridae genomes. In every case the contig consensus sequences for all time points clustered closely together (Fig. 3B), arguing against immigration of related Microviridae and supporting the model of continuous substitution in long-term viral residents.
Fig. 3.
Longitudinal DNA substitution in Microviridae. (A) Substitution rates in the four Microviridae genomes with the highest values measured. Because many pairwise comparisons are possible between the time points at which each virus was detected, the plot shows distances between time points on the x-axis and the percent substitution on the y-axis. The percent substitution values within each time point were subtracted from the between-time point values before the plot was constructed. Colors differentiate the four viruses studied. (Inset) The genome with the highest substitution rate (contig 122_321). (B) Phylogenetic tree of microphages detected in this and other studies. The four microphage contigs with the highest substitution rates observed in this study are shown in large black lettering. Database microphages are shown in red, microphages from ref. 6 are shown in green, and additional microphages identified in this study are shown in blue. (Scale bar: the proportion of amino acid substitutions within the 919-aa major coat protein, which was aligned to make the tree.) Longitudinal maps of substitution accumulation are shown to the right. Note that all of the variations shown in the sequences to the right are plotted in the phylogenetic tree but are not visible because of the comparatively low divergence. Only time points with high-quality complete-genome assemblies are shown.
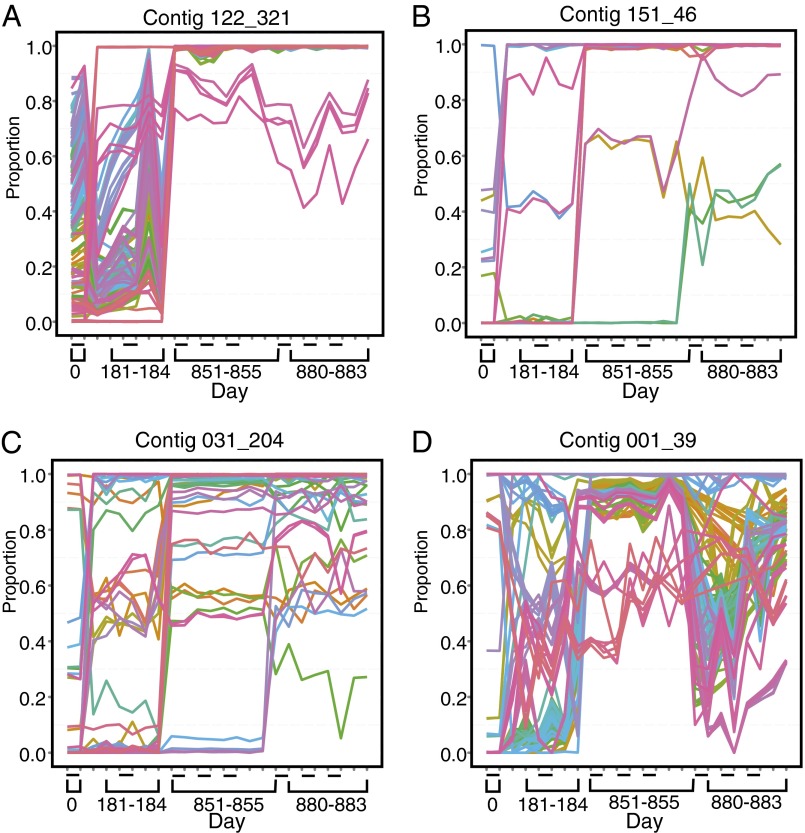
A detailed analysis of the longitudinal change of each SNP detected (Fig. 4) showed that a complex community of variants was present at most time points and that new SNPs accumulated on this background. Substitutions could accumulate either at a steady rate or in an episodic fashion, for example in response to a change in selective pressure. Linear modeling of substitution rates versus time showed correlation coefficients of 0.91–0.99, consistent with generally steady substitution rates, although with considerable sample-specific fluctuations. Longitudinal sequence divergence in major variants predicted from the Illumina data were confirmed using Sanger sequencing for two of the Microviridae (described in SI Methods).
Fig. 4.
Relative abundance of SNPs in four Microviridae genomes analyzed longitudinally. Contigs studied are marked above each figure panel. The x-axis shows elapsed time since the start of the study. The y-axis shows the relative proportion of each variant in the population. The dashes on the x-axis show replicate analysis of single time points, allowing assessment of within-time point variability. Only positions with SNPs that transitioned from minor (<0.5) to major (>0.5) are plotted. The colors are used to make the different positions easier to visualize. Panel labels A–D show data for the contigs indicated at the top of each panel.
Clustered Regularly Interspaced Short Palindromic Repeats Targeting Phage Genomes.
One force driving phage sequence variation is the bacterial Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) system (23–26). DNA sequences from invaders such as bacteriophage or plasmids are incorporated as spacers into arrays in the bacterial genome. Transcription of such arrays allows the CRISPR spacer RNAs to be incorporated into nucleoprotein effector complexes that target the destruction of sequence-complementary invaders. Thus, bacteriophages are under pressure to mutate to evade degradation by the CRISPR system, as has been documented in model systems (23–25, 27). The deep analysis of viral sequences presented here, together with the shotgun metagenomic analysis of host bacterial sequences, allowed the influence of the CRISPR system in vivo to be studied in detail. A total of 34 types of CRISPR repeat sequences and their associated spacers were identified in the bacterial metagenomic sequence. Table 1 shows that several of these spacers targeted contigs from the virome sequence data. Up to 28 spacers could be identified targeting a single viral contig.
Table 1.
CRISPR arrays from bacterial metagenomic sequence targeting viral contigs detected in this study
| CRISPR | Organism hosting CRISPR | No. of spacers associated with repeat | Median spacer length (bp) | Viral contig targeted | No. of spacers matching viral contig |
| CRISPR-2 | Ruminococcus bromii L2-63 (temperate phage) | 64 | 30 (29–31) | 232_308 | 1 |
| CRISPR-3 | Unknown | 38 | 30 (21–33) | 112_6 | 2 |
| CRISPR-7 | Unknown | 64 | 36 (22–40) | 051_116 | 1 |
| 75 | 1 | ||||
| CRISPR-21 | Unknown | 59 | 35 (30–38) | 111_52 | 4 |
| CRISPR-31 | Eubacterium siraeum V10Sc8a | 110 | 37 (25–40) | 132_57 | 1 |
| CRISPR-32 | Eubacterium siraeum V10Sc8a | 230 | 37 (22–46) | 132_57 | 27 |
| CRISPR-37 | Bacteroides fragilis NCTC 9343 | 32 | 30 (29–30) | 111_52 | 1 |
The CRISPR-targeted viral contigs were analyzed for their relative abundance over time. No simple pattern was seen relating the presence of CRISPR spacers to the relative abundance over all of the targeted viruses. In one case, a viral contig accumulated a base substitution in a CRISPR target site, and the mutant contig increased in abundance while the original contig declined, suggestive of CRISPR evasion by mutation (Fig. S3).
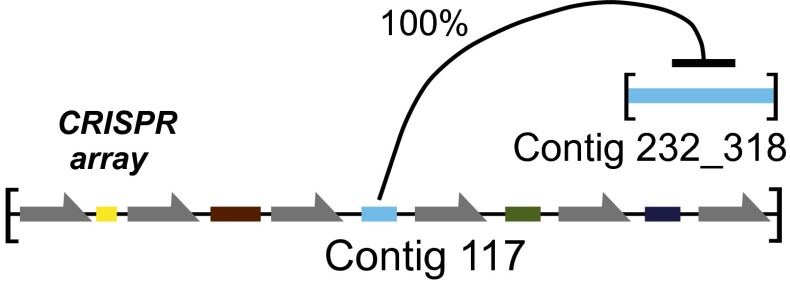
Of the CRISPR arrays identified, four appeared to be encoded by temperate phage. Several previous reports also have documented phage-encoded CRISPR arrays (5, 28, 29). An analysis of longitudinal variation in phage CRISPR arrays would be useful, but uncertainties in reconstructing arrays from short read data precluded a detailed analysis. For the CRISPR array with the most sequence coverage (contig 117), we found that the entire collection of spacers was replaced over the time series studied.
The phage-encoded CRISPR array on phage contig 117 encoded spacers that targeted four different phage contigs from our study (Fig. 5 shows one example). We previously reported another example from a different subject of a phage-encoded CRISPR spacer targeting a different phage in the same virome sample (5). Evidently phages commonly use CRISPR systems to compete with one another.
Fig. 5.
A phage-encoded CRISPR array targeting another phage. The array shown (contig 117) was detected in the viral contig collection. Gray indicates CRISPR repeats, and colors indicate CRISPR spacers. The target contig (contig 102) also was identified and observed to be present at some of the same time points; three other contigs also were targeted by the CRISPR array in contig 117. The CRISPR array in viral contig 117 is closely similar to CRISPR-2 detected in the total stool metagenomic sequencing.
Identifying Phage Hosts.
Characterization of bacteriophage populations by sequencing typically does not specify the host bacterial species, leaving important gaps in our understanding of phage–host interactions. Analysis of CRISPRs, however, provides a means of connecting phage–host pairs (Table 1). Three previously sequenced bacterial genomes, from Ruminococcus bromii, Eubacterium siraeum, and Bacteriodes fragilis, contain CRISPR repeats that were found here linked to spacers matching virome contigs from this study (contig 232_308, contig 132_57, and contig 111_52, respectively), allowing us to infer that these phages infect these three bacteria in the subject studied. In another approach to associating phage–host pairs, phage sequences annotated as integrated prophages in sequenced bacterial genomes could be recognized that resembled our newly sequenced phage contigs, thereby also specifying potential hosts (4–6). Bacterial lineages identified as harboring phage from the virome analysis included Bacteroides fragilis, Eubacterium siraeum, Ruminococcus bromii, Blautia hansenii, and Lachnospiraceae, all of which were found to be present in metagenomic sequence analysis of total stool DNA (Fig. S2). Overall, 19 of the phage contigs sequenced here could be associated with bacterial hosts by at least one of the two approaches (Table S2), although for the great majority the hosts remain unknown.
Longitudinal Sequence Variation Driven by Diversity-Generating Retroelements.
Another force diversifying bacteriophage genomes are diversity-generating retroelements (DGRs), which are reverse transcriptase-based systems that introduce mutations at adenines in specific repeated sequences using a copy–paste targeting mechanism (6, 30–33). We analyzed the viral contigs described here to investigate whether DGRs were detectably active within the human gut. DGRs were identified by searching contigs for regions that matched three criteria: (i) they contained protein-coding regions resembling reverse transcriptases, (ii) they encompassed short repeat regions containing mismatches in adenine positions, and (iii) they contained hypervariable regions. Of the 20 contigs with both a reverse transcriptase and an adenine-mismatched repeat, six were associated with hypervariable regions (located no more than 100 bp away; Table S3) and were selected for further study. As was found previously, hypervariation was directed toward asparagine AAY codons in genes encoding either predicted C-type lectin or Ig-superfamily proteins (6, 30–33).
We next asked whether any of the DGRs were detectably active over the time series studied. The longest gap between sample collections was 22 mo, so to maximize sensitivity we asked whether the hypervariable regions had evolved to become clearly different over this time interval. Of the two hypervariable regions with sufficient longitudinal coverage for analysis, one (contig 42) showed change over the 22-mo time period, and change was greater than for samples closer together in time (P < 0.0001) or for pairs of samples from the same time point (P < 0.0001). For the second (contig d03-2), we did not obtain evidence for longitudinal variation. We conclude that one of our DGRs was active in the human gut. For the others, it is unclear whether they were inactive or whether we did not have enough sequence coverage to detect activity. Analysis showed that DGR-containing contigs were not among the most variable, highlighting the local nature of DGR variation and emphasizing the contributions of other mechanisms. The possibility that some of the DGRs were inactive raises the question of whether the mutagenic activity might be regulated in the human host.
Discussion
Here we report a study of longitudinal variation in the human gut virome and some of the mechanisms responsible for change over time. Loss and acquisition of viral types was uncommon: Fully ∼80% of viral forms persisted over the 2.5-y time course studied, as is consistent with previous studies of shorter duration (4–6). Most viral contigs showed diversity within each time point and accumulated variation over time. Temperate DNA phages showed relatively modest rates of variation compared with lytic phage, as is consistent with temperate phage DNA replication by accurate bacterial polymerases in the prophage state, and potentially fewer total rounds of replication. In contrast, the strictly lytic ssDNA Microviridae showed up to 4% substitutions in the major variants present over the time period studied. DGRs showed high diversity in variable repeat regions, and one was detectably active over the time series studied. CRISPR arrays encoded in viral genomes also were associated with longitudinal variation. Thus, multiple mechanisms contributed to viral sequence variation, and our data provide a detailed picture of their relative contributions.
This study did not yield any clear examples of known DNA viruses infecting animal cells. Rare reads did align to genomes of animal cell viruses, but it is uncertain whether these alignments represent true detection of these viruses or rare regions of homology between animal cell viruses and phages. In contrast, several studies have reported frequent detection of animal cell viruses in metagenomic analysis of stool DNA from humans and other primates, raising the question of how these studies differed. One observation is that samples from sick individuals (34, 35) or SIV-infected macaques (36) have yielded animal cell viruses more frequently than samples from healthy controls. Some of these studies did not attempt to analyze bacterial viruses, instead using bioinformatic filters to extract animal cell viruses from complex sequence mixtures, potentially leading to an under-appreciation of the size of the phage populations. Thus, our data emphasize that in the healthy human gut bacterial viruses are much more numerous than animal cell viruses, although it remains possible that some of our contigs with no database matches correspond to previously unknown viruses infecting human cells.
Given the findings reported here, we can return to the question of why human gut viromes differ so greatly among human individuals. One factor must be the differences in bacterial populations in the guts of different humans. Many metagenomic studies emphasize that, although the human gut typically contains bacteria from only a few phyla, the bacterial strains are mostly different between individuals (11–13). Phages can be highly selective for different bacterial lineages—indeed, phage sensitivity is used clinically to distinguish some bacterial strains (e.g., refs. 37 and 38)—likely explaining some of the differences in phage populations in different individuals.
However, a second basis for the differences among individuals, highlighted in data reported here, is rapid within-host viral evolution. Microviridae lineages showed up to 4% substitution in the main variant over the 2.5-y period studied, consistent with laboratory experiments also showing high mutation rates for Microviridae (39). There is no single threshold of sequence identity accepted for splitting related viruses into separate species (40), but different Microviridae species specified by the International Committee on Taxonomy of Viruses show as little as 3.1% divergence (Table S4). Evidently the divergence seen here for Microviridae contigs 122_321 and 001_39 approaches the level sufficient for designation as speciation events. Extrapolating from these rates, our data suggest that multiple new viral species commonly will arise in the gut of a typical human over the course of a human life. Thus, part of the explanation for the extremely large populations of gut viruses inferred from sequence information and for the extreme differences among individual humans appears to be rapid within-individual evolution of long-term viral residents.
Methods
Longitudinal stool samples were collected from a single healthy male adult under a protocol approved by the Internal Review Board of the Perelman School of Medicine at the University of Pennsylvania. Samples of viral particles were purified by filtration, Centricon ultrafiltration, and nuclease treatment, and then total DNA was extracted using the QIAamp DNA Stool kit. Sequence information was acquired using Illumina paired-end technology. Sequences were assembled by iterative deBruijn graph assembly using MetaIDBA, and contigs were combined using Minimo. Taxonomy was assigned using Blastp, ORFs were predicted using Glimmer, and bacterial taxa were called using Metaphlan. Oligonucleotides used in this study are presented in Table S5. All sequence information has been deposited at the National Center for Biotechnology Information. For further details see SI Methods.
Supplementary Material
Acknowledgments
We thank members of the F.D.B. laboratory for help and suggestions. This work was supported by Human Microbiome Roadmap Demonstration Project Grant UH2DK083981 (to G.D.W., F.D.B., and J.D.L.), the Penn Genome Frontiers Institute, and the University of Pennsylvania Center for AIDS Research Grant P30 AI 045008. S.M. was supported by National Institutes of Health Training Grant T32AI060516.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The sequences reported in this paper have been deposited in the National Center for Biotechnology (NCBI) database, www.ncbi.nlm.nih.gov (accession no. SRP021107).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1300833110/-/DCSupplemental.
References
- 1.Rohwer F. Global phage diversity. Cell. 2003;113(2):141. doi: 10.1016/s0092-8674(03)00276-9. [DOI] [PubMed] [Google Scholar]
- 2.Schoenfeld T, et al. Functional viral metagenomics and the next generation of molecular tools. Trends Microbiol. 2010;18(1):20–29. doi: 10.1016/j.tim.2009.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Suttle CA. Viruses in the sea. Nature. 2005;437(7057):356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 4.Reyes A, et al. Viruses in the faecal microbiota of monozygotic twins and their mothers. Nature. 2010;466(7304):334–338. doi: 10.1038/nature09199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Minot S, et al. The human gut virome: inter-individual variation and dynamic response to diet. Genome Res. 2011;21(10):1616–1625. doi: 10.1101/gr.122705.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Minot S, Grunberg S, Wu GD, Lewis JD, Bushman FD. Hypervariable loci in the human gut virome. Proc Natl Acad Sci USA. 2012;109(10):3962–3966. doi: 10.1073/pnas.1119061109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Minot S, Wu GD, Lewis JD, Bushman FD. Conservation of gene cassettes among diverse viruses of the human gut. PLoS ONE. 2012;7(8):e42342. doi: 10.1371/journal.pone.0042342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Reyes A, Semenkovich NP, Whiteson K, Rohwer F, Gordon JI. Going viral: next-generation sequencing applied to phage populations in the human gut. Nat Rev Microbiol. 2012;10(9):607–617. doi: 10.1038/nrmicro2853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O’Brien AD, et al. Shiga-like toxin-converting phages from Escherichia coli strains that cause hemorrhagic colitis or infantile diarrhea. Science. 1984;226(4675):694–696. doi: 10.1126/science.6387911. [DOI] [PubMed] [Google Scholar]
- 10.Waldor MK, Mekalanos JJ. Lysogenic conversion by a filamentous phage encoding cholera toxin. Science. 1996;272(5270):1910–1914. doi: 10.1126/science.272.5270.1910. [DOI] [PubMed] [Google Scholar]
- 11.Turnbaugh PJ, et al. A core gut microbiome in obese and lean twins. Nature. 2009;457(7228):480–484. doi: 10.1038/nature07540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yatsunenko T, et al. Human gut microbiome viewed across age and geography. Nature. 2012;486(7402):222–227. doi: 10.1038/nature11053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wu GD, et al. Linking long-term dietary patterns with gut microbial enterotypes. Science. 2011;334(6052):105–108. doi: 10.1126/science.1208344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rodriguez-Valera F, et al. Explaining microbial population genomics through phage predation. Nat Rev Microbiol. 2009;7(11):828–836. doi: 10.1038/nrmicro2235. [DOI] [PubMed] [Google Scholar]
- 15.Zhang T, et al. RNA viral community in human feces: prevalence of plant pathogenic viruses. PLoS Biol. 2006;4(1):e3. doi: 10.1371/journal.pbio.0040003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peng Y, Leung HC, Yiu SM, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–1428. doi: 10.1093/bioinformatics/bts174. [DOI] [PubMed] [Google Scholar]
- 17.Sauvage V, et al. Identification of the first human gyrovirus, a virus related to chicken anemia virus. J Virol. 2011;85(15):7948–7950. doi: 10.1128/JVI.00639-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Breitbart M, et al. Viral diversity and dynamics in an infant gut. Res Microbiol. 2008;159(5):367–373. doi: 10.1016/j.resmic.2008.04.006. [DOI] [PubMed] [Google Scholar]
- 19.Breitbart M, et al. Metagenomic analyses of an uncultured viral community from human feces. J Bacteriol. 2003;185(20):6220–6223. doi: 10.1128/JB.185.20.6220-6223.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Segata N, et al. Metagenomic microbial community profiling using unique clade-specific marker genes. Nat Methods. 2012;9(8):811–814. doi: 10.1038/nmeth.2066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Domingo-Calap P, Sanjuán R. Experimental evolution of RNA versus DNA viruses. Evolution. 2011;65(10):2987–2994. doi: 10.1111/j.1558-5646.2011.01339.x. [DOI] [PubMed] [Google Scholar]
- 22.Berman L, et al. Defining surgical therapy for pseudomembranous colitis with toxic megacolon. J Clin Gastroenterol. 2008;42(5):476–480. doi: 10.1097/MCG.0b013e31804bbe12. [DOI] [PubMed] [Google Scholar]
- 23.Brouns SJ, et al. Small CRISPR RNAs guide antiviral defense in prokaryotes. Science. 2008;321(5891):960–964. doi: 10.1126/science.1159689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sorek R, Kunin V, Hugenholtz P. CRISPR—a widespread system that provides acquired resistance against phages in bacteria and archaea. Nat Rev Microbiol. 2008;6(3):181–186. doi: 10.1038/nrmicro1793. [DOI] [PubMed] [Google Scholar]
- 25.Karginov FV, Hannon GJ. The CRISPR system: small RNA-guided defense in bacteria and archaea. Mol Cell. 2010;37(1):7–19. doi: 10.1016/j.molcel.2009.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Horvath P, Barrangou R. CRISPR/Cas, the immune system of bacteria and archaea. Science. 2010;327(5962):167–170. doi: 10.1126/science.1179555. [DOI] [PubMed] [Google Scholar]
- 27.Semenova E, et al. Interference by clustered regularly interspaced short palindromic repeat (CRISPR) RNA is governed by a seed sequence. Proc Natl Acad Sci USA. 2011;108(25):10098–10103. doi: 10.1073/pnas.1104144108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sebaihia M, et al. Genome sequence of a proteolytic (Group I) Clostridium botulinum strain Hall A and comparative analysis of the clostridial genomes. Genome Res. 2007;17(7):1082–1092. doi: 10.1101/gr.6282807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Seed KD, Lazinski DW, Calderwood SB, Camilli A. A bacteriophage encodes its own CRISPR/Cas adaptive response to evade host innate immunity. Nature. 2013;494(7438):489–491. doi: 10.1038/nature11927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.McMahon SA, et al. The C-type lectin fold as an evolutionary solution for massive sequence variation. Nat Struct Mol Biol. 2005;12(10):886–892. doi: 10.1038/nsmb992. [DOI] [PubMed] [Google Scholar]
- 31.Miller JL, et al. Selective ligand recognition by a diversity-generating retroelement variable protein. PLoS Biol. 2008;6(6):e131. doi: 10.1371/journal.pbio.0060131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dai W, et al. Three-dimensional structure of tropism-switching Bordetella bacteriophage. Proc Natl Acad Sci USA. 2010;107(9):4347–4352. doi: 10.1073/pnas.0915008107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Doulatov S, et al. Tropism switching in Bordetella bacteriophage defines a family of diversity-generating retroelements. Nature. 2004;431(7007):476–481. doi: 10.1038/nature02833. [DOI] [PubMed] [Google Scholar]
- 34.Gevers D, et al. The Human Microbiome Project: a community resource for the healthy human microbiome. PLoS Biol. 2012;10(8):e1001377. doi: 10.1371/journal.pbio.1001377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ursell LK, et al. The interpersonal and intrapersonal diversity of human-associated microbiota in key body sites. J Allergy Clin Immunol. 2012;129(5):1204–1208. doi: 10.1016/j.jaci.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Handley SA, et al. Pathogenic simian immunodeficiency virus infection is associated with expansion of the enteric virome. Cell. 2012;151(2):253–266. doi: 10.1016/j.cell.2012.09.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sell TL, Schaberg DR, Fekety FR. Bacteriophage and bacteriocin typing scheme for Clostridium difficile. J Clin Microbiol. 1983;17(6):1148–1152. doi: 10.1128/jcm.17.6.1148-1152.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Mahony DE, Clow J, Atkinson L, Vakharia N, Schlech WF. Development and application of a multiple typing system for Clostridium difficile. Appl Environ Microbiol. 1991;57(7):1873–1879. doi: 10.1128/aem.57.7.1873-1879.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Cuevas JM, Domingo-Calap P, Sanjuán R. The fitness effects of synonymous mutations in DNA and RNA viruses. Mol Biol Evol. 2012;29(1):17–20. doi: 10.1093/molbev/msr179. [DOI] [PubMed] [Google Scholar]
- 40. Cantalupo PG, et al. (2011) Raw sewage harbors diverse viral populations. MBio 2(5):1–11. [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.