Abstract
Specification of organelle size is crucial for cell function, yet we know little about the molecular mechanisms that report and regulate organelle growth and steady-state dimensions. The biflagellated green alga Chlamydomonas requires continuous-length feedback to integrate the multiple events that support flagellar assembly and disassembly and at the same time maintain the sensory and motility functions of the organelle. Although several length mutants have been characterized, the requisite molecular reporter of length has not been identified. Previously, we showed that depletion of Chlamydomonas aurora-like protein kinase CALK inhibited flagellar disassembly and that a gel-shift–associated phosphorylation of CALK marked half-length flagella during flagellar assembly. Here, we show that phosphorylation of CALK on T193, a consensus phosphorylation site on the activation loop required for kinase activity, is distinct from the gel-shift–associated phosphorylation and is triggered when flagellar shortening is induced, thereby implicating CALK protein kinase activity in the shortening arm of length control. Moreover, CALK phosphorylation on T193 is dynamically related to flagellar length. It is reduced in cells with short flagella, elevated in the long flagella mutant, lf4, and dynamically tracks length during both flagellar assembly and flagellar disassembly in WT, but not in lf4. Thus, phosphorylation of CALK in its activation loop is implicated in the disassembly arm of a length feedback mechanism and is a continuous and dynamic molecular marker of flagellar length during both assembly and disassembly.
Keywords: cilia and flagella, flagellar length control, cilia length, organelle size control, aurora kinase
Understanding control of organelle size is a fundamental problem in cell biology. Eukaryotic flagella and cilia have been used as simple systems to address this question because their size can be simply defined as length and can be easily tracked and measured (1). After flagellar amputation in Chlamydomonas, rapid flagellar elongation occurs followed by a decreased elongation rate as the organelles approach their full length (∼12 μm) (2). Flagellar precursors delivered by intraflagellar transport (IFT) to the flagellar tip drive assembly of the microtubule-rich axoneme (3, 4). Steady-state length is established when the rate of assembly comes into balance with the basal rate of disassembly (5–7).
The relatively constant amount of IFT complexes within flagella that supports elongation is controlled in the cytoplasm (5, 6). The frequency of injection of groups of IFT complexes (trains) into the flagellum increases with increasing length, whereas IFT train size shows a corresponding decrease with increasing length (7). In addition, the rate of transcription of flagellar genes and transcript stability change during elongation (reviewed in ref. 8). When flagellar shortening is triggered, the number of IFT complexes entering increases two- to fourfold, but the complexes carry much less anterograde cargo (9). In addition, a depolymerizing kinesin moves into the flagella and the rate of flagellar disassembly increases over the basal rate (5, 9, 10). Thus, during flagellar assembly and disassembly as well as at steady state, length must be directly translated into changes in a molecular reporter in the cytoplasm that can be used to modulate and integrate the many parameters of the assembly and disassembly arms of this complex feedback system (11–13). To date, however, not a single protein in any system has been shown to change its properties continuously with length during both flagellar growth and shortening.
Elegant genetic studies over many years have identified several mutants with defects in flagellar length control in Chlamydomonas (13, 14). Of the five long flagella mutant genes identified (none of the short flagella genes is known yet), LF2, LF4, and LF5 encode protein kinases. LF2 (a cyclin-dependent protein kinase) is a cytoplasmic protein that interacts with the two nonkinase LF proteins LF1 and LF3 (15). LF4 (a MAPK) and LF5 (also a cyclin-dependent protein kinase) are in the flagella (16, 17). Homologs of LF2 and LF4 (CCRK and MAK, respectively) also influence ciliary length in vertebrate cells (18–20).
Several other signaling proteins and pathways are known to influence assembly/disassembly and length control of cilia and flagella in Chlamydomonas and other organisms, including the Notch developmental signaling pathway (21), gamma secretase activity (22), MAP4-septin interactions (23), and the target-of-rapamycin (TOR) pathway (24). The full list of proteins/signaling molecules that influence ciliary and flagellar length is long and includes a mammalian adenylyl cyclase (25); NIMA-related protein kinases in Tetrahymena and Chlamydomonas (26, 27); protein kinase GSK3 in Chlamydomonas and vertebrate cells (25, 28); phosphatase CDC14B in zebrafish (29), actin (30), and regulators of gene transcription (24, 30, 31). Perhaps the best-characterized ciliary disassembly protein in vertebrates is Aurora A, which regulates HDAC6 and is essential for ciliary shortening (32). Whether these several molecules and pathways sense length as part of a feedback loop or act directly as effectors for assembly or disassembly is largely unknown.
Our previous work has placed the Chlamydomonas aurora-like protein kinase CALK at the nexus of pathways essential for flagellar assembly and disassembly. Cells depleted of CALK by RNAi methods were inhibited in flagellar shortening, and a gel shift-associated phosphorylation on an uncharacterized residue of CALK responded to changes in the state of flagellation of cells during flagellar assembly (33, 34). The gel shift-associated phosphorylation of CALK failed to meet the criteria of a length reporter/modulator in that its relation to protein kinase activity was unknown, and the phosphorylation did not track length during the entire assembly process, but only marked a stage (half-length) during assembly. Moreover, once shortening was triggered, the phosphorylation state did not change as the flagella shortened.
Here, we show that CALK T193, a consensus phosphorylation site in the conserved activation loop whose phosphorylation is known to be essential for regulating the activity of most protein kinase family members (35, 36), is essential for CALK protein kinase activity. Our findings in cells undergoing flagellar shortening implicate T193 phosphorylation in flagellar disassembly. Experiments with cells undergoing flagellar shortening and flagellar elongation show that the cellular level of CALK phosphorylated on T193 is a dynamic reporter of flagellar length in both. Moreover, CALK phosphorylation on T193 is elevated in the long flagella mutant, lf4, and dysregulated in lf4 cells during elongation and shortening. Thus, CALK is implicated as an effector in the disassembly arm of flagellar length control and is the first dynamic molecular reporter of organelle size.
Results
CALK Phosphorylation on T193 in the Activation Loop Is Required for Protein Kinase Activity.
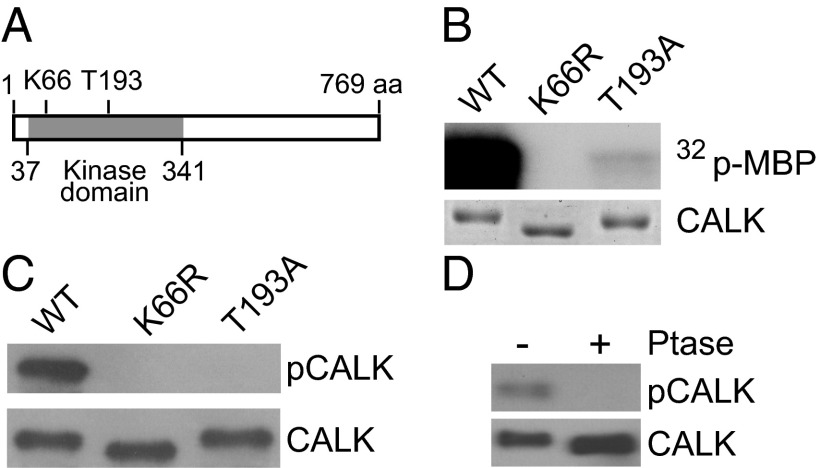
Phosphorylation of a conserved threonine in a domain termed the activation loop (or T-loop) is required for full activity of many protein kinases, including closely related CALK family members, the aurora protein kinases (32, 35). Activation-loop phosphorylation, mediated by autocatalytic activity or upstream protein kinases, stimulates kinase activity by several orders of magnitude (36). The protein kinase domain of CALK is in the N-terminal half of this 769-residue protein (residues 37–341) (Fig. 1A) (37). To characterize more fully the properties of CALK linked to flagellar status, we tested whether the conserved T193 in the activation loop of CALK was important for protein kinase activity. In vitro kinase activity assays of WT and several mutant forms of recombinant CALK expressed in bacteria using myelin basic protein (MBP) as substrate showed that, whereas WT CALK was an active protein kinase, mutation of the residue K66 to R in the catalytic domain abolished CALK kinase activity (Fig. 1B). As expected, the protein kinase activity of bacterially expressed T193A CALK also was reduced substantially compared with WT, indicating that T193 phosphorylation was essential for full protein kinase activity.
Fig. 1.
T-loop T193 phosphorylation is required for CALK activity. (A) A diagram of CALK domain structures showing the protein kinase domain and residues critical for CALK activity. (B) T193 is required for CALK activity. Bacterially expressed WT and mutated forms of CALK were assayed for kinase activity in vitro with MBP as substrate. SDS/PAGE analysis with Coomassie blue staining showed equal loading. (C) T193 is phosphorylated in WT CALK but not in kinase-dead and T-loop mutants. Bacterially expressed WT and mutated forms of CALK were analyzed by immunoblotting with anti-pCALK and CALK antibodies. pCALK stains CALK phosphorylated at T193, whereas CALK stains total CALK. (D) Endogenous CALK T193 is phosphorylated at steady state. Steady-state cell lysates were incubated with phosphatase followed by immunoblot analysis with anti-pCALK and CALK antibodies.
We generated a polyclonal antibody (pCALK antibody) that recognized phosphorylated T19. Immunoblot analysis showed that our original CALK antibody recognized bacterially expressed WT K66R and T193A CALK. In contrast, the pCALK antibody reacted with bacterially expressed WT CALK (which underwent autophosphorylation in the bacteria), but not with the K66R kinase-dead CALK or the T193A activation loop mutant CALK (Fig. 1C). Furthermore, the absence of T193 phosphorylation in the K66R kinase-dead form expressed in bacteria indicated that phosphorylation of T193 in the WT recombinant protein was due to autophosphorylation (38). In related experiments with endogenous CALK in Chlamydomonas, immunoblotting showed that Chlamydomonas cells with full-length flagella possessed phosphorylated T193, and that phosphatase treatment abolished this phosphorylation (Fig. 1D). Taken together, these data demonstrated the specificity of the antibody, they indicated that T193 phosphorylation was required for CALK activity, and they showed that a portion of CALK was active in cells with flagella of steady-state length.
T193 Phosphorylation Is Distinct from the Gel Shift-Associated CALK Phosphorylation(s).
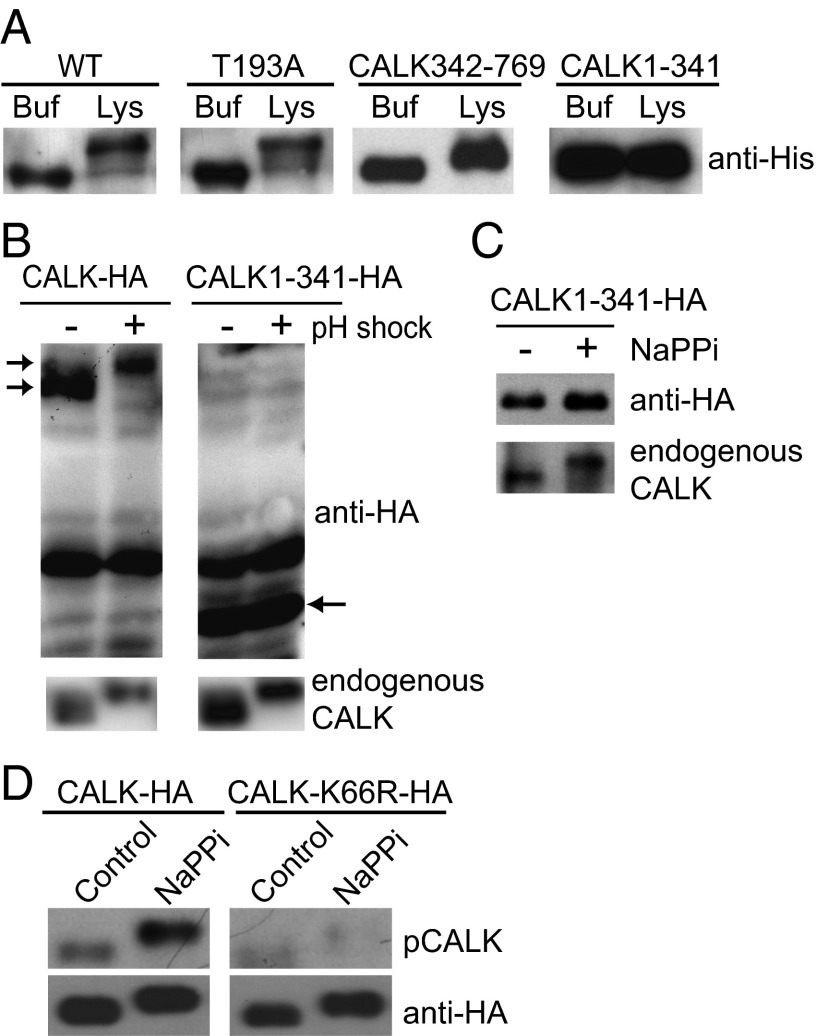
Multiple, independent sets of experiments indicated that T193 phosphorylation was distinct from the previously reported gel shift-associated phosphorylations (33, 34). First, upon incubation with ATP and Chlamydomonas cell lysates from aflagellate cells obtained by pH shock, both WT bacterially expressed, His-tagged CALK and bacterially expressed T193A CALK underwent the gel shift-associated phosphorylation (Fig. 2A). Second, in similar experiments, only the nonkinase domain-containing, C-terminal half of bacterially expressed CALK (CALK342-769) underwent the gel shift when incubated with Chlamydomonas lysates in the presence of ATP; the bacterially expressed N-terminal domain (CALK1-341) failed to undergo the gel shift (Fig. 2A).
Fig. 2.
The CALK gel shift is associated with C-terminal phosphorylation, but not with T193 phosphorylation. (A) The C-terminal domain is required for CALK gel shift-associated phosphorylations. Bacterially expressed and His-tagged WT and mutated forms of CALK were incubated with buffer or with a lysate from deflagellated Chlamydomonas cells in the presence of ATP. After incubation for 15 min at 30 °C, samples were analyzed by immunoblot analysis with anti-His antibody. Buf, buffer; Lys, cell lysate. (B) CALK C-terminal region is required for the gel-shift–associated phosphorylation that occurs upon flagellar detachment. Chlamydomonas cells expressing HA-tagged WT CALK or C-terminally truncated CALK (CALK1-341-HA) were deflagellated and subjected to immunoblot analysis with anti-HA and anti-CALK antibodies. Endogenous CALK is shown as a control. Left arrows indicate CALK-HA and right arrow, CALK1-341-HA. (C) The gel shift-associated phosphorylation during flagellar shortening occurs on the C-terminal region of CALK. Chlamydomonas cells expressing HA-tagged, C-terminally truncated CALK were exposed to 20 mM NaPPi for 5 min to trigger activation of the flagellar shortening pathway and analyzed by immunoblotting. (D) T193 phosphorylation does not cause a CALK gel shift. Chlamydomonas cells expressing HA-tagged WT CALK or kinase-dead mutant K66R-HA CALK were treated with NaPPi for 5 min, followed by immunoprecipitation with HA antibody and immunoblot analysis.
Third, treatments of Chlamydomonas [pH shock or incubation in sodium pyrophosphate (NaPPi)] that triggered the gel shift in endogenous WT and in ectopically expressed, epitope-tagged CALK (CALK-HA) in Chlamydomonas cells, failed to trigger the shift in the ectopically expressed CALK1-341-HA (Fig. 2 B and C). Fourth, only WT CALK (CALK-HA) and not kinase-dead CALK (CALK-K66R-HA) immunoprecipitated from NaPPi-treated Chlamydomonas cells was phosphorylated on T193, but both underwent the gel shift-associated phosphorylation (Fig. 2D). Taken together, these results demonstrated that the previously described gel shift-associated phosphorylations were distinct from T193 phosphorylation and that uncharacterized protein kinases were likely responsible for the C-terminal phosphorylation.
The Amount of CALK Phosphorylated on T193 Increases When Flagellar Shortening Is Triggered.
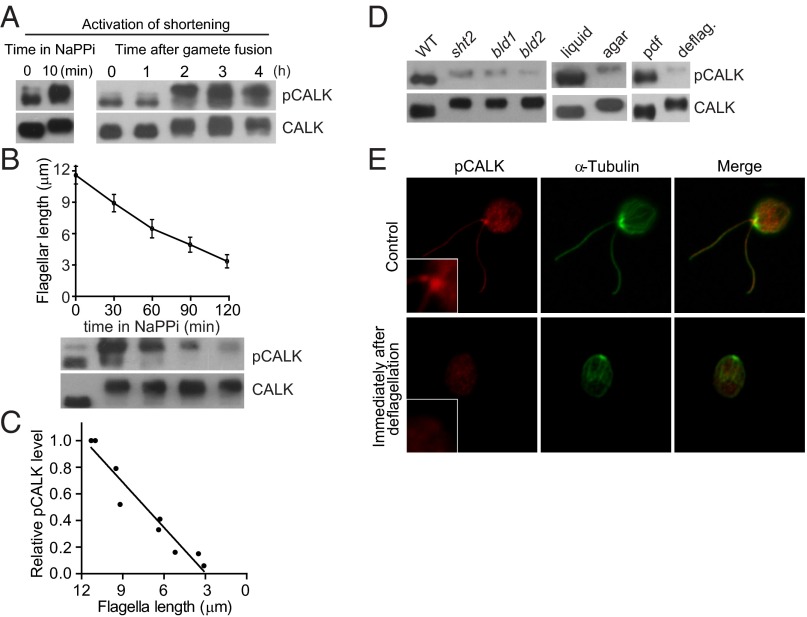
Our previous inhibitor studies showing that protein kinase activity was required for flagellar shortening along with our earlier RNAi results showing that depletion of CALK inhibited shortening (33) led us to test whether T193 phosphorylation was induced when shortening was triggered. First, we used an environmental cue to induce shortening via depletion of divalent cations by suspension of cells in medium containing 20 mM NaPPi (39). Within minutes after the shortening pathway was activated, immunoblotting with the anti-CALK antibody showed that CALK underwent the expected gel shift-associated phosphorylation in its C terminus (Fig. 3A). Moreover, activation of shortening also sparked a rapid, dramatic increase in the level of CALK phosphorylated on T193 (Fig. 3A). Because it was possible that the NaPPi treatment brought about changes in cells unrelated to shortening that triggered T193 phosphorylation, we also examined T193 phosphorylation in cells undergoing shortening activated by a developmental trigger, zygote development. Within 2 h after gametes of opposite mating types had fused to form a quadriflagellated zygote, the flagella began the expected shortening (33). Confirming the NaPPi results, the cellular level of CALK phosphorylated on T193 increased several fold (Fig. 3A, Right). These results, that two independent methods for inducing flagella shortening also triggered phosphorylation of the residue in CALK that is required for its protein kinase activity, implicated T193 phosphorylation in the disassembly arm of flagellar length regulation.
Fig. 3.
Activation of the flagellar shortening pathway induces a rapid increase in T193 phosphorylation, and shortening is accompanied by a concomitant decrease in levels of CALK phosphorylated on T193. (A) The level of CALK phosphorylated on T193 increases when flagellar shortening is triggered by NaPPi and during zygotic development. Samples harvested at the indicated times after NaPPi treatment or after mixing gametes of opposite mating types were analyzed by immunoblotting. (B) Dynamic changes of CALK T193 phosphorylation during flagellar shortening induced by NaPPi. Cells were induced to shorten their flagella by adding 20 mM NaPPi. At the times indicated during flagellar shortening, samples were analyzed for flagellar length (Upper) and for CALK phosphorylation by immunoblotting (Lower). Flagellar length data shown here and below are represented as mean ± SD). (C) The relative amount of CALK phosphorylated on T193 is proportional to flagellar length during flagellar shortening. The relative amounts of pCALK as determined by densitometry from immunoblotting with pCALK and CALK antibodies from two experiments are plotted against flagellar length during shortening. (D) CALK T193 phosphorylation in cells with short or absent flagella. Aflagellate mutants bld1 and bld2, aflagellate cells grown on agar plates, aflagellate cells generated by pH shock, and the short flagella mutant shf2 were analyzed by immunoblotting with anti-CALK and anti-pCALK antibodies. deflag, deflagellation by pH shock; pdf, predeflagellation. (E) Localization of CALK T193 phosphorylated form at the basal body region. Control cells and cells deflagellated by pH shock were immunostained with pCALK and anti–α-tubulin antibodies. (Insets) Higher magnification views of basal body regions.
T193-Phosphorylated CALK Is Enriched in the Basal Body Region and the Relative Level of CALK Phosphorylated on T193 Decreases as Flagella Shorten.
A surprising observation from the shortening experiments was that after the cellular levels of T193 phosphorylated CALK had undergone the increase upon triggering of the shortening pathway, the level of CALK phosphorylated on T193 began to decrease. Remarkably, the level continued to fall concomitantly with flagellar length (Fig. 3 B and C), and when the flagella were less than 30% of their original length, relatively low levels of CALK phosphorylated on T193 were detected. To test whether low T193 phosphorylation was a consistent feature of cells with short flagella, we analyzed CALK in short flagella mutants, in agar-grown cells, and in cells immediately after experimental deflagellation. The shf2 strain harbors an unknown mutation that leads to cells that possess flagella shorter than half length (14, 34). The IFT52 and bld1 mutants; the epsilon tubulin mutant, bld2; and cells grown on agar plates are aflagellate (40, 41). Consistent with the shortening results, and compared with control cells with flagella of normal length, the level of CALK phosphorylated on T193 in each of these cell types was dramatically reduced (Fig. 3D). We found the same result when we experimentally detached flagella by pH shock. Almost immediately after cells were deflagellated, the amount of CALK phosphorylated on T193 fell dramatically (Fig. 3D). Immunofluorescence of cells before and after deflagellation confirmed the immunoblotting results. As shown in Fig. 3E, T193 CALK was detectable with the pCALK antibody in cells before deflagellation, where it was enriched in the basal body region and became much reduced after deflagellation. Immunodepletion experiments showed that the basal body localization of pCALK was specific (Fig. S1). No difference between control and deflagellated cells was observed with CALK by using anti-HA antibody to recognize HA-tagged CALK, which was detected throughout the cytoplasm (Fig. S2). Immunostaining of CALK T193 during flagellar shortening also identified basal body localization of pCALK, and its changes were consistent with the immunoblotting results (Fig. S3). Taken together, these results demonstrated that the relative amount of CALK phosphorylated on T193 fell as flagella shortened and that low levels of T193 CALK were a property of cells with short or absent flagella, independently of the conditions responsible for their short flagellar status.
The Relative Amount of CALK Phosphorylated on T193 Increases as Flagella Increase in Length.
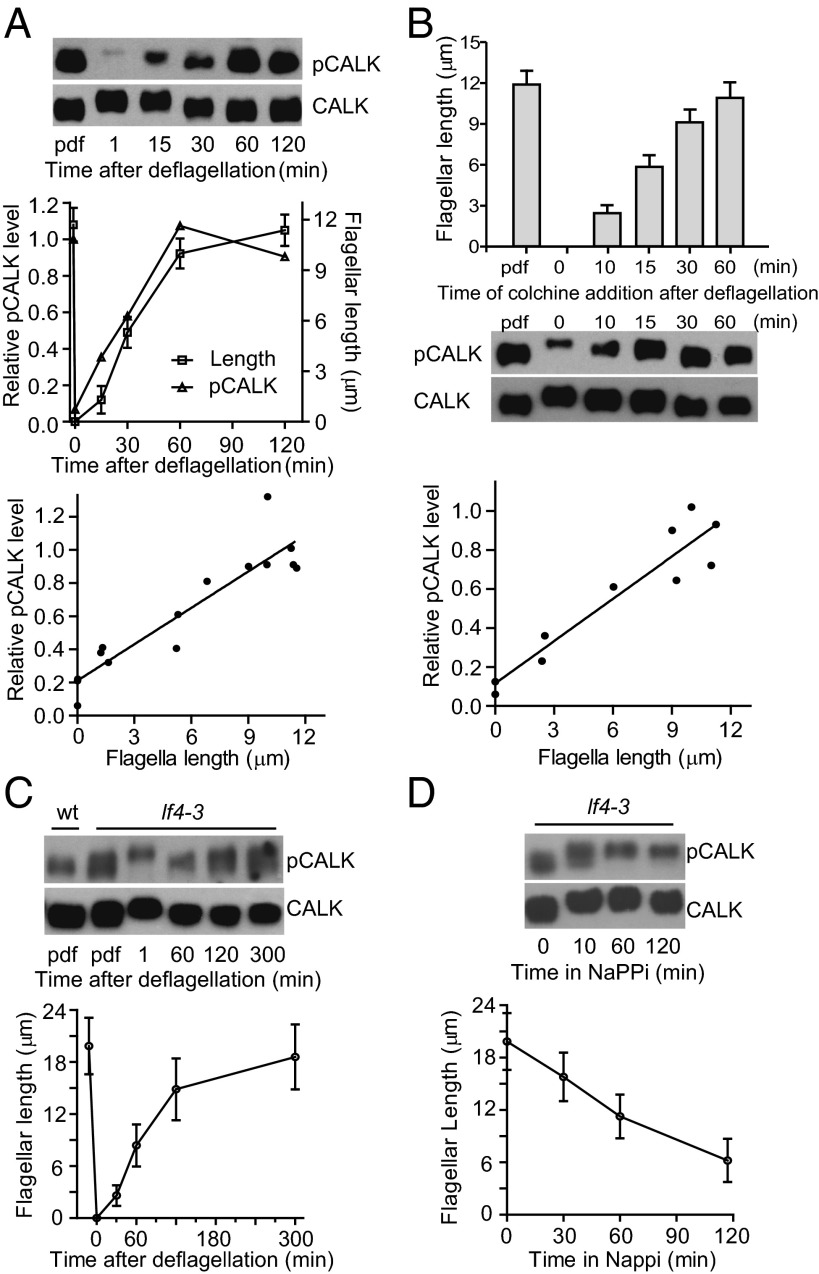
If the relative amount of CALK phosphorylated on T193 indeed is a dynamic reporter of flagellar length, then as cells begin to regenerate the organelles after detachment, the relative amount of CALK phosphorylated on T193 should increase. WT cells were deflagellated and allowed to regenerate flagella (Fig. 4A, Middle). Immediately after flagellar loss, CALK underwent the expected gel shift associated phosphorylation and the amount of T193-phosphorylated CALK decreased dramatically (Fig. 4A, Top). In a striking mirror of the shortening data, the cellular levels of T193-phosphorylated CALK increased concomitantly with flagellar length, reaching predeflagellation amounts when the flagella reached their steady-state length. Quantification by densitometry of T193-phosphorylated CALK during flagellar growth demonstrated that the relative amounts of T193-phosphorylated CALK were directly related to flagellar length (Fig. 4A, Middle and Bottom).
Fig. 4.
Length-dependent regulation of phosphorylation state of CALK T193. (A) Change in levels of CALK phosphorylated on T193 during flagellar elongation. At the indicated times during flagellar regeneration after pH shock, samples were analyzed for T193 phosphorylation state by immunoblotting (Top) and for flagellar length (Middle). The ratio of CALK phosphorylated on T193 (pCALK) versus total CALK determined by densitometry is plotted against time (Middle) and flagellar length (Bottom). Data shown are from three independent experiments. (B) Levels of T193 phosphorylated CALK in cells whose flagellar length was fixed by addition of colchicine during flagellar regeneration. Colchicine was added at the indicated times after deflagellation; 2 h later, samples were analyzed for flagellar lengths (Top) and by immunoblot analysis (Middle). The relative amounts of T193 phosphorylated CALK from two independent experiments were quantified by densitometry (Bottom). (C) Analysis of T193 phosphorylation at steady state and during flagellar elongation in the long flagellar mutant lf4-3. lf4-3 cells were deflagellated by mechanical shearing and allowed to regenerate flagella. Cell samples pdf and at the indicated times after deflagellation were analyzed by immunoblotting (Upper) and for flagellar length (Lower). An immunoblot of a WT control sample is also shown. (D) Analysis of T193 phosphorylation during flagellar shortening in lf4-3. Flagellar lengths and pCALK and CALK immunoblot analysis of lf4-3 cells during NaPPi-induced shortening.
To confirm that the relative amount of CALK phosphorylated on T193 was related to length and not simply to time after deflagellation, we generated cells with flagella of different lengths by adding colchicine at the indicated times after deflagellation (Fig. 4B, Top) and 2 h later collected samples for immunoblotting and length measurements (2, 34). As shown in Fig. 4B (Middle and Bottom), the relative level of T193-phosphorylated CALK was also directly related to flagellar length in these cells. Thus, the cellular levels of T193-phosphorylated CALK dynamically report flagellar length during both flagellar assembly and flagellar disassembly.
The Steady-State Level of CALK Phosphorylated on T193 Is Elevated in the Long Flagella Mutant, lf4, and T193 Phosphorylation Is Dysregulated During Flagellar Elongation and Shortening.
If the cellular mechanisms for length regulation are linked to the relative amount of CALK phosphorylated on T193, then T193 phosphorylation should be altered in a cell with aberrantly long flagella. Several of the lf mutants have flagella that indeed are longer than WT, but most exhibit substantial variability in length (42). The lf4 mutant (flagella approximately two times longer than WT) is the most robust and reliable of the long flagella mutants (16). Immunoblotting showed that lf4 steady-state levels of CALK phosphorylated on T193 were substantively higher than WT (Fig. 4C). On the other hand, after deflagellation, even though flagellar elongation began almost immediately, there was a lag before the amount of CALK phosphorylated on T193 began to increase. During the first 60 min of flagellar regeneration, when the lf4 flagella reached nearly WT length, T193 phosphorylation changed little, if at all, whereas in WT cells the amount of CALK phosphorylated on T193 increased around fivefold during the same time. Thereafter, the amount of phosphorylated T193 CALK in the lf4 cells increased with increasing length (Fig. 4C). In addition to the apparent inability of cells lacking the LF4 protein to sense when their flagella were of WT length as assessed by T193 phosphorylation, they also failed to sense length during shortening. The relative amount of CALK T193 phosphorylation fell proportionately with length in WT cells undergoing shortening, but the amounts in the lf4 cells were essentially unchanged during shortening (Fig. 4D). Thus, the link between T193 phosphorylation and flagellar length during growth and shortening of the organelles depends on the length-controlling protein, LF4.
Discussion
An attractive model for flagellar length control is that the activity of a reporter molecule is directly related to length and at the center of a feedback system that controls and integrates the multiple pathways required for orderly flagellar assembly, steady-state length, and flagellar disassembly (11, 12, 42). We believe that the results here firmly establish that the phosphorylation of CALK on the activity-regulating T193 dynamically reports flagellar length. Furthermore, previous RNAi results (33, 34) along with the several-fold increase in CALK T193 phosphorylation upon activation of the shortening pathway strongly implicate CALK protein kinase activity in modulating disassembly to control flagellar length. In vertebrate cells, phosphorylation of Aurora A on the threonine in its activation loop regulates its activity. One of its downstream effectors is the tubulin deacetylase HDAC6, whose activity is essential for regulated ciliary shortening in vertebrate cells (32). It will be interesting to determine if a similar process occurs in Chlamydomonas.
Our result that this protein kinase with a role in flagellar disassembly is phosphorylated on its activation loop in cells with flagella at steady-state length attests to the complexity of length regulation and confirms that length indeed is not a fixed condition, but rather a balance between active assembly and disassembly. At steady-state length, even in the lf4 cells with very long flagella, cells can tolerate or balance the “pressure” for disassembly driven by active CALK with the pressure for assembly provided by IFT.
Notwithstanding the importance of regulating length, simply assembling such a complex organelle as the flagellum, all of the while maintaining its motility and sensory functions, is a major feat of cellular coordination of trafficking of IFT complexes and their cargo, including soluble proteins (e.g., tubulin), large protein complexes, and membrane vesicles. Because cargo loading at the base is reduced when disassembly is activated (9), it is likely that cargo loading during assembly also is regulated. The rates of injection and train size of IFT complexes are known to be regulated by length (7), synthesis of new flagellar precursors is reduced as flagella reach full length (39), and flagellar membrane lipids and proteins must be added coordinately with flagellar elongation. The length-dependent, continuously changing levels of activation loop-phosphorylated CALK provides a potential regulator. The enrichment of CALK phosphorylated on T193 in the basal body region, where entry of IFT complexes and cargo loading occurs, raises the possibility that this length reporter could participate in regulation of the IFT machinery.
These results demonstrating that the relative level of CALK phosphorylated on T193 reports length raises the question of why the RNAi strains described previously did not show an obvious length phenotype, even though they did show a shortening phenotype (33). One possibility is that it is not total amounts of CALK phosphorylated on T193, but the ratio of inactive to T193 phosphorylated CALK that is important for length regulation. According to this idea, the small amounts of CALK that remained in the RNAi strains might have been sufficient for length regulation.
The inverse relationship between CALK T193 and C-terminal phosphorylation during the first half of flagellar elongation is intriguing. In aflagellate cells, the CALK C terminus was phosphorylated when the cellular level of CALK phosphorylated on T193 was at its lowest. The flagellar sensing system might activate a protein kinase when flagella are lost that phosphorylates CALK at the C terminus. A specific phosphatase at the same time could dephosphorylate T193. The phosphorylation state of the CALK homolog Aurora A on the activation loop is regulated by protein phosphatase 6 (43). Thus, the phosphatase that participates in regulating T193 phosphorylation also could be regulated by flagellar length, given that CALK remains phosphorylated on its C terminus as T193 phosphorylation gradually decreases during disassembly. The kinases that phosphorylate CALK are unknown.
The ability of cells to independently phosphorylate CALK on T913 and on the C terminus provides a potential mechanism for fine-tuning CALK action on assembly/disassembly. Once growing flagella reach approximately half-length, the C terminus becomes dephosphorylated (34) and levels of T193 CALK continue to increase with increasing length. When flagellar shortening is induced, however, levels of T193 CALK increase even further, and CALK becomes phosphorylated on its C terminus. One consequent model is that the C-terminal phosphorylation itself influences CALK’s ability to regulate assembly/disassembly. Or, C-terminal phosphorylation could influence CALK interaction with a binding partner to regulate CALK action on its effectors.
Several mechanisms could underlie the link between flagellar length and CALK T193 phosphorylation state (11). In a TOF model, a signaling molecule that moves through the flagella by IFT might undergo a time- and therefore length-dependent change. Such a protein could modulate CALK phosphorylation in the cell body where most CALK is localized (33). In the Hedgehog signaling pathway in vertebrates, regulation of transcription by Gli transcription factors depends on their transit through the cilium (44). Signals generated by ion channels in the flagellar membrane (12) or gradients generated by flagellar tip-localized protein kinase(s) could also change as a function of flagellar length (45).
In summary, we provide evidence that cells possess a feedback system that regulates the relative amounts of CALK phosphorylated on its activation loop. To our knowledge, CALK T193 phosphorylation state is the first reporter of organelle size to be uncovered in any system. The dynamic relationship between CALK T193 phosphorylation and flagellar length provides a novel platform for dissecting the molecular networks controlling ciliary and flagellar length.
Materials and Methods
Rabbit anti-pCALK antibody was made against peptide Cys-QERPVT(P)RVGT-amide (PhosphoSolutions). The phosphorylated threonine is at 193 in CALK. The antibody was affinity-purified using nonphosphorylated peptide. The other antibodies used are as follows: rabbit anti-CALK (1:5,000) (33), anti-pCALK (1:150), anti-GST (1:2,500) (Abmart), mouse anti–α-tubulin (1:5,000) (Sigma), anti-Histidine (1:500) (Qiagen), and rat anti-HA (1:1,000) (Roche). The relative amount of total CALK versus CALK phosphorylated on T193 was quantified by densitometry based on immunoblotting by anti-CALK and anti-pCALK antibodies.
Supplementary Material
Acknowledgments
This work was supported by National Basic Research Program of China (973 Program) Grants 2012CB945003 and 2013CB910700, National Natural Science Foundation of China Grants 30830057 and 30988004 (to J.P.), and by National Institutes of Health Grant GM25661 (to W.J.S.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1302364110/-/DCSupplemental.
References
- 1.Wemmer KA, Marshall WF. Flagellar length control in chlamydomonas—paradigm for organelle size regulation. Int Rev Cytol. 2007;260:175–212. doi: 10.1016/S0074-7696(06)60004-1. [DOI] [PubMed] [Google Scholar]
- 2.Rosenbaum JL, Moulder JE, Ringo DL. Flagellar elongation and shortening in Chlamydomonas. The use of cycloheximide and colchicine to study the synthesis and assembly of flagellar proteins. J Cell Biol. 1969;41(2):600–619. doi: 10.1083/jcb.41.2.600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Qin H, Diener DR, Geimer S, Cole DG, Rosenbaum JL. Intraflagellar transport (IFT) cargo: IFT transports flagellar precursors to the tip and turnover products to the cell body. J Cell Biol. 2004;164(2):255–266. doi: 10.1083/jcb.200308132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hao L, et al. Intraflagellar transport delivers tubulin isotypes to sensory cilium middle and distal segments. Nat Cell Biol. 2011;13(7):790–798. doi: 10.1038/ncb2268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Marshall WF, Rosenbaum JL. Intraflagellar transport balances continuous turnover of outer doublet microtubules: Implications for flagellar length control. J Cell Biol. 2001;155(3):405–414. doi: 10.1083/jcb.200106141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marshall WF, Qin H, Rodrigo Brenni M, Rosenbaum JL. Flagellar length control system: Testing a simple model based on intraflagellar transport and turnover. Mol Biol Cell. 2005;16(1):270–278. doi: 10.1091/mbc.E04-07-0586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Engel BD, Ludington WB, Marshall WF. Intraflagellar transport particle size scales inversely with flagellar length: Revisiting the balance-point length control model. J Cell Biol. 2009;187(1):81–89. doi: 10.1083/jcb.200812084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lefebvre PA, Rosenbaum JL. Regulation of the synthesis and assembly of ciliary and flagellar proteins during regeneration. Annu Rev Cell Biol. 1986;2:517–546. doi: 10.1146/annurev.cb.02.110186.002505. [DOI] [PubMed] [Google Scholar]
- 9.Pan J, Snell WJ. Chlamydomonas shortens its flagella by activating axonemal disassembly, stimulating IFT particle trafficking, and blocking anterograde cargo loading. Dev Cell. 2005;9(3):431–438. doi: 10.1016/j.devcel.2005.07.010. [DOI] [PubMed] [Google Scholar]
- 10.Piao T, et al. A microtubule depolymerizing kinesin functions during both flagellar disassembly and flagellar assembly in Chlamydomonas. Proc Natl Acad Sci USA. 2009;106(12):4713–4718. doi: 10.1073/pnas.0808671106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chan YH, Marshall WF. How cells know the size of their organelles. Science. 2012;337(6099):1186–1189. doi: 10.1126/science.1223539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosenbaum J. Organelle size regulation: Length matters. Curr Biol. 2003;13(13):R506–R507. doi: 10.1016/s0960-9822(03)00440-8. [DOI] [PubMed] [Google Scholar]
- 13.Asleson CM, Lefebvre PA. Genetic analysis of flagellar length control in Chlamydomonas reinhardtii: A new long-flagella locus and extragenic suppressor mutations. Genetics. 1998;148(2):693–702. doi: 10.1093/genetics/148.2.693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kuchka MR, Jarvik JW. Short-flagella mutants of Chlamydomonas reinhardtii. Genetics. 1987;115(4):685–691. doi: 10.1093/genetics/115.4.685. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tam LW, Wilson NF, Lefebvre PA. A CDK-related kinase regulates the length and assembly of flagella in Chlamydomonas. J Cell Biol. 2007;176(6):819–829. doi: 10.1083/jcb.200610022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Berman SA, Wilson NF, Haas NA, Lefebvre PA. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol. 2003;13(13):1145–1149. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- 17.Tam LW, Ranum PT, Lefebvre PA. CDKL5 regulates flagellar length and localizes to the base of the flagella in Chlamydomonas. Mol Biol Cell. 2013;24(5):588–600. doi: 10.1091/mbc.E12-10-0718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Omori Y, et al. Negative regulation of ciliary length by ciliary male germ cell-associated kinase (Mak) is required for retinal photoreceptor survival. Proc Natl Acad Sci USA. 2010;107(52):22671–22676. doi: 10.1073/pnas.1009437108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ozgül RK, et al. European Retinal Disease Consortium Exome sequencing and cis-regulatory mapping identify mutations in MAK, a gene encoding a regulator of ciliary length, as a cause of retinitis pigmentosa. Am J Hum Genet. 2011;89(2):253–264. doi: 10.1016/j.ajhg.2011.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ko HW, et al. Broad-minded links cell cycle-related kinase to cilia assembly and hedgehog signal transduction. Dev Cell. 2010;18(2):237–247. doi: 10.1016/j.devcel.2009.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lopes SS, et al. Notch signalling regulates left-right asymmetry through ciliary length control. Development. 2010;137(21):3625–3632. doi: 10.1242/dev.054452. [DOI] [PubMed] [Google Scholar]
- 22.Jurisch-Yaksi N, et al. Rer1p maintains ciliary length and signaling by regulating γ-secretase activity and Foxj1a levels. J Cell Biol. 2013;200(6):709–720. doi: 10.1083/jcb.201208175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Ghossoub R, et al. (2013) Septins 2, 7, and 9 and MAP4 co-localize along the axoneme in the primary cilium and control ciliary length. J Cell Sci, April 9. [DOI] [PMC free article] [PubMed]
- 24.Yuan S, et al. Target-of-rapamycin complex 1 (Torc1) signaling modulates cilia size and function through protein synthesis regulation. Proc Natl Acad Sci USA. 2012;109(6):2021–2026. doi: 10.1073/pnas.1112834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ou Y, et al. Adenylate cyclase regulates elongation of mammalian primary cilia. Exp Cell Res. 2009;315(16):2802–2817. doi: 10.1016/j.yexcr.2009.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wloga D, et al. Members of the NIMA-related kinase family promote disassembly of cilia by multiple mechanisms. Mol Biol Cell. 2006;17(6):2799–2810. doi: 10.1091/mbc.E05-05-0450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bradley BA, Quarmby LM. A NIMA-related kinase, Cnk2p, regulates both flagellar length and cell size in Chlamydomonas. J Cell Sci. 2005;118(Pt 15):3317–3326. doi: 10.1242/jcs.02455. [DOI] [PubMed] [Google Scholar]
- 28.Wilson NF, Lefebvre PA. Regulation of flagellar assembly by glycogen synthase kinase 3 in Chlamydomonas reinhardtii. Eukaryot Cell. 2004;3(5):1307–1319. doi: 10.1128/EC.3.5.1307-1319.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Clément A, Solnica-Krezel L, Gould KL. The Cdc14B phosphatase contributes to ciliogenesis in zebrafish. Development. 2011;138(2):291–302. doi: 10.1242/dev.055038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J, et al. Functional genomic screen for modulators of ciliogenesis and cilium length. Nature. 2010;464(7291):1048–1051. doi: 10.1038/nature08895. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458(7238):651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pugacheva EN, Jablonski SA, Hartman TR, Henske EP, Golemis EA. HEF1-dependent Aurora A activation induces disassembly of the primary cilium. Cell. 2007;129(7):1351–1363. doi: 10.1016/j.cell.2007.04.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Pan J, Wang Q, Snell WJ. An aurora kinase is essential for flagellar disassembly in Chlamydomonas. Dev Cell. 2004;6(3):445–451. doi: 10.1016/s1534-5807(04)00064-4. [DOI] [PubMed] [Google Scholar]
- 34.Luo M, et al. The phosphorylation state of an aurora-like kinase marks the length of growing flagella in Chlamydomonas. Curr Biol. 2011;21(7):586–591. doi: 10.1016/j.cub.2011.02.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Nolen B, Taylor S, Ghosh G. Regulation of protein kinases; controlling activity through activation segment conformation. Mol Cell. 2004;15(5):661–675. doi: 10.1016/j.molcel.2004.08.024. [DOI] [PubMed] [Google Scholar]
- 36.Narayanan A, Jacobson MP. Computational studies of protein regulation by post-translational phosphorylation. Curr Opin Struct Biol. 2009;19(2):156–163. doi: 10.1016/j.sbi.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 37.Pan J, Snell WJ. Regulated targeting of a protein kinase into an intact flagellum. An aurora/Ipl1p-like protein kinase translocates from the cell body into the flagella during gamete activation in chlamydomonas. J Biol Chem. 2000;275(31):24106–24114. doi: 10.1074/jbc.M002686200. [DOI] [PubMed] [Google Scholar]
- 38.Tsai MY, et al. A Ran signalling pathway mediated by the mitotic kinase Aurora A in spindle assembly. Nat Cell Biol. 2003;5(3):242–248. doi: 10.1038/ncb936. [DOI] [PubMed] [Google Scholar]
- 39.Lefebvre PA, Nordstrom SA, Moulder JE, Rosenbaum JL. Flagellar elongation and shortening in Chlamydomonas. IV. Effects of flagellar detachment, regeneration, and resorption on the induction of flagellar protein synthesis. J Cell Biol. 1978;78(1):8–27. doi: 10.1083/jcb.78.1.8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Dutcher SK, Morrissette NS, Preble AM, Rackley C, Stanga J. Epsilon-tubulin is an essential component of the centriole. Mol Biol Cell. 2002;13(11):3859–3869. doi: 10.1091/mbc.E02-04-0205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Brazelton WJ, Amundsen CD, Silflow CD, Lefebvre PA. The bld1 mutation identifies the Chlamydomonas osm-6 homolog as a gene required for flagellar assembly. Curr Biol. 2001;11(20):1591–1594. doi: 10.1016/s0960-9822(01)00485-7. [DOI] [PubMed] [Google Scholar]
- 42.Barsel SE, Wexler DE, Lefebvre PA. Genetic analysis of long-flagella mutants of Chlamydomonas reinhardtii. Genetics. 1988;118(4):637–648. doi: 10.1093/genetics/118.4.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zeng K, Bastos RN, Barr FA, Gruneberg U. Protein phosphatase 6 regulates mitotic spindle formation by controlling the T-loop phosphorylation state of Aurora A bound to its activator TPX2. J Cell Biol. 2010;191(7):1315–1332. doi: 10.1083/jcb.201008106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Tukachinsky H, Lopez LV, Salic A. A mechanism for vertebrate Hedgehog signaling: Recruitment to cilia and dissociation of SuFu-Gli protein complexes. J Cell Biol. 2010;191(2):415–428. doi: 10.1083/jcb.201004108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Martin SG, Berthelot-Grosjean M. Polar gradients of the DYRK-family kinase Pom1 couple cell length with the cell cycle. Nature. 2009;459(7248):852–856. doi: 10.1038/nature08054. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.