Abstract
When host cells are infected by an RNA virus, pattern-recognition receptors (PRRs) recognize the viral RNA and induce the antiviral innate immunity. Toll-like receptor 7 (TLR7) detects the genomic RNA of incoming murine leukemia virus (MLV) in endosomes and mediates the antiviral response. However, the RNA-sensing PRR that recognizes the MLV in the cytosol is not fully understood. Here, we definitively demonstrate that zinc-finger antiviral protein (ZAP) acts as a cytosolic RNA sensor, inducing the degradation of the MLV transcripts by the exosome, an RNA degradation system, on RNA granules. Although the retinoic acid inducible gene I (RIG-I)–like receptors (RLRs) RIG-I and melanoma differentiation-associated protein 5 detect various RNA viruses in the cytosol and induce the type I IFN-dependent antiviral response, RLR loss does not alter the replication efficiency of MLV. In sharp contrast, the loss of ZAP greatly enhances the replication efficiency of MLV. ZAP localizes to RNA granules, where the processing-body and stress-granule proteins assemble. ZAP induces the recruitment of the MLV transcripts and exosome components to the RNA granules. The CCCH-type zinc-finger domains of ZAP, which are RNA-binding motifs, mediate its localization to RNA granules and MLV transcripts degradation by the exosome. Although ZAP was known as a regulator of RIG-I signaling in a human cell line, ZAP deficiency does not affect the RIG-I–dependent production of type I IFN in mouse cells. Thus, ZAP is a unique member of the cytosolic RNA-sensing PRR family that targets and eliminates intracellular RNA viruses independently of TLR and RLR family members.
Keywords: host defense, retrovirus, ZC3HAV1
Innate immunity is induced after the recognition of viral RNAs by pattern-recognition receptors (PRRs) and is the first line of the host defenses against a variety of RNA viruses (1, 2). Among the PRRs, the Toll-like receptor (TLR) and retinoic acid-inducible gene I (RIG-I)-like receptor (RLR) families play major roles in the recognition of viral RNAs. The RLR’s RIG-I [also called DEAD (Asp-Glu-Ala-Asp) box polypeptide 58 (DDX58)] and melanoma differentiation-associated protein 5 [MDA5, also called interferon induced with helicase C domain 1 (IFIH1)] are RNA helicases that sense the ds form of viral RNAs in the cytosol (3, 4). After sensing dsRNA, the RLRs trigger a signaling pathway that activates interferon (IFN) regulatory factor 3 (IRF3) and IRF7, transcription factors that induce IFN stimulation-responsive, element-dependent transcription (5, 6). This results in the production of type I IFN and the expression of IFN-inducible antiviral proteins. The sensing of viral RNAs by TLR family members also induces the IRF3- and IRF7-dependent type I IFN response (1, 2). In epithelial cells, TLR3, a sensor of dsRNA, detects the incoming RNA virus genomes in endosomes and induces the activation of IRF3, leading to the production of type I IFN (7, 8). In plasmacytoid dendritic cells, TLR7, a sensor of single-stranded (ss) RNA, detects incoming RNA virus genomes in endo-lysosomes and triggers the activation of IRF7, leading to the robust production of type I IFN (9–13). Thus, TLRs and RLRs play major roles in the establishment of an antiviral state by mediating the production of type I IFN.
Murine leukemia virus (MLV), a retrovirus belonging to the gammaretroviral genus of the family Retroviridae, is a causative agent of cancer in murine hosts (14, 15). Although type I IFN is essential for the protection of hosts from lethal infection with a variety of RNA viruses, such as influenza A virus (IAV) and vesicular stomatitis virus (VSV), type I IFN is not essential for induction of the antiviral state against MLV (16–18). Therefore, a different type of innate immune system has been proposed to protect hosts from MLV infection. Although TLR7 has been shown to induce virus-neutralizing immunity after MLV genomic RNA is detected in endosomes (16), the RNA sensor responsible for the elimination of MLV in the cytosol has not been fully understood. RLRs are candidate RNA sensors of intracellular MLV. RLRs might mediate the antiviral response to MLV after the viral RNA is detected, independently of type I IFN because RLRs stimulate not only IRF3/IRF7, but also other transcription factors, such as NF-κB and activator protein 1, which are responsible for the production of inflammatory cytokines and chemokines (19). Another candidate sensor is zinc-finger antiviral protein [ZAP, also called zinc finger CCCH-type, antiviral 1 (ZC3HAV1)]. ZAP was originally identified with an expression cloning method as one of the antiviral proteins directed against MLV (20). ZAP reduces the level of MLV transcripts in the cytosol to suppress MLV infection at the posttranscriptional stage, whereas ZAP does not inhibit the early stage of the MLV infection. ZAP recognizes the MLV transcripts via its CCCH-type zinc-finger domains and binds with RNA helicases and the components of the exosome (an RNA degradation system) to induce the degradation of the MLV transcripts (21–25). However, it is unclear whether endogenous ZAP is involved in the antiviral response to replication-competent MLV in primary cells. In the present study, we examined the roles of these two types of cytosolic RNA sensors and demonstrated the spatial regulation of the innate immune response directed against intracellular MLV.
Results
RLRs Do Not Regulate the Antiviral Response to MLV in Primary Mouse Embryonic Fibroblasts.
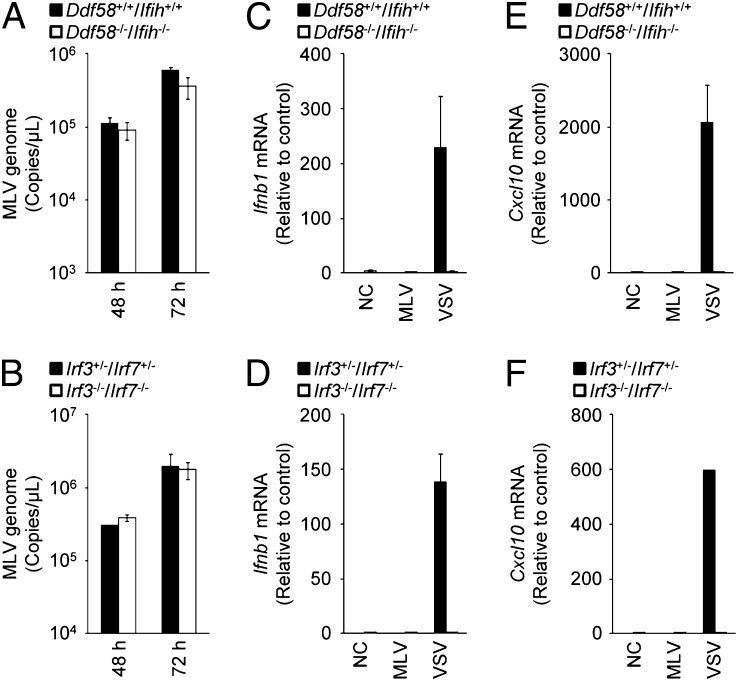
We first examined the involvement of RLRs in the antiviral response to MLV in mouse embryonic fibroblasts (MEFs). The replication efficiency of MLV in Ddx58−/−/Ifih1−/− MEFs was similar to that in Ddx58+/+/Ifih1+/+ MEFs (Fig. 1A). Furthermore, the replication efficiency of MLV in Irf3−/−/Irf7−/− MEFs was similar to that in Irf3+/−/Irf7+/− MEFs (Fig. 1B). Consistent with this, the levels of Ifnb1 and chemokine (C-X-C motif) ligand 10 (Cxcl10) mRNAs did not change during MLV infection (Fig. 1 C–F). The RLR–IRF3/7 signaling axis is essential for the up-regulation of Ifnb1 and Cxcl10 mRNAs during VSV infection. R848, a ligand of TLR7, failed to stimulate MEFs isolated from C57BL/6 mice (Fig. S1), indicating that no RNA-sensing TLR family member recognizes MLV in the extracellular space of MEFs. Therefore, MLV evades the RLR and TLR systems and does not induce the type I IFN response in MEFs.
Fig. 1.
RIG-I–like receptors are not essential for the antiviral response to MLV in primary MEFs. (A and B) Ddx58+/+/Ifih1+/+ and Ddx5−/−/Ifih1−/− MEFs (A) or Irf3+/−/Irf7+/− and Irf−/−/Irf7−/− MEFs (B) were infected with MLV (2 × 1010 copies per μL) for 48 or 72 h. The copy numbers of the MLV genome in the culture supernatants were measured by quantitative RT-PCR. (C–F) Ddx58+/+/Ifih1+/+ and Ddx58−/−/Ifih1−/− MEFs (C and E) or Irf3+/−/Irf7+/− and Irf3−/−/Irf7−/− MEFs (D and F) were infected with MLV (2 × 1010 copies per μL) or VSV [multiplicity of infection (MOI) = 1] for 12 h. The levels of Ifnb1 (C and D) and Cxcl10 (E and F) mRNAs were measured by quantitative RT-PCR. The results shown are means ± SD (n = 3).
Endogenous ZAP Limits the Replication of MLV in Primary MEFs.
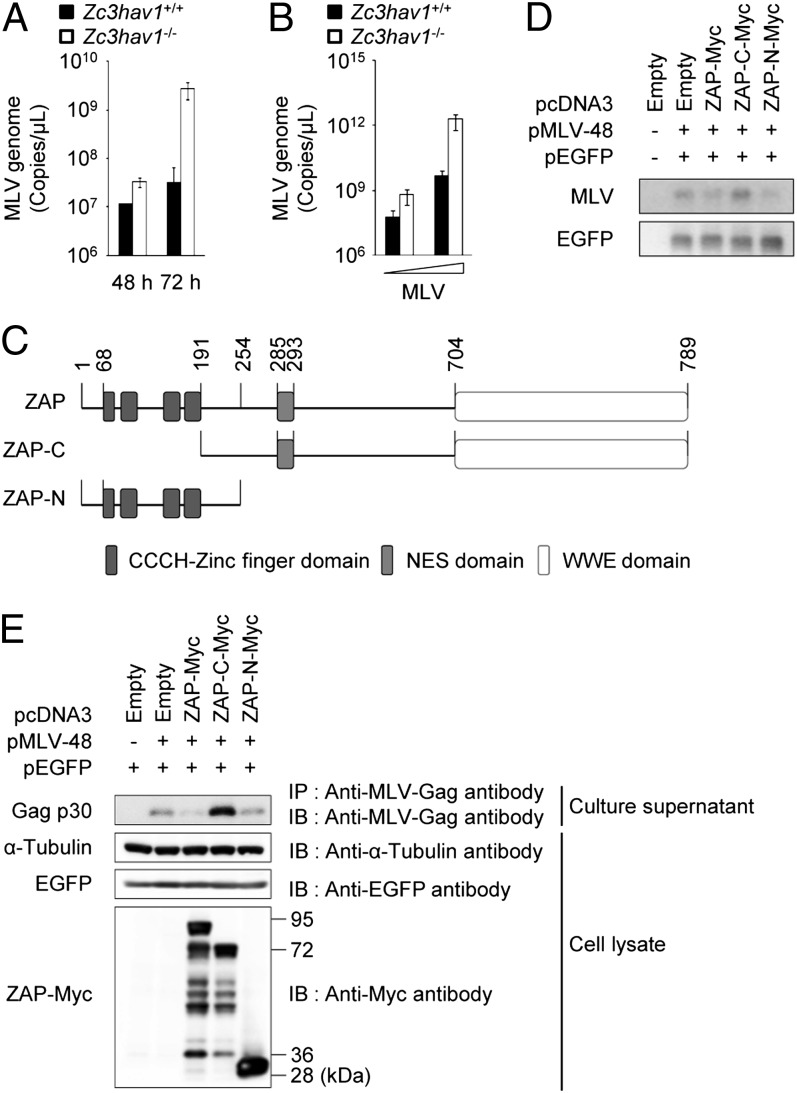
We next investigated the role of ZAP, another cytosolic sensor of viral RNA, in the antiviral response to MLV. Previous studies have demonstrated that the ectopic expression of ZAP potently inhibits replication-incompetent MLV in the cytoplasm of various types of cell lines (20). Therefore, we generated Zc3hav1−/− mice to examine whether endogenous ZAP controls the replication of MLV in primary cells (Fig. S2). Detectable levels of ZAP protein were expressed in Zc3hav1+/+ MEFs before and after MLV infection (Fig. S2D). Whereas ZAP deficiency did not alter the replication efficiency of VSV in MEFs (Fig. S3), ZAP deficiency greatly enhanced the replication efficiency of MLV (Fig. 2 A and B). These findings indicate that endogenous ZAP is responsible for the antiviral response to replication-competent MLV in primary mouse cells.
Fig. 2.
ZAP inhibits MLV replication in primary MEFs. (A) Zc3hav1+/+ and Zc3hav1−/− MEFs were infected with MLV (2 × 1010 copies per μL). Viral RNA was isolated at the indicated time points. The copy numbers of the MLV genome in the culture supernatants were measured by quantitative RT-PCR. (B) Zc3hav1+/+ and Zc3hav1−/− MEFs were infected with increasing doses of MLV (2 × 108 and 2 × 109 copies per μL) for 96 h. The copy numbers of the MLV genome in the culture supernatants were measured by quantitative RT-PCR. (C) Domain architecture of ZAP. (D and E) 293T cells were transfected with pMLV-48 and pEGFP-N1 together with the indicated ZAP expression plasmids for 48 h. Cytoplasmic RNA was subjected to Northern blotting analysis of the indicated RNAs (D). The culture supernatants were subjected to immunoprecipitation coupled to immunoblotting to detect the indicated proteins (E). The results shown are means ± SD (n = 3). NES, nuclear export signal.
The CCCH-type zinc-finger domains of ZAP are known to recognize the MLV transcripts and to induce its degradation (21, 25). Consistent with this, the ectopic expression of the N-terminal portion of ZAP, which contains the CCCH-type zinc-finger domains, but not the ectopic expression of the C-terminal portion of ZAP, which lacks CCCH-type zinc-finger domains, reduced the level of MLV transcripts in the cytosol (Fig. 2 C and D). The ectopic expression of the CCCH-type zinc-finger domains of ZAP also suppressed the expression of the Gag protein of MLV (Fig. 2E). Therefore, the CCCH-type zinc-finger domains of ZAP are essential for its antiviral action against MLV.
CCCH-Type Zinc-Finger Domains of ZAP Mediate Its Localization to the RNA Granules.
The involvement of ZAP in the antiviral response to MLV prompted us to determine the mechanism underlying the ZAP-dependent degradation of the MLV transcripts. Although a previous study showed that ZAP acts in the cytosol (20), it was still unclear where in the cytosol ZAP eliminates the MLV transcripts. Therefore, we examined whether ZAP localizes to a cytosolic compartment, such as in the processing bodies (P-bodies) (26). When it was ectopically expressed, ZAP localized to cytoplasmic dot-like structures in a manner that was dependent on its CCCH-type zinc-finger domains (Fig. 3A). The ZAP-positive dot-like structures colocalized with marker proteins for P-bodies, such as DCP1 decapping enzyme homolog A (Saccharomyces cerevisiae; DCP1A) and DDX6 (Fig. 3B). ZAP induced the enlargement of the DCP1A- and DDX6-positive dot-like structures, suggesting that the ZAP-positive dot-like structures are not conventional P-bodies. ZAP also colocalized with marker proteins for stress granules, such as GTPase-activating protein (SH3 domain) binding protein 1 (G3BP1) and cytotoxic granule-associated RNA binding protein (TIA-1) (Fig. S4). Furthermore, the RNA helicase DEAH (Asp-Glu-Ala-His) box polypeptide 30 (DHX30), which binds to ZAP to facilitate its antiviral action against MLV (24), colocalized with ZAP to the DCP1A-positive dot-like structures (Fig. S5). By contrast, ZAP did not colocalize with mitochondrial preprotein translocases of the outer membrane 20 (TOM20), 70-kDa peroxisomal membrane protein (PMP70), early endosome antigen 1 (EEA1), or lysosomal-associated membrane protein 1 (LAMP1), marker proteins for the mitochondria, peroxisomes, endosomes, and lysosomes, respectively (Fig. 3C). These findings indicate that ZAP localizes to the RNA granules, where the marker proteins for P-bodies and stress granules assemble.
Fig. 3.
ZAP localizes to DCP1A- and DDX6-positive RNA granules. (A–C) 293T cells were transfected with the indicated vectors for 48 h and then fixed. The samples were immunostained with the indicated antibodies and then observed by confocal laser scanning microscopy. The data are representative of three independent experiments. (Scale bars, 10 μm.)
ZAP Recruits the MLV Transcripts and Exosome Components to RNA Granules.
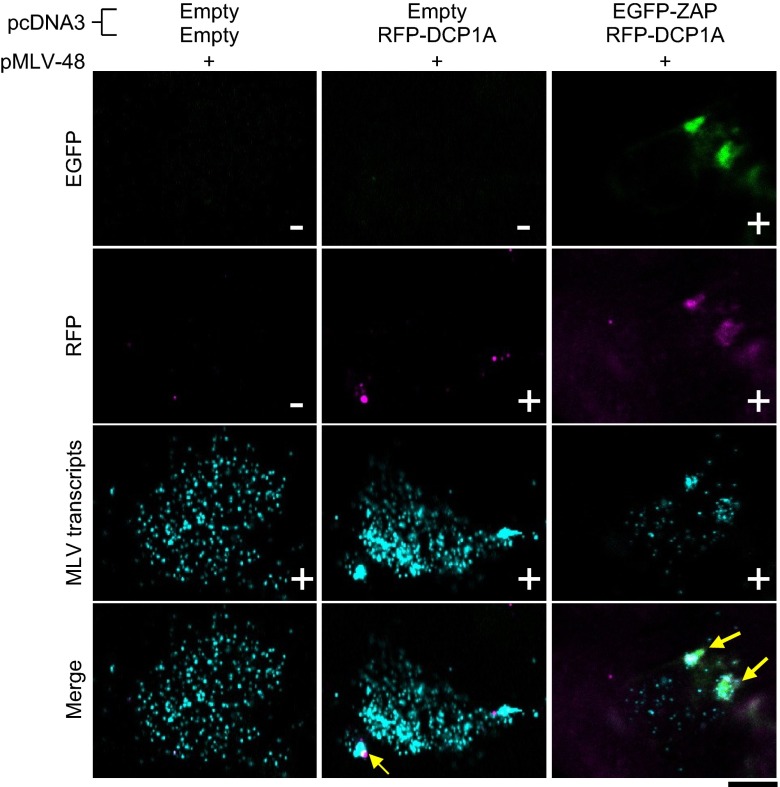
The localization of the MLV transcripts has been poorly understood. We used an improved RNA FISH method to visualize the subcellular localization of viral RNA and identified the cytosolic compartments in which ZAP acts on the MLV transcripts. The MLV transcripts mainly localize in the cytosol and colocalize with DCP1A-positive RNA granules at low frequency (Fig. 4). However, the ectopic expression of ZAP reduced the level of MLV transcripts in the cytosol and dramatically altered its localization from the cytosol to ZAP- and DCP1A-positive RNA granules (Fig. 4 and Fig. S6). Therefore, ZAP tethers the MLV transcripts and transfers it to the RNA granules.
Fig. 4.
ZAP recruits the MLV transcripts to RNA granules. 293T cells were transfected with the indicated plasmids for 48 h and then fixed. The samples were subjected to in situ hybridization analysis with a fluorescent probe for MLV transcripts and then observed by confocal laser scanning microscopy. (Scale bar, 10 μm.)
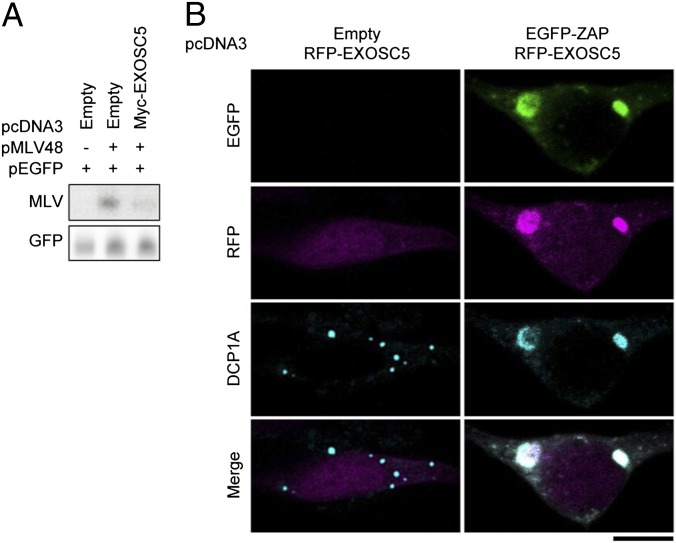
Because ZAP is not a ribonuclease, it requires the support of an RNA degradation system to destabilize the MLV transcripts. Consistent with this, previous studies have shown that exosome components and RNA helicases interact with ZAP to mediate the antiviral response to MLV (22–24). Therefore, we focused on the localization of exosome component 5 (EXOSC5, also known as RRP46) (27). The ectopic expression of EXOSC5 reduced the level of MLV transcripts in the cytosol (Fig. 5A). Under normal conditions, EXOSC5 localized in the cytosol and nuclei, and colocalized with the DCP1A-positive RNA granules at low frequency (Fig. 5B). However, when ZAP was ectopically expressed, EXOSC5 moved from the cytosol to the ZAP- and DCP1A-positive RNA granules (Fig. 5B). These findings indicate that ZAP recruits the exosome component to the RNA granules to induce the degradation of MLV transcripts.
Fig. 5.
EXOSC5 colocalizes with ZAP on RNA granules. (A) 293T cells were transfected with pMLV-48 and pEGFP-N1 together with the indicated ZAP expression plasmids for 48 h. Cytoplasmic RNA was subjected to Northern blotting analysis to detect the indicated RNAs. (B) 293T cells were transfected with the indicated plasmids and then fixed. The samples were immunostained with anti-DCP1A antibody and then observed by confocal laser scanning microscopy. The data are representative of three independent experiments. (Scale bar, 10 μm.)
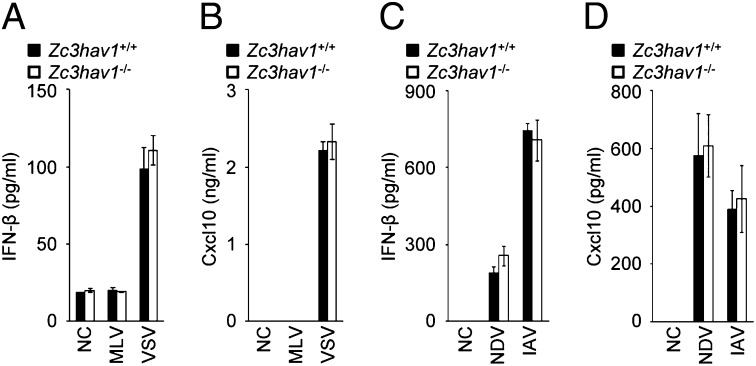
ZAP Does Not Regulate the RIG-I–Dependent Type I IFN Response in Primary Mouse Cells.
A recent study showed that ZAP positively regulated RIG-I signaling during RNA virus infection in a human cell line (28). Therefore, we examined the involvement of ZAP in the RIG-I–dependent type I IFN response in primary mouse cells. In Zc3hav1−/− primary MEFs, the IFN-β and Cxcl10 proteins were produced normally in response to VSV, an RNA virus recognized by RIG-I (Fig. 6 A and B). Although ZAP deficiency greatly enhanced the replication of MLV (Fig. 2 A and B), no IFN-β or Cxcl10 protein was produced in Zc3hav1−/− MEFs infected with MLV. In Zc3hav1−/− mouse primary dendritic cells, IFN-β and Cxcl10 were also normally produced in response to Newcastle disease virus (NDV) and IAV, RNA viruses recognized by RIG-I (Fig. 6 C and D). Furthermore, ZAP deficiency did not affect the production of IFN-β in MEFs stimulated with the RIG-I ligand, 5′ triphosphate dsRNA (3pRNA) (Fig. S7 A and B), the MDA5 ligand poly(rI–rC), and a synthetic dsDNA poly(dA–dT) (Fig. S7C). These findings indicate that ZAP is not a regulator of the RIG-I–dependent type I IFN response in primary mouse cells and strengthen our conclusion that ZAP eliminates MLV independently of the RLR–IRF3/7 signaling axis.
Fig. 6.
ZAP is not essential for the RIG-I–mediated type I IFN response. (A and B) Zc3hav1+/+ and Zc3hav1−/− MEFs were infected with MLV (2 × 1010 copies per μL) or VSV (MOI = 1) for 12 h. The levels of IFN-β (A) and Cxcl10 (B) proteins in the culture supernatants were measured with ELISAs. (C and D) Zc3hav1+/+ and Zc3hav1−/− bone marrow-derived dendritic cells were infected with NDV (2.5 × 105 pfu/mL) or IAV (PR8, 100 Hematoglutinin) for 24 h. The levels of IFN-β (C) and Cxcl10 (D) proteins in the culture supernatants were measured with ELISAs. The results shown are means ± SD (n = 3).
Discussion
In this study, we showed that endogenous ZAP suppresses the replication of MLV in MEFs. This raises the issue of whether endogenous ZAP suppresses the replication of other types of RNA viruses, including human retroviruses. The RNAi-mediated knockdown of ZC3HAV1 mRNA enhanced the replication of xenotropic MLV-related virus, an artificial retrovirus belonging to the gammaretroviral genus of the family Retroviridae (29), in 293T cells (Fig. S8 A and B), whereas the knockdown of ZC3HAV1 mRNA did not enhance the replication of human T-cell leukemia virus type I, a retrovirus belonging to the deltaretroviral genus of the family Retroviridae (30), in MT-2 cells (Fig. S8 C and D). In a previous study, the knockdown of ZC3HAV1 mRNA enhanced the replication of HIV-1, a retrovirus belonging to the lentiviral genus of the family Retroviridae (31), in HOS-CD4 cells expressing chemokine (C-C motif) receptor 5 (32). Therefore, ZAP functions in human cells to target not all but certain types of retroviruses. ZAP is also known to suppress the replication of RNA viruses belonging to the families Filoviridae and Togaviridae (33, 34). Although ZAP has been shown to recognize the viral RNA of RNA viruses belonging to the families Filoviridae, Togaviridae, and Retroviridae via, its CCCH-type zinc-finger domains, the common features that are recognized by these domains, such as specific sequences or structural characteristics, have not been determined. Further studies are required to identify the RNA ligand of ZAP that induces the destabilization of the viral RNA by the RNA degradation machinery.
Although accumulating evidence indicates that ZAP counters a variety of RNA viruses under in vitro experimental conditions (20, 33, 34), it is still unclear whether ZAP protects hosts from RNA viral infections in vivo. RNA-sensing TLRs and the ssDNA cytosine deaminase apolipoprotein B mRNA-editing, enzyme-catalytic, polypeptide-like 3 are other antiviral systems that affect mouse retroviruses, and also control the replication of endogenous retroviruses (ERVs) (16, 35–37). Therefore, ZAP might also contribute to the antiviral response to ERVs and prevent the ERV-induced generation of tumors in vivo. To assess this, we are now establishing a colony of Zc3hav1−/− mice in the C57BL/6 genetic background. In a future study, we will attempt to determine the in vivo role of ZAP in the host defense responses to endogenous and exogenous microbes.
The CCCH-type zinc-finger–domain-containing protein family regulates RNA synthesis, splicing, and degradation, and is involved in a variety of cellular events, including cell growth, cell death, the inflammatory response, and the antimicrobial response (38, 39). To date, more than 50 CCCH-type zinc-finger-domain–containing proteins have been identified (40). Although various CCCH-type zinc-finger-domain-containing proteins, including tristetraprolin, roquin, and regnase-1, have been shown to be regulators of cytokine mRNA stability, ZAP is the only CCCH-type zinc-finger–domain-containing protein known to promote the destabilization of viral RNA (20, 41–43). Therefore, it will be interesting to identify a CCCH-type zinc-finger–domain-containing protein capable of mediating an antiviral response to RNA viruses that have evaded ZAP and the other RNA-sensing PRRs.
Materials and Methods
Reagents.
Anti-MLV-Gag antibody (ABIN457547) was purchased from Antibodies-online. Anti–α-tubulin antibody (T6199) was purchased from Sigma. Anti-GFP antibody (598) was purchased from MBL. Chicken anti–avian myelocytomatosis viral oncogene homolog (Myc) antibody (A190-103A) for the immunostaining assay was purchased from Bethyl Laboratories. Mouse anti–Myc-tag antibody (2276S) for immunoblotting was purchased from Cell Signaling. Anti-DDX6 (ab40684), anti-PMP70 (ab3421), and anti-LAMP1 (ab24170) antibodies were purchased from Abcam. Anti-DCP1A antibody (H00055802-M06) was purchased from Abnova. Anti-TOM20 antibody (SC-11415) was purchased from Santa Cruz Biotechnology. Anti-EEA1 antibody (610456) was purchased from BD Biosciences. The ELISA kit for mouse IFN-β was purchased from Pestka Biomedical Laboratories Interferon Source. The ELISA kit for mouse Cxcl10 was purchased from R&D Systems.
Plasmids.
pMLV-48 (GenBank accession no. J02255.1) was previously described (44) and kindly donated by H. Fan (University of California, Irvine, CA). pcDNA3.1(+) was purchased from Invitrogen. To generate the ZAP expression constructs, NheI/NotI cDNA fragments encoding full-length mouse ZAP (GenBank accession no. NM_028864.2) and the C-terminal portion of ZAP and a BamHI/NotI cDNA fragment encoding the N-terminal portion of ZAP were amplified from pCMV–SPORT6–Zc3hav1 (MMM1013-7511214, Open Biosystems) by PCR and cloned into the corresponding restriction sites of pcDNA3 to produce pcDNA3–ZAP, pcDNA3–ZAP–C, and pcDNA3–ZAP–N, respectively. To generate the expression construct for the EGFP–ZAP fusion protein, an NheI/SpeI cDNA fragment encoding EGFP was amplified from pEGFP-N1 (Clontech) by PCR and cloned into the NheI site of pcDNA3–ZAP to produce pcDNA3–EGFP–ZAP. To generate the red fluorescent protein (RFP) expression construct, a BamHI/EcoRI cDNA fragment of RFP was amplified from pTagRFP-N1 (Evrogen) by PCR and cloned into the BamHI/EcoRI sites of pcDNA3 to produce pcDNA3–RFP. To generate the expression constructs for the RFP–DCP1A and RFP–EXOSC5 fusion proteins, EcoRI/NotI cDNA fragments of human DCP1A and human EXOSC5 were amplified from a 293T cDNA library by PCR, and cloned into the EcoRI/NotI sites of pcDNA3–RFP to produce pcDNA3–RFP–DCP1A and pCDNA3–RFP–EXOSC5.
Mice, Cells, and Viruses.
C57BL/6 mice were purchased from CLEA Japan, Inc. Irf3−/−/Irf7−/− mice were kindly donated by T. Taniguchi (The University of Tokyo, Tokyo, Japan). The Ddx58−/−/Ifih1−/− mice have been described previously (45). The mice were maintained in our animal facility and treated in accordance with the guidelines of Osaka University. Primary MEFs were prepared from pregnant female mice on embryonic day 13.5, as described previously (4). To prepare bone marrow-derived dendritic cells, mouse bone marrow cells were cultured in the presence of 10 ng/mL GM-CSF (PeproTech) for 6 d, during which time the culture medium was replaced with medium containing GM-CSF every 2 d. The 293T cells have been described previously (46). Replication-competent MLV was produced by 293T cells transfected with pMLV-48. To induce infection, MLV was incubated with MEFs for 2 h in the presence of 10 μg/mL Polybrene (Millipore). VSV, IAV (A/Puerto Rico/8/34, H1N1 strain), and NDV have been described elsewhere (3, 4).
Quantitative RT-PCR.
Total RNA was isolated using the ZR RNA MicroPrep kit (Zymo Research), according the manufacturer’s instructions. Viral RNA was isolated from the culture supernatants using the ZR Viral RNA kit (Zymo Research), according the manufacturer’s instructions. RT was performed using random primers and Verso reverse transcriptase (Thermo Scientific) according to the manufacturer’s instructions. For quantitative PCR, the cDNA fragments were amplified from the RT products with Real-Time PCR Master Mix (Toyobo) according to the manufacturer’s instructions. The fluorescence from the TaqMan probe for each cytokine was detected with a 7500 Real-Time PCR System (Applied Biosystems). To determine the relative induction of cytokine mRNAs, the level of mRNA expressed from each gene was normalized to the expression of 18S RNA. The copy number of the MLV genomic RNA was determined with the dsDNA copy number calculator program. The experiments were repeated at least three times, with reproducible results.
ELISAs.
The levels of IFN-β and Cxcl10 in the culture supernatants were measured with ELISAs in accordance with the manufacturer’s instructions. The experiments were repeated at least three times, with reproducible results.
Northern Blotting.
Cytoplasmic RNA was extracted using the Cytoplasmic and Nuclear RNA Purification Kit (Norgen) according the manufacturer’s instructions. The RNA obtained was separated electrophoretically, transferred to nylon membranes, and hybridized with the indicated probes. An RNA probe was designed to hybridize specifically to the Gag region from nucleotide 1291 to nucleotide 1472 of the MLV transcripts. The experiments were repeated at least three times, with reproducible results.
Immunoblotting.
Immunoblotting was performed as described previously (47). The experiments were repeated at least three times, with reproducible results.
Immunostaining Assay.
Cells cultured in microscopy chambers (ibidi) were fixed with 3% (wt/vol) paraformaldehyde and then processed for immunostaining as described previously (47). The samples were examined under an LSM 780 confocal laser scanning microscope (Carl Zeiss). The experiments were repeated at least three times, with reproducible results.
Detection of the MLV Transcripts with FISH.
The cells were fixed with 4% paraformaldehyde. FISH was performed using the QuantiGene ViewRNA ISH Cell Assay kit (Veritas) according to the manufacturer’s instructions. A Cy5-labeled FISH probe was designed to hybridize specifically to the Gag region from nucleotide 607 to nucleotide 1833 of the MLV transcripts. The samples were examined under an LSM780 confocal laser scanning microscope. The experiments were repeated at least three times, with reproducible results.
Supplementary Material
Acknowledgments
We thank Drs. H. Fan, D. Trono, H. Miyoshi, and T. Taniguchi for providing invaluable materials and the members of the Laboratory of Host Defenses for their assistance. This work was supported by a Japan Society for the Promotion of Science Grant-in-Aid for Challenging Exploratory Research (to T. Saitoh); the Cabinet Office, Government of Japan, and the Japan Society for the Promotion of Science Funding Program for World-Leading Innovative Research and Development on Science and Technology “FIRST Program” (to S.A.); and National Institutes of Health Grant P01-AI070167 (to S.A.).
Footnotes
The authors declare no conflict of interest.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1310604110/-/DCSupplemental.
References
- 1.Kawai T, Akira S. The roles of TLRs, RLRs and NLRs in pathogen recognition. Int Immunol. 2009;21(4):317–337. doi: 10.1093/intimm/dxp017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Iwasaki A. A virological view of innate immune recognition. Annu Rev Microbiol. 2012;66:177–196. doi: 10.1146/annurev-micro-092611-150203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yoneyama M, et al. The RNA helicase RIG-I has an essential function in double-stranded RNA-induced innate antiviral responses. Nat Immunol. 2004;5(7):730–737. doi: 10.1038/ni1087. [DOI] [PubMed] [Google Scholar]
- 4.Kato H, et al. Differential roles of MDA5 and RIG-I helicases in the recognition of RNA viruses. Nature. 2006;441(7089):101–105. doi: 10.1038/nature04734. [DOI] [PubMed] [Google Scholar]
- 5.Yoneyama M, et al. Direct triggering of the type I interferon system by virus infection: Activation of a transcription factor complex containing IRF-3 and CBP/p300. EMBO J. 1998;17(4):1087–1095. doi: 10.1093/emboj/17.4.1087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Honda K, et al. IRF-7 is the master regulator of type-I interferon-dependent immune responses. Nature. 2005;434(7034):772–777. doi: 10.1038/nature03464. [DOI] [PubMed] [Google Scholar]
- 7.Alexopoulou L, Holt AC, Medzhitov R, Flavell RA. Recognition of double-stranded RNA and activation of NF-kappaB by Toll-like receptor 3. Nature. 2001;413(6857):732–738. doi: 10.1038/35099560. [DOI] [PubMed] [Google Scholar]
- 8.Yamamoto M, et al. Role of adaptor TRIF in the MyD88-independent toll-like receptor signaling pathway. Science. 2003;301(5633):640–643. doi: 10.1126/science.1087262. [DOI] [PubMed] [Google Scholar]
- 9.Cella M, et al. Plasmacytoid monocytes migrate to inflamed lymph nodes and produce large amounts of type I interferon. Nat Med. 1999;5(8):919–923. doi: 10.1038/11360. [DOI] [PubMed] [Google Scholar]
- 10.Diebold SS, Kaisho T, Hemmi H, Akira S, Reis e Sousa C. Innate antiviral responses by means of TLR7-mediated recognition of single-stranded RNA. Science. 2004;303(5663):1529–1531. doi: 10.1126/science.1093616. [DOI] [PubMed] [Google Scholar]
- 11.Heil F, et al. Species-specific recognition of single-stranded RNA via toll-like receptor 7 and 8. Science. 2004;303(5663):1526–1529. doi: 10.1126/science.1093620. [DOI] [PubMed] [Google Scholar]
- 12.Kawai T, et al. Interferon-alpha induction through Toll-like receptors involves a direct interaction of IRF7 with MyD88 and TRAF6. Nat Immunol. 2004;5(10):1061–1068. doi: 10.1038/ni1118. [DOI] [PubMed] [Google Scholar]
- 13.Honda K, et al. Spatiotemporal regulation of MyD88-IRF-7 signalling for robust type-I interferon induction. Nature. 2005;434(7036):1035–1040. doi: 10.1038/nature03547. [DOI] [PubMed] [Google Scholar]
- 14.Ihle JN, Rein A, Mural R. Immunological and virological mechanisms in retrovirus-induced murine leukemogenesis. In: Klein G, editor. Advances in Viral Oncology. New York: Raven Press; 1984. pp. 95–137. [Google Scholar]
- 15.Schiff RD, Oliff A. The pathophysiology of murine retrovirus-induced leukemias. Crit Rev Oncol Hematol. 1986;5(3):257–323. doi: 10.1016/s1040-8428(86)80041-5. [DOI] [PubMed] [Google Scholar]
- 16.Kane M, et al. Innate immune sensing of retroviral infection via Toll-like receptor 7 occurs upon viral entry. Immunity. 2011;35(1):135–145. doi: 10.1016/j.immuni.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Everitt AR, et al. GenISIS Investigators MOSAIC Investigators IFITM3 restricts the morbidity and mortality associated with influenza. Nature. 2012;484(7395):519–523. doi: 10.1038/nature10921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fensterl V, et al. Interferon-induced Ifit2/ISG54 protects mice from lethal VSV neuropathogenesis. PLoS Pathog. 2012;8(5):e1002712. doi: 10.1371/journal.ppat.1002712. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Goubau D, Deddouche S, Reis E Sousa C. Cytosolic sensing of viruses. Immunity. 2013;38(5):855–869. doi: 10.1016/j.immuni.2013.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gao G, Guo X, Goff SP. Inhibition of retroviral RNA production by ZAP, a CCCH-type zinc finger protein. Science. 2002;297(5587):1703–1706. doi: 10.1126/science.1074276. [DOI] [PubMed] [Google Scholar]
- 21.Guo X, Carroll JW, Macdonald MR, Goff SP, Gao G. The zinc finger antiviral protein directly binds to specific viral mRNAs through the CCCH zinc finger motifs. J Virol. 2004;78(23):12781–12787. doi: 10.1128/JVI.78.23.12781-12787.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Guo X, Ma J, Sun J, Gao G. The zinc-finger antiviral protein recruits the RNA processing exosome to degrade the target mRNA. Proc Natl Acad Sci USA. 2007;104(1):151–156. doi: 10.1073/pnas.0607063104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chen G, Guo X, Lv F, Xu Y, Gao G. p72 DEAD box RNA helicase is required for optimal function of the zinc-finger antiviral protein. Proc Natl Acad Sci USA. 2008;105(11):4352–4357. doi: 10.1073/pnas.0712276105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ye P, Liu S, Zhu Y, Chen G, Gao G. DEXH-Box protein DHX30 is required for optimal function of the zinc-finger antiviral protein. Protein Cell. 2010;1(10):956–964. doi: 10.1007/s13238-010-0117-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang X, Lv F, Gao G. Mutagenesis analysis of the zinc-finger antiviral protein. Retrovirology. 2010;7:19. doi: 10.1186/1742-4690-7-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Reineke LC, Lloyd RE. Diversion of stress granules and P-bodies during viral infection. Virology. 2013;436(2):255–267. doi: 10.1016/j.virol.2012.11.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu Q, Greimann JC, Lima CD. Reconstitution, activities, and structure of the eukaryotic RNA exosome. Cell. 2006;127(6):1223–1237. doi: 10.1016/j.cell.2006.10.037. [DOI] [PubMed] [Google Scholar]
- 28.Hayakawa S, et al. ZAPS is a potent stimulator of signaling mediated by the RNA helicase RIG-I during antiviral responses. Nat Immunol. 2011;12(1):37–44. doi: 10.1038/ni.1963. [DOI] [PubMed] [Google Scholar]
- 29.Paprotka T, et al. Recombinant origin of the retrovirus XMRV. Science. 2011;333(6038):97–101. doi: 10.1126/science.1205292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamamoto N, Hinuma Y. Viral aetiology of adult T-cell leukaemia. J Gen Virol. 1985;66(Pt 8):1641–1660. doi: 10.1099/0022-1317-66-8-1641. [DOI] [PubMed] [Google Scholar]
- 31.Haseltine WA. Replication and pathogenesis of the AIDS virus. J Acquir Immune Defic Syndr. 1988;1(3):217–240. [PubMed] [Google Scholar]
- 32.Zhu Y, et al. Zinc-finger antiviral protein inhibits HIV-1 infection by selectively targeting multiply spliced viral mRNAs for degradation. Proc Natl Acad Sci USA. 2011;108(38):15834–15839. doi: 10.1073/pnas.1101676108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Müller S, et al. Inhibition of filovirus replication by the zinc finger antiviral protein. J Virol. 2007;81(5):2391–2400. doi: 10.1128/JVI.01601-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bick MJ, et al. Expression of the zinc-finger antiviral protein inhibits alphavirus replication. J Virol. 2003;77(21):11555–11562. doi: 10.1128/JVI.77.21.11555-11562.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Okeoma CM, Lovsin N, Peterlin BM, Ross SR. APOBEC3 inhibits mouse mammary tumour virus replication in vivo. Nature. 2007;445(7130):927–930. doi: 10.1038/nature05540. [DOI] [PubMed] [Google Scholar]
- 36.Santiago ML, et al. Apobec3 encodes Rfv3, a gene influencing neutralizing antibody control of retrovirus infection. Science. 2008;321(5894):1343–1346. doi: 10.1126/science.1161121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yu P, et al. Nucleic acid-sensing Toll-like receptors are essential for the control of endogenous retrovirus viremia and ERV-induced tumors. Immunity. 2012;37(5):867–879. doi: 10.1016/j.immuni.2012.07.018. [DOI] [PubMed] [Google Scholar]
- 38.Chen CY, et al. AU binding proteins recruit the exosome to degrade ARE-containing mRNAs. Cell. 2001;107(4):451–464. doi: 10.1016/s0092-8674(01)00578-5. [DOI] [PubMed] [Google Scholar]
- 39.Hurt JA, et al. A conserved CCCH-type zinc finger protein regulates mRNA nuclear adenylation and export. J Cell Biol. 2009;185(2):265–277. doi: 10.1083/jcb.200811072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liang J, Song W, Tromp G, Kolattukudy PE, Fu M. Genome-wide survey and expression profiling of CCCH-zinc finger family reveals a functional module in macrophage activation. PLoS ONE. 2008;3(8):e2880. doi: 10.1371/journal.pone.0002880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lai WS, et al. Evidence that tristetraprolin binds to AU-rich elements and promotes the deadenylation and destabilization of tumor necrosis factor alpha mRNA. Mol Cell Biol. 1999;19(6):4311–4323. doi: 10.1128/mcb.19.6.4311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu D, et al. Roquin represses autoimmunity by limiting inducible T-cell co-stimulator messenger RNA. Nature. 2007;450(7167):299–303. doi: 10.1038/nature06253. [DOI] [PubMed] [Google Scholar]
- 43.Matsushita K, et al. Zc3h12a is an RNase essential for controlling immune responses by regulating mRNA decay. Nature. 2009;458(7242):1185–1190. doi: 10.1038/nature07924. [DOI] [PubMed] [Google Scholar]
- 44.Bacheler L, Fan H. Isolation of recombinant DNA clones carrying complete integrated proviruses of Moloney murine leukemia virus. J Virol. 1981;37(1):181–190. doi: 10.1128/jvi.37.1.181-190.1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kato H, et al. Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med. 2008;205(7):1601–1610. doi: 10.1084/jem.20080091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Saitoh Y, et al. Overexpressed NF-kappaB-inducing kinase contributes to the tumorigenesis of adult T-cell leukemia and Hodgkin Reed-Sternberg cells. Blood. 2008;111(10):5118–5129. doi: 10.1182/blood-2007-09-110635. [DOI] [PubMed] [Google Scholar]
- 47.Saitoh T, et al. Loss of the autophagy protein Atg16L1 enhances endotoxin-induced IL-1beta production. Nature. 2008;456(7219):264–268. doi: 10.1038/nature07383. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.