Significance
Plasmodium parasites have two distinct intracellular growth stages inside the mammalian host—the first stage, which is clinically silent, in liver hepatocytes, and the second, which causes the symptoms of malaria, in red blood cells. This study reports the discovery of a class of antimalarial compounds called torins, which are extremely potent inhibitors of both intracellular stages of Plasmodium. We show that torins block trafficking of liver stage parasite proteins to the physical host–parasite interface, called the parasitophorous vacuole membrane (PVM), and that without continuous trafficking of PVM-resident proteins, the parasite is subject to elimination by its host hepatocyte.
Keywords: host-parasite interactions, malaria, protein trafficking, P. falciparum
Abstract
Residence within a customized vacuole is a highly successful strategy used by diverse intracellular microorganisms. The parasitophorous vacuole membrane (PVM) is the critical interface between Plasmodium parasites and their possibly hostile, yet ultimately sustaining, host cell environment. We show that torins, developed as ATP-competitive mammalian target of rapamycin (mTOR) kinase inhibitors, are fast-acting antiplasmodial compounds that unexpectedly target the parasite directly, blocking the dynamic trafficking of the Plasmodium proteins exported protein 1 (EXP1) and upregulated in sporozoites 4 (UIS4) to the liver stage PVM and leading to efficient parasite elimination by the hepatocyte. Torin2 has single-digit, or lower, nanomolar potency in both liver and blood stages of infection in vitro and is likewise effective against both stages in vivo, with a single oral dose sufficient to clear liver stage infection. Parasite elimination and perturbed trafficking of liver stage PVM-resident proteins are both specific aspects of torin-mediated Plasmodium liver stage inhibition, indicating that torins have a distinct mode of action compared with currently used antimalarials.
The population at risk for developing malaria is vast, comprising some 3.3 billion people particularly in sub-Saharan Africa and Southeast Asia, with mortality estimates ranging from 655,000 to 1,200,000 (1). Widespread resistance has limited the therapeutic utility of most existing antimalarial drugs, and artemisinin, the highly efficacious cornerstone of artemisinin combination therapies, appears to be at risk for the same fate (2). The need for new antimalarial chemotherapeutic strategies is thus acute.
Plasmodium spp., the causative agents of malaria, have a complex life cycle with alternating motile-nonreplicative and sessile-replicative forms in both mammal and mosquito. In the mammalian host, Plasmodium invades and replicates inside two very distinct cell types: hepatocytes and red blood cells (RBCs). In mammals, the Plasmodium life cycle is initiated by a motile sporozoite that invades a hepatocyte, where it resides for 2–14 d, multiplying into >10,000 merozoites in a single cycle (3). Once released into the bloodstream, each of these motile merozoites will infect an RBC and, within 1–3 d, generate 10–30 new merozoites, which will contribute to the continuous cycle of blood stage infection that causes the symptoms, morbidity, and mortality of malaria.
These two stages of mammalian infection, despite taking place in distinct cell types and having an orders-of-magnitude difference in parasite replication, do share common features. In both, the motile “zoite” invades the host cell through formation of a parasitophorous vacuole (PV). Both stages grow and replicate exclusively within the confines of the PV, and the parasitophorous vacuole membrane (PVM), which is populated with parasite proteins, constitutes the physical host–parasite interface throughout development. Unlike the vacuoles of many intracellular pathogens including Leishmania, Chlamydia, Mycobacteria, and Legionella (4, 5), the Plasmodium vacuole, like that of Toxoplasma gondii, does not fuse with host lysosomes and is not acidified (6). This is not unsurprising in the context of Plasmodium development in an RBC, which lacks endomembrane system trafficking and, indeed, lysosomes. The highly polarized hepatocyte, however, has extensive vesicular transport networks (7) and can target intracellular pathogens residing in a vacuole (8), suggesting that the exoerythrocytic form (EEF) may need to resist host cell attack.
Although the PVM is thought to be critical for Plasmodium growth in both the hepatocyte and the RBC contexts, its cellular roles remain elusive. The importance of several Plasmodium PVM-resident proteins, however, has been conclusively demonstrated in both blood and liver stages. Attempts to generate exported (exp)1 and Plasmodium translocon of exported protein (ptex)150 knockout parasites in Plasmodium falciparum failed (9, 10), revealing that these are both essential proteins for the blood stage, whereas Plasmodium berghei and Plasmodium yoelii mutants lacking up-regulated in sporozoites (uis)3 or uis4 fail to complete liver stage development (11, 12). These PVM-resident proteins, and thus the PVM itself, are performing functions that are crucial for Plasmodium growth, but delineating the functions of individual PVM-resident proteins has proven as difficult as identifying the cellular processes mediated by the PVM.
The one process in which both the centrality of the PVM is known and evidence for the participation of specific PVM proteins exists is the export of parasite proteins to the RBC. A cohort of parasite proteins that are involved in extensive physiological and structural modifications of the infected RBC (iRBC) is exported into the iRBC cytoplasm and beyond (13). Five proteins have been identified as components of PTEX, the proposed export machinery at the iRBC PVM (9). Although liver stage protein export has been shown for the Circumsporozite (CS) protein (14) and PTEX components are expressed in P. falciparum EEFs (15), a role for parasite protein export into the hepatocyte remains speculative; the host hepatocyte may not require the extensive structural remodeling that the iRBC does.
Conversely, however, the hepatocyte, with its extensive vesicular transport network, intuitively constitutes a more hostile host environment than the RBC, and there is evidence that the liver stage PVM may play a crucial role in preventing host cell-mediated parasite killing, as it does in Toxoplasma gondii (16). Support for a protective role for the liver stage PVM comes from knockout parasites that fail in the earliest steps of PVM formation and remodeling. Sporozoites lacking the p52/p36 gene pair invade hepatocytes successfully, but fail in PVM formation (17, 18) and are severely reduced in abundance midway through liver stage development. Parasites lacking slarp/sap (19, 20), a regulator of early liver stage development, fail to express UIS4 and exported protein 1 (EXP-1), along with other parasite proteins, and are also eliminated at the beginning of infection.
Acquisition of resources from the host-cell environment, an unambiguous requirement for an obligate intracellular parasite like Plasmodium, is a function ascribed to the PVM in both mammalian stages. The PVM allows the free passage of molecules (21, 22), presumably through proteinaceous pores, which may contribute to acquisition of host nutrients and disposal of parasite waste products. Members of the early transcribed membrane protein (ETRAMP) family, single-pass transmembrane proteins conserved among Plasmodium spp., which are highly expressed and developmentally regulated in both blood and liver stage parasites (23, 24), could be candidates for mediating uptake of host resources. Such a role in lipid uptake has indeed been proposed for the P. berghei ETRAMP UIS3 on the basis of its interaction with host-cell L-FABP (liver fatty acid binding protein) (25).
Although Plasmodium parasites must use host resources to support their own growth in both mammalian stages, the single cycle replicative output of the liver stage parasite is vastly greater than that of the blood stage, which may reflect a similarly increased need for host resources. In this respect, the hepatocyte constitutes far superior “raw material” compared with the RBC; hepatocytes are not only metabolically active, but also highly versatile cells, which are capable of altering uptake, storage, production, and degradation of a wide array of macromolecules in response to cellular and organismal requirements. The presence of a growing Plasmodium parasite is sensed by the host hepatocyte, which responds with activation of cellular stress responses and altered metabolism (26, 27). The mammalian target of rapamycin (mTOR) kinase integrates signals from amino acids, stress, oxygen, energy, and growth factors and responds by altering cellular protein and lipid synthesis, as well as autophagy (28). As such, we sought to determine how inhibition of host mTOR signaling would affect Plasmodium liver stage development. Here we show that torins, a single structural class of mTOR inhibitors, are highly potent antiplasmodial compounds targeting both mammalian stages in vitro and in vivo. Independent of host-cell mTOR, torins impair trafficking of Plasmodium liver stage PVM-resident proteins, revealing the fast turnover of these proteins at the liver stage PVM, and provoke elimination of liver stage parasites.
Results
Torins Potently Inhibit Plasmodium Liver and Blood Stages.
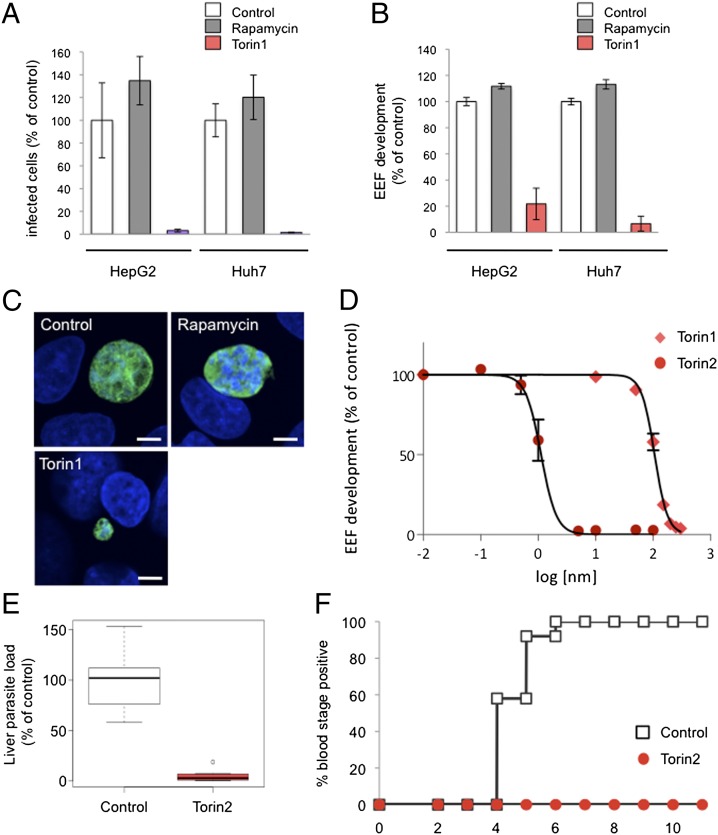
We first tested whether Plasmodium infection could be modulated by inhibition of host mTOR signaling using two unrelated mTOR inhibitors (see Table S1 for structures, reviewed in ref. 29): rapamycin, a naturally occurring macrolide that is an allosteric mTOR inhibitor, and Torin1, a tricyclic benzonaphthyridinone developed through medicinal chemistry efforts as an ATP-competitive inhibitor (30). We plated two human hepatoma cell lines, Huh7 and HepG2, and infected the cells with GFP-expressing P. berghei sporozoites. Two hours after infection, by which time sporozoite invasion is completed, cells were switched into medium containing 250 nM of either rapamycin or Torin1. Control cells were treated with DMSO alone (vehicle control). Infection parameters were quantified by flow cytometry 48 h after sporozoite addition. In both cell lines, the effects of the two mTOR inhibitors were strikingly different. Treatment with rapamycin led to a modest increase in the proportion of infected cells (number of GFP+ cells) (Fig. 1A), or parasite development (Fig. 1B), as indicated by the geometric mean of the GFP signal intensity, which correlates with parasite development (31). Treatment with Torin1, however, eliminated the vast majority of parasites in both cells lines (Fig. 1A; HepG2 P < 0.0001) and blocked the development of those few that remained (Fig. 1B; HepG2 P < 0.0001). These results were confirmed by microscopy, and representative examples of control, rapamycin-treated, and the rare remaining Torin1-treated EEFs are shown in Fig. 1C.
Fig. 1.
Torins are antiplasmodial compounds with nanomolar potency against Plasmodium liver stages and are capable of curing infection. (A–C) Effect of mTOR inhibitors, rapamycin, and Torin1 on P. berghei liver stage infection. HepG2 or Huh7 cells infected with P. berghei-GFP sporozoites on the day after seeding. Torin1, rapamycin, or DMSO (vehicle as control) were added 2 h after infection and remained present until 48 h after sporozoite addition, when samples were processed for flow cytometry (A and B) or microscopy (C). Data from technical triplicates of the DMSO control were averaged, and the mean was set to 100%; all samples were then normalized to this value. Mean ± SD from one representative experiment is shown. (A) The number of GFP+ events detected represents the number of infected cells. (B) The geometric mean of the GFP signal, known to correlate with parasite growth, represents EEF development. (C) Representive confocal images. Huh7 cells were fixed and labeled with anti-PbHSP70 (P. berghei heat shock protein 70) (EEF, green) and DAPI (nuclei, blue). (D) Dose-dependent effects of Torin1 and Torin2 on P. berghei liver stage infection. HepG2 cell infection analyzed by flow cytometry 48 h later, as in B. Each data point represents n = 3 biological triplicates. Four-variable curve fitting was carried out using GraphPad Prism. (E) Effect of a single Torin2 oral dose on parasite liver load. C57BL/6 mice were infected i.v. with 10,000 Pb-GFP sporozoites and given 10 mg/kg Torin2 suspended in sunflower oil or an equivalent dose of sunflower oil alone 2 h after infection. Each point represents a single animal (n = 3 independent experiments); mean ± SD is indicated for each group. (F) Effect of a single Torin2 oral dose on prepatency period. C57BL/6 mice were infected i.v. with 500 Pb-GFP sporozoites and given 10 mg/kg Torin2 suspended in sunflower oil or an equivalent dose of sunflower oil alone 2 h after infection. Animals were monitored daily for appearance of parasitaemia in the blood by flow cytometry. Data are from three independent experiments with a total of 12 animals in each group.
A Torin1 analog with properties more amenable to large-scale synthesis and in vivo use was recently reported (32). We next synthesized and tested this analog, Torin2, for antiplasmodial activity. Dose–response analysis revealed that Torin2 is an ∼100-fold more potent an inhibitor of Plasmodium liver stage growth than Torin1; the calculated cellular EC50 of Torin1 and Torin2 for parasite development was 106 nM 95% confidence interval (CI) 101–107 nM and 1.1 nM (95% CI 0.95–1.33 nM), respectively (Fig. 1D). Furthermore, a single dose of 10 mg/kg Torin2, administered to mice 2 h after infection with 10,000 sporozoites, led to a highly significant reduction in Plasmodium liver load (Fig. 1E, P < 0.0001) 40 h postinfection. In a controlled physiological infection model with infection initiated by 500 P. berghei-GFP sporozites, the same 10 mg/kg dose of Torin2 was curative. Control mice became blood stage positive by flow cytometry detection of GFP from day 4 to 6 postinfection, but none of the Torin2-treated animals ever developed blood parasitaemia by day 11 (Fig. 1F). The flow cytometry data were confirmed by microscopic examination of thin blood smears for all of the Torin2-treated animals.
The discrepancy between the extremely potent inhibition of P. berghei EEFs by the torins and the slight (and opposing) effects of rapamycin led us to wonder whether the antiplasmodial activity of the torins was really mediated by host mTOR. As a first step in addressing this, we tested if the inhibitory effects of the torins would extend to P. falciparum asexual blood stages, as the mature enucleate RBCs in which P. falciparum replicates have no catabolic capacity for mTOR to stimulate. Torin1 (200 nM) and Torin2 (10 nM) were added to synchronized P. falciparum 3D7 ring stages, and parasite replication and reinvasion were assessed by flow cytometry 48 h later. iRBCs were identified based on SYBR green labeling of P. falciparum DNA. Strikingly, both Torin1 and Torin2 blocked parasite development and completely prevented reinvasion, as evidenced by the static parasitemia, compared with the DMSO-treated control (Fig. 2A, P < 0.0001 for both Torin1 and Torin2 vs. control). Using a different cytometry-based assay and the P2G12 clone of P. falciparum 3D7 (33), we performed dose–response assays and determined the Torin2 EC50 for asexual blood stages to be 1.4 nM (95% CI 1.31–1.59 nM) (Fig. 2B). Torin2 is also highly potent against early gametocytes, with a slightly lower EC50 of 6.62 nM (95% CI 4.59–9.54 nM). (Table S1). We next tested whether Torin2 would be capable of antimalarial activity against the blood stage of P. berghei in vivo. C57BL/6 mice infected with P. berghei-GFP iRBCs were treated with a single oral dose of 10 mg/kg Torin2 on day 4 after infection when the parasitaemia had reached 3%. All control mice succumbed to experimental cerebral malaria by day 7, whereas the majority of the Torin2-treated group did not (Fig. 2C), a highly significant difference in survival (P < 0.0001). The single Torin2 dose also led to a highly significant blunting of parasitaemia (Fig. 2D, P < 0.0001). Overall, our data demonstrate that torins are similarly effective and potent against both blood and liver stage Plasmodium parasites in vitro and in vivo. Thus, the mediator of the antimalarial activity must be present in both of these host–parasite settings.
Fig. 2.
Torins potently inhibit Plasmodium blood stage infection. (A) Effect of Torin1 and Torin2 in P. falciparum blood stage in vitro cultures. Synchronized P. falciparum 3D7 ring stage parasites were treated with 200 nM Torin1, 10 nM Torin2, or DMSO (vehicle control); reinvasion was analyzed after SybrGreen labeling of parasite DNA 48 h later. Data represent mean ± SD of three independent experiments. Starting parasitaemia are represented by dotted line. (B) Dose-dependent effect of Torin2 on P. falciparum blood growth. Data represent mean ± SD of three independent experiments. (C and D) Effect of a single 10-mg/kg oral dose of Torin2 administered on day 4 after infection of C57BL/6 mice with 1 × 106 Pb-GFP–parasitized RBC on survival (C) and parasitemia (D). Animals were monitored daily for malaria-associated pathology (experimental cerebral malaria symptoms), and parasitaemia was analyzed by flow cytometry. (C) Cumulative survival curve for three independent experiments; n = 10 mice for control and Torin2-treated groups. (D) Parasitaemia; each point represents the mean ± SD from three independent experiments.
Inhibition of Host mTOR Signaling by Other Means Does Not Phenocopy the Antiplasmodial Effects of Torins.
The ability of torins to potently target both liver and blood stages of Plasmodium infection suggested a direct effect on the parasite and not on host mTOR. We sought to further test this hypothesis using chemical and genetic means to reduce host mTOR activity in hepatoma cells.
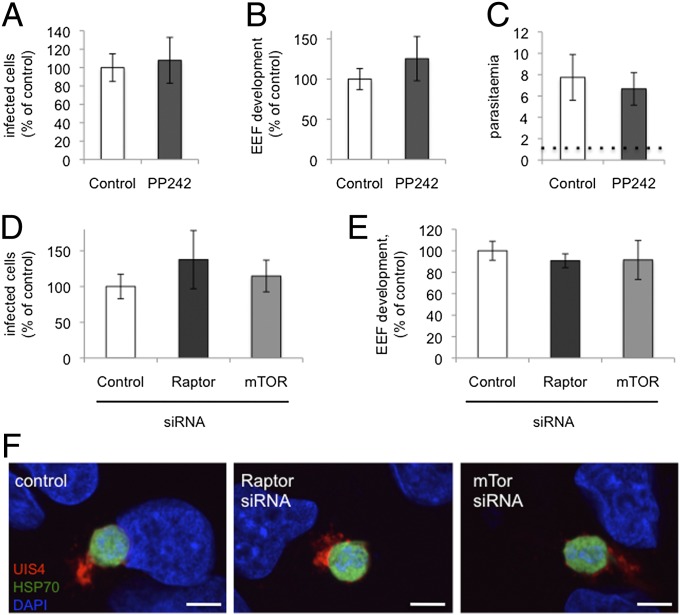
First, we evaluated the ability of PP242, another highly specific ATP-competitive mTOR inhibitor that is structurally unrelated to the torins (34), to inhibit Plasmodium liver stages. Treatment with 2.5 μM PP242 [concentration showing complete inhibition of mTORC1 and mTORC2 signaling in cellular assays (34)] did not reduce either infected hepatocyte number or Plasmodium liver stage growth (Fig. 3 A and B). Likewise, addition of PP242 to P. falciparum-synchronized 3D7 ring stages had no impact on blood stage development and reinvasion (Fig. 3C).
Fig. 3.
mTOR inhibition by other means does not phenocopy the antiplasmodial effect of torins. (A and B) Effect of PP242 on inhibition of liver stage infection. Infected HepG2 cells were treated with 2.5 uM PP242 2 h after spozoroite addition, and the infection was analyzed by flow cytometry 48 h later, as in Fig. 1. Data represent mean ± SD three independent experiments. (C) Effect of PP242 on blood stage infection. Synchronized P. falciparum 3D7 rings were treated with 2.5 μM PP242 or DMSO (identical control data set as in Fig. 2A), and reinvasion was analyzed after SybrGreen labeling of parasite DNA 48 h later. Data represent mean ± SD of three independent experiments. (D–F) Effect of siRNA-mediated knockdown of raptor or mTOR on liver stage infection. Validated siRNA pools were reverse-transfected into HepG2 cells, which were infected with sporozoites 2 d later and analyzed by flow cytometry as described above or with immunofluorescence analysis (IFA) 24 h after sporozoite addition. (F) Representative images with PbHSP7 (green), UIS4 (red), and DAPI (blue).
We next turned to siRNA knockdown of mTORC1 components in hepatoma cells. siRNA oligonucleotide pools targeting human mTor or raptor transcripts were introduced by reverse transfection and the knockdown cells infected with GFP-expressing P. berghei sporozoites 48 h later. siRNA-mediated knockdown of neither mTOR nor raptor could phenocopy the dramatic reduction of infected cells or parasite development observed with Torin2 treatment (Fig. 3 D and E). EEF morphology 24 h after infection was also comparable across the control, mTOR, and raptor knockdown cells (Fig. 3F). Taken together, our data strongly suggest that the target mediating the antiplasmodial effects of torins is not the host cell mTOR kinase, but rather is parasite-encoded.
Torin2 Is Not Permissive to Generation of Drug-Resistant Parasite Lines in Vitro.
To identify the target of Torin2, we attempted to generate drug-resistant mutants in P. falciparum blood stage parasites. We first determined that the multidrug-resistant Dd2 strain of P. falciparum retained Torin2 sensitivity, with an EC50 of 0.7 nM (95% CI 0.48–1.02 nM) in asexual blood stage growth assays (35). We then performed resistance selection with clonal Dd2 ring stage parasites, exposing them to 10× EC50 (7 nM) Torin2 for 8 d. When parasites failed to recrudesce 60 d after this treatment, we varied the length of selection to optimize the mutation-selection window. We achieved parasite recrudescence in 4 of 32 independent selection attempts and only with exposure times of 48 h or less (summarized in Table 1). Remarkably, the four selected strains all failed to display any resistance phenotype by SYBR dose–response or decrease in time to recrudescence in subsequent repetitions of the selection protocol (Table 1). We, and others, have used these selection methodologies to raise resistance to antimalarials that act on a variety of targets within the parasite’s cytosol and mitochondria (36, 37). In contrast, Torin2 pressure has thus far failed to induce a stable resistance phenotype after varied, repeated attempts, suggesting that additional unbiased approaches will be necessary to elucidate the drug target(s).
Table 1.
Torin2 is refractory to repeated attempts at resistance selection in vitro
| Selection strategy | Recrudescence of parasites | Days to recrudescence (first/second/third round)* | Fold EC50 (nM)** |
| 10× EC50, 192 h | 0/8 | NA | NA |
| 10x EC50, 144 h | 0/4 | NA | NA |
| 20x EC50, 48 h pulsed three times | 0/4 | NA | NA |
| 20x EC50, 48 h | 1/7 | 26/24/30 | 1.04 |
| 20x EC50, 36 h | 1/3 | 26/23/30 | 0.93 |
| 20x EC50, 24 h | 2/4 | 22/24/31 | 1.14 |
| 20x EC50, 12 h | 0/2 | NA | NA |
Asexual Dd2 parasites were subjected to varied step-wise intermittent selection protocol with10x EC50 Torin2.
Number of days for each independent selection to return to 1% parasitemia. The time to recrudescence for subsequent rounds of selection is expressed separately for the first/second/third round of selections and expressed as the average of the number of days when multiple independent selections were performed with identical protocols.
Parental Dd2 strain EC50 is 0.7 nM (0.48,1.02). Values expressed as fold change over Dd2 EC50.
Plasmodium Growth Is Inhibited by Torin2 Throughout Liver Stage Development.
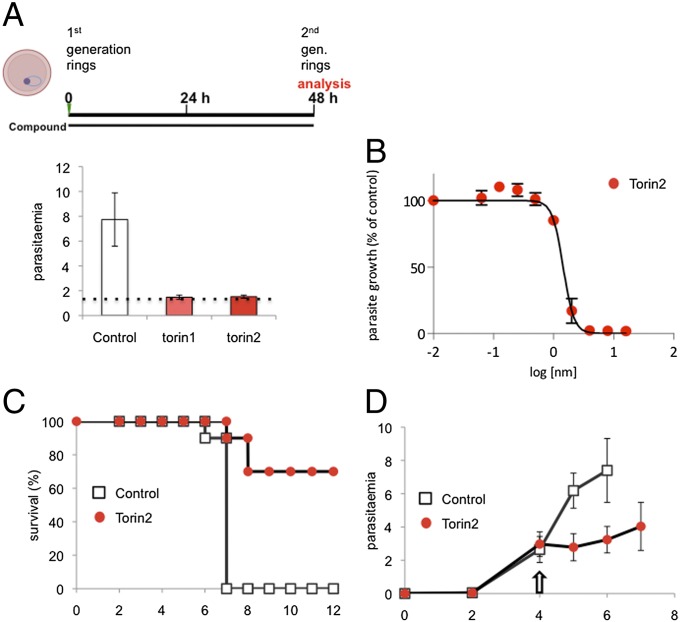
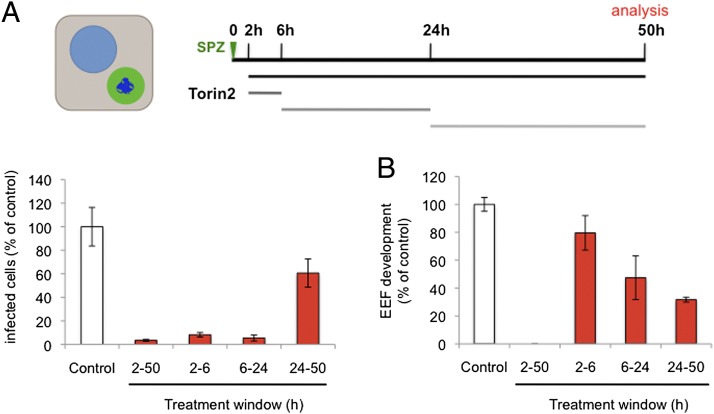
With the evidence leaning toward torins acting directly on Plasmodium spp., we sought to gain insight into the killing mechanism by asking when the target(s) of Torin2 antiplasmodial activity are present in liver stages. We first varied compound treatment windows to target different stages of parasite invasion and development during liver stage infection. Initially, we checked whether Torin2 treatment of HepG2 cells 2 h before sporozoite addition could impact either sporozoite invasion or EEF development. Torin2 pretreatment did not alter either parasite numbers or development, as assayed 24 h after infection (Fig. S1A). We next tested if Torin2 treatment concomitant with sporozoite addition would affect parasite invasion and again found no effect; comparable amounts of cells were infected (GFP+) after 2 h, demonstrating that sporozoite invasion occurs normally in the presence of Torin2 (Fig. S1B). Next, sporozoites were allowed to complete invasion of HepG2 cells, and then 10 nM Torin2 was added for varying time periods (as schematized in Fig. 4A) corresponding roughly to “early PVM remodeling” (2 h), trophozoite (6 h), and schizont (24 h) stages of intrahepatocyte development. Infection was analyzed 50 h after sporozoite addition by flow cytometry, and none of the Torin2 treatment periods was found to significantly increase HepG2 cell death (Fig. S1C). As previously shown, continuous incubation of infected cells with 10 nM Torin2 after sporozoite invasion results in near-complete parasite elimination (Fig. 4A). Remarkably, a mere 4-h incubation with 10 nM Torin2 from 2 to 6 h after infection was also capable of eliminating more than 90% of all parasites (Fig. 4A); Torin2 treatment from 6 to 24 h postinfection was similarly effective (Fig. 4A). The very few developing EEFs under the 2- to 6- or 6- to 24-h conditions showed slightly reduced development compared with the control (Fig. 4B). Interestingly, when Torin2 treatment was initiated only after the start of schizogony (24–50 h), fewer parasites were eliminated, with parasite numbers about 60% of the control (Fig. 4B). However, in this treatment group, EEF development showed the strongest inhibition—to 30% of control levels (Fig. 4B); on an individual level, these parasites show aberrant development and fail to form merozoites (Fig. S1D). Our data demonstrate that Torin2 is a potent inhibitor of all phases of Plasmodium EEF development through late schizogony. Torin2-treated parasite elimination, however, occurred efficiently only when EEFs were exposed to Torin2 before the onset of schizogony.
Fig. 4.
Plasmodium growth is inhibited by Torin2 throughout liver stage development. (A and B) Effects of Torin2 throughout P. berghei liver stage development in vitro. Infected HepG2 cells were treated with 10 nM Torin2 or DMSO (vehicle control) as indicated in the schematic illustration, and the infection analyzed by flow cytometry 50 h after sporozoite addition, as in Fig. 1. Data represent mean ± SD of quadruplicates from one representative experiment with all conditions processed in parallel.
Torin2 Treatment Leaves the PVM Structurally Intact, but Lacking PVM-Resident Proteins.
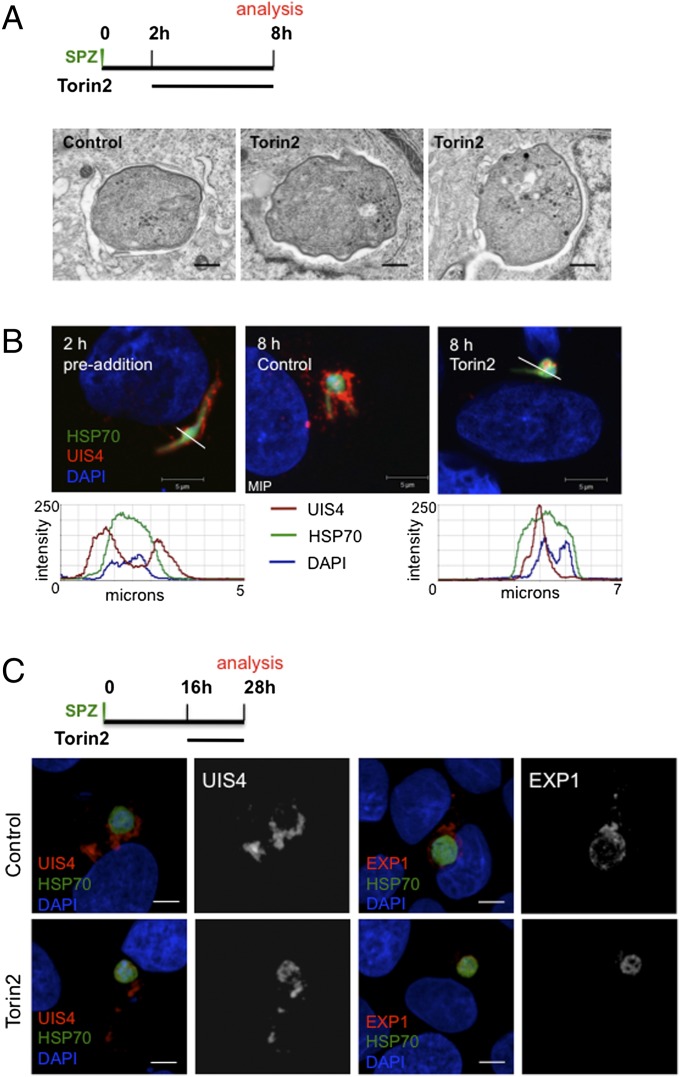
A number of gene-knockout parasite lines that successfully invade hepatocytes exist, but fail during PVM establishment or remodeling and are rapidly eliminated (11, 12, 17–20, 38). Given this phenotypic parallel to the effects of Torin treatment, which also does not affect the invasion process itself, but leads to parasite elimination, we wondered if Torin2 treatment might be altering the parasite PVM, or alternatively, if infected cells were selectively rendered nonviable by a brief Torin2 treatment. To this end, we used a live imaging setup to address two questions: (i) Are infected cells viable after Torin2 treatment? and (ii) Is the PVM maintained intact after Torin2 treatment? To that end, we infected HepG2 cells with P. berghei-GFP sporozoites and used the vital dye tetramethylrhodamine, ethyl ester (TMRE), which labels mitochondria with an intact membrane potential, to assess host-cell mitochondrial activity and unambiguously identify intracellular EEFs. Additionally, we used the vital DNA dye Hoechst 33258, which freely labels host-cell DNA or free sporozoites, which lack a PVM, but is unable to label the DNA of developing EEFs until maturation-induced PVM changes occur late in liver stage development (39). Torin2 was added 2 h after infection, and the cells were imaged 6 h later, after TMRE and Hoechst 33258 labeling. Based on our earlier data (Fig. 4B), the vast majority of Torin2-treated EEFs are ultimately nonviable after such a treatment. In both control and Torin2-treated cells, the 8-h EEFs developed in host cells with normal nuclear morphology and active mitochondria (Fig. S2A). Importantly, the EEF nuclei are not labeled by Hoechst 33258 in either drug-treated or control cells (Fig. S2A), although the nuclei of extracellular sporozoites that have failed to invade clearly are an indication that Torin2-treated EEFs reside in an intact PVM. To confirm this interpretation, we used transmission electron microscopy (TEM) to investigate PVM integrity in EEFs that were exposed to Torin2 from 2 to 8 h after infection. As expected, the 8-h control EEFs were uniformly PVM-positive (Fig. 5A). Torin2-treated parasites were also clearly surrounded by a PVM in all cases (Fig. 5A) and tended to have more parasite PM waviness and intracellular complexity (e.g., membranous whorls) than the control parasites observed.
Fig. 5.
Torin2 treatment leaves the PVM structurally intact but lacking PVM-resident proteins. (A and B) Effect of Torin2 treatment on the PVM and UIS4 localization in young liver stage trophozoites. Schematic illustrates experimental setup. (A) Representative images of TEM of 8-h EEFs after Torin2 or DMSO (control) treatment. (B) IFA at 2 h (control, single confocal slice) and at 8 h in control [maximum intensity projection (MIP)] and Torin2-treated cells [single confocal slice (HSP70, green; DAPI, blue; UIS4, red)]. Fluorescence intensity graphs of the trajectory indicated by the white lines in the preaddition and Torin2-treated images show the relative spatial intensity peaks of the three fluorophores. (C) Effect of Torin2 treatment on UIS4 and EXP1 during early liver stage schizogony. Schematic illustrates experimental setup. MIPs of the entire EEF are shown for A–C.
Given that the PVM appeared intact in Torin2-treated infected cells, we next checked whether Plasmodium proteins known to localize to the PVM were altered after exposure to Torin2. UIS4, which is already transcribed in mature salivary gland sporozoites (40), is localized to the PVM throughout development inside the hepatocyte (11). Using UIS4-specific antiserum, we confirmed that 2 h after infection UIS4 was already localized to the PVM; the peak UIS4 signal is clearly outside of the parasite soma delineated by P. berghei heat shock protein 70 (HSP70) antibody staining (Fig. 5B). Torin2 treatment initiated 2 h after infection is thus acting upon developing parasites already enclosed in UIS4-positive PVMs. Surprisingly, we found that 6 h of Torin2 treatment completely abolished the PVM localization of UIS4. In stark contrast to both the pretreated 2-h parasites and the 8-h control parasites, UIS4 is found exclusively within the parasite soma in Torin2-treated parasites (Fig. 5B).
Torins are potent antiplasmodials throughout liver stage development, so we next investigated whether or not Torin2 treatment could induce mislocalization of PVM-resident proteins in more mature parasites. Infected HepG2 cells were treated with 10 nM Torin2 during a 12-h window starting 16 h after infection, and localization of both UIS4 and EXP1, another known PVM-resident protein [also called Hep17 (41)], was assessed in 28 h EEFs. As expected, UIS4 and EXP1 were localized almost completely outside of the parasite soma in control 28-h EEFs (Fig. 5C). Again, Torin2 treatment led to a dramatic mislocalization of UIS4, with signal largely found in the parasite soma in 28-h EEFs (Fig. 5C), although some residual UIS4 could also be observed in the host-cell cytoplasm. EXP1 localization was also completely perturbed by Torin2 treatment, with a PVM (control) to soma (Torin2-treated) shift paralleling that of UIS4 (Fig. 5C). Additionally, Torin1 treatment also induces mislocalization of UIS4 to the parasite soma (Fig. S2B). These data imply that torins act against Plasmodium by altering the localization of PVM-resident proteins.
EEF Elimination and PVM-Resident Protein Mislocalization Are Not General Aspects of Liver Stage Inhibition by Antimalarials, Indicating a Distinct Mechanism of Action for Torins.
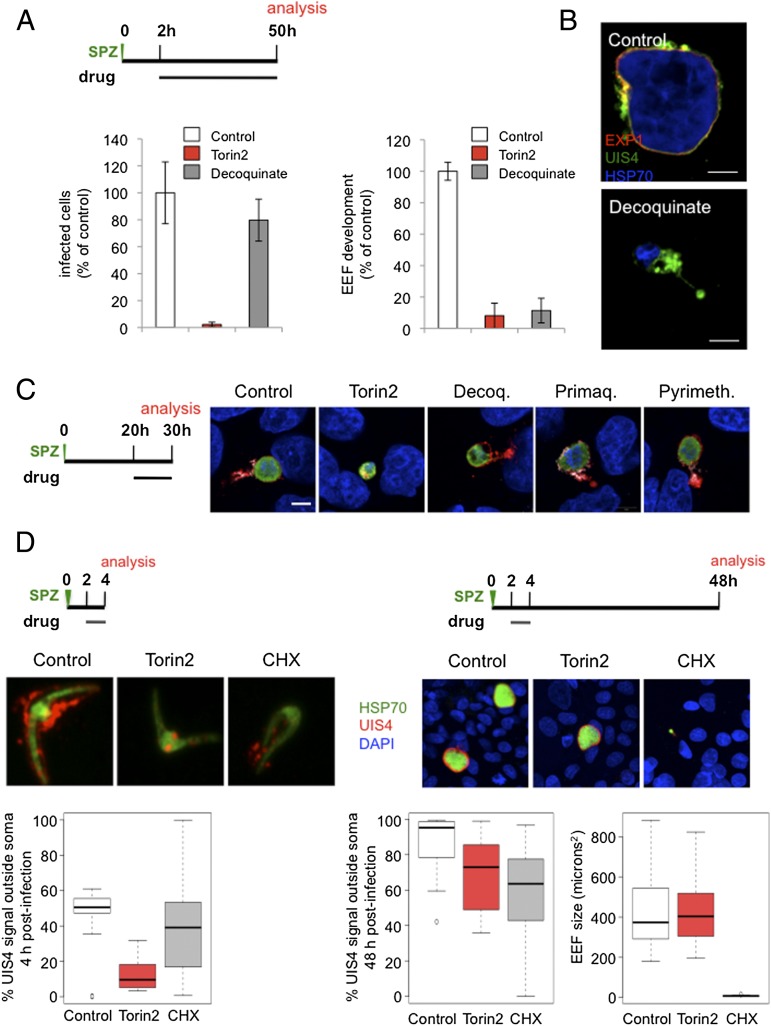
Although a comprehensive study of the activity of currently used antimalarials against rodent liver stage parasites has been carried out (42), we lack such information on a phenotypic level, leading us to wonder whether parasite clearance is the inevitable outcome for EEFs that are rendered nonviable during the first hours of intrahepatocyte development. To address this, we first confirmed the elimination of Torin2-treated parasites in HepG2 cells by microscopy; continuous exposure to Torin2 initiated after sporozoite invasion results in a complete absence of developing EEFs 48 h later. We then compared Torin2 side by side with the recently identified antiplasmodial decoquinate, which has the same mechanism of action (MoA) as atovaquone (43, 44), the most potent antimalarial in clinical use effective in the liver stage (42). We chose to focus on decoquinate as its potency is more similar to that of Torin2 than that of atovaquone. Treatment of infected HepG2 cells 2 h after sporozoite invasion with either 10 nM Torin2 or 26 nM decoquinate (10× EC50) resulted in complete Plasmodium inhibition. Torin2 eliminated EEFs, as expected (Fig. 6A, P < 0.0001), whereas decoquinate led to a modest reduction in the number of EEFs present 50 h after infection (Fig. 6A, P < 0.05). Furthermore, a 6-h treatment with decoquinate from 2 to 8 h after infection was phenotypically equivalent to the 2- to 50-h treatment (Fig. S3). Despite the persistence of decoquinate-treated EEFs, they appear to be arrested very early in development (Fig. 6A, P < 0.0001), a phenotype we have confirmed by microscopy in primary mouse hepatocytes (Fig. 6B). EEF elimination is thus not a default outcome of parasite nonviability early in development, but rather reflects a specific aspect of the torin-treated EEF phenotype. This provides an intriguing parallel to the phenotypes described for those mutant parasites that successfully invade hepatocytes, but fail during PVM establishment or remodeling and are rapidly eliminated (17–20).
Fig. 6.
EEF elimination and PVM-resident protein-trafficking defects are torin-specific phenotypes, not general aspects of liver stage inhibition by antimalarials. (A and B) Effects of decoquinate and Torin2 on EEF numbers and development. (A) Schematic of treatment and analysis. Infected HepG2 cells analyzed by flow cytometry at 50 h after sporozoite addition. Mean of n = 3 biological experiments, each condition in triplicate. (B) Mouse primary hepatocytes infected ex vivo with EXP1 (red), UIS4 (green), and HSP70 (blue) labeling. (C) Effect of current antimalarial classes active against the liver stage at the onset of schizogony. Schematic illustrates experimental setup; representative confocal images shown with UIS4 (red), EXP1 (white), PbHSP70 (green), and DAPI (blue) labeling. (D) Effects of 2 h Torin2 or cycloheximide treatment on UIS4 localization and EEF development. Schematic illustrates experimental setup. Representative confocal images of control (DMSO), Torin2-, and cycloheximide-treated EEFs labeled with anti-UIS4 (red), anti-PbHSP70 (green), and DAPI (blue) 4 and 48 h postinfection. Quantification of the proportion of UIS4+ pixels not overlapping PbHSP70 in 20 EEFs from each condition at 4 and 48 h postinfection; quantification of EEF area 48 h postinfection.
We next checked if the mislocalization of PVM-resident proteins induced by the torins could be a previously unnoted feature of pharmacological inhibition/killing of Plasmodium liver stages, also provoked by known antimalarials active against the liver stages. We tested this hypothesis by evaluating UIS4 and EXP1 localization in infected cells treated with representative members of the classes, in terms of both chemical structure and MoA, of currently known antimalarials (42). Primaquine, pyrimethamine, and decoquinate were individually added to HepG2 cells 20 h after infection at 10× EC50 concentrations. Ten hours later, coverslips were fixed and processed for immunofluorescence. Compared with the control and Torin2 conditions (Fig. 6C), decoquinate-treated parasites retained robust anti-UIS4 labeling of the PVM but, notably, EXP1 staining was nearly abolished (Fig. 6C), a feature we also noted in mouse primary hepatocytes (Fig. 6B). In the primaquine- and pyrimethamine-treated cells, both PVM-resident proteins were properly localized (Fig. 6C). Additionally, PVM-resident proteins remain properly localized in cells treated with the known liver stage inhibitors genistein (45), lopinavir (46), and cyclosporin A (47), as well as with the PI3K inhibitor LY294002 and rapamycin (Fig. S4). Furthermore, Torin2 treatment from 2 to 4 h postinfection is sufficient to mislocalize UIS4, but is reversible, in terms of both parasite growth and UIS4 localization at 48 h postinfection (Fig. 6D). Additionally, cycloheximide treatment (10 µg/mL), which blocks translation and is a potent antiplasmodial compound (48), during the same 2 h window is uniformly lethal for the developing EEFs, which do not grow after this time period; this lethality is not accompanied by a penetrant defect in UIS4 localization as assayed at either 4 or 50 h after infection (Fig. 6D). Thus, PVM-protein mislocalization is definitively not a consequence of parasite death.
As such, we conclude that both parasite elimination and altered localization of PVM resident proteins are specific phenotypes of torin-mediated Plasmodium liver stage inhibition, which strongly indicates that torins have a distinct MoA from currently used antimalarials.
Plasmodium Liver Stage Parasites Require Replenishment of PVM-Resident Proteins for Viability.
As the alteration of PVM-resident protein localization was not a general consequence of antimalarial activity against EEFs, we sought to determine the mechanism by which Torin2 provokes the mislocalization of these proteins.
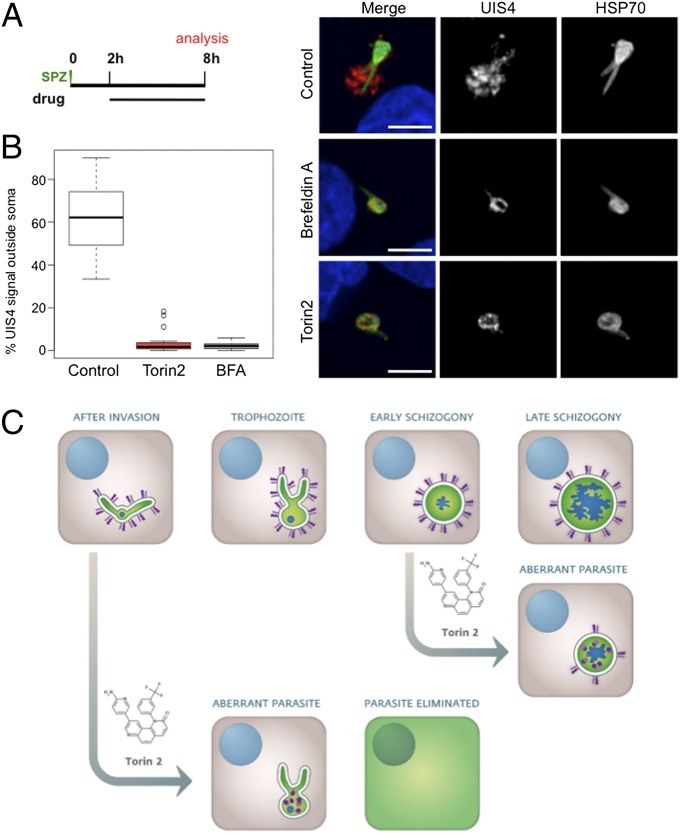
Trafficking of blood stage P. falciparum proteins to specific organelles, the vacuolar space, PVM, and beyond into the iRBC itself has been a subject of intense study (49). Brefeldin A (BFA), an inhibitor of eukaryotic ADP-ribosylation factor (ARF) GTPases and the retrograde Golgi-ER trafficking that they mediate in many species, including Plasmodium (50), blocks export of knob-associated histidine-rich protein (KAHRP), P. falciparum erythrocyte membrane protein 1 (PfEMP1) (51), and EXP1 (52) among other proteins in blood stage parasites. Nothing is known about the trafficking pathways used by the liver stage parasite, so we tested whether UIS4 trafficking to the PVM proceeds through a BFA-sensitive pathway. We added BFA to HepG2 cells 2 h after infection, when UIS4 is already present in the PVM. After BFA (5 µM) treatment for 6 h postsporozoite invasion, UIS4 is found inside the parasite soma as in Torin2-treated parasites (Fig. 7A). Compared with Torin2 treatment, which causes UIS4 to accumulate in discrete intracellular puncta (Fig. 7A) during the same window, BFA causes UIS4 to accumulate in what appears to be a more continuous distribution within the parasite soma. As UIS4 colocalizes with the microneme marker thrombospondin-related anonymous protein (TRAP) in mature salivary gland sporozoites (40) and as TRAP is maintained in puncta throughout the majority of liver stage development (53), we tested whether UIS4 relocalized into TRAP-positive structures upon Torin2 or BFA treatment. The data clearly show that UIS4 and TRAP do not substantially colocalize in control, BFA-, or Torin2-treated cells (Fig. S5A).
Fig. 7.
Plasmodium liver stage parasites require replenishment of PVM-resident proteins for viability. (A and B) Comparative effects of Torin2 and BFA on UIS4 trafficking to the PVM in young liver stage trophozoites. Schematic illustrates experimental setup. (A) Representative confocal images of control (methanol, BFA vehicle), Torin2-, and BFA-treated EEFs labeled with anti-UIS4 (red), anti-PbHSP70 (green), and DAPI (blue). (B) Quantification of the proportion of UIS4+ pixels not overlapping PbHSP70 in 20 EEFs from each condition. DMSO and methanol, the vehicle controls for Torin2 and BFA, respectively, are grouped together as control. (C) Model of Torin2 liver stage MoA.
Most strikingly, BFA treatment, which blocks only protein secretion, phenocopied the complete loss of PVM-localized UIS4 seen in the Torin2-treated parasites (Fig. 7A). We quantified the distribution of UIS4 and found that, on average, 62% of the UIS4 signal is present outside of the parasite soma in 8-h control EEFs, whereas BFA or Torin2 treatment resulted in >95% overlap of UIS4 and HSP70 in the parasite soma (Fig. 7B). We extended these results to P. berghei EEFs developing inside murine primary hepatocytes ex vivo (Fig. S5B); UIS4 and EXP1 trafficking to the PVM is BFA- and Torin2-sensitive in young liver stage schizonts (Fig. S5B), and both PVM-resident proteins are also concomitantly depleted from the PVM.
Taken together, our data support a model (Fig. 7C) in which Torin2 treatment prevents secretion of liver stage PVM-resident proteins, leading to their accumulation inside the EEF soma. This block in secretion causes the PVM to be left devoid of PVM-resident proteins, particularly in trophozoites, which leads to parasite elimination by the host hepatocyte.
Discussion
This study reports that torins, developed as ATP-competitive mammalian mTOR kinase inhibitors, are extremely potent inhibitors of both the liver and blood stages of Plasmodium parasites. Although we first tested Torin1 (together with rapamycin) to investigate the role of host hepatocyte mTOR signaling in liver stage Plasmodium infection, our data as a whole provide extremely strong support for a parasite-encoded target mediating the antiplasmodial activity of the torins. We present three lines of evidence that suggest that host-cell mTOR is not the mediator of the antiplasmodial activity of the torins. First, unrelated small-molecule mTOR inhibitors (rapamycin and PP242) or siRNA-mediated reduction of mTOR signaling both fail to inhibit Plasmodium liver stage infection. Strong evidence that host mTOR is uninvolved comes from the nonequivalence of PP242 and torins. Both cause complete inhibition of mTORC1 and mTORC2 activity (30, 32, 34). The chemical structures of the two compounds are not at all similar although, as we show here, neither are their effects on liver or blood stage Plasmodium infection. Second, torins inhibit both liver and blood stage parasites, with Torin2 maintaining a near identical EC50 for the two stages and for different Plasmodium species. Although it is easy to envisage mechanisms by which reduction of host hepatocyte mTOR signaling could alter liver stage Plasmodium development, this is not the case for the iRBC. The mature RBC lacks the capacity for both protein and lipid synthesis, as well as gene expression, and although mTOR has been found in the RBC “hidden proteome,” no evidence of functional protein exists (54). Third, torins are able to alter the localization of Plasmodium proteins during liver stage infection, an effect very unlikely to be mediated by a host protein. As a whole, our data point unequivocally to a Plasmodium-encoded molecule or molecules, conserved across the genus and expressed in all mammalian stages, as the target of the antiplasmodial activity of Torin1 and Torin2.
Sequence homology queries of current Plasmodium genome assemblies show that no Plasmodium orthologs of mTOR exist, and indeed the PI3K-like protein kinase family, of which mTOR is a member, is not found (55, 56). The P. berghei proteins showing the highest sequence similarity to human mTOR are the single predicted phosphatidylinositol-3-kinase (PI3K) (PBANKA_111490) and the two predicted phosphatidylinositol-4 kinases (PI4Ks) (PBANKA_110940 and PBANKA_072200), which show conservation primarily in the kinase catalytic domain. Although little is known about the activity of the two predicted Plasmodium PI4Ks, PbPI3K is an essential gene (57), and treatment with the known PI3K inhibitors wortmannin and LY294002 reduced PI3P production by PfPI3K in vitro and inhibited blood stage parasite growth (58). LY294002 does not phenocopy Torin2 inhibition of P. berghei liver stages, however, and the inhibition of P. falciparum replication by wortmannin and LY294002 (58) is much more modest than what we observed with either Torin1 or Torin2.
Our data clearly show that Torin2 is a potent antimalarial with in vivo activity against both liver and blood stages, capable of curing liver stage infection with a single, well-tolerated oral dose. Still, given its demonstrated potent inhibition of human mTOR, we cannot, and do not, see Torin2 itself as a lead compound; further medicinal chemistry elaboration will be necessary to realize a torin analog as a suitable antimalarial lead. Nevertheless, torins represent a new antimalarial chemotype that certainly warrants further medicinal chemistry investigation due to the following highly desirable properties: (i) versatility: Torin2 targets both liver and blood stages, and is active against immature gametocytes; (ii) speed of action: torins render P. falciparum parasites nonviable in a single blood stage cycle and short treatment windows are sufficient to kill liver stage parasites; (iii) distinct MoA from currently utilized antimalarials: none of the compounds in current clinical use which are active against Plasmodium liver stages provoke PVM trafficking defects; and (iv) Torin2 retains potency against the multidrug resistant Dd2 strain of P. falciparum, and is not amenable to in vitro resistance generation, in contrast to most antimalarials in current clinical use and leading candidates (36, 37, 59–63). Additionally, the mediator(s) of the antiplasmodial activity of the torins is clearly druggable in vivo during both mammalian stages.
Although the value of torin analogs for human antimalarial use will rely on future medicinal chemistry elaboration, the utility of torins for probing PVM function and protein-trafficking pathways in the Plasmodium EEF is immediate. Plasmodium parasites contain many unusual cellular compartments and have evolved strategies to direct proteins not only to these but also to destinations outside of the parasite soma—from the PV to the host-cell membrane itself. The transport pathways that the blood stage parasite uses to direct proteins to these diverse locations has been a subject of intense study; whereas specific trafficking pathways and molecular determinants ultimately mediate protein movement to discrete locations—i.e., the vacuolar space, the Maurer’s clefts in the iRBC cytoplasm, the digestive vacuole, or the PVM—most secreted proteins seem to initially follow a common, canonical BFA-sensitive ER-Golgi route in the blood stage parasites (49). As such, it is perhaps not surprising that UIS4 and EXP1 trafficking to the liver stage PVM requires the canonical BFA-sensitive ER-Golgi transport route, as we have demonstrated. Still, it highlights the fact that the parasite does use this, and undoubtedly other core pathways, throughout all mammalian stages. It remains to be determined whether the EEF uses the same parasite-encoded molecules for acquisition of host hepatocyte resources that the blood stage parasite uses in remodelling the “inert” RBC or whether components of the host hepatocyte vesicular trafficking networks are co-opted by the parasite.
Comparing BFA- and Torin2-treated EEFs, we find clear commonalities and subtle differences in phenotype. When either drug is added after sporozoite invasion and intial UIS4-positive PVM establishment, both treatments result in complete loss of UIS4 at the PVM 6 h later, as well as intracellular accumulation of UIS4. BFA blocks ER to Golgi trafficking by inhibiting the P. falciparum orthologue of Arf1 (64). BFA-treated blood stage parasites form a hybrid ER- Golgi compartment (65) that accumulates proteins utilizing the canonical secretory pathway. We interpret the similarity of BFA- and Torin2-treated EEF phenotypes as highly suggestive that Torin2, like BFA, causes a failure in anterograde protein trafficking. The compartment in which UIS4 is retained appears qualitatively different, however, with Torin2 treatment leading to the appearance of small puncta of UIS4 and BFA treatment leading to a more continuous distribution of UIS4 inside the parasite soma. Future characterization of the intracellular location in which PVM-resident proteins accumulate after Torin2 treatment may help shed light on which trafficking step is inhibited by Torin2.
Localization of a membrane protein to a specific compartment can be achieved by either its retention at, or continuous transport to, the compartment. Most intriguingly, our data illustrate that UIS4 and EXP1 must be continuously transported to the PVM throughout at least the first 30 h of liver stage development. Several fascinating questions arise from this: Are UIS4 and EXP1 “lost” to the host cell during EEF development? Are UIS4 and EXP1 subject to retrograde trafficking back into the PV or parasite soma? Are these dynamics generalizable to all liver stage PVM-resident proteins, or do they reflect the specific functions of EXP1 and UIS4? Whether or not torin treatment alters protein trafficking in Plasmodium asexual and sexual blood stages also remains to be established.
Proteins that are known to populate the nascent PVM of the invading P. falciparum merozoite are synthesized during the preceding schizont stage and stored in the apical organelles of the merozoite before release in the invasion process (66). The translocon components HSP101, PTEX150, and EXP2 are examples of this; they are found in the dense granules of merozoites, but will be associated with the PVM throughout the subsequent cycle postinvasion (67). The sporozoite invasion process is assumed to be analogous to that of the merozoite. During the transit of the Plasmodium sporozoite from the bite site to the hepatocytes, UIS4 is stored inside the sporozoite where it colocalizes with TRAP (40), apparently in the micronemes, and is discharged only once the sporozoite is in the process of hepatocyte invasion. We clearly demonstrate that the pool of UIS4 that initially populates the young EEF PVM is gone after 6 h of either Torin2 or BFA treatment. As BFA is not thought to affect retrograde trafficking, it is very unlikely that this indicates a block in UIS4 recycling from the parasite soma to the PVM, suggesting that this pool of UIS4 has been degraded either by the host cell or the parasite itself. However, this is not the case with the translocon components in the iRBC, in which the initial pool of proteins released from the dense granules appears to be stably retained at the PVM throughout blood stage development, with further synthesis and trafficking not required (67).
Although it is clear that the blood stage parasite is actively secreting proteins, such as the stage-specific ETRAMPS (24), to the PVM throughout development, more investigation will be required to determine if some blood PVM proteins show dynamics like those of UIS4, and indeed whether EXP1 itself is similarly dynamically localized to the blood stage PVM. The hypothesis that PVM protein turnover would be related to protein function is attractive, but as the liver stage PVM also must contend with extensive interactions with hepatocyte components (22, 68), it remains possible that the host cell itself dictates the turnover of the liver stage PVM-resident proteins that we have examined.
Regardless of whether host or parasite ultimately drives the turnover of the liver stage PVM-resident proteins that we have studied, their replenishment at the PVM via continued expression and secretion through the early schizont stage, at the least, is crucial for parasite viability. Clearly, the trafficking route to the liver stage PVM and the molecular players that mediate it, as well as the protective role of the PVM, constitute fascinating avenues for future research into Plasmodium biology, as well as antimalarial drug development.
Experimental Procedures
See detailed version in SI Experimental Procedures.
Plasmodium Liver Stage Assays.
GFP-expressing P. berghei sporozoites were added to HepG2 or Huh7 cells cultured in 24-well plates. Infected cells were processed and analyzed by flow cytometry as described in ref. 31.
A total of 10,000 P. berghei-GFP sporozoites were injected i.v. into C57BL6 mice; 2 h later, a 10-mg/kg dose of Torin2 was given orally as a sunflower oil slurry. Control animals received an equal dose of oil. Livers were harvested 44 h after infection, mRNA was extracted, and liver parasite load was determined by quantitative RT-PCR of P. berghei 18s rRNA.
Plasmodium Blood Stage Assays.
P. falciparum strains were cultured in vitro, and parasite proliferation was determined by flow cytometry.
To test the antimalarial properties of Torin2 in vivo, 1 × 106 P. berghei-GFP iRBCs were injected intraperitoneally into C57BL/6 mice, and parasitaemia was monitored by flow cytometry. Torin2 (10 mM) in DMSO was diluted in PBS, and 10 mg/kg was given orally on day 4 postinfection when parasitaemia was above 3%. Control animals received equal doses of DMSO in PBS.
For resistance selection, ∼109 parasites were subject to a stepwise intermittent selection protocol beginning with 10× EC50 for various exposure-time windows. Optimal resistance-selection conditions were obtained that selected against the majority of parasites, but allowed recrudescence of resistant parasites within 60 d. Upon recrudescence, additional selection rounds were conducted to optimally obtain clones uninhibited by Torin2.
Immunofluorescence and Microscopy.
Infected cells were fixed in 4% paraformaldehyde (wt/vol) for 10 min at room temperature, permeabilized, blocked in 2% BSA (wt/vol), and incubated with 1° antibodies. After washing, appropriate 2° antibodies were added, and coverslips were mounted in Fluoromount. All images were acquired on Zeiss confocal microscopes.
Supplementary Material
Acknowledgments
We thank Ana Parreira for mosquito production and infection; Fernanda Baptista for laboratory support; and Liliana Mancio, Vanessa Luis, and Ghislain Cabal for advice and reagents. Additionally, we are grateful to Volker Heussler, Stefan Kappe, and Miguel Seabra for providing antisera, and to David Sabatini for providing Torin1. This work was supported by Fundação para a Ciência e Tecnologia (FCT, Portugal) Grants PTDC/SAU-GMG/100313/2008 and EXCL/IMI-MIC/0056/2012, and European Research Council funding (to M.M.M.). K.K.H. was supported by funds from the European Community’s Seventh Framework Programme (FP7/2007-2013) Marie Curie IntraEuropean Fellowship Grant PIEF-GA-2008-221854 and FCT Grant SFRH/BPD/40989/2007.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1306097110/-/DCSupplemental.
References
- 1.Murray CJL, et al. Global malaria mortality between 1980 and 2010: A systematic analysis. Lancet. 2012;379(9814):413–431. doi: 10.1016/S0140-6736(12)60034-8. [DOI] [PubMed] [Google Scholar]
- 2.O’Brien C, Henrich PP, Passi N, Fidock DA. Recent clinical and molecular insights into emerging artemisinin resistance in Plasmodium falciparum. Curr Opin Infect Dis. 2011;24(6):570–577. doi: 10.1097/QCO.0b013e32834cd3ed. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prudêncio M, Rodriguez A, Mota MM. The silent path to thousands of merozoites: The Plasmodium liver stage. Nat Rev Microbiol. 2006;4(11):849–856. doi: 10.1038/nrmicro1529. [DOI] [PubMed] [Google Scholar]
- 4.Moradin N, Descoteaux A. Leishmania promastigotes: Building a safe niche within macrophages. Front Cell Infect Microbiol. 2012;2:121. doi: 10.3389/fcimb.2012.00121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kumar Y, Valdivia RH. Leading a sheltered life: Intracellular pathogens and maintenance of vacuolar compartments. Cell Host Microbe. 2009;5(6):593–601. doi: 10.1016/j.chom.2009.05.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lingelbach K, Joiner KA. The parasitophorous vacuole membrane surrounding Plasmodium and Toxoplasma: An unusual compartment in infected cells. J Cell Sci. 1998;111(Pt 11):1467–1475. doi: 10.1242/jcs.111.11.1467. [DOI] [PubMed] [Google Scholar]
- 7.Wang L, Boyer JL. The maintenance and generation of membrane polarity in hepatocytes. Hepatology. 2004;39(4):892–899. doi: 10.1002/hep.20039. [DOI] [PubMed] [Google Scholar]
- 8.Bast A, et al. Defense mechanisms of hepatocytes against Burkholderia pseudomallei. Front Microbiol. 2011;2:277. doi: 10.3389/fmicb.2011.00277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.de Koning-Ward TF, et al. A newly discovered protein export machine in malaria parasites. Nature. 2009;459(7249):945–949. doi: 10.1038/nature08104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Maier AG, et al. Exported proteins required for virulence and rigidity of Plasmodium falciparum-infected human erythrocytes. Cell. 2008;134(1):48–61. doi: 10.1016/j.cell.2008.04.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mueller AK, et al. Plasmodium liver stage developmental arrest by depletion of a protein at the parasite-host interface. Proc Natl Acad Sci USA. 2005;102(8):3022–3027. doi: 10.1073/pnas.0408442102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mueller AK, Labaied M, Kappe SH, Matuschewski K. Genetically modified Plasmodium parasites as a protective experimental malaria vaccine. Nature. 2005;433(7022):164–167. doi: 10.1038/nature03188. [DOI] [PubMed] [Google Scholar]
- 13.Maier AG, Cooke BM, Cowman AF, Tilley L. Malaria parasite proteins that remodel the host erythrocyte. Nat Rev Microbiol. 2009;7(5):341–354. doi: 10.1038/nrmicro2110. [DOI] [PubMed] [Google Scholar]
- 14.Singh AP, et al. Plasmodium circumsporozoite protein promotes the development of the liver stages of the parasite. Cell. 2007;131(3):492–504. doi: 10.1016/j.cell.2007.09.013. [DOI] [PubMed] [Google Scholar]
- 15.Vaughan AM, et al. Complete Plasmodium falciparum liver-stage development in liver-chimeric mice. J Clin Invest. 2012;122(10):3618–3628. doi: 10.1172/JCI62684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hunn JP, Feng CG, Sher A, Howard JC. The immunity-related GTPases in mammals: A fast-evolving cell-autonomous resistance system against intracellular pathogens. Mamm Genome. 2011;22(1–2):43–54. doi: 10.1007/s00335-010-9293-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Labaied M, et al. Plasmodium yoelii sporozoites with simultaneous deletion of P52 and P36 are completely attenuated and confer sterile immunity against infection. Infect Immun. 2007;75(8):3758–3768. doi: 10.1128/IAI.00225-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.van Dijk MR, et al. Genetically attenuated, P36p-deficient malarial sporozoites induce protective immunity and apoptosis of infected liver cells. Proc Natl Acad Sci USA. 2005;102(34):12194–12199. doi: 10.1073/pnas.0500925102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Silvie O, Goetz K, Matuschewski K. A sporozoite asparagine-rich protein controls initiation of Plasmodium liver stage development. PLoS Pathog. 2008;4(6):e1000086. doi: 10.1371/journal.ppat.1000086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aly AS, et al. Targeted deletion of SAP1 abolishes the expression of infectivity factors necessary for successful malaria parasite liver infection. Mol Microbiol. 2008;69(1):152–163. doi: 10.1111/j.1365-2958.2008.06271.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Desai SA, Rosenberg RL. Pore size of the malaria parasite’s nutrient channel. Proc Natl Acad Sci USA. 1997;94(5):2045–2049. doi: 10.1073/pnas.94.5.2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bano N, Romano JD, Jayabalasingham B, Coppens I. Cellular interactions of Plasmodium liver stage with its host mammalian cell. Int J Parasitol. 2007;37(12):1329–1341. doi: 10.1016/j.ijpara.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 23.MacKellar DC, Vaughan AM, Aly AS, DeLeon S, Kappe SH. A systematic analysis of the early transcribed membrane protein family throughout the life cycle of Plasmodium yoelii. Cell Microbiol. 2011;13(11):1755–1767. doi: 10.1111/j.1462-5822.2011.01656.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Spielmann T, Fergusen DJ, Beck HP. etramps, a new Plasmodium falciparum gene family coding for developmentally regulated and highly charged membrane proteins located at the parasite-host cell interface. Mol Biol Cell. 2003;14(4):1529–1544. doi: 10.1091/mbc.E02-04-0240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mikolajczak SA, Jacobs-Lorena V, MacKellar DC, Camargo N, Kappe SH. L-FABP is a critical host factor for successful malaria liver stage development. Int J Parasitol. 2007;37(5):483–489. doi: 10.1016/j.ijpara.2007.01.002. [DOI] [PubMed] [Google Scholar]
- 26.Albuquerque SS, et al. Host cell transcriptional profiling during malaria liver stage infection reveals a coordinated and sequential set of biological events. BMC Genomics. 2009;10:270. doi: 10.1186/1471-2164-10-270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chattopadhyay R, et al. Early transcriptional responses of HepG2-A16 liver cells to infection by Plasmodium falciparum sporozoites. J Biol Chem. 2011;286(30):26396–26405. doi: 10.1074/jbc.M111.240879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laplante M, Sabatini DM. mTOR signaling in growth control and disease. Cell. 2012;149(2):274–293. doi: 10.1016/j.cell.2012.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guertin DA, Sabatini DM. The pharmacology of mTOR inhibition. Sci Signal. 2009;2(67):pe24. doi: 10.1126/scisignal.267pe24. [DOI] [PubMed] [Google Scholar]
- 30.Thoreen CC, et al. An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J Biol Chem. 2009;284(12):8023–8032. doi: 10.1074/jbc.M900301200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Prudêncio M, Rodrigues CD, Ataíde R, Mota MM. Dissecting in vitro host cell infection by Plasmodium sporozoites using flow cytometry. Cell Microbiol. 2008;10(1):218–224. doi: 10.1111/j.1462-5822.2007.01032.x. [DOI] [PubMed] [Google Scholar]
- 32.Liu Q, et al. Discovery of 9-(6-aminopyridin-3-yl)-1-(3-(trifluoromethyl)phenyl)benzo[h][1,6]naphthyridin-2(1H)-one (Torin2) as a potent, selective, and orally available mammalian target of rapamycin (mTOR) inhibitor for treatment of cancer. J Med Chem. 2011;54(5):1473–1480. doi: 10.1021/jm101520v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buchholz K, et al. A high-throughput screen targeting malaria transmission stages opens new avenues for drug development. J Infect Dis. 2011;203(10):1445–1453. doi: 10.1093/infdis/jir037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Feldman ME, et al. Active-site inhibitors of mTOR target rapamycin-resistant outputs of mTORC1 and mTORC2. PLoS Biol. 2009;7(2):e38. doi: 10.1371/journal.pbio.1000038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Van Tyne D, et al. Identification and functional validation of the novel antimalarial resistance locus PF10_0355 in Plasmodium falciparum. PLoS Genet. 2011;7(4):e1001383. doi: 10.1371/journal.pgen.1001383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Dharia NV, et al. Use of high-density tiling microarrays to identify mutations globally and elucidate mechanisms of drug resistance in Plasmodium falciparum. Genome Biol. 2009;10(2):R21. doi: 10.1186/gb-2009-10-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rottmann M, et al. Spiroindolones, a potent compound class for the treatment of malaria. Science. 2010;329(5996):1175–1180. doi: 10.1126/science.1193225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vera IM, Beatty WL, Sinnis P, Kim K. Plasmodium protease ROM1 is important for proper formation of the parasitophorous vacuole. PLoS Pathog. 2011;7(9):e1002197. doi: 10.1371/journal.ppat.1002197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sturm A, et al. Alteration of the parasite plasma membrane and the parasitophorous vacuole membrane during exo-erythrocytic development of malaria parasites. Protist. 2009;160(1):51–63. doi: 10.1016/j.protis.2008.08.002. [DOI] [PubMed] [Google Scholar]
- 40.Kaiser K, Matuschewski K, Camargo N, Ross J, Kappe SH. Differential transcriptome profiling identifies Plasmodium genes encoding pre-erythrocytic stage-specific proteins. Mol Microbiol. 2004;51(5):1221–1232. doi: 10.1046/j.1365-2958.2003.03909.x. [DOI] [PubMed] [Google Scholar]
- 41.Doolan DL, et al. Identification and characterization of the protective hepatocyte erythrocyte protein 17 kDa gene of Plasmodium yoelii, homolog of Plasmodium falciparum exported protein 1. J Biol Chem. 1996;271(30):17861–17868. doi: 10.1074/jbc.271.30.17861. [DOI] [PubMed] [Google Scholar]
- 42.Delves M, et al. The activities of current antimalarial drugs on the life cycle stages of Plasmodium: A comparative study with human and rodent parasites. PLoS Med. 2012;9(2):e1001169. doi: 10.1371/journal.pmed.1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Nam TG, et al. A chemical genomic analysis of decoquinate, a Plasmodium falciparum cytochrome b inhibitor. ACS Chem Biol. 2011;6(11):1214–1222. doi: 10.1021/cb200105d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.da Cruz FP, et al. Drug screen targeted at Plasmodium liver stages identifies a potent multistage antimalarial drug. J Infect Dis. 2012;205(8):1278–1286. doi: 10.1093/infdis/jis184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cunha-Rodrigues M, et al. Genistein-supplemented diet decreases malaria liver infection in mice and constitutes a potential prophylactic strategy. PLoS ONE. 2008;3(7):e2732. doi: 10.1371/journal.pone.0002732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hobbs CV, et al. HIV protease inhibitors inhibit the development of preerythrocytic-stage plasmodium parasites. J Infect Dis. 2009;199(1):134–141. doi: 10.1086/594369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Meister S, et al. Imaging of Plasmodium liver stages to drive next-generation antimalarial drug discovery. Science. 2011;334(6061):1372–1377. doi: 10.1126/science.1211936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gershon PD, Howells RE. Mitochondrial protein synthesis in Plasmodium falciparum. Mol Biochem Parasitol. 1986;18(1):37–43. doi: 10.1016/0166-6851(86)90048-4. [DOI] [PubMed] [Google Scholar]
- 49.Deponte M, et al. Wherever I may roam: Protein and membrane trafficking in P. falciparum-infected red blood cells. Mol Biochem Parasitol. 2012;186(2):95–116. doi: 10.1016/j.molbiopara.2012.09.007. [DOI] [PubMed] [Google Scholar]
- 50.Crary JL, Haldar K. Brefeldin A inhibits protein secretion and parasite maturation in the ring stage of Plasmodium falciparum. Mol Biochem Parasitol. 1992;53(1–2):185–192. doi: 10.1016/0166-6851(92)90020-k. [DOI] [PubMed] [Google Scholar]
- 51.Wickham ME, et al. Trafficking and assembly of the cytoadherence complex in Plasmodium falciparum-infected human erythrocytes. EMBO J. 2001;20(20):5636–5649. doi: 10.1093/emboj/20.20.5636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nacer A, Berry L, Slomianny C, Mattei D. Plasmodium falciparum signal sequences: Simply sequences or special signals? Int J Parasitol. 2001;31(12):1371–1379. doi: 10.1016/s0020-7519(01)00253-3. [DOI] [PubMed] [Google Scholar]
- 53.Jayabalasingham B, Bano N, Coppens I. Metamorphosis of the malaria parasite in the liver is associated with organelle clearance. Cell Res. 2010;20(9):1043–1059. doi: 10.1038/cr.2010.88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.D’Alessandro A, Righetti PG, Zolla L. The red blood cell proteome and interactome: An update. J Proteome Res. 2010;9(1):144–163. doi: 10.1021/pr900831f. [DOI] [PubMed] [Google Scholar]
- 55.Brown JR, Auger KR. Phylogenomics of phosphoinositide lipid kinases: Perspectives on the evolution of second messenger signaling and drug discovery. BMC Evol Biol. 2011;11:4. doi: 10.1186/1471-2148-11-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ward P, Equinet L, Packer J, Doerig C. Protein kinases of the human malaria parasite Plasmodium falciparum: The kinome of a divergent eukaryote. BMC Genomics. 2004;5:79. doi: 10.1186/1471-2164-5-79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tawk L, et al. Phosphatidylinositol 3-phosphate, an essential lipid in Plasmodium, localizes to the food vacuole membrane and the apicoplast. Eukaryot Cell. 2010;9(10):1519–1530. doi: 10.1128/EC.00124-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Vaid A, Ranjan R, Smythe WA, Hoppe HC, Sharma P. PfPI3K, a phosphatidylinositol-3 kinase from Plasmodium falciparum, is exported to the host erythrocyte and is involved in hemoglobin trafficking. Blood. 2010;115(12):2500–2507. doi: 10.1182/blood-2009-08-238972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Oduola AM, Milhous WK, Weatherly NF, Bowdre JH, Desjardins RE. Plasmodium falciparum: Induction of resistance to mefloquine in cloned strains by continuous drug exposure in vitro. Exp Parasitol. 1988;67(2):354–360. doi: 10.1016/0014-4894(88)90082-3. [DOI] [PubMed] [Google Scholar]
- 60.Ritchie GY, et al. In vitro selection of halofantrine resistance in Plasmodium falciparum is not associated with increased expression of Pgh1. Mol Biochem Parasitol. 1996;83(1):35–46. doi: 10.1016/s0166-6851(96)02746-6. [DOI] [PubMed] [Google Scholar]
- 61.Eastman RT, Dharia NV, Winzeler EA, Fidock DA. Piperaquine resistance is associated with a copy number variation on chromosome 5 in drug-pressured Plasmodium falciparum parasites. Antimicrob Agents Chemother. 2011;55(8):3908–3916. doi: 10.1128/AAC.01793-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Korsinczky M, et al. Mutations in Plasmodium falciparum cytochrome b that are associated with atovaquone resistance are located at a putative drug-binding site. Antimicrob Agents Chemother. 2000;44(8):2100–2108. doi: 10.1128/aac.44.8.2100-2108.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Barnes DA, Foote SJ, Galatis D, Kemp DJ, Cowman AF. Selection for high-level chloroquine resistance results in deamplification of the pfmdr1 gene and increased sensitivity to mefloquine in Plasmodium falciparum. EMBO J. 1992;11(8):3067–3075. doi: 10.1002/j.1460-2075.1992.tb05378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Baumgartner F, Wiek S, Paprotka K, Zauner S, Lingelbach K. A point mutation in an unusual Sec7 domain is linked to brefeldin A resistance in a Plasmodium falciparum line generated by drug selection. Mol Microbiol. 2001;41(5):1151–1158. doi: 10.1046/j.1365-2958.2001.02572.x. [DOI] [PubMed] [Google Scholar]
- 65.Elmendorf HG, Haldar K. Identification and localization of ERD2 in the malaria parasite Plasmodium falciparum: Separation from sites of sphingomyelin synthesis and implications for organization of the Golgi. EMBO J. 1993;12(12):4763–4773. doi: 10.1002/j.1460-2075.1993.tb06165.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Cowman AF, Berry D, Baum J. The cellular and molecular basis for malaria parasite invasion of the human red blood cell. J Cell Biol. 2012;198(6):961–971. doi: 10.1083/jcb.201206112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bullen HE, et al. Biosynthesis, localization, and macromolecular arrangement of the Plasmodium falciparum translocon of exported proteins (PTEX) J Biol Chem. 2012;287(11):7871–7884. doi: 10.1074/jbc.M111.328591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gomes-Santos CS, et al. Highly dynamic host actin reorganization around developing Plasmodium inside hepatocytes. PLoS ONE. 2012;7(1):e29408. doi: 10.1371/journal.pone.0029408. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.