Viruses of marine bacteria (bacteriophage) have been characterized since the mid-20th century (1). However, it took several additional decades before electron micrographs of seawater revealed that particles of virus-like morphology could exceed concentrations of 1 million per milliliter of seawater (2, 3), and that phage infection could cause substantial mortality in marine bacterial communities (4). Subsequent research in the field of marine viral ecology has focused on the characterization of the diversity of the virioplankton (community of extracellular viruses), elucidating how viruses influence the community dynamics and evolution of their hosts and ultimately the role viruses play in biogeochemical cycling. Critical to the understanding of viral ecology is the identification of the constituents of the viral community and whom they infect. This is a formidable challenge because there are thousands of different types of viruses in every liter of seawater (5) that are mostly distantly related to the catalog of known viruses, and whose morphology and genotype do not identify the host that they infect. A clear, albeit challenging, path forward to gaining a greater understanding of viral ecology is the isolation and characterization of viruses that infect hosts who play important roles in the environment. In PNAS, Kang et al. (6) make a significant contribution to the field with a report describing the isolation and characterization of a bacteriophage, HMO-2011, which infects a member of the SAR116 clade, a group of marine bacteria that are abundant and ubiquitous in the surface ocean (7, 8). The isolation of a phage that lyses a SAR116 strain demonstrates that viruses are an additional source of mortality for these organisms, and analyses based on the sequence of the HMO-2011 genome suggest that viruses related to this phage comprise a substantial fraction of the virioplankton.
The isolation of a virus–host system from the marine environment can be a demanding process; nevertheless, the payoff for this often frustrating and exacting work has been substantial. The characterization of the modest number of cultivated marine phage–host systems has resulted in data critical in the modeling of marine viral dynamics, understanding the extent of gene transfer between virus and host, and the compilation of a reference database crucial to the interpretation of sequences generated from ostensibly cultivation-independent approaches. An excellent example of the benefit of cultivation-based research is the line of investigation founded on the isolation of viruses that infect the cyanobacterium Prochlorococcus (9), the most abundant primary producer in the largest regions of the ocean (10). One highlight was the discovery that these viruses encode genes that form the core components of the cyanobacterial photosynthetic apparatus (11). These genes may enable the virus to coax more energy from the host cell to complete its replication cycle, even as the virus is simultaneously destroying the cell from within. The HMO-2011 genome contains genes that suggest that the virus is capable of altering its host’s metabolism as well (6), raising the possibility that future research based on this virus–host system may bear similar fruit.
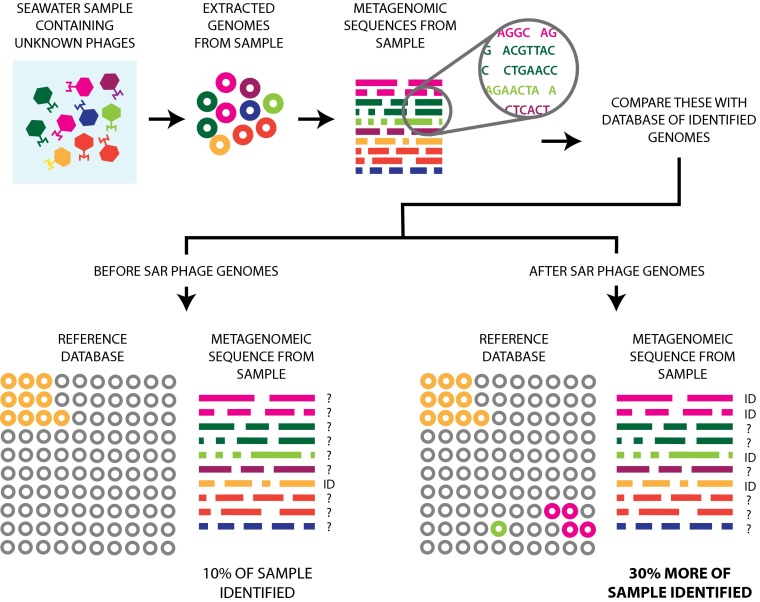
Bringing thousands of marine prokaryotes and tens of thousands of viruses into culture to produce an accurate assessment of the composition of a virus community is presently intractable. Therefore, a cultivation-independent, metagenomic approach has been the most commonly used method to characterizing diversity (e.g., ref. 12). In this method, the genomes from a community of viruses are extracted from seawater, sequenced in toto, and analyzed (Fig. 1). Of the billions of nucleotide sequences generated from metagenomic studies of the marine virus community, often 90% of the sequences have no similarity to reference databases that include the genome sequences of known viruses (13). These results are particularly problematic because they indicate that the identity of a significant majority of the virioplankton (and therefore their hosts) remains unknown. However, one of the remarkable findings of the research by Kang et al. (6) is that when the genome sequence from HMO-2011, as well as four recently described phages that infect a strain of SAR11 (14) (another important group of marine bacteria), are included in similarity searches with sequences from viral metagenomes generated from the Indian and Pacific Ocean, an average of 30% more of the total sequences assigned to viruses are identified than if these phages were not included (Fig. 1). These results suggest that viruses related to SAR phages comprise a substantial fraction of the virioplankton. Nevertheless, the mosaic nature of double-stranded DNA (dsDNA) phage genomes and the high frequency of exchange between them means that viruses that infect disparate hosts can have sections of genomes that are highly related (15). Thus, the apparent prevalence of SAR phages based on sequence similarity must be confirmed independently. Beyond providing greater insight into the unknown viral majority, the SAR phages should provide excellent model systems to better
Fig. 1.
A simplified depiction of how the addition of the SAR116 and SAR11 phages to the catalog of known viruses results in the identification of substantially more of the marine phage community. A seawater sample containing a community of viruses is harvested from the ocean, the viral genomes are extracted, and a metagenome is generated through random sequencing. To identify the viruses in the sample, the metagenomic sequences are compared with a reference library of known viruses. The identification of these viruses is ultimately dependent on their similarity to viruses in the reference database. The addition of the SAR phages results in the identification of ∼30% more metagenomic sequences (6). In the reference database, the orange, light green, and pink circles represent known, SAR116, and SAR11 phages, respectively. A question mark next to a row of sequences indicates the sequences remain unidentified, and an ID indicates the sequences are known. Graphics by Amanda Toperoff.
Analyses based on the sequence of the HMO-2011 genome suggest that viruses related to this phage comprise a substantial fraction of the virioplankton.
understand the influence of viruses on these key groups of organisms in the ocean.
Although these exciting findings represent substantial progress toward identifying the dominant marine viruses, the virioplankton appears to be more diverse than previously appreciated. Whereas dsDNA phage are undoubtedly major contributors to the virioplankton, diverse communities of largely uncharacterized viruses with single-stranded (ss)DNA (16), ssRNA and dsRNA genomes are present as well (17). In fact, there are data that suggest that at times the abundance of RNA viruses can exceed that of dsDNA phage (18). Moreover sequences closely related to a group of viruses with massive dsDNA genomes have been identified in marine microbial metagenomic libraries (19), suggesting that these viruses are also persistent constituents of the virus community. Integrating the diverse and complex groups that comprise the virioplankton into a cohesive ecological picture presents a substantial challenge. Fortunately, increased sequencing capabilities, advances in analytical tools, more refined modeling efforts, and innovative new methodologies promise to expedite this process. Nevertheless, the importance of cultivation, as exemplified in the work of Kang et al. (6), is incontrovertible. Expanding this collection is of paramount importance.
Footnotes
The author declares no conflict of interest.
See companion article on page 12343.
References
- 1.Spencer R. Indigenous marine bacteriophages. J Bacteriol. 1960;79(4):614. doi: 10.1128/jb.79.4.614-614.1960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torrella F, Morita RY. Evidence by electron micrographs for a high incidence of bacteriophage particles in the waters of Yaquina Bay, Oregon: Ecological and taxonomical implications. Appl Environ Microbiol. 1979;37(4):774–778. doi: 10.1128/aem.37.4.774-778.1979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bergh Ø, Børsheim KY, Bratbak G, Heldal M. High abundance of viruses found in aquatic environments. Nature. 1989;340(6233):467–468. doi: 10.1038/340467a0. [DOI] [PubMed] [Google Scholar]
- 4.Proctor L, Fuhrman J. Viral mortality of marine bacteria and cyanobacteria. Nature. 1990;343(6253):60–62. [Google Scholar]
- 5.Edwards RA, Rohwer F. Viral metagenomics. Nat Rev Microbiol. 2005;3(6):504–510. doi: 10.1038/nrmicro1163. [DOI] [PubMed] [Google Scholar]
- 6.Kang I, Oh H-M, Kang D, Cho J-C. Genome of a SAR116 bacteriophage shows the prevalence of this phage type in the oceans. Proc Natl Acad Sci USA. 2013;110:12343–12348. doi: 10.1073/pnas.1219930110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mou X, Hodson RE, Moran MA. Bacterioplankton assemblages transforming dissolved organic compounds in coastal seawater. Environ Microbiol. 2007;9(8):2025–2037. doi: 10.1111/j.1462-2920.2007.01318.x. [DOI] [PubMed] [Google Scholar]
- 8.Rappé MS, Vergin K, Giovannoni SJ. Phylogenetic comparisons of a coastal bacterioplankton community with its counterparts in open ocean and freshwater systems. FEMS Microbiol Ecol. 2000;33(3):219–232. doi: 10.1111/j.1574-6941.2000.tb00744.x. [DOI] [PubMed] [Google Scholar]
- 9.Sullivan MB, Waterbury JB, Chisholm SW. Cyanophages infecting the oceanic cyanobacterium Prochlorococcus. Nature. 2003;424(6952):1047–1051. doi: 10.1038/nature01929. [DOI] [PubMed] [Google Scholar]
- 10.Chisholm SW, et al. A novel free-living prochlorophyte abundant in the oceanic euphotic zone. Nature. 1988;334(6154):340–343. [Google Scholar]
- 11.Mann NH, Cook A, Millard A, Bailey S, Clokie M. Marine ecosystems: Bacterial photosynthesis genes in a virus. Nature. 2003;424(6950):741. doi: 10.1038/424741a. [DOI] [PubMed] [Google Scholar]
- 12.Breitbart M, et al. Genomic analysis of uncultured marine viral communities. Proc Natl Acad Sci USA. 2002;99(22):14250–14255. doi: 10.1073/pnas.202488399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hurwitz BL, Sullivan MB. The Pacific Ocean virome (POV): A marine viral metagenomic dataset and associated protein clusters for quantitative viral ecology. PLoS ONE. 2013;8(2):e57355. doi: 10.1371/journal.pone.0057355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zhao Y, et al. Abundant SAR11 viruses in the ocean. Nature. 2013;494(7437):357–360. doi: 10.1038/nature11921. [DOI] [PubMed] [Google Scholar]
- 15.Hendrix RW, Smith MCM, Burns RN, Ford ME, Hatfull GF. Evolutionary relationships among diverse bacteriophages and prophages: All the world’s a phage. Proc Natl Acad Sci USA. 1999;96(5):2192–2197. doi: 10.1073/pnas.96.5.2192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Angly FE, et al. The marine viromes of four oceanic regions. PLoS Biol. 2006;4(11):e368. doi: 10.1371/journal.pbio.0040368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Culley AI, Lang AS, Suttle CA. Metagenomic analysis of coastal RNA virus communities. Science. 2006;312(5781):1795–1798. doi: 10.1126/science.1127404. [DOI] [PubMed] [Google Scholar]
- 18.Steward GF, et al. Are we missing half of the viruses in the ocean? ISME J. 2013;7(3):672–679. doi: 10.1038/ismej.2012.121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hingamp P, et al. Exploring nucleo-cytoplasmic large DNA viruses in Tara Oceans microbial metagenomes. ISME J. 2013 doi: 10.1038/ismej.2013.59. 10.1038/ismej.2013.59. [DOI] [PMC free article] [PubMed] [Google Scholar]