Abstract
Major depressive disorder (MDD) is a complex psychiatric condition with strong genetic predisposition. The association of MDD with genetic polymorphisms, such as Val66Met (rs6265), in the brain derived neurotrophic factor (BDNF), have been reported in many studies and the results were conflicting. In this study, we performed a systematic literature search and conducted random-effects meta-analysis to evaluate genetic variants in BDNF with MDD. A gene-based analysis was also conducted to investigate the cumulative effects of genetic polymorphisms in BDNF. A total of 28 studies from 26 published articles were included in our analysis. Meta-analysis yielded an estimated odds ratio (OR) of 0.96 (95% CI: 0.89–1.05; P= 0.402) for Val66Met (rs6265), 0.83 (95% CI: 0.67–1.04; P= 0.103) for 11757C/G, 1.16 (95% CI: 0.74–1.82; P= 0.527) for 270T/C, 1.03 (95% CI: 0.18–5.75; P= 0.974) for 712A/G and 0.98 (95% CI: 0.85–1.14; P= 0.831) for rs988748. The gene-based analysis indicated that BDNF is not associated with MDD (P>0.21). Our updated meta- and novel gene-based analyses provide no evidence of the association of BDNF with major depression.
Keywords: major depressive disorder, BDNF, polymorphism, meta-analysis, gene-based analysis
INTRODUCTION
Major depressive disorder (MDD) is a complex psychiatric condition [Sullivan et al., 2000] defined by nearly constant depressed mood and/or anhedonia lasting for weeks [American Psychiatric Association, 2000]. MDD is a huge and growing public health problem around the world [Kessler et al., 2003; Ustun et al., 2004]. Although lifestyle and environmental factors are significant contributors to the etiology of MDD, genetic predisposition has been found to play a strong role [Sullivan et al., 2000]. Elucidating genetic factors involved in the etiology of depression will facilitate the design and implementation of novel treatments for this under-treated and often poorly managed condition.
The brain derived neurotrophic factor (BDNF) gene was discovered for the role of its protein product in promoting neuron survival [Hofer and Barde, 1988; Jones and Reichardt, 1990; Maisonpierre et al., 1991]. Investigation into the common genetic variants of the gene in the human population revealed a single nucleotide polymorphism (SNP) that changes the 66th amino acid in the protein from the ancestral valine to methionine (rs6265 or Val66Met). Initial tests for association between MDD and Val66-Met in case control studies were conflicting, with many published reports in favor of the hypothesis [Jiang et al., 2005; Schumacher et al., 2005; Hwang et al., 2006; Iga et al., 2007] and opposed [Hong et al., 2003; Tsai et al., 2003; Oswald et al., 2005; Choi et al., 2006; Surtees et al., 2007; Kim et al., 2008]. The first meta-analyses of this single allele found no net effect of the Val66Met allele [Chen et al., 2008; Lopez-Leon et al., 2008]. Recent genome-wide association studies (GWAS) on MDD also indicate there are no large or consistent effects of any common BDNF single-nucleotide polymorphisms [Sullivan et al., 2009; Lewis et al., 2010; Muglia et al., 2010; Shi et al., 2011]. However, new reports continuously emerge suggesting that the Val66Met polymorphism plays a role in processes that might influence prevalence of MDD, such as response rate for common treatments [Kato and Serretti, 2010; Zou et al., 2010].
Meanwhile, a growing number of studies have tested for association of other genetic variants in BDNF with MDD. Data has accumulated to make it possible to conduct meta-analyses on some minor variants with modest prevalence in the population. This also makes it possible to pool the effects of multiple SNPs for a gene-based analysis [Neale and Sham, 2004]. Thus, we conducted an updated meta-analysis of the Val66Met polymorphisms and other genetic variants in BDNF gene, and also performed a gene-based analysis to examine the cumulative association of BDNF with MDD.
METHODS
Search Strategy
We did an extensive literature search in July 2012 in MEDLINE, Cochrane Library and Web of Science for studies on the association of genetic variants in BDNF with major depressive disorder. Search terms included “major depression,” “depressive disorder,” “genetic variant,” “polymorphism,” “brain-derived neurotrophic factor,” and “BDNF.” References of all relevant publications were also hand searched for additional studies missed by the database search.
Inclusion Criteria
Studies were included in our analysis if they met the following criteria: (1) studies on human subjects; (2) outcome measures include major depression; and (3) case–control studies reporting genotype or allele frequencies of individual SNPs in BNDF of subjects with MDD and non-depressed controls. Case status was defined as having diagnosis of MDD assessed by psychiatric interviews. We include studies of any ethnic group or any age category. We did not include subsyndromal depression in our analysis. Case only studies were excluded, as well as studies only reporting the effect of BDNF genetic variants on depression-related phenotypes, such as anxiety or anti-depressant treatments. We did not specify Hardy–Weinberg equilibrium as an inclusion criterion. All potentially relevant publications were retrieved and evaluated for inclusion. Only studies published in the English language were included. Two authors (W.Y. and J.Y.) performed the search independently. Disagreement over eligibility of a study was resolved by discussion until a consensus was reached.
Data Extraction
Reviewers (W.Y. and J.Y.) independently extracted the following data according to a pre-specified protocol: first author’s name, year of publication, characteristics (sample size, age, and prevalence of MDD) of the study populations, criteria for diagnosing MDD, genotype data for subjects with MDD and controls. No efforts were made to contact the authors for additional information. Extracted data were entered into a computerized spreadsheet for analysis and compared between reviewers. Discrepancies were resolved by discussion.
Statistical Analysis
Odds ratio (OR) was used as a measure of the association of the genetic variants in BDNF with MDD. In the presence of heterogeneity among the studies, we used random-effects models to calculate the OR and the corresponding 95% confidence interval (CI); otherwise, a fixed-effect model was used. The Z-test was used to calculate the P-value of the overall effect for the meta-analysis. We used a forest plot to graphically present the calculated pooled ORs and the 95% CIs. Each study was represented by a square in the plot, and the area of the square is proportional to the weight of the study. In the random-effects meta-analysis, we used the inverse of the variance of each study as the weight for the study. The overall effect from meta-analysis is represented by a diamond, with its width representing the 95% CI for the estimate. We used a Chisquared heterogeneity test for assessment of between-study heterogeneity. Publication bias was assessed visually using a funnel plot and tested with Egger’s regression test [Egger et al., 1997].
In order to assess the overall association of BDNF with MDD, we conducted a gene-based analysis using the P-values of the association of genetic variants in BDNF with MDD calculated from published literature and from our meta-analysis. We used four popular P-value combination methods to assess this association: the Fisher’s method, [Fisher, 1932] the Simes method, [Simes, 1986] the modified inverse normal method [Hartung, 1999] and the truncated product method (TPM) [Zaykin et al., 2002; Sheng and Yang, 2012]. To deal with differences in sample size in studies assessing the association of each individual SNP, we calculated unweighted and weighted TPM. Un-weighted TPM disregards the difference in sample size and weighted TPM employs the sample size as its weight, allowing studies with larger sample size to play a larger role in the calculation [Zaykin et al., 2002] A detailed description of the four methods was reported elsewhere [Sheng and Yang, 2012; Wang et al., 2012a]. Because these P-values are most likely to be dependent, we used 100,000 simulations to estimate the P-value for TPM.
Meta-analysis was performed using Stata 11.2 (StataCorp LP, College Station, TX). All the other analyses were performed using SAS version 9.2 (SAS Institute Inc., Cary, NC) and Matlab 7.10.0.499 (The MathWorks, Inc., Natick, MA).
RESULTS
Literature Search and Eligible Studies
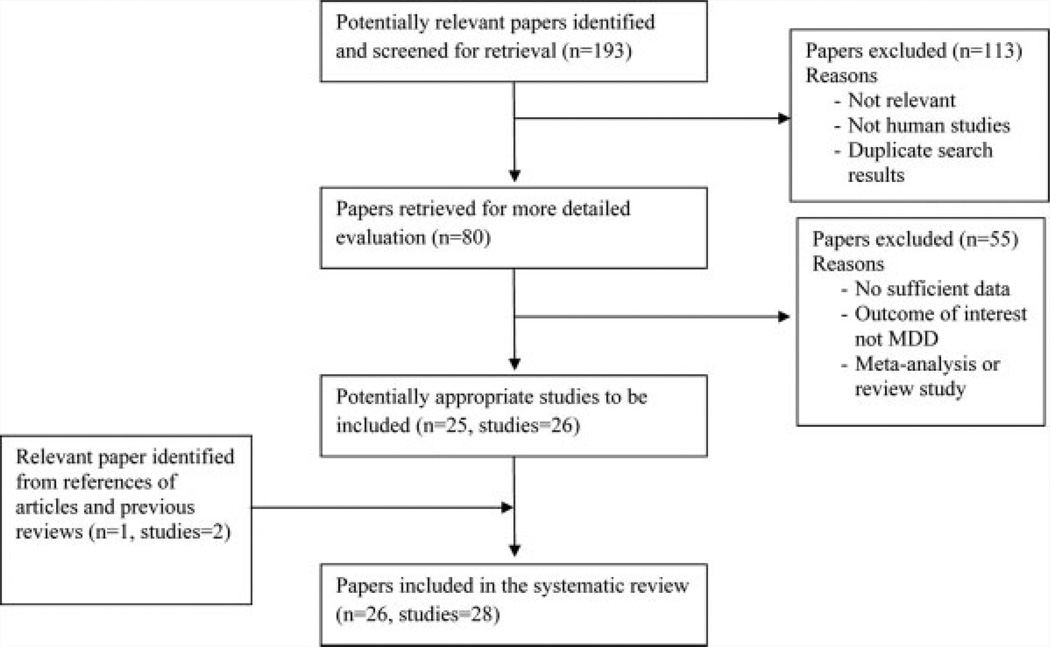
Figure 1 is the flow diagram showing the process of selection of studies included in our analysis. Using our pre-defined search strategy, we identified a total of 193 potential studies through our initial search. After screening of the abstracts of these studies, 113 were excluded either because they were irrelevant,the study was not about human subjects, or the record was a duplicate search result. The remaining 80 studies were retrieved for more detailed evaluations, which excluded an additional 55 studies because no sufficient genotype data were reported, the outcome of interest is not MDD or a review or meta-analysis study, leaving 25 potentially relevant papers (total studies = 26) to be included in our analysis. A further review of the references of these studies and review article identified one more article with two studies. As a result, a total of 28 studies from 26 articles met the eligibility criteria and were included in our analyses [Hong et al., 2003; Tsai et al., 2003; Oswald et al., 2005; Schumacher et al., 2005; Choi et al., 2006; Hwang et al., 2006; Anttila et al., 2007; Frodl et al., 2007; Iga et al., 2007; Ribeiro et al., 2007; Surtees et al., 2007; Enoch et al., 2008; Gratacos et al., 2008; Taylor et al., 2008; Chi et al., 2010, 2011; Kim et al., 2010; Ozan et al., 2010; Zhang et al., 2010; Chen et al., 2011; Cole et al., 2011; Kanellopoulos et al., 2011; Sun et al., 2011; Pae et al., 2012; Quinn et al., 2012; Wang et al., 2012b].
FIG. 1.
Flow diagram of studies included in the systematic review
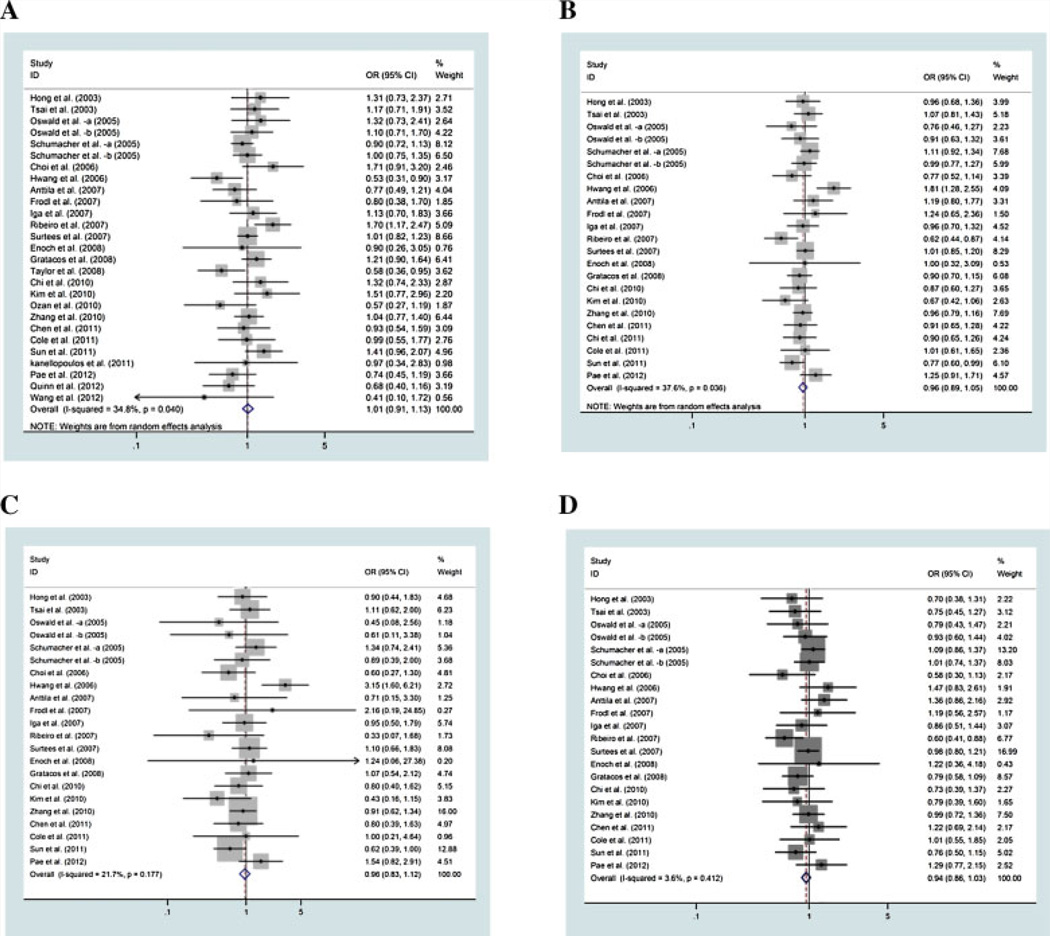
All qualified articles included in this study were published since 2003 and had sample sizes ranging from 38 to 7,173 participants (Table I). Prevalence of MDD ranges from 6% to 72%. Of these 28 studies, 23 reported a summary of genotype data for Val66Met, three studies for 11757C/G [Oswald et al., 2005; Chen et al., 2011], and two studies for 270T/C [Chen et al., 2011; Sun et al., 2011], 712A/G [Chen et al., 2011; Sun et al., 2011] and rs988748 [Oswald et al., 2005], respectively, and they were included in the corresponding meta-analysis for each of the five SNPs (Tables II and III). In addition to these five SNPs, three other studies also reported summaries of genotype data of a total of 12 other SNPs in BDNF [Gratacos et al., 2008; Zhang et al., 2010; Pae et al., 2012]. We computed the association of these SNPs with MDD and the results were included in our gene-based analysis. Different genetic models were used in the calculation of the association of genetic variants in BDNF with MDD. For simplicity, we only report the results of minor allele versus major allele. Results for other genetic comparisons show a similar pattern (Fig. 2).
TABLE 1.
Basic Characteristics of All Studies
| MDD |
Control |
|||||
|---|---|---|---|---|---|---|
| Study (author, year) | n | Mean age | n | Mean age | Definition of MDD | Diagnosis method |
| Wang et al. [2012a, b] | 18 | NA | 20 | NA | DSM-IV | — |
| Quinn et al. [2012] | 128 | 40.1 | 128 | 40.1 | DSM-IV | Structured interview using MINI by research officers trained and supervised by psychiatrists |
| Pae et al. [2012] | 145 | 41.4 ±14.1 | 170 | 38.8 ±12.8 | DSM-IV | Using MINI |
| Chi et al. [2011] | 198 | 40.7 ±14.2 | 106 | 34.0 ±12.7 | DSM-IV | Structured interview by experienced psychiatrists using MINI |
| Cole et al. [2011] | 84 | 33.0±9.2 | 111 | 48.8 ±8.9 | DSM-IV | Structured interview using SCAN |
| Chen et al. [2011] | 83 | 57.9 ± 11.1 | 400 | 64.5 ±5.8 | DSM-IV | — |
| Kanellopoulos et al. [2011] | 33 | 72.3 | 23 | 70.9 | DSM-IV | Structured interview |
| Sun et al. [2011] | 202 | NA | 346 | NA | DSM-IV | Structured interview by two psychiatrists |
| Chi et al. [2010] | 117 | 36.2 ±12.6 | 106 | 34.0 ±12.7 | DSM-IV | Structured interview using a Chinese version of MINI |
| Zhang et al. [2010] | 447 | 27.8±8.0 | 432 | 28.3 ±8.7 | DSM-IV | Structured interview by at least two consultant psychiatrists using the Chinese version of SCID-I/P |
| Kim et al. [2010] | 42 | NA | 349 | NA | DSM-IV | Structured interview by board-certified psychiatrists |
| Ozan et al. [2010] | 66 | 34.0 | 56 | 33.0 | DSM-IV | Structured interview using SCID-I/P |
| Taylor et al. [2008] | 245 | 69.7±7.5 | 94 | 69.8 ±5.8 | NIMH DIS plus DSM-IV | A self-report questionnaire plus an interview with a geriatric psychiatrist |
| Enoch et al. [2008] | 15 | NA | 101 | NA | DSM-III-R | — |
| Ribeiro et al. [2007] | 278 | 39.0 ±9.9 | 320 | NA | DSM-IV | Structured interview by experienced clinical investigators using SCID-I/P |
| Iga et al. [2007] | 154 | 45.1 ±14.2 | 154 | 45.0 ±14.0 | DSM-IV | Consensus by at least two trained psychiatrists |
| Gratacos et al. [2008] | 374 | 57.2 ± 15.3 | 342 | 39.8 ±11.9 | DSM-IV | Structured interview by experienced psychiatrists using SCID-I/P |
| Frodl et al. [2007] | 60 | 44.2 ±11.8 | 60 | 41.6 ±12.3 | DSM-IV | Consensus of at least two psychiatrists based on structured interview |
| Anttila et al. [2007] | 119 | 57.5 ±14.0 | 392 | 44.4 ±11.1 | DSM-IV | — |
| Choi et al. [2006] | 83 | 53.9 ±11.7 | 128 | 40.7 ±15.5 | DSM-IV plus K-DIGS | Structured interview by trained psychiatrists |
| Hwang et al. [2007] | 110 | 75.0 ±5.3 | 171 | 76.0 ±5.5 | DSM-IV | — |
| Surtees et al. [2007] | 429 | 57.0 ±8.6 | 6,744 | 60.4 ±9.2 | HLEQ | Structured self-assessment |
| Schumacher et al. [2005]—a | 312 | 52.2 ±13.4 | 444 | 50.5 ±12.8 | DSM-IV | Combination of multiple sources of information including a personal structured interview using SCID-I/P, medical records, and the family history; assessed by least two experienced psychiatrists or psychologists |
| Schumacher et al. [2005]—b | 446 | 47.4 ±13.7 | 1,084 | 44.6 ±15.9 | DSM-IV | Structured interview using SCAN |
| Tsai et al. [2003] | 152 | 45.3 ±17.0 | 164 | 45.7 ±13.1 | DSM-IV | — |
| Hong et al. [2003] | 84 | 47.9 ±17.4 | 392 | 44.8 ±21.4 | DSM-IV | Combination of interview, clinical observation, medical records, past history and re-assessed by a psychiatrist |
| Oswad et al. [2005]—a | 66 | 25.7 ±3.5 | 66 | 32.2±9.7 | DSM-IV | Semi-structured interview by a trained psychiatrist using MINI |
| Oswad et al. [2005]—b | 92 | 50.2 ±12.1 | 92 | 40.1 ±7.8 | DSM-IV | Semi-structured interview by a trained psychiatrist using MINI |
DSM-IV, diagnosis and statistical manual of mental health disorders, fourth edition.
MINI, the mini international neuropsychological interview, a structured psychiatric interview based on DSM-IV.
SCID-I/P, the modified structured clinical interview for DSM-IV TR Axis I disorders, patient edition.
K-GIGS, the Korean version of diagnostic interview for genetic studies.
HLEQ, a structured self-assessment version of DSM-IV.
SCAN, schedule for clinical assessment in neuropsychiatry.
TABLE II.
Meta-Analysis of the Association Val66Met With Major Depression
| Case | Controls | OR (95% CI) | P | Pfor heterogeneity | |
|---|---|---|---|---|---|
| Val/val versus met carrier | 4,447 | 12,979 | 1.01 (0.91–1.13) | 0.823 | 0.040 |
| Met versus val | 4,173 | 12,747 | 0.96 (0.89–1.05) | 0.402 | 0.036 |
| Met/met versus val/val | 3,975 | 12,641 | 0.96 (0.83–1.12) | 0.618 | 0.177 |
| Met/val versus val/val | 3,975 | 12,641 | 0.94 (0.86–1.03) | 0.189 | 0.412 |
TABLE III.
Meta-Analysis of Other SNPs in BDNF With Major Depression
| SNP | Study | Cases | Controls | OR (95% CI) | P |
|---|---|---|---|---|---|
| 11757C/G | Chen et al. | 83 | 400 | 0.93 (0.66–1.29) | 0.654 |
| Oswald et al.—a | 92 | 92 | 0.62 (0.38–1.02) | 0.060 | |
| Oswald et al.—b | 156 | 196 | 0.86 (0.59–1.24) | 0.410 | |
| Total | 331 | 688 | 0.83 (0.67–1.04) | 0.103 | |
| 270T/C | Chen et al. | 83 | 400 | 0.94 (0.43–2.04) | 0.870 |
| Sun et al. | 202 | 346 | 1.29 (0.74–2.24) | 0.372 | |
| Total | 285 | 746 | 1.16 (0.74–1.82) | 0.527 | |
| 712A/G | Chen et al. | 83 | 400 | 0.39 (0.12–1.28) | 0.121 |
| Sun et al. | 202 | 346 | 2.28 (1.40–3.71) | 0.001 | |
| Total | 285 | 746 | 1.03 (0.18–5.75) | 0.974 | |
| rs988748 | Oswald et al.—a | 465 | 1,097 | 1.02 (0.85–1.23) | 0.797 |
| Oswald et al.—b | 312 | 444 | 0.92 (0.72–1.17) | 0.483 | |
| Total | 777 | 1,541 | 0.98 (0.85–1.14) | 0.831 |
FIG. 2.
Odds ratios of Val66Met (rs6265) with major depressive disorder (A: Val/val vs. met;B:Met vs. val; C: Met/met vs. val/val; D:Met/val vs. val/val).
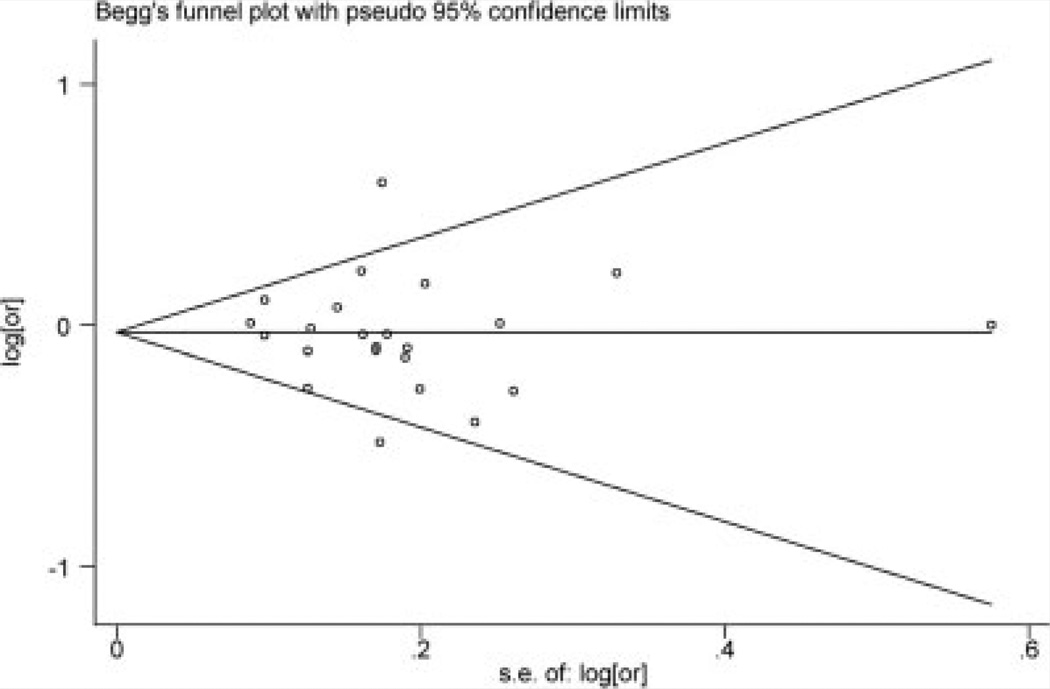
Assessment of Publication Bias
Both funnel plot and Egger’s test were used to assess publication bias (Fig. 3). The funnel plot for the meta-analysis of Val66Met seems symmetrical, suggesting no evidence of publication bias. Egger’s test also shows no publication bias (α=−0.38, 95% CI: −1.99, 1.23; P =0.63). There was significant publication bias for the metaanalysis of 11757C/G (α =−4.79, 95% CI: −6.95, −2.63; P = 0.023). The Egger’s test was not available for meta-analysis of 270T/C, 712A/G and rs988748 due to too few studies.
FIG. 3.
Funnel plot for meta-analysis of Val66Met.
Association of Individual SNPs With MDD
Twenty-three studies provided summaries of genotype data of Val66Met (minor allele vs. major allele). Random-effects metaanalysis indicates no association of the SNP with MDD (OR = 0.96 for Met vs. Val; 95% CI: 0.89–1.05; P = 0.402), (Fig. 2b, Table II). There was significant between-study heterogeneity (χ2 =35.26, P=0.036).
Three studies provided summaries of genotype data of 11757C/G with MDD [Oswald et al., 2005; Chen et al., 2011]. Fixed-effect meta-analysis gives an estimated odds ratio of 0.83 (95% CI: 0.67–1.04; P= 0.103), indicating no significant association with MDD (Table III). There was no significant between-study heterogeneity (χ2 = 1.76, P=0.415).
Two studies provided summaries of genotype data of 270T/C [Chen et al., 2011; Sun et al, 2011]. Fixed-effect meta-analysis gives an estimated odds ratio of 1.16 (95% CI: 0.74–1.82; P= 0.527), indicating no significant association with MDD (Table III). There was no significant between-study heterogeneity (χ2 = 0.43, P=0.515).
Two studies provided summaries of genotype data of 712A/G [Chen et al., 2011;Sun et al., 2011]. Random-effects meta-analysis gives an estimated odds ratio of 1.03 (95% CI: 0.18–5.75; P = 0.974), indicating no significant association with MDD (Table III). There was significant between-study heterogeneity (χ2 = 7.22, P = 0.007).
Two studies provided summaries of genotype data of rs988748 with MDD [Oswald et al., 2005]. Fixed-effect meta-analysis gives an estimated odds ratio of 0.98 (95% CI: 0.85–1.14; P = 0.831), indicating no association with MDD (Table III). There was no significant between-study heterogeneity (χ2 = 0.51, P = 0.474).
In addition to the above five SNPs, three other studies [Gratacos et al., 2008; Zhang et al., 2010; Pae et al., 2012] reported genotype information of 12 additional SNPs in BDNF. We calculated the association (minor allele vs. major allele) of these SNPs with MDD and summarized them in Table IV with results from the meta-analyses.
TABLE IV.
Summary of Association of Individual SNPs in BDNF With Major Depression*
| SNP | Allelesa | OR (95% CI) | P |
|---|---|---|---|
| Val66Met (rs6265) | A/G | 0.96(0.89–1.05) | 0.402 |
| 11757C/G | C/G | 0.83 (0.67–1.04) | 0.103 |
| 270T/C | T/C | 1.16 (0.74–1.82) | 0.527 |
| 712A/G | A/G | 1.03 (0.18–5.75) | 0.974 |
| rs988748 | C/G | 0.98 (0.85–1.14) | 0.831 |
| rs10501087 | C/T | 1.01 (0.79–1.29) | 0.929 |
| rs11030096 | C/T | 1.12 (0.91–1.38) | 0.295 |
| rs12273363 | C/T | 1.03 (0.78–1.36) | 0.845 |
| rs1491850 | C/T | 1.00 (0.81–1.23) | 0.990 |
| rs1491851 | T/C | 1.18 (0.96–1.46) | 0.122 |
| rs908867 | A/G | 0.78 (0.53–1.16) | 0.224 |
| rs925946 | T/G | 0.93 (0.73–1.20) | 0.598 |
| rs10835210 | A/C | 0.78 (0.56–1.10) | 0.156 |
| rs11030101 | T/A | 0.78 (0.56–1.10) | 0.156 |
| rs2030324 | T/C | 0.77 (0.56–1.05) | 0.102 |
| rs7103873 | G/C | 0.76 (0.56–1.04) | 0.088 |
| rs7124442 | T/C | 1.34 (0.91–1.96) | 0.138 |
Major/minor
Calculated based on meta-analysis and published literature
Gene-Based Analysis
Using the P-values obtained above; we performed a gene-based association study to examine the cumulative association of these genetic variants with MDD. All the methods indicate no significant association of BDNF with MDD (all P>0.21; Table V). We examined whether the association between BDNF and MDD was influenced by Val66Met, the most widely reported genetic variant in BDNF. After removing this SNP, the gene-based association did not change dramatically (all P> 0.19; Table V).
TABLE V.
Gene-Based Analysis of Genetic Variants in BDNF With Major Depression
| Fisher | Simes | Inverse | TPM |
||
|---|---|---|---|---|---|
| BDNF | 0.212 | 0.379 | 0.291 | Un-weighted | 0.627 |
| Weighted | 0.625 | ||||
| BDNFa | 0.199 | 0.357 | 0.309 | Un-weighted | 0.603 |
| Weighted | 0.603 | ||||
Excluding Val66Met
DISCUSSION
In this article, we did a systematic literature search of studies on the association between genetic variants in BDNF and MDD. Out of the five individual SNPs evaluated, our meta-analyses found no significant association of genetic polymorphisms in BDNF with major depression. Gene-based analysis also indicates that BDNF shows no significant cumulative association with MDD. To the best of our knowledge, this is the first study on the association of polymorphisms in BDNF with MDD through a gene-based approach.
Most studies on the effect of genetic polymorphisms in BDNF on MDD focus on Val66Met, with conflicting results being reported. Among the 23 studies on Val66Met, only three studies show significant association of this SNP with MDD, with one study [Sun et al., 2011] showing marginal association (OR = .77,95% CI: 0.60–0.99; P= 0.039) and two other studies [Hwang et al, 2006; Ribeiro et al, 2007] reporting stronger association (both P < 0.006) but in different directions. All other studies fail to detect a significant association. Similarly, the study by Sun et al. [2011] found a significant association of 712A/G with MDD in 548 participants in China (OR = 2.28, 95% CI: 1.40–3.71; P= 9.57 × 10−4 ). However, a study by Chen et al. [2011] failed to detect a strong association in 483 participants in China (OR = 0.39; 95% CI: 0.12–1.28; P = 0.121). It is unclear what factors contribute to the conflicting results reported in these studies. Differences in genetic structure, sample size, gender distribution and definition and measurement of MDD might contribute to the disparity in results. Of course, other factors, such as environment and diet, might play roles in these differences as well.
It appears that BDNF’s status as a candidate gene for MDD risk is waning. The hypothesis that genetic variants in the BDNF gene are associated with risk of MDD emerged from studies in animal models suggesting functional relationships between BDNF and stressful experiences [Smith et al., 1995] and antidepressant treatment response [Nibuya et al., 1995]. In vitro studies suggested that theVal66Met polymorphism could influence the function of BDNF in neurons [Egan et al., 2003; Chen et al., 2004], and in early candidate gene studies was associated with reduced hippocampal size, [Pezawas et al., 2004] which is a neurological feature believed to be shared with MDD [Videbech and Ravnkilde, 2004]. However, new experimental evidence has emerged indicating BDNF’s effect in brain regions outside the hippocampus plays opposing roles in anxiety and depression that could directly counteract and thus nullify BDNF’s overall role in MDD [Groves, 2007].
The meta-analyses to date, including our own, indicate that the initial reports of a main effect of Val66Met on MDD were probably false positives. The issue of heterogeneity between studies could be mediated by a variety of factors, but the possibility remains that the Val66Met does influence depression risk by certain mechanisms that remain unclear. The Val66Met knock-in mouse model appears to confirm a role of this genetic variant in certain aspects of anxietylike behavior [Chen et al., 2006; Spencer et al., 2010]. However, when findings in mutant mice do not necessarily translate between strains of the same species [Phillips et al., 1999], it remains difficult to interpret how these particular findings—even for a knock-in of the human mutation—are directly relevant to the human condition.
In our meta-analysis, we performed tests for publication bias where a sufficient number of observations were available. However, in three out of the five SNPs used in meta-analyses and in the 12 additional SNPs also included in the gene-based analysis, we could not test for or rule out publication bias. This might lead to bias in the resulting meta-analytic data influencing the validity of the overall gene-based analysis. In two out of the five meta-analyses, we found significant between-study heterogeneities (meta-analysis of Val66-Met and 712A/G). The heterogeneity can be due to various clinical and methodological factors. For example, diagnostic criteria for MDD vary across studies (Table I). Variation in the accuracy and sensitivity of these approaches can lead to differences in the estimation of the effects [Glasziou and Sanders, 2002]. Differences in the study designs can also contribute to the heterogeneity between studies. For example, some studies used age and sex-matched controls [Ribeiro et al., 2007], while most other studies did not match by age and sex in recruiting non-depressed controls. Some studies recruited elderly or geriatric MDD participants [Taylor et al., 2008], while many other studies did not consider age as an inclusion criterion in recruiting. To handle the heterogeneity between the studies, we used random-effects meta-analyses when statistical analysis indicates the presence of heterogeneity.
Future gene-based analyses on increasingly large and well-characterized groups of people needs to consider pooling individuals based on alleles that confer dramatic reductions or complete absence of BDNF protein to test for effects of the most extreme variants in functional signaling of this conserved protein. But considering that MDD is a highly complex disorder and BDNF is part of redundant signaling network regulated by multiple negative feedback pathways, there is much less reason to expect a substantial main effect in population-level analyses [McClearn 2006; Plomin et al., 2010]. To be considered trustworthy, follow-up work probably needs to await the availability of sequence data from a very large number of MDD phenotyped subjects.
This study has some limitations. First, although the meta-analysis of Val66Met involves a large number of studies, the number of studies involved in the meta-analysis of other genetic variants in BDNF is relatively small, due to limited availability of published results. Moreover, we could only perform meta-analysis for five SNPs in BDNF. The association of the remaining 12 SNPs was based on the results from single studies. We expect that a more accurate estimation of the cumulative association of BDNF with MDD could be obtained when more studies become available. Second, the definition of MDD is not consistent across the studies for the meta- and gene-based analyses (Table I). Third, due to limited data, we did not stratify analysis by ethnicity and gender. A recent meta-analysis found no significant association of Val66Met in the total sample, nor in a stratified analysis by ethnicity (Asian vs. Caucasian), but did detect a significant effect of this genetic polymorphism in men, [Verhagen et al., 2010] perhaps due to the gender-related differences in the etiology of MDD. Future studies on the effects of genetic polymorphisms in BDNF should take into account gender and/or ethnicity differences, including planned analyses of sex by gene interactions and similar tests for empirically observed genetic stratification in the populations. Fourth, due to lack of consistently available information on comorbidities of MDD, we were unable to control for them in our analyses, which might influence the conclusions of this study.
In summary, we conducted a systematic literature search and performed meta- and gene-based analyses to assess the association of genetic variants in BDNF with major depressive disorder. Our updated meta-analysis and novel gene-based analysis did not detect any SNP in BDNF showing significant association with MDD. The gene-based analysis indicated that, based on current evidence from published studies, the cumulative effect of polymorphisms in BDNF is not significantly associated with MDD. Further investigation is warranted on the association between genetic polymorphisms in BDNF with MDD, particularly studies with larger sample size that resequence the gene to fully evaluate rare variants while taking into account potential interactions between gender and ethnicity.
ACKNOWLEDGMENTS
This research was supported by award number P50DA010075-16 from the National Institute on Drug Abuse. The content of this research is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute on Drug Abuse or the National Institute of Health.
REFERENCES
- American Psychiatric Association. Diagnostic and statistical manual of mental disorders. Washington, DC: American Psychiatric Association Press; 2000. [Google Scholar]
- Anttila S, Huuhka K, Huuhka M, Rontu R, Hurme M, Leinonen E, Lehtimaki T. Interaction between 5-HT1A and BDNF genotypes increases the risk of treatment-resistant depression. J Neural Transm. 2007;114:1065–1068. doi: 10.1007/s00702-007-0705-9. [DOI] [PubMed] [Google Scholar]
- Chen L, Lawlor DA, Lewis SJ, Yuan W, Abdollahi MR, Timpson NJ, Day IN, Ebrahim S, Smith GD, Shugart YY. Genetic association study of BDNF in depression: Finding from two cohort studies and a meta-analysis. Am J Med Genet Part B. 2008;147B:814–821. doi: 10.1002/ajmg.b.30686. [DOI] [PubMed] [Google Scholar]
- Chen L, Wang Y, Xiao H, Wang L, Wang C, Guo S, Zhao Y, Hua P, Liu W, Zhang N. The 712A/G polymorphism of brain-derived neurotrophic factor is associated with Parkinson’s disease but not major depressive disorder in a Chinese Han population. Biochem Biophys Res Commun. 2011;408:318–321. doi: 10.1016/j.bbrc.2011.04.030. [DOI] [PubMed] [Google Scholar]
- Chen ZY, Jing D, Bath KG, Ieraci A, Khan T, Siao CJ, Herrera DG, Toth M, Yang C, McEwen BS, et al. Genetic variant BDNF (Val66Met) polymorphism alters anxiety-related behavior. Science. 2006;314:140–143. doi: 10.1126/science.1129663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen ZY, Patel PD, Sant G, Meng CX, Teng KK, Hempstead BL, Lee FS. Variant brain-derived neurotrophic factor (BDNF) (Met66) alters the intra-cellular trafficking and activity-dependent secretion of wild-type BDNF in neurosecretory cells and cortical neurons. J Neurosci. 2004;24:4401–4411. doi: 10.1523/JNEUROSCI.0348-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chi MH, Chang HH, Lee SY, Lee IH, Gean PW, Yang YK, Lu RB, Chen PS. Brain derived neurotrophic factor gene polymorphism (Val66Met) and short-term antidepressant response in major depressive disorder. J Affect Disord. 2010;126:430–435. doi: 10.1016/j.jad.2010.07.006. [DOI] [PubMed] [Google Scholar]
- Chi MH, Lee SY, Chang HH, Yang YK, Lin E, Chen PS. Comparison of antidepressant efficacy-related SNPs among Taiwanese and four populations in the HapMap database. J Formos Med Assoc. 2011;110:478–482. doi: 10.1016/S0929-6646(11)60071-5. [DOI] [PubMed] [Google Scholar]
- Choi MJ, Kang RH, Lim SW, Oh KS, Lee MS. Brain-derived neurotrophic factor gene polymorphism (Val66Met) and citalopram response in major depressive disorder. Brain Res. 2006;1118:176–182. doi: 10.1016/j.brainres.2006.08.012. [DOI] [PubMed] [Google Scholar]
- Cole J, Weinberger DR, Mattay VS, Cheng X, Toga AW, Thompson PM, Powell-Smith G, Cohen-Woods S, Simmons A, McGuffin P, et al. No effect of 5HTTLPR or BDNF Val66Met polymorphism on hippocampal morphology in major depression. Genes Brain Behav. 2011;10:756–764. doi: 10.1111/j.1601-183X.2011.00714.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Egan MF, Kojima M, Callicott JH, Goldberg TE, Kolachana BS, Bertolino A, Zaitsev E, Gold B, Goldman D, Dean M, et al. The BDNF val66met polymorphism affects activity-dependent secretion of BDNF and human memory and hippocampal function. Cell. 2003;112:257–269. doi: 10.1016/s0092-8674(03)00035-7. [DOI] [PubMed] [Google Scholar]
- Egger M, Davey Smith G, Schneider M, Minder C. Bias in meta-analysis detected by a simple, graphical test. BMJ. 1997;315:629–634. doi: 10.1136/bmj.315.7109.629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Enoch MA, White KV, Waheed J, Goldman D. Neurophysiological and genetic distinctions between pure and comorbid anxiety disorders. Depress Anxiety. 2008;25:383–392. doi: 10.1002/da.20378. [DOI] [PubMed] [Google Scholar]
- Fisher RA. Statistical methods for research workers. xiii. Edinburgh: Oliver and Boyd; 1932. p. 1. l. [Google Scholar]
- Frodl T, Schule C, Schmitt G, Born C, Baghai T, Zill P, Bottlender R, Rupprecht R, Bondy B, Reiser M, et al. Association of the brain-derived neurotrophic factor Val66Met polymorphism with reduced hippocampal volumes in major depression. Arch Gen Psychiatry. 2007;64:410–416. doi: 10.1001/archpsyc.64.4.410. [DOI] [PubMed] [Google Scholar]
- Glasziou PP, Sanders SL. Investigating causes of heterogeneity in systematic reviews. Stat Med. 2002;21:1503–1511. doi: 10.1002/sim.1183. [DOI] [PubMed] [Google Scholar]
- Gratacos M, Soria V, Urretavizcaya M, Gonzalez JR, Crespo JM, Bayes M, de Cid R, Menchon JM, Vallejo J, Estivill X. A brain-derived neurotrophic factor (BDNF) haplotype is associated with antidepressant treatment outcome in mood disorders. Pharmacogenomics J. 2008;8:101–112. doi: 10.1038/sj.tpj.6500460. [DOI] [PubMed] [Google Scholar]
- Groves JO. Is it time to reassess the BDNF hypothesis of depression? Mol Psychiatry. 2007;12:1079–1088. doi: 10.1038/sj.mp.4002075. [DOI] [PubMed] [Google Scholar]
- Hartung J. A note on combining dependent tests of significance. Biometrical J. 1999;41:849–855. [Google Scholar]
- Hofer MM, Barde YA. Brain-derived neurotrophic factor prevents neuronal death in vivo. Nature. 1988;331:261–262. doi: 10.1038/331261a0. [DOI] [PubMed] [Google Scholar]
- Hong CJ, Huo SJ, Yen FC, Tung CL, Pan GM, Tsai SJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and mood disorders, age of onset and suicidal behavior. Neuropsychobiology. 2003;48:186–189. doi: 10.1159/000074636. [DOI] [PubMed] [Google Scholar]
- Hwang JP, Tsai SJ, Hong CJ, Yang CH, Lirng JF, Yang YM. The Val66Met polymorphism of the brain-derived neurotrophic-factor gene is associated with geriatric depression. Neurobiol Aging. 2006;27:1834–1837. doi: 10.1016/j.neurobiolaging.2005.10.013. [DOI] [PubMed] [Google Scholar]
- Iga J, Ueno S, Yamauchi K, Numata S, Tayoshi-Shibuya S, Kinouchi S, Nakataki M, Song H, Hokoishi K, Tanabe H, et al. The Val66Met polymorphism of the brain-derived neurotrophic factor gene is associated with psychotic feature and suicidal behavior in Japanese major depressive patients. Am J Med Genet Part B. 2007;144B:1003–1006. doi: 10.1002/ajmg.b.30520. [DOI] [PubMed] [Google Scholar]
- Jiang X, Xu K, Hoberman J, Tian F, Marko AJ, Waheed JF, Harris CR, Marini AM, Enoch MA, Lipsky RH. BDNF variation and mood disorders: a novel functional promoter polymorphism and Val66Met are associated with anxiety but have opposing effects. Neuropsychopharma-cology. 2005;30:1353–1361. doi: 10.1038/sj.npp.1300703. [DOI] [PubMed] [Google Scholar]
- Jones KR, Reichardt LF. Molecular cloning of a human gene that is a member of the nerve growth factor family. Proc Natl Acad Sci USA. 1990;87:8060–8064. doi: 10.1073/pnas.87.20.8060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanellopoulos D, Gunning FM, Morimoto SS, Hoptman MJ, Murphy CF, Kelly RE, Glatt C, Lim KO, Alexopoulos GS. Hippocampal volumes and the brain-derived neurotrophic factor val66met polymorphism in geriatric major depression. Am J Geriatr Psychiatry. 2011;19:13–22. doi: 10.1097/jgp.0b013e3181f61d62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kato M, Serretti A. Review and meta-analysis of antidepressant pharmacogenetic findings in major depressive disorder. Mol Psychiatry. 2010;15:473–500. doi: 10.1038/mp.2008.116. [DOI] [PubMed] [Google Scholar]
- Kessler RC, Berglund P, Demler O, Jin R, Koretz D, Merikangas KR, Rush AJ, Walters EE, Wang PS. The epidemiology of major depressive disorder: Results from the National Comorbidity Survey Replication (NCS-R) JAMA. 2003;289:3095–3105. doi: 10.1001/jama.289.23.3095. [DOI] [PubMed] [Google Scholar]
- Kim JM, Stewart R, Kim SW, Yang SJ, Shin IS, Kim YH, Yoon JS. BDNF genotype potentially modifying the association between incident stroke and depression. Neurobiol Aging. 2008;29:789–792. doi: 10.1016/j.neurobiolaging.2006.11.021. [DOI] [PubMed] [Google Scholar]
- Kim SJ, Cho SJ, Jang HM, Shin J, Park PW, Lee YJ, Cho IH, Choi JE, Lee HJ. Interaction between brain-derived neurotrophic factor Val66Met polymorphism and recent negative stressor in harm avoidance. Neuropsychobiology. 2010;61:19–26. doi: 10.1159/000258639. [DOI] [PubMed] [Google Scholar]
- Lewis CM, Ng MY, Butler AW, Cohen-Woods S, Uher R, Pirlo K, Weale ME, Schosser A, Paredes UM, Rivera M, et al. Genome-wide association study of major recurrent depression in the U.K population. Am J Psychiatry. 2010;167:949–957. doi: 10.1176/appi.ajp.2010.09091380. [DOI] [PubMed] [Google Scholar]
- Lopez-Leon S, Janssens AC, Gonzalez-Zuloeta Ladd AM, Del-Favero J, Claes SJ, Oostra BA, van Duijn CM. Meta-analyses of genetic studies on major depressive disorder. Mol Psychiatry. 2008;13:772–785. doi: 10.1038/sj.mp.4002088. [DOI] [PubMed] [Google Scholar]
- Maisonpierre PC, Le Beau MM, Espinosa R, III, Ip NY, Belluscio L, de la Monte SM, Squinto S, Furth ME, Yancopoulos GD. Human and rat brain-derived neurotrophic factor and neurotrophin-3: Gene structures, distributions, and chromosomal localizations. Genomics. 1991;10:558–568. doi: 10.1016/0888-7543(91)90436-i. [DOI] [PubMed] [Google Scholar]
- McClearn GE. Contextual genetics. Trends Genet. 2006;22:314–319. doi: 10.1016/j.tig.2006.04.005. [DOI] [PubMed] [Google Scholar]
- Muglia P, Tozzi F, Galwey NW, Francks C, Upmanyu R, Kong XQ, Antoniades A, Domenici E, Perry J, Rothen S, et al. Genome-wide association study of recurrent major depressive disorder in two European case–control cohorts. Mol Psychiatry. 2010;15:589–601. doi: 10.1038/mp.2008.131. [DOI] [PubMed] [Google Scholar]
- Neale BM, Sham PC. The future of association studies: Gene-based analysis and replication. Am J Hum Genet. 2004;75:353–362. doi: 10.1086/423901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nibuya M, Morinobu S, Duman RS. Regulation of BDNF and trkB mRNA in rat brain by chronic electroconvulsive seizure and antidepressant drug treatments. J Neurosci. 1995;15:7539–7547. doi: 10.1523/JNEUROSCI.15-11-07539.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oswald P, Del-Favero J, Massat I, Souery D, Claes S, Van Broeckhoven C, Mendlewicz J. Noimplication of brain-derived neurotrophic factor (BDNF) gene in unipolar affective disorder: Evidence from Belgian first and replication patient-control studies. Eur Neuropsychopharmacol. 2005;15:491–495. doi: 10.1016/j.euroneuro.2005.01.001. [DOI] [PubMed] [Google Scholar]
- Ozan E, Okur H, Eker C, Eker OD, Gonul AS, Akarsu N. The effect of depression, BDNF gene val66met polymorphism and gender on serum BDNF levels. Brain Res Bull. 2010;81:61–65. doi: 10.1016/j.brainresbull.2009.06.022. [DOI] [PubMed] [Google Scholar]
- Pae CU, Chiesa A, Porcelli S, Han C, Patkar AA, Lee SJ, Park MH, Serretti A, De Ronchi D. Influence of BDNF variants on diagnosis and response to treatment in patients with major depression, bipolar disorder, and schizophrenia. Neuropsychobiology. 2012;65:1–11. doi: 10.1159/000327605. [DOI] [PubMed] [Google Scholar]
- Pezawas L, Verchinski BA, Mattay VS, Callicott JH, Kolachana BS, Straub RE, Egan MF, Meyer-Lindenberg A, Weinberger DR. The brain-derived neurotrophic factor val66met polymorphism and variation in human cortical morphology. J Neurosci. 2004;24:10099–10102. doi: 10.1523/JNEUROSCI.2680-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Phillips TJ, Hen R, Crabbe JC. Complications associated with genetic background effects in research using knockout mice. Psychopharmacology (Berl) 1999;147:5–7. doi: 10.1007/s002130051128. [DOI] [PubMed] [Google Scholar]
- Plomin R, Haworth CM, Davis OS. Genetics of learning abilities and disabilities: Recent developments from the UK and possible directions for research in China 2008. Behav Genet. 2010;40:297–305. doi: 10.1007/s10519-010-9355-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quinn CR, Dobson-Stone C, Outhred T, Harris A, Kemp AH. The contribution of BDNF and 5-HTT polymorphisms and early life stress to the heterogeneity of major depressive disorder: A preliminary study. Aust N Z J Psychiatry. 2012;46:55–63. doi: 10.1177/0004867411430878. [DOI] [PubMed] [Google Scholar]
- Ribeiro L, Busnello JV, Cantor RM, Whelan F, Whittaker P, Deloukas P, Wong ML, Licinio J. The brain-derived neurotrophic factor rs6265 (Val66Met) polymorphism and depression in Mexican-Americans. Neuroreport. 2007;18:1291–1293. doi: 10.1097/WNR.0b013e328273bcb0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schumacher J, Jamra RA, Becker T, Ohlraun S, Klopp N, Binder EB, Schulze TG, Deschner M, Schmal C, Hofels S, et al. Evidence for a relationship between genetic variants at the brain-derived neurotrophic factor (BDNF) locus and major depression. Biol Psychiatry. 2005;58:307–314. doi: 10.1016/j.biopsych.2005.04.006. [DOI] [PubMed] [Google Scholar]
- Sheng X, Yang J. Truncated product methods for panel unit root tests. Oxf Bull Econ Stat. 2012 doi: 10.1111/j.1468-0084.2012.00705.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shi J, Potash JB, Knowles JA, Weissman MM, Coryell W, Scheftner WA, Lawson WB, DePaulo JR, Jr., Gejman PV, Sanders AR, et al. Genome-wide association study of recurrent early-onset major depressive disorder. Mol Psychiatry. 2011;16:193–201. doi: 10.1038/mp.2009.124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Simes RJ. An improved bonferroni procedure for multiple tests of significance. Biometrika. 1986;73:751–754. [Google Scholar]
- Smith MA, Makino S, Kvetnansky R, Post RM. Stress and gluco-corticoids affect the expression of brain-derived neurotrophic factor and neurotrophin-3 mRNAs in the hippocampus. J Neurosci. 1995;15:1768–1777. doi: 10.1523/JNEUROSCI.15-03-01768.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spencer JL, Waters EM, Milner TA, Lee FS, McEwen BS. BDNF variant Val66Met interacts with estrous cycle in the control of hippocampal function. Proc Natl Acad Sci USA. 2010;107:4395–4400. doi: 10.1073/pnas.0915105107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, de Geus EJ, Willemsen G, James MR, Smit JH, Zandbelt T, Arolt V, Baune BT, Blackwood D, Cichon S, et al. Genome-wide association for major depressive disorder: A possible role for the presynaptic protein piccolo. Mol Psychiatry. 2009;14:359–375. doi: 10.1038/mp.2008.125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sullivan PF, Neale MC, Kendler KS. Genetic epidemiology of major depression: Review and meta-analysis. Am J Psychiatry. 2000;157:1552–1562. doi: 10.1176/appi.ajp.157.10.1552. [DOI] [PubMed] [Google Scholar]
- Sun RF, Zhu YS, Kuang WJ, Liu Y, Li SB. The G-712A polymorphism of brain-derived neurotrophic factor is associated with major depression but not schizophrenia. Neurosci Lett. 2011;489:34–37. doi: 10.1016/j.neulet.2010.11.061. [DOI] [PubMed] [Google Scholar]
- Surtees PG, Wainwright NW, Willis-Owen SA, Sandhu MS, Luben R, Day NE, Flint J. No association between the BDNF Val66Met polymorphism and mood status in a non-clinical community sample of 7389 older adults. J Psychiatr Res. 2007;41:404–409. doi: 10.1016/j.jpsychires.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Taylor WD, Zuchner S, McQuoid DR, Steffens DC, Blazer DG, Krishnan KR. Social support in older individuals: The role of the BDNF Val66Met polymorphism. Am J Med Genet Part B. 2008;147B:1205–1212. doi: 10.1002/ajmg.b.30754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai SJ, Cheng CY, Yu YW, Chen TJ, Hong CJ. Association study of a brain-derived neurotrophic-factor genetic polymorphism and major depressive disorders, symptomatology, and antidepressant response. Am J Med Genet Part B. 2003;123B:19–22. doi: 10.1002/ajmg.b.20026. [DOI] [PubMed] [Google Scholar]
- Ustun TB, Ayuso-Mateos JL, Chatterji S, Mathers C, Murray CJ. Global burden of depressive disorders in the year 2000. Br J Psychiatry. 2004;184:386–392. doi: 10.1192/bjp.184.5.386. [DOI] [PubMed] [Google Scholar]
- Verhagen M, van der Meij A, van Deurzen PA, Janzing JG, Arias-Vasquez A, Buitelaar JK, Franke B. Meta-analysis of the BDNF Val66Met polymorphism in major depressive disorder: Effects of gender and ethnicity. Mol Psychiatry. 2010;15:260–271. doi: 10.1038/mp.2008.109. [DOI] [PubMed] [Google Scholar]
- Videbech P, Ravnkilde B. Hippocampal volume and depression: A meta-analysis of MRI studies. Am J Psychiatry. 2004;161:1957–1966. doi: 10.1176/appi.ajp.161.11.1957. [DOI] [PubMed] [Google Scholar]
- Wang H, Dong S, Xu H, Qian J, Yang J. Genetic variants in FTO associated with metabolic syndrome: A meta- and gene-based analysis. Mol Biol Rep. 2012a;39:5691–5698. doi: 10.1007/s11033-011-1377-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ashley-Koch A, Steffens DC, Krishnan KR, Taylor WD. Impact of BDNF Val66Met and 5-HTTLPR polymorphism variants on neural substrates related to sadness and executive function. Genes Brain Behav. 2012b;11:352–359. doi: 10.1111/j.1601-183X.2012.00764.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaykin DV, Zhivotovsky LA, Westfall PH, Weir BS. Truncated product method for combining P-values. Genet Epidemiol. 2002;22:170–185. doi: 10.1002/gepi.0042. [DOI] [PubMed] [Google Scholar]
- Zhang K, Yang C, Xu Y, Sun N, Yang H, Liu J, Xu Q, Shen Y. Genetic association of the interaction between the BDNF and GSK3B genes and major depressive disorder in a Chinese population. J Neural Transm. 2010;117:393–401. doi: 10.1007/s00702-009-0360-4. [DOI] [PubMed] [Google Scholar]
- Zou YF, Ye DQ, Feng XL, Su H, Pan FM, Liao FF. Meta-analysis of BDNF Val66Met polymorphism association with treatment response in patients with major depressive disorder. Eur Neuropsychopharmacol. 2010;20:535–544. doi: 10.1016/j.euroneuro.2009.12.005. [DOI] [PubMed] [Google Scholar]