Abstract
Nuclease sensitive element binding protein 1 (NSEP1) is a member of the EFIA/NSEP1/YB-1 family of DNA-binding proteins whose members share a cold shock domain; it has also been termed DNA-binding protein B and Y box binding protein-1 because of its recognition of transcriptional regulatory elements. In addition, NSEP1 functions in the translational regulation of renin, ferritin, and interleukin 2 transcripts, and our laboratory has reported that it plays a role in the biosynthesis of selenium-containing proteins. To test the functional importance of NSEP1 in murine embryonic development, we have utilized a clone of ES cells in which the NSEP1 gene had been disrupted by integration of a plasmid gene-trapping vector into the seventh exon. Injection of these cells into C57BL/6 blastocysts resulted in 11 high percentage chimeric mice; crosses to wild type C57BL/6 mice generated 82 F1 agouti mice, indicating germ line transmission of the ES cell clone, but genotyping showed no evidence of the disrupted allele in any of these agouti offspring even though spermatozoa from four of five tested mice contained the targeted allele. Embryos harvested after timed matings of chimeric male mice demonstrated only the wildtype allele in 27 embryos tested at E7.5, E12.5, and E18.5. These results suggest that gene targeting of NSEP1 induces a lethal phenotype in early embryos, due to either haploinsufficiency of NSEP1 or formation of a dominant negative form of the protein. In either case, these data indicate the functional importance of the NSEP1 gene in murine early embryonic development.
Keywords: nuclease sensitive element binding protein 1, gene disruption, embryonic, lethality, Y-box binding protein 1, DNA-binding protein B, RNA-binding proteins, selenium
Nuclease sensitive element binding protein 1 (NSEP1) (GenBank locus NM_004559; OMIM #154030) is a member of the EFIA/NSEP1/YB-1 family of DNA-binding proteins whose members share a highly conserved region of 100 amino acids highly homologous to the E. coli cold shock domain [Goldstein et al., 1990; Kudo et al., 1995]. This molecule was originally named DNA-binding protein B because it was first identified by screening a human placenta cDNA expression library for binding to double-stranded DNA fragments derived from enhancer and promoter regions [Sakura et al., 1988]. The molecule has also been termed Y-box binding protein-1 in recognition of its role as a transcription factor that recognizes the Y-box element in HLA class II gene promoters [Didier et al., 1988; Kohno et al., 2003]. NSEP1 also binds to regulatory sites in the leukosialin (CD43) promoter, human gamma-globin gene upstream regulatory region, and promoter regions of pathogenic viruses including HIV-1, HTLV-1, and polyoma virus JC [Horwitz et al., 1994; Swamynathan et al., 1998]. It plays a functionally significant role in the transcriptional regulation of the human genes encoding Fas, gelatinase A, collagen α1(I), and multidrug resistance 1 [Mertens et al., 1998; Ohga et al., 1998; Lasham et al., 2000; Norman et al., 2001].
In addition, NSEP1 protein binds to RNA and functions in the translational regulation of renin, ferritin, and interleukin 2 transcripts [Chenetal.,2000; Ashizuka etal.,2002; Skalweit et al., 2003]. The bifunctional activities of NSEP1 are typical of many other nucleic acid-binding proteins that serve as both transcriptional and translational regulators or even as metabolic enzymes [Nakagawa et al., 1995; Matsumoto and Wolffe, 1998; Kim and Dang, 2005].
Our laboratory has previously reported that NSEP1 also plays a role in the biosynthesis of selenium-containing proteins [Shen et al., 1998]. This small but important class of proteins, comprised of 25 selenoproteins in human cells [Kryukov et al., 2003], includes important antioxidants such as the glutathione peroxidase family [Chambers et al., 1986; Esworthy et al., 1991; Schuckelt et al., 1991; Chu et al., 1993]. Selenoproteins incorporate selenium by translational insertion of selenocysteine (SeCys) at a UGA codon [Sunde, 1990; Böck et al., 1991], which otherwise serves as a termination signal. Translation of this codon in eukaryotic selenoprotein mRNA depends upon a SeCys insertion sequence (SECIS) in the 3′-untranslated region [Berry et al., 1993; Shen et al., 1993]. The mammalian SeCys translation apparatus includes multiple proteins involved in SECIS recognition and binding. The best-established of these constituents are SECIS-binding protein 2 [Lesoon et al., 1997; Copeland and Driscoll, 1999; Copeland et al., 2000] and mSelB [Fagegaltier et al., 2000b; Tujebajeva et al., 2000], the mammalian homolog of the prokaryotic selenoprotein-specific translation factor SelB [Forchhammer et al., 1989].
NSEP1 contains RNA-binding elements including four characteristic arginine-rich motifs [Burd and Dreyfuss, 1994] and specifically binds to the SECIS element of human cellular glutathione peroxidase mRNA [Shen et al., 1998]. However, Krol’s group—which independently identified NSEP1 as a SECIS-binding protein by 3-hybrid screening—did not detect SECIS binding in electromobility shift assays [Fagegaltier et al., 2000a], so the protein’s activity in selenoprotein translation has remained controversial.
Our current study of NSEP1 gene targeting provides the first in vivo evidence that the protein plays an essential role in mammalian embryonic development.
MATERIALS AND METHODS
NSEP1 Gene Targeting
The German Gene Trap Consortium (http://www.genetrap.de/) provided a clone of murine 129/SvP ES cells in which the NSEP1 gene had been disrupted by integration of a plasmid gene-trapping vector [Hansen et al., 2003]. The size and location of the integrated vector was determined by PCR and sequencing. Injection of this clone into C57BL/6 blastocysts generated eleven high percentage chimeric mice, identified by coat color. The chimeras were then crossed to wildtype C57BL/6 mice to produce F1 offspring, including 82 agouti mice. PCR genotyping was carried out by analysis of DNA from mouse tail snips, using three pairs of primers:
-
primers targeting only the inserted vector
-
1
(pT1F) 5′-GTAAGTGAAGCGACCCGCATTGA
-
2
(pT1R) 5′-TCAAGAAGGCGATAGAAGGCGATG
-
1
-
primers targeting both genomic DNA and the inserted vector
-
3
(EX7F) 5′-CTCGCCAAAGACAGCCTAGAGAG
-
4
(lacZR) 5′-GGATTGACCGTAATGGGATAGGTC
-
3
-
primers targeting genomic DNA bracketing the inserted vector
-
3
(EX7F, above)
-
5
(IN7R) 5′-CAGAGGGCAAAAAGCAAGCAC.
-
3
Southern blotting was performed by standard techniques, using HindIII digestion of genomic DNA and hybridization with a 32P-labeled probe formed by PCR amplification of mouse genomic DNA with the following primers:
NDBPBKOF1, 5′-CGGTATCGCCGAAACTTCAATTA
NDBPBKOR1, 5′-CAACACTGTCTTCAGAGCACAGGA.
NSEP1-targeted chimeric males were sacrificed, and the epididymis and vas deferentia were harvested and minced with an 18-gauge syringe needle in HTF media (Chemicon). The sperm was concentrated by brief microcentrifugation in an Eppendorf tube and flash frozen in liquid nitrogen. DNA was prepared from these samples and other harvested organs using standard methods. All animals were maintained and used in accordance with the University of Massachusetts Animal Care and Use Committee.
Embryo Analysis
NSEP1 +/− males were mated with NSEP1 +/+ females, and embryos were flushed from the uterine horns of females at days 7.5, 12.5, and 18.5 post coitum. Genomic DNA was prepared by placing individual blastocycts into10 μl of NSPK buffer (20 nmM Tris (pH 8.0), 100 mM KCl, 4 mM MgCl2, 0.9% Triton-X 100, 0.9% NP-40, with 300 μg/ml Proteinase K) for 4 h at 60°C. One half of the reaction was used for genotyping by a standard PCR protocol.
RESULTS AND DISCUSSION
Murine embryonic stem cell clone W094B04, derived from the TBV2 (129/SvP) ES cell line, was obtained from the German Gene Trap Consortium (http://www.genetrap.de/). Its position in the NSEP1 gene was mapped by PCR amplification from the inserted gene trap plasmid and sequencing of the murine genomic flanking regions. As shown schematically in Figure 1, exon 7 of the NSEP1 gene was interrupted by insertion of the gene trapping vector pT1βgeo, which includes an upstream 3′ splice consensus sequence, a promoterless fusion gene formed from the beta-galactosidase (lacZ) reporter and the neomycin resistance selector, and signals for chain termination and polyadenylation [Hansen et al., 2003]. In the resulting transcript, endogenous splicing acceptor of exon 7 is being used, with transcription through the upstream region of exon 7 then through the targeting vector (T. Floss, German Gene Trap Consortium, personal communication). The vector fusion gene’s chain termination signal results in loss of all downstream NSEP1 sequences. The clone was expanded and cells injected into C57BL/6 blastocysts, generating eleven high percentage chimeric mice, identified by coat color. Estimated percent chimerism ranged from 50% to 95%.
Fig. 1.
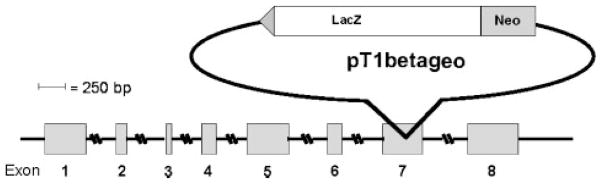
Schematic diagram of NSEP1 gene disruption by the exon-trapping vector pT1βgeo. The fusion gene insert is represented by the upstream splice acceptor site (triangle) and the open reading frames for beta galactosidase (LacZ) and neomycin resistance (Neo).
The 11 high percentage chimeras were crossed to wild type C57BL/6 mice and six of them generated 82 F1 agouti mice, indicating germ line transmission of the ES cell clone in these F1 founders. Genotyping of these F1 mice by PCR showed no evidence of the disrupted allele in any of the agouti offspring of the chimeric founders when tail snip DNA was amplified with primer pairs 1–2 or 3–5. Figure 2 illustrates the results from a representative set of F1 tail snip DNA samples tested with the internal plasmid primer pair 1–2, located entirely within the targeting vector. These primers amplified a 1499 bp product from DNA extracted from the W094B04 targeted cell line (lane 2) but not from control AB2.2 ES cells (lane 1) or any of the F1 tail snip DNA samples (lanes 3–11).
Fig. 2.

PCR genotyping of F1 agouti mice, using primers directed to the targeting vector. Lane 1, AB2.2 negative control; lane 2, W094B04 positive control; lanes 3–11, F1 agouti mouse tail snip DNA samples.
Figure 3 illustrates the results from a representative set of F1 tail snip DNA samples tested with the bracketing primer pair 3–5, which generates a 924 bp product from the wild type allele and an 8 kb product from the disrupted allele containing the targeting vector. The control ES cell line AB2.2 (lane 1) demonstrates the wild type allele; the W094B04 targeted ES cell line (lane 2) shows both the wildtype and targeted alleles; and the tail snip samples (lanes 2–11) show only the wild type, indicating lack of transmission of the targeted allele.
Fig. 3.

PCR genotyping of F1 agouti mice, using exon 7 and intron 7 primers that bracket the insertion. Lane 1, AB2.2 wildtype control; lane 2, W094B04 positive control; lanes 3–11, F1 agouti mouse tail snip DNA samples.
To confirm the presence of ES-derived germ cells the potential for transmission of the targeted NSEP1 allele, we performed PCR genotyping of spermatozoa, as well as somatic tissues, from five chimeric male founders, all of which had produced agouti F1 offspring. As shown in Figure 4, spermatozoa from four of five tested mice showed detectable levels of the targeted allele, as did their other tissues tested (not shown).
Fig. 4.

PCR genotyping of spermatozoa from chimeric founder males, using primers directed to the targeting vector (upper panel) and beta-actin (lower panel). Lanes 1–5, sperm DNA samples from individual chimeric founder mice; lane 6, template-free control.
In view of the absence of the targeted allele in live births, we sought to determine whether the targeted allele was present in embryos. PCR genotyping was performed on DNA extracted from embryos harvested at E7.5–E18.5 after timed matings of chimeric male mice to wild type C57BL/6 females. Figure 5 illustrates the results from a representative set of embryo DNA samples tested with the bracketing primer pair 3–5; all appeared homozygous for the wildtype NSEP1 allele. As shown in Table I, only the wildtype allele was present in all 27 embryos tested at E7.5, E12.5, and E18.5.
Fig. 5.

PCR genotyping of F1 E12.5 embryos, using exon 7 and intron 7 primers that bracket the insertion. Lane 1, AB2.2 wildtype control; lane 2, W094B04 positive control; lanes 3–12, embryo DNA samples.
TABLE I.
Genotyping of F1 Embryos for the Targeted NSEP1 Allele
| Day of development | Genotype
|
||
|---|---|---|---|
| Number tested | wt/wt | wt/KO | |
| E7.5 | 9 | 9 | 0 |
| E12.5 | 10 | 10 | 0 |
| E18.5 | 8 | 8 | 0 |
Embryos were harvested and DNA extracted as described in “Methods.” DNA samples were analyzed by PCR using the bracketing primer pair 3–5 for detection of the wild type (“wt”) and targeted (“KO”) alleles of the NSEP1 gene, as illustrated in Figure 4.
These results suggest that haploinsufficiency of NSEP1 induces a lethal phenotype in early embryos. Alternatively, the gene-targeting event could have created a dominant negative form of the protein. The gene trap vector inserted into the seventh exon of the gene, leaving intact the first six exons, which contain the RNA-binding cold shock domain [Toh et al., 1998], and eliminating all or most of the less-conserved seventh and eighth exons, which probably contain sites for protein–protein interactions. Thus, the protein product, a fusion of NSEP1 exons 1–6 with a reporter/selector fusion gene, is likely to bind to the SECIS. However, the absence of normal C-terminal domains and steric hindrance by the gene trap-encoded fusion region could prevent protein–protein interactions and thus result in a dominant negative effect. Another, less likely, possibility would be that NSEP1 gene disruption produced a functional defect in mature spermatozoa, either by diminished transcription of an essential gene with a critical Y-box promoter element or by inhibiting translation of an essential selenoprotein. Elongating spermatids express high levels of the selenoprotein thioredoxin–glutatione reductase [Su et al., 2005] and mature sperm utilize the selenoenzyme glutathione peroxidase 4 as the major structural protein of the mitochondrial mid-piece [Ursini et al., 1999].
In either case, the current data indicate the functional importance of the NSEP1 gene, which has heretofore been considered a controversial participant in selenoprotein translation and a transcription factor with many functions, none of which had been proven critical for survival of a cell or organism. The phenotype is consistent with, but hardly specific to, the embryonic lethal effect of disrupting translation by knockout of the Trsp gene encoding the selenocysteine carrier tRNA[Kumaraswamy et al., 2003]. Alternatively, or additionally, the lethal phenotype could result from decreased expression of a Y-box gene such as leukosialin, Fas, gelatinase A, collagen a1(I), or multidrug resistance 1 [Mertens et al., 1998; Ohga et al., 1998; Swamynathan et al., 1998; Lasham et al., 2000; Norman et al., 2001].
Acknowledgments
Grant sponsor: National Research Initiative of the USDA Cooperative State Research, Education and Extension Service; Grant number: 2001-35200-10692; Grant sponsor: John H. Pierce Pediatric Oncology Research Fund; Grant sponsor: Institute of Diabetes and Digestive and Kidney Diseases; Grant number: P30DK32520; Grant sponsor: National Cancer Institute; Grant numbers: R01CA95216, R01CA77735.
We thank Judith Gallant and Jane Carlson for technical assistance.
References
- Ashizuka M, Fukuda T, Nakamura T, Shirasuna K, Iwai K, Izumi H, Kohno K, Kuwano M, Uchiumi T. Novel translational control through an iron-responsive element by interaction of multifunctional protein YB-1 and IRP2. Mol Cell Biol. 2002;22:6375–6383. doi: 10.1128/MCB.22.18.6375-6383.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Berry MJ, Banu L, Harney JW, Larsen PR. Functional characterization of the eukaryotic SECIS elements which direct selenocysteine insertion at UGA codons. EMBO J. 1993;12:3315–3322. doi: 10.1002/j.1460-2075.1993.tb06001.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burd CG, Dreyfuss G. Conserved structures and diversity of functions of RNA-binding proteins. Science. 1994;265:615–621. doi: 10.1126/science.8036511. [DOI] [PubMed] [Google Scholar]
- Böck A, Forchhammer K, Heider J, Baron C. Selenoprotein synthesis: An expansion of the genetic code. Trends Biochem Sci. 1991;16:463–467. doi: 10.1016/0968-0004(91)90180-4. [DOI] [PubMed] [Google Scholar]
- Chambers I, Frampton J, Goldfarb P, Affara N, McBain W, Harrison PR. The structure of the mouse glutathione peroxidase gene: The selenocysteine in the active site is encoded by the termination codon, TGA. EMBO J. 1986;5:1221–1227. doi: 10.1002/j.1460-2075.1986.tb04350.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen CY, Gherzi R, Andersen JS, Gaietta G, Jürchott K, Royer HD, Mann M, Karin M. Nucleolin and YB-1 are required for JNK-mediated interleukin-2 mRNA stabilization during T-cell activation. Genes Dev. 2000;14:1236–1248. [PMC free article] [PubMed] [Google Scholar]
- Chu F-F, Doroshow JH, Esworthy RS. Expression, characterization, and tissue distribution of a new cellular selenium-dependent glutathione peroxidase, GSHPx-GI. J Biol Chem. 1993;268:2571–2576. [PubMed] [Google Scholar]
- Copeland PR, Driscoll DM. Purification, redox sensitivity, and RNA binding properties of SECIS-binding protein 2, a protein involved in selenoprotein biosynthesis. J Biol Chem. 1999;274:25447–25454. doi: 10.1074/jbc.274.36.25447. [DOI] [PubMed] [Google Scholar]
- Copeland PR, Fletcher JE, Carlson BA, Hatfield DL, Driscoll DM. A novel RNA binding protein, SBP2, is required for the translation of mammalian selenoprotein mRNAs. EMBO J. 2000;19:306–314. doi: 10.1093/emboj/19.2.306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Didier DK, Schiffenbauer J, Woulfe SL, Zacheis M, Schwartz BD. Characterization of the cDNA encoding a protein binding to the major histocompatibility complex class II Y box. Proc Natl Acad Sci U S A. 1988;85:7322–7326. doi: 10.1073/pnas.85.19.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Esworthy RS, Chu F-F, Paxton RJ, Akman S, Doroshow JH. Characterization and partial amino acid sequence of human plasma glutathione peroxidase. Arch Biochem Biophys. 1991;286:330–336. doi: 10.1016/0003-9861(91)90048-n. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Carbon P, Krol A. The selenocysteine insertion sequence binding protein SBP is different from the Y-box protein dbpB. Biochimie. 2000a;82:117–122. doi: 10.1016/s0300-9084(00)00192-9. [DOI] [PubMed] [Google Scholar]
- Fagegaltier D, Hubert N, Yamada K, Mizutani T, Carbon P, Krol A. Characterization of mSelB, a novel mammalian elongation factor for selenoprotein translation. EMBO J. 2000b;19:4796–4805. doi: 10.1093/emboj/19.17.4796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forchhammer K, Leinfelder W, Böck A. Identification of a novel translation factor necessary for the incorporation of selenocysteine into protein. Nature. 1989;342:453–456. doi: 10.1038/342453a0. [DOI] [PubMed] [Google Scholar]
- Goldstein J, Pollitt NS, Inouye M. Major cold shock protein of Escherichia coli. Proc Natl Acad Sci U S A. 1990;87:283–287. doi: 10.1073/pnas.87.1.283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen J, Floss T, Van Sloun P, Fuchtbauer EM, Vauti F, Arnold HH, Schnutgen F, Wurst W, von Melchner H, Ruiz P. A large-scale, gene-driven mutagenesis approach for the functional analysis of the mouse genome. Proc Natl Acad Sci USA. 2003;100:9918–9922. doi: 10.1073/pnas.1633296100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horwitz EM, Maloney KA, Ley TJ. A human protein containing a “cold shock” domain binds specifically to H-DNA upstream from the human gamma-globin genes. J Biol Chem. 1994;269:14130–14139. [PubMed] [Google Scholar]
- Kim JW, Dang CV. Multifaceted roles of glycolytic enzymes. Trends Biochem Sci. 2005;30:142–150. doi: 10.1016/j.tibs.2005.01.005. [DOI] [PubMed] [Google Scholar]
- Kohno K, Izumi H, Uchiumi T, Ashizuka M, Kuwano M. The pleiotropic functions of the Y-box-binding protein, YB-1. BioEssays. 2003;25:691–698. doi: 10.1002/bies.10300. [DOI] [PubMed] [Google Scholar]
- Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigo R, Gladyshev VN. Characterization of mammalian selenoproteomes. Science. 2003;300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- Kudo S, Mattei MG, Fukuda M. Characterization of the gene for dbpA, a family member of the nucleic- acid-binding proteins containing a cold-shock domain. Eur J Biochem. 1995;231:72–82. doi: 10.1111/j.1432-1033.1995.tb20672.x. [DOI] [PubMed] [Google Scholar]
- Kumaraswamy E, Carlson BA, Morgan F, Miyoshi K, Robinson GW, Su D, Wang S, Southon E, Tessarollo L, Lee BJ, Gladyshev VN, Hennighausen L, Hatfield DL. Selective removal of the selenocysteine tRNA [Ser]Sec gene (Trsp) in mouse mammary epithelium. Mol Cell Biol. 2003;23:1477–1488. doi: 10.1128/MCB.23.5.1477-1488.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lasham A, Lindridge E, Rudert F, Onrust R, Watson J. Regulation of the human fas promoter by YB-1, Purα and AP-1 transcription factors. Gene. 2000;252:1–13. doi: 10.1016/s0378-1119(00)00220-1. [DOI] [PubMed] [Google Scholar]
- Lesoon A, Mehta A, Singh R, Chisolm GM, Driscoll DM. An RNA-binding protein recognizes a mammalian selenocysteine insertion sequence element required for cotranslational incorporation of selenocysteine. Mol Cell Biol. 1997;17:1977–1985. doi: 10.1128/mcb.17.4.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto K, Wolffe AP. Gene regulation by Y-box proteins: Coupling control of transcription and translation. Trends Cell Biol. 1998;8:318–323. doi: 10.1016/s0962-8924(98)01300-2. [DOI] [PubMed] [Google Scholar]
- Mertens PR, Alfonso-Jaume MA, Steinmann K, Lovett DH. A synergistic interaction of transcription factors AP2 and YB-1 regulates gelatinase A enhancer-dependent transcription. J Biol Chem. 1998;273:32957–32965. doi: 10.1074/jbc.273.49.32957. [DOI] [PubMed] [Google Scholar]
- Nakagawa J, Waldner H, Meyer-Monard S, Hofsteenge J, Jenö P, Moroni C. AUH, a gene encoding an AU-specific RNA binding protein with intrinsic enoyl-CoA hydratase activity. Proc Natl Acad Sci U S A. 1995;92:2051–2055. doi: 10.1073/pnas.92.6.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Norman JT, Lindahl GE, Shakib K, En-Nia A, Yilmaz E, Mertens PR. The Y-box binding protein YB-1 suppresses collagen α1(I) gene transcription via an evolutionarily conserved regulatory element in the proximal promoter. J Biol Chem. 2001;276:29880–29890. doi: 10.1074/jbc.M103145200. [DOI] [PubMed] [Google Scholar]
- Ohga T, Uchiumi T, Makino Y, Koike K, Wada M, Kuwano M, Kohno K. Direct involvement of the Y-box binding protein YB-1 in genotoxic stress-induced activation of the human multidrug resistance 1 gene. J Biol Chem. 1998;273:5997–6000. doi: 10.1074/jbc.273.11.5997. [DOI] [PubMed] [Google Scholar]
- Sakura H, Maekawa T, Imamoto F, Yasuda K, Ishii S. Two human genes isolated by a novel method encode DNA-binding proteins containing a common region of homology. Gene. 1988;73:499–507. doi: 10.1016/0378-1119(88)90514-8. [DOI] [PubMed] [Google Scholar]
- Schuckelt R, Brigelius-Flohé R, Maiorino M, Roveri A, Reumkens J, Strassburger W, Ursini F, Wolf B, Flohé L. Phospholipid hydroperoxide glutathione peroxidase is a selenoenzyme distinct from the classical glutathione peroxidase as evident from cDNA and amino acid sequencing. Free Radic Res Commun. 1991;14:343–361. doi: 10.3109/10715769109093424. [DOI] [PubMed] [Google Scholar]
- Shen Q, Chu F-F, Newburger PE. Sequences in the 3′ untranslated region of the human cellular glutathione peroxidase gene are necessary and sufficient for seleno-cysteine incorporation at the UGA codon. J Biol Chem. 1993;268:11463–11469. [PubMed] [Google Scholar]
- Shen Q, Wu R, Leonard JL, Newburger PE. Identification and molecular cloning of a human seleno-cysteine insertion sequence-binding protein. A bifunctional role for DNA binding protein B. J Biol Chem. 1998;273:5443–5446. doi: 10.1074/jbc.273.10.5443. [DOI] [PubMed] [Google Scholar]
- Skalweit A, Doller A, Huth A, Kahne T, Persson PB, Thiele BJ. Posttranscriptional control of renin synthesis: Identification of proteins interacting with renin mRNA 3′-untranslated region. Circ Res. 2003;92:419–427. doi: 10.1161/01.RES.0000059300.67152.4E. [DOI] [PubMed] [Google Scholar]
- Su D, Novoselov SV, Sun QA, Moustafa ME, Zhou Y, Oko R, Hatfield DL, Gladyshev VN. Mammalian seleno-protein thioredoxin-glutathione reductase - Roles in disulfide bond formation and sperm maturation. J Biol Chem. 2005;280:26491–26498. doi: 10.1074/jbc.M503638200. [DOI] [PubMed] [Google Scholar]
- Sunde RA. Molecular biology of selenoproteins. Annu Rev Nutr. 1990;10:451–474. doi: 10.1146/annurev.nu.10.070190.002315. [DOI] [PubMed] [Google Scholar]
- Swamynathan SK, Nambiar A, Guntaka RV. Role of single-stranded DNA regions and Ybox proteins in transcriptional regulation of viral and cellular genes. FASEB J. 1998;12:515–522. doi: 10.1096/fasebj.12.7.515. [DOI] [PubMed] [Google Scholar]
- Toh S, Nakamura T, Ohga T, Koike K, Uchiumi T, Wada M, Kuwano M, Kohno K. Genomic organization of the human Y-box protein (YB-1) gene. Gene. 1998;206:93–97. doi: 10.1016/s0378-1119(97)00570-2. [DOI] [PubMed] [Google Scholar]
- Tujebajeva RM, Copeland PR, Xu X-M, Carlson BA, Harney JW, Driscoll DM, Hatfield DL, Berry MJ. Decoding apparatus for eukaryotic selenocyteine insertion. EMBO Rep. 2000;1:158–163. doi: 10.1093/embo-reports/kvd033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ursini F, Heim S, Kiess M, Maiorino M, Roveri A, Wissing J, Flohe L. Dual function of the selenoprotein PHGPx during sperm maturation. Science. 1999;285:1393–1396. doi: 10.1126/science.285.5432.1393. [DOI] [PubMed] [Google Scholar]


