Abstract
Non-coding RNAs fold into precise base-pairing patterns to carry out critical roles in genetic regulation and protein synthesis, but determining RNA structure remains difficult. Here, we show that coupling systematic mutagenesis with high-throughput chemical mapping enables accurate base-pair inference of domains from ribosomal RNA, ribozymes and riboswitches. For a six-RNA benchmark that has challenged previous chemical/computational methods, this ‘mutate-and-map’ strategy gives secondary structures that are in agreement with crystallography (helix error rates, 2%), including a blind test on a double-glycine riboswitch. Through modelling of partially ordered states, the method enables the first test of an interdomain helix-swap hypothesis for ligand-binding cooperativity in a glycine riboswitch. Finally, the data report on tertiary contacts within non-coding RNAs, and coupling to the Rosetta/FARFAR algorithm gives nucleotide-resolution three-dimensional models (helix root-mean-squared deviation, 5.7 Å) of an adenine riboswitch. These results establish a promising two-dimensional chemical strategy for inferring the secondary and tertiary structures that underlie non-coding RNA behaviour.
The transcriptomes of living cells and viruses continue to reveal novel classes of non-coding RNA (ncRNA) with critical functions in gene regulation, metabolism and pathogenesis1–3. The functional behaviours of these molecules are intimately tied to specific base-pairing patterns, and these patterns are challenging to identify using existing strategies based on phylogenetic analysis4,5, nuclear magnetic resonance (NMR)6,7, crystallography8–14, molecular rulers15,16 or functional mutation/rescue experiments17,18. A more facile approach to the characterization of RNA structure involves high-throughput chemical mapping at single-nucleotide resolution. This method is applicable to RNAs as large as the ribosome as well as entire viruses, both in vitro and in their cellular milieu19–21. Measurements of the accessibility of every nucleotide to solution chemical modification can guide or filter structural hypotheses from computational models22,23. Nevertheless, approximations in computational models and in correlating structure to chemical accessibility limit the inherent accuracy of this approach22–25.
This Article presents a strategy to expand the information content of chemical mapping by means of a two-dimensional ‘mutate-and-map’ methodology26. Here, sequence mutation acts as a second dimension in a manner analogous to initial perturbation steps in multidimensional NMR pulse sequences for structure determination7 or pump–probe experiments in other spectroscopic fields27. Based on elegant precedents in group I intron studies28,29, we reasoned that if one nucleotide involved in a base pair is mutated, its partner might become more exposed and thus be readily detectable by chemical mapping. In practice, some mutations might not lead to the desired ‘release’ of the pairing partners, and some mutations might produce larger perturbations, such as the unfolding of an entire helix. Nevertheless, if even a subset of the probed mutations leads to precise release of interacting nucleotides, the base-pairing pattern of the RNA could potentially be read out from this extensive data set. Indeed, our recent proof-of-concept studies have demonstrated systematic inference of Watson–Crick base pairs in a 20-base-pair DNA/RNA duplex26 and a 35-nucleotide RNA hairpin30. However, these artificial systems were designed to include single long helices and thus may not adequately represent natural, functional non-coding RNAs with many shorter helices, extensive non-canonical interactions and multiple solution states.
We therefore sought to apply the mutate-and-map strategy to a diverse set of non-coding RNAs with available crystal structures for some states and unknown structures for other states. The benchmark, which includes ribozymes, riboswitches and ribosomal RNA domains (Supplementary Table S1), is challenging; an earlier (one-dimensional) chemical/computational approach missed and mis-predicted ~20% of the benchmark’s helices24. We can report that our mutate-and-map strategy achieves 98% accuracy in inferring Watson–Crick base-pairing patterns and gives useful confidence estimates through bootstrap analysis. Furthermore, the method permits the generation and falsification of structural hypotheses about partially ordered RNA states, as highlighted by new results on a glycine-sensing riboswitch. We focus predominantly on the basic but unsolved problem of RNA secondary structure inference24,25,31 from biochemical data. Extensions of the method and advances in computational modelling may permit robust tertiary contact inference and three-dimensional models, and we present one such case as a proof-of-concept.
Results
Proof-of-concept for an adenine-binding riboswitch
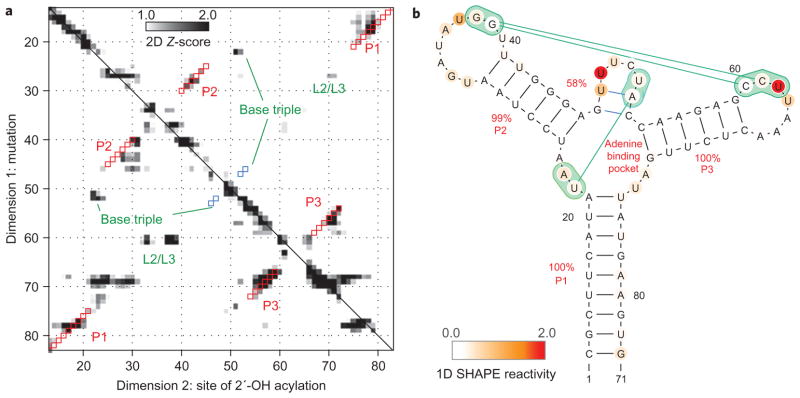
We first established the information content and accuracy of the strategy for the 71-nucleotide adenine-sensing add riboswitch from Vibrio vulnificus, which has been studied extensively6,17,32–35 and solved in the adenine-bound state using crystallography9. The add secondary structure is incorrectly modelled by the RNAstructure algorithm alone, but can be recovered through the inclusion of standard one-dimensional SHAPE (selective 2′ hydroxyl acylation with primer extension) data24; this RNA therefore serves as a well-characterized control. We prepared 71 variants of the RNA, mutating each base to its complement using high-throughput polymerase chain reaction (PCR) assembly, in vitro transcription and magnetic bead purification methods in a 96-well format, as discussed previously30. SHAPE data for all mutants were collected in a single afternoon. For a detailed view, Fig. 1a compares electrophoretic traces for the starting sequence and the C18G variant in the presence of 5 mM adenine. For a global view, the electropherograms for all constructs are given in Fig. 1b. As in earlier studies26,30, Z-scores (Fig. 2a, number of standard deviations from the mean accessibility; see Methods) highlight the most significant features of the data.
Figure 1. The mutate-and-map method gives an information-rich picture of RNA structure.
a, Mutating a nucleotide and mapping chemical accessibility reveals interactions in the three-dimensional structure of the RNA. The traces are for wild-type (blue) and C18G-mutated (red) variants of the adenine-binding domain of the add riboswitch. These 2′-OH acylation (SHAPE) data were read out by reverse transcription with fluorescently labelled primers and capillary electrophoresis; peaks (left to right) correspond to nucleotides from the 5′ to 3′ end of the RNA. Arrows mark exposure of the mutation site (C18) and of sequence-distant regions brought near this nucleotide by base-pairing (partner G78). b, Entire mutate-and-map data set across 71 single mutations, plotted in grey scale, revealing numerous elements of riboswitch structure. Dark features highlight: (I) the main diagonal stripe showing localized perturbations following C18G mutation; (II–IV) punctate features marking base pairs C18–G78, C26–G44 and C54–G72 in three different helices; (V–VII) more delocalized effects upon helix mutations G14C, G44C and G59C; (VIII) large-scale changes from C69G mutation due to secondary structure rearrangement; (IX) perturbations consistent with loss of adenine binding in A52U variant; (X) evidence for long-range tertiary contact between L2 and L3 upon mutation of C60 and C61 in L3; (XI) ‘symmetric’ mutations in L2 that affect L3; (XII) evidence for U22–A52 base pair in the adenine binding site.
Figure 2. Mutate-and-map data and secondary structure.
a, Strong features of mutate-and-map data isolated by Z-score analysis (number of standard deviations from mean at each residue). Squares show secondary structure model guided by mutate-and-map data (red, match to crystallographic Watson–Crick stems; blue, match to non-Watson–Crick stem). b, Secondary structure derived from incorporating Z-scores into the RNAstructure modelling algorithm; bootstrap confidence estimates given as red percentage values. Additional tertiary contacts inferred from a separate clustering analysis are given in green. Nucleotides are coloured according to SHAPE reactivity.
As with simpler model systems, the add mutate-and-map data demonstrate perturbations near mutation sites (marked at nucleotide 18 in Fig. 1a; distinct diagonal stripes (I) in Fig. 1b). More importantly, the data show numerous features corresponding to interacting pairs of sequence-separated nucleotides. For example, mutation C18G led to increased exposure of nucleotides 74–79, with the strongest effect at G78 (Fig. 1a; marked II in Fig. 1b). This observation strongly supports a C18–G78 base pair in the P1 helix. Such ‘punctate’ single-nucleotide-resolution base-pair features are also visible for the other helical stems of this RNA (for example, C26–G44 and C54–G72, marked III and IV in Fig. 1b). Several mutations led to more delocalized perturbations (V–VII, Fig. 1b; stems marked in Fig. 2a) due to disruption of multiple consecutive base pairs. Although not punctate, these features confirm interactions at helix-level resolution. Some single mutations produce larger-scale changes, reflecting shifts in the secondary structure (for example, C69G, VIII in Fig. 1b; see also U25A, G46C) or loss of adenine binding (for example, A52U, IX in Fig. 1b; see also Supplementary Figs S1 and S2). Interestingly, within each helix, some mutations gave strong signals, whereas others led to minimal perturbations (for example, the 3′ segment of P2 in Fig. 2a)26,30, underscoring the need to survey all possible mutations.
To assess the predictive power of these data for structure modelling, we applied the measured Z-scores as energetic bonuses in the RNAstructure secondary structure prediction algorithm. We further estimated confidence values for all inferred helices by bootstrapping the mutate-and-map data and repeating the secondary structure calculation36. As expected from the visual analysis above, the crystallographic secondary structure was robustly recovered (Fig. 2a, b), with ≥99% bootstrap values for helices P1, P2 and P3. An ‘extra’ two-base-pair helix was also found, with a weak bootstrap value (58%); these nucleotides are in fact base-paired in the add riboswitch crystallographic model9, but one pairing is a non-canonical Watson–Crick/Hoogsteen pair and part of a base triple. Together with additional mutate-and-map signals (X–XII, Fig. 1b), these data were sufficient for determining the global tertiary fold of the RNA, as described in the following.
A challenging benchmark of base pair inference
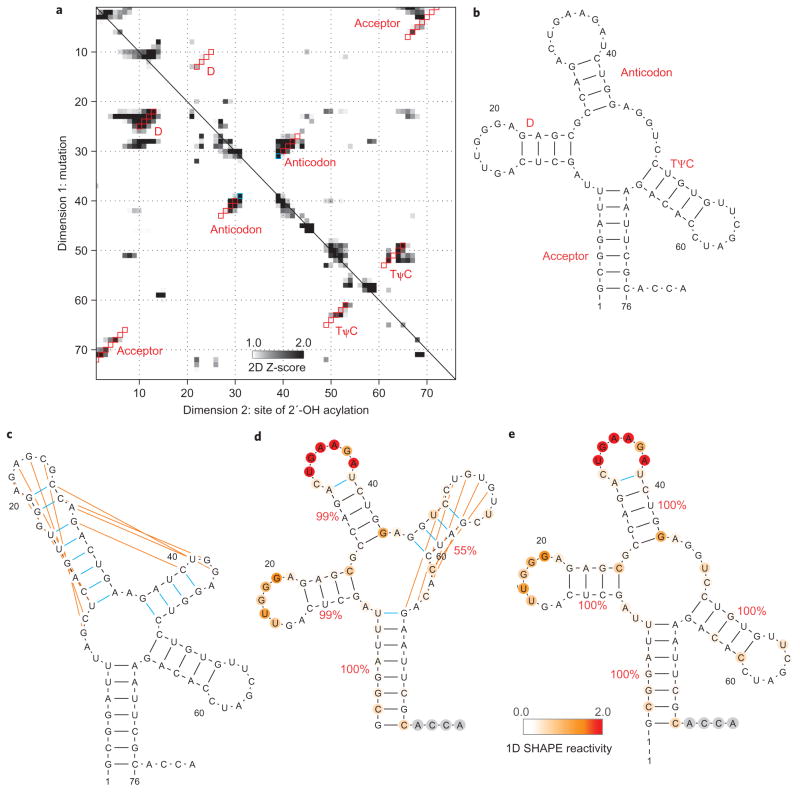
To complete our benchmark of the mutate-and-map strategy, we applied the method to RNAs for which base-pairing patterns have been more challenging to recover. The smallest of these, unmodified tRNAphe from Escherichia coli10, offers a simple illustration of the information content of the new method (Fig. 3a). The RNAstructure algorithm mispredicted two of the four helices of the tRNA ‘cloverleaf’ (the D and anticodon helices; cf. Fig. 3b, c). Inclusion of one-dimensional SHAPE reactivities corrected these errors, but introduced an additional error, mispredicting the TψC helix (Fig. 3d); protection of the loop of this helix by tertiary contacts renders its modelling uncertain with one-dimensional data alone. The mutate-and-map SHAPE data for this tRNA (Fig. 3a) gave clear signals for all four helices. Applying the two-dimensional mutate-and-map data set to RNAstructure corrected the inherent inaccuracies of the algorithm and recovered the entire four-helix secondary structure (>99% bootstrap values; Fig. 3e). One additional edge base pair was predicted for the anticodon arm; this and other fine-scale errors are discussed in the following.
Figure 3. Comparison of chemical/computational modelling approaches on tRNAphe.
a, Mutate-and-map Z-score data for tRNAphe from E. coli. b–e, Secondary structure models of this RNA from crystallography (b), the RNAstructure algorithm without data (c), calculations guided by one-dimensional SHAPE data (d) and calculations guided by the two-dimensional mutate-and-map data (e). Red squares (a) give Watson–Crick base pairs from the mutate-and-map model that match the crystallographic secondary structure. Blue squares (a) or lines (b–e) give model Watson–Crick base pairs not present in the crystallographic secondary structure. Orange lines give crystallographic Watson–Crick base pairs missed in each model. Helix confidence estimates from bootstrapping one-dimensional (d) or two-dimensional (e) data are given as red percentage values; nucleotides are coloured according to SHAPE reactivity.
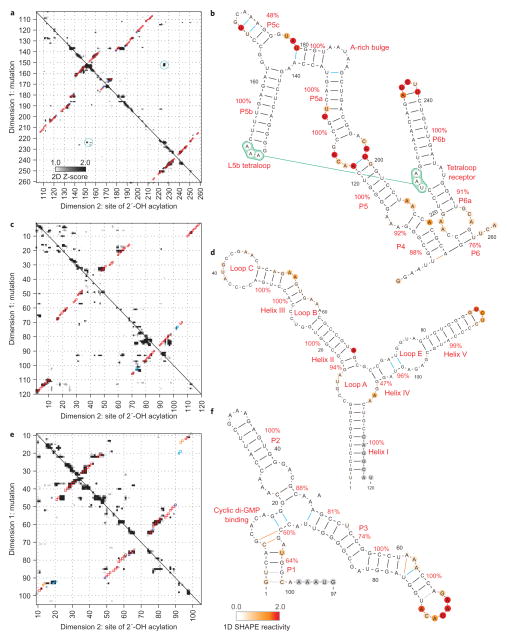
The remaining RNAs in our benchmark exceeded 100 nucleotides in length. As in the tRNAphe case, earlier chemical/computational methods assigned incorrect secondary structures to these sequences, but the mutate-and-map strategy led to accurate base-pairing patterns. The mutate-and-map data for a widely studied model RNA, the P4–P6 domain of the group I Tetrahymena ribozyme, gave visible features corresponding to all helices in the RNA14 (Fig. 4a) and led to correct recovery of the secondary structure (Fig. 4b). One of the helices, P5c, was correctly modelled but with a weak bootstrap value (48%); this low score is consistent with conformational fluctuations in P5c identified in previous biochemical and NMR studies37,38.
Figure 4. Accurate secondary structure models for non-coding RNAs.
a–f, Mutate-and-map Z-score data and resulting secondary structure models for the P4–P6 domain of the Tetrahymena group I ribozyme (a, b), the 5S ribosomal RNA from E. coli (c, d) and the domain that binds cyclic di-guanosine monophosphate from the V. cholerae VC1722 riboswitch (in the presence of 10 μM ligand; e, f). Colouring of squares (a, c, e) and lines and nucleotides (b, d, f) are as in Fig. 3.
As a more stringent test of the mutate-and-map strategy, we applied the method to the E. coli 5S ribosomal RNA, a notable problem case for earlier chemical/computational approaches22,39. In particular, the segments around the non-canonical loop E motif have been mispredicted in all previous studies, including the most recent (one-dimensional) SHAPE-directed approach24. By providing pairwise information on interacting nucleotides (Fig. 4c), the mutate-and-map method recovered the entire secondary structure with high confidence (>90%; Fig. 4d). One extra helix (blue in Fig. 4d) corresponds to a segment that in fact forms non-canonical base pairs within the loop E motif.
The ligand-binding domain of the cyclic di-GMP riboswitch from Vibrio cholerae provided an additional challenge; helix P1 of this RNA was not found in the original phylogenetic analysis18, but was instead later revealed by crystallography. Based on measurements in the presence of 10 μM ligand, the mutate-and-map strategy (Fig. 4e) recovered nearly the entire secondary structure (7 of 8 helices), including P1 (Fig. 4f).
Blind prediction for the glycine riboswitch
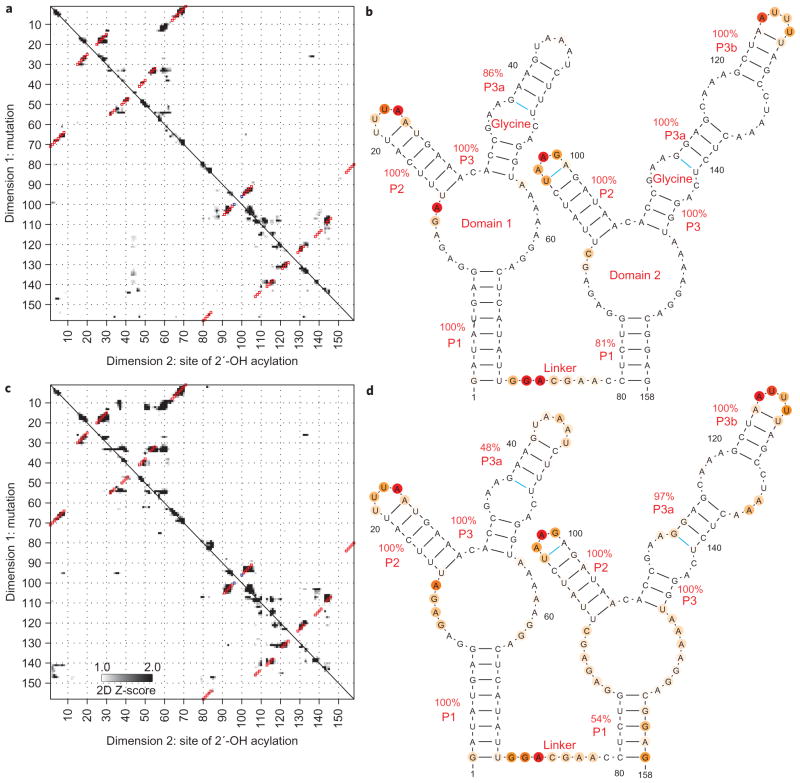
As a final rigorous test, we acquired mutate-and-map data for an RNA for which a crystallographic model was not available at the time of modelling: the ligand-binding domain of the glycine-binding riboswitch from Fusobacterium nucleatum40,41. The mutate-and-map data in the presence of 10 mM glycine gave a secondary structure with nine helices (Fig. 5a); the model agreed with the nine helices that were identified by phylogeny. The secondary structure was confirmed by a crystallographic model released at the time of the submission of this Article43.
Figure 5. Two states of a glycine-binding riboswitch.
a–d, Mutate-and-map Z-score data and resulting secondary structure models for the double-ligand-binding domain of the F. nucleatum glycine riboswitch with 10 mM glycine (a, b) and without glycine (c, d), indicating no inter-domain helix swap upon glycine binding. Colouring of squares (a, c) and lines and nucleotides (b, d) are as in Fig. 3.
Overall accuracy of secondary structure modelling
Overall, the mutate-and-map method demonstrated high accuracy in secondary structure inference for a benchmark of six diverse RNAs including 661 nucleotides in 42 helices (Table 1). As a baseline, an earlier method, using RNAstructure directed by one-dimensional SHAPE data, gave a false negative rate and false discovery rate of 17% and 21%, respectively, on this benchmark24. The mutate-and-map method recovered 41 of 42 helices, giving a sensitivity of 98% and a false negative rate of 2%, nearly an order of magnitude less than the previous method. The only missing helix was a two-base-pair helix in the cyclic diGMP riboswitch (see below). At a finer resolution, a small number (<6%) of base pairs in mutate-and-map calculated helices were either missed or added relative to the crystallographic secondary structures (1 and 11 of 197 base pairs, respectively; Supplementary Table S2). All these errors were either G–U or A–U pairs at the edges of otherwise correct helices (Figs 2– 5 and Table S2). Variation of the assumed coefficient of the two-dimensional Z-scores in the RNAstructure energy bonus or addition/subtraction of an offset did not improve the recovery at the level of base pairs or helices (data not shown).
Table 1.
Accuracy of RNA secondary structure models.
| RNA | Length★ | Number of helices†
|
||||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Cryst. | No data
|
1D
|
1D + 2D
|
2D
|
||||||
| TP | FP | TP | FP | TP | FP | TP | FP | |||
| Adenine riboswitch‡ | 71 | 3 | 2 | 3 | 3 | 0 (1) | 3 | 0 (1) | 3 | 0 (1) |
| tRNAphe | 76 | 4 | 2 | 3 | 3 | 1 | 4 | 0 | 4 | 0 |
| P4–P6 RNA | 158 | 11 | 10 | 1 | 9 | 2 | 9 | 2 | 11 | 0 |
| 5S rRNA | 118 | 7 | 1 | 9 | 6 | 3 | 7 | 0 (1) | 7 | 0 (1) |
| c-di-GMP riboswitch‡ | 80 | 8 | 6 | 2 | 6 | 2 | 7 | 1 | 7 | 1 |
| Glycine riboswitch‡ | 158 | 9 | 5 | 3 | 8 | 1 | 9 | 0 | 9 | 0 |
| Total | 661 | 42 | 26 | 21 | 35 | 9 (10) | 39 | 3 (5) | 41 | 1 (3) |
| False negative rate§ | 38.1% | 16.7% | 7.1% | 2.4% | ||||||
| False discovery rate|| | 44.7% | 20.4 (22.2)% | 7.1 (11.4)% | 2.3 (6.8)% | ||||||
Length of RNA in nucleotides.
Cryst, number of helices in crystallographic model; TP, true positive helices; FP, false positive helices; 1D, models using one-dimensional SHAPE chemical mapping data; 2D, models using mutate-and-map data. For FP, a helix was considered incorrect if its base pairs did not match the majority of base pairs in a crystallographic helix. Numbers in parentheses required that the matching crystallographic base pairs have Watson–Crick geometry.
Ligand-binding riboswitches were probed in the presence of small-molecule partners (5 mM adenine, 10 μM cyclic di-guanosine-monophosphate or 10 mM glycine). All experiments were carried out with 10 mM MgCl2, 50 mM Na-HEPES, pH 8.0.
False negative rate = (Cryst–TP)/TP.
False discovery rate = FP/(FP + TP). Numbers in parentheses count matches of model base pairs to non-Watson–Crick crystallographic base pairs as false discoveries.
In terms of the false discovery rate, the mutate-and-map method gave only three extra helices, all of which were the smallest possible length (2 bp). As discussed above, two of these extra helices in fact correspond to non-canonical stems observed in crystallographic models. The remaining false helix gave a weak bootstrap value (60%) and may correspond to a stem sampled in the ligand-free conformation of the cyclic diGMP riboswitch (see below). The overall positive predictive value was 93–98% depending on whether the non-canonical helices are counted as correct. The false discovery rate was 2–7%, nearly an order of magnitude less than the earlier one-dimensional SHAPE-directed method (21%). Somewhat surprisingly, using both the one-dimensional SHAPE data and two-dimensional mutate-and-map data gave slightly worse accuracy than using the two-dimensional data alone (false negative rate of 7% compared with 2%); this result may reflect inaccuracies in interpreting absolute SHAPE reactivity, as opposed to Z-score changes in reactivity induced by mutations. We conclude that secondary structures derived from the mutate-and-map method are accurate (~2% error rates) for structured non-coding RNAs.
Testing an ‘inter-domain helix swap’ hypothesis for glycine riboswitch cooperativity
Beyond recovering known information about non-coding RNA secondary structure, we sought to generate or falsify novel hypotheses that would be difficult to explore using standard structural methods. The three riboswitch ligand-binding domains for adenine, cyclic di-GMP and glycine provide interesting test cases because their ligand-free states will generally be partially ordered and thus difficult to crystallize. First, application of the mutate-and-map strategy indicated that the secondary structure of the add riboswitch ligand-binding domain remains the same in adenine-free and adenine-bound states (Supplementary Fig. S3), consistent with biophysical data from other approaches6,35. In contrast, mutate-and-map data indicate that the cyclic di-GMP riboswitch shifts its secondary structure near P1 on ligand binding (Supplementary Fig. S4). This shift is potentially involved in the mechanism of the riboswitch12,13,18 and may account for the weak phylogenetic signature of the P1 helix.
Among these ‘non-crystallographic’ targets, we were most interested in the glycine-binding riboswitch, which exhibits cooperative binding of two glycines to separate domains and is under intense investigation by several groups40–44. Analogous to the tense/relaxed equilibrium in the Monod–Wyman–Changeaux model for haemoglobin45, we considered that cooperativity might stem from an inter-domain helix swap. In this model, an alternative (‘tense’) secondary structure involving non-native interactions between the two domains would be rearranged upon glycine binding. Although one-dimensional mapping experiments (Fig. 5 and refs 40–42) show changes in the RNA following glycine binding, these data are consistent with a range of secondary structures and do not provide stringent tests of the inter-domain swap model. Furthermore, the model is not easily testable by crystallography43,44, which, if successful, is biased towards more structured conformations.
Application of the mutate-and-map strategy (Fig. 5b, d) gave a strong test of the hypothesis; the data in the absence of glycine gave the same domain-separated secondary structure as under conditions with glycine bound (Fig. 5a, c). Any changes in secondary structure for these constructs are thus either at edge base pairs or are negligible. We note that additional 5′ and 3′ flanking elements are likely to play critical roles in the modes of genetic regulation of these RNAs2; these longer segments are now under investigation.
Tertiary structure and cooperative fluctuations
The analysis described above focused on the first level of RNA structure, the Watson–Crick base-pairing pattern. Nevertheless, many non-coding RNAs use tertiary contacts and ordered junctions to position Watson–Crick helices into intricate three-dimensional structures. Qualitatively, we found evidence for numerous such tertiary interactions in the mutate-and-map data of these RNAs. For example, the add riboswitch is stabilized by tertiary interactions between the loops L2 (nucleotides 32–38) and L3 (nucleotides 60–66). In the presence of 5 mM adenine, mutations at G37 and G38 resulted in exposure of their partners C61 and C60 (X, Fig. 1b), and vice versa (XI, Fig. 1b; L2/L3 pseudoknot marked in Fig. 1b). Nevertheless, other mutations led to longer range effects (VIII, IX in Fig. 1b) due to cooperative unfolding of subdomains of tertiary structure or loss of adenine binding. For example, mutation of nucleotide A52 (VIII) gave chemical accessibilities that were different from the adenine-bound wild type RNA throughout the sequence, but consistent with the adenine-unbound state (Supplementary Fig. S1).
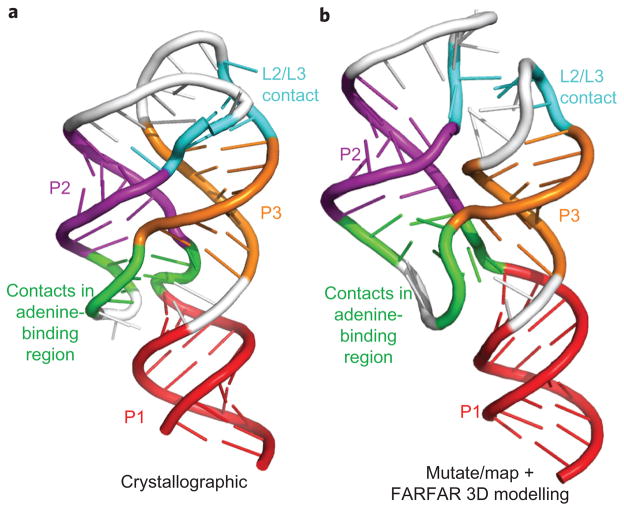
To extract tertiary base-pairing information, we could not use the RNAstructure method above, as it focuses on Watson–Crick base pairs. We therefore implemented filters enforcing strong, punctate signals and symmetry but not A–U, G–C or G–U pairing (see ref. 30, Methods and Supplementary Fig. S5). This analysis, independent of any computational models of RNA structure, recovered the majority of Watson–Crick helices in this benchmark. The analysis also recovered three tertiary contacts: the L2/L3 interaction of the add riboswitch (X and XI in Fig. 1b) and a U22–A52 base pair in the adenine binding pocket (XII, Fig. 1b); and a tetraloop/receptor interaction in the P4–P6 RNA (Fig. 4a, b). These features are accurate, but, in most test cases, their number is significantly less than the number of helices, precluding effective three-dimensional modelling. For the one case in which multiple tertiary contacts could be determined, the add adenine-sensing riboswitch, we carried out three-dimensional modelling using the FARFAR de novo assembly method. The algorithm gave a structural ensemble with helix RMSD of 5.7 Å and overall RMSD of 7.7 Å to the crystallographic model9 (Fig. 6a, b). This resolution is comparable to the average distance between nearest nucleotides (5.9 Å) and significantly better than model accuracy without mutate-and-map data (helix RMSD 8.9 Å; overall RMSD 16.9 Å) or values expected by chance (P < 1 × 10−3 for modelling a 71-nt RNA with secondary structure information46). These results on a favourable case suggest that rapid inference of three-dimensional structure for general RNAs might be achievable with other chemical probes that discriminate non-canonical interactions (for example, dimethyl sulfate30,47 for A-minor interactions) or more sophisticated methods for mining tertiary information or ligand-binding sites from mutate-and-map data. We note also that features corresponding to cooperative changes in chemical accessibility, while not reporting on specific tertiary contacts, can reveal ‘excited’ states in the folding landscapes of the RNA that may be functional48,49. We are making the information-rich data sets acquired for this Article publicly available in the Stanford RNA Mapping Database (http://rmdb.stanford.edu) to encourage the development of novel analysis methods to explore tertiary contact extraction and landscapes.
Figure 6. Three-dimensional modelling from mutate-and-map data.
a, b, Models of the (ligand-bound) adenine riboswitch derived from X-ray crystallography (a) and from the chemical/computational protocol introduced here (b). De novo modelling (using the Rosetta FARFAR algorithm) was carried out with secondary structure (P1, P2, P3) and tertiary contacts (L2/L3 and two contacts in adenine-binding region) inferred from solution mutate-and-map data. The mutate-and-map model agrees with the crystallographic model at nucleotide resolution (helix root-mean-squared deviation (RMSD) of 5.7 Å; overall RMSD of 7.7 Å).
Discussion
We have demonstrated that a mutate-and-map strategy permits the high-throughput inference of non-coding RNA base-pairing patterns. With error rates of ~2% and confidence estimates via bootstrapping, the method determines the secondary structures of riboswitch, ribosomal and ribozyme domains for which earlier chemical/computational approaches gave incorrect models. In addition to recovering known structures, the mutate-and-map data permit the rapid generation and falsification of hypotheses for structural rearrangements in three ligand-binding RNAs in partially ordered ligand-free states, including a cooperative glycine riboswitch with a poorly understood mechanism. Finally, the data yield rapid information on tertiary contacts of ncRNAs. Although not sufficient to yield crystallographic-quality structure models, in an adenine-sensing riboswitch, the data permit the modelling of the three-dimensional helix arrangement of the RNA at nucleotide resolution (5.7 Å). Further insights will come from detailed biophysical modelling of secondary and tertiary structure fluctuations induced by mutations; the public availability of these information-rich data sets should promote such analyses.
The mutate-and-map method only requires commercially available reagents, widely accessible capillary electrophoresis sequencers and freely available software. Further, each data set was acquired and analysed in a week or less. Therefore, for non-coding RNA domains up to ~300 nucleotides in length, the technology should be applicable as a front-line structural tool. The combined expense of the mutagenesis and mapping grows as the square of the RNA length. Thus, characterization of transcripts with thousands of nucleotides is presently challenging but may be facilitated by next-generation sequencing strategies50.
Expanding experimental technologies from one to multiple dimensions has transformed fields ranging from NMR to infrared spectroscopy. We propose that the mutate-and-map strategy will be analogously enabling for chemical mapping approaches, permitting the confident secondary structure determination and tertiary contact characterization of non-coding RNAs that are difficult or intractable for previous experimental methods. Applications to full-length RNA messages in vitro or in extract, to complex ribo-nucleoprotein systems, and even to full viral RNA genomes appear feasible and are exciting frontiers for this high-throughput approach.
Methods
Mutate-and-map experimental protocol and data processing
Preparation of DNA templates, in vitro transcription of RNAs, SHAPE chemical mapping, and capillary electrophoresis were carried out in a 96-well format, accelerated through the use of magnetic bead purification steps, as has been described previously26,30,51. Data were analysed with the HiTRACE52 software package, and Z-scores were computed in MATLAB. A complete protocol is given in the Supplementary Methods. Code for analysing mutate-and-map data is being made available as part of HiTRACE. Z-scores were used for secondary structure inference and sequence-independent feature analysis by single-linkage clustering, as described in the Supplementary Methods.
Secondary structure inference
The Fold executable of the RNAstructure package (v5.3) was used to infer secondary structures. The entire RNA sequences (Supplementary Table S1), including added flanking sequences, were used for all calculations. The flag ‘–T 297.15’ set the temperature to match our experimental conditions (24 °C). The flags ‘–sh’ and ‘–x’ were used to input (one-dimensional) SHAPE data files and (two-dimensional) base-pair energy bonuses (equal to −1 kcal mol−1 times the Z-scores), respectively. In the RNAstructure implementation, the pseudoenergies were applied to each nucleotide forming an edge base pair, and doubly applied to each nucleotide forming an internal base pair23. Additional flags ‘–xs’ and ‘–xo’ permitted scaling and offset of the Z-score bonuses, but default values of 1.0 kcal mol−1 and 0.0 kcal mol−1, respectively, were found to be optimal. For bootstrap analyses, mock SHAPE data replicates were generated by randomly choosing mutants with replacement36. The analysis is being made available as a server at http://rmdb.stanford.edu/structureserver. Secondary structure images were prepared in VARNA53.
Assessment of secondary structure accuracy
A crystallographic helix was considered correctly recovered if more than 50% of its base pairs were observed in a helix by the computational model. (In practice, 40 of 41 such helices in models based on mutate-and-map data retained all crystallographic base pairs.) Helix slips of ±1 were not considered correct (that is, the pairing (i, j) was not allowed to match the pairings (i, j–1) or (i, j + 1)).
Three-dimensional modelling with Rosetta
Three-dimensional models were acquired using the Fragment Assembly of RNA with Full Atom Refinement (FARFAR) methodology51 in the Rosetta framework. Briefly, ideal A-form helices were created for each helix greater than two base pairs in length in the modelled secondary structure. Then, remaining nucleotides were modelled by FARFAR as separate motifs interconnecting these ideal helices, generating up to 4,000 potential structures. Finally, these motif conformations were assembled in a Monte Carlo procedure, optimizing the FARNA low-resolution potential and tertiary constraint potentials defined by the sequence-independent clustering analysis of mutate-and-map data. Runs without mutate-and-map data used the one-dimensional SHAPE-directed secondary structure (which agrees with crystallography for the add riboswitch) and constraints only for the two-base-pair non-canonical helix (G47–C54, U48–A53). Explicit command lines and example files are given in the Supplementary Information. The code, as well as a Python job-setup script setup_rna_assembly_ jobs. py and documentation, are being incorporated into Rosetta release 3.4, which is freely available to academic users at http://www.rosettacommons.org. Before release, the code is available on request from the authors. The P-value for the add riboswitch was estimated by comparing the all-atom RMSD (7.7 Å) to the range expected by chance (13.5±1.8 Å), as described in ref. 46.
Supplementary Material
Acknowledgments
The authors thank A. Laederach and J. Lucks for comments on the manuscript and the authors of RNAstructure for making their source code freely available. This work was supported by the Burroughs-Wellcome Foundation (CASI to R.D.), the National Institutes of Health (T32 HG000044 to C.C.V.) and a Stanford Graduate Fellowship (to P.C.).
Footnotes
Author contributions
R.D. conceived and designed the experiments. W.K., C.C.V. and R.D. performed the experiments. C.C.V., P.C. and R.D. analysed the data. R.D. wrote the paper. All authors discussed the results and commented on the manuscript.
The authors declare no competing financial interests.
Supplementary information accompanies this paper at www.nature.com/naturechemistry.
Reprints and permission information is available online at http://www.nature.com/reprints.
References
- 1.Yanofsky C. The different roles of tryptophan transfer RNA in regulating trp operon expression in E. coli versus B. subtilis. Trends Genet. 2004;20:367–374. doi: 10.1016/j.tig.2004.06.007. [DOI] [PubMed] [Google Scholar]
- 2.Winkler WC, Breaker RR. Genetic control by metabolite-binding riboswitches. Chembiochem. 2003;4:1024–1032. doi: 10.1002/cbic.200300685. [DOI] [PubMed] [Google Scholar]
- 3.Zaratiegui M, Irvine DV, Martienssen RA. Noncoding RNAs and gene silencing. Cell. 2007;128:763–776. doi: 10.1016/j.cell.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 4.Levitt M. Detailed molecular model for transfer ribonucleic acid. Nature. 1969;224:759–763. doi: 10.1038/224759a0. [DOI] [PubMed] [Google Scholar]
- 5.Lehnert V, Jaeger L, Michel F, Westhof E. New loop-loop tertiary interactions in self-splicing introns of subgroup IC and ID: a complete 3D model of the Tetrahymena thermophila ribozyme. Chem Biol. 1996;3:993–1009. doi: 10.1016/s1074-5521(96)90166-0. [DOI] [PubMed] [Google Scholar]
- 6.Lee MK, Gal M, Frydman L, Varani G. Real-time multidimensional NMR follows RNA folding with second resolution. Proc Natl Acad Sci USA. 2010;107:9192–9197. doi: 10.1073/pnas.1001195107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wuthrich K. NMR studies of structure and function of biological macromolecules (Nobel lecture) Angew Chem Int Ed. 2003;42:3340–3363. doi: 10.1002/anie.200300595. [DOI] [PubMed] [Google Scholar]
- 8.Cruz JA, Westhof E. The dynamic landscapes of RNA architecture. Cell. 2009;136:604–609. doi: 10.1016/j.cell.2009.02.003. [DOI] [PubMed] [Google Scholar]
- 9.Serganov A, et al. Structural basis for discriminative regulation of gene expression by adenine- and guanine-sensing mRNAs. Chem Biol. 2004;11:1729–1741. doi: 10.1016/j.chembiol.2004.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Byrne RT, Konevega AL, Rodnina MV, Antson AA. The crystal structure of unmodified tRNAPhe from Escherichia coli. Nucleic Acids Res. 2010;38:4154–4162. doi: 10.1093/nar/gkq133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Correll CC, Freeborn B, Moore PB, Steitz TA. Metals, motifs, and recognition in the crystal structure of a 5S rRNA domain. Cell. 1997;91:705–712. doi: 10.1016/s0092-8674(00)80457-2. [DOI] [PubMed] [Google Scholar]
- 12.Smith KD, Lipchock SV, Livingston AL, Shanahan CA, Strobel SA. Structural and biochemical determinants of ligand binding by the c-di-GMP riboswitch. Biochemistry. 2010;49:7351–7359. doi: 10.1021/bi100671e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kulshina N, Baird NJ, Ferre-D’Amare AR. Recognition of the bacterial second messenger cyclic diguanylate by its cognate riboswitch. Nature Struct Mol Biol. 2009;16:1212–1217. doi: 10.1038/nsmb.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cate JH, et al. Crystal structure of a group I ribozyme domain: principles of RNA packing. Science. 1996;273:1678–1685. doi: 10.1126/science.273.5282.1678. [DOI] [PubMed] [Google Scholar]
- 15.Lemay JF, Penedo JC, Mulhbacher J, Lafontaine DA. Molecular basis of RNA-mediated gene regulation on the adenine riboswitch by single-molecule approaches. Methods Mol Biol. 2009;540:65–76. doi: 10.1007/978-1-59745-558-9_6. [DOI] [PubMed] [Google Scholar]
- 16.Das R, et al. Structural inference of native and partially folded RNA by high-throughput contact mapping. Proc Natl Acad Sci USA. 2008;105:4144–4149. doi: 10.1073/pnas.0709032105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mandal M, Breaker RR. Adenine riboswitches and gene activation by disruption of a transcription terminator. Nature Struct Mol Biol. 2004;11:29–35. doi: 10.1038/nsmb710. [DOI] [PubMed] [Google Scholar]
- 18.Sudarsan N, et al. Riboswitches in eubacteria sense the second messenger cyclic di-GMP. Science. 2008;321:411–413. doi: 10.1126/science.1159519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Culver GM, Noller HF. In vitro reconstitution of 30S ribosomal subunits using complete set of recombinant proteins. Methods Enzymol. 2000;318:446–460. doi: 10.1016/s0076-6879(00)18069-3. [DOI] [PubMed] [Google Scholar]
- 20.Adilakshmi T, Lease RA, Woodson SA. Hydroxyl radical footprinting in vivo: mapping macromolecular structures with synchrotron radiation. Nucleic Acids Res. 2006;34:e64. doi: 10.1093/nar/gkl291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wilkinson KA, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6:e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mathews DH, et al. Incorporating chemical modification constraints into a dynamic programming algorithm for prediction of RNA secondary structure. Proc Natl Acad Sci USA. 2004;101:7287–7292. doi: 10.1073/pnas.0401799101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Deigan KE, Li TW, Mathews DH, Weeks KM. Accurate SHAPE-directed RNA structure determination. Proc Natl Acad Sci USA. 2009;106:97–102. doi: 10.1073/pnas.0806929106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kladwang W, VanLang CC, Cordero P, Das R. Understanding the errors of SHAPE-directed RNA modeling. Biochemistry. 2011;50:8049–8056. doi: 10.1021/bi200524n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Quarrier S, Martin JS, Davis-Neulander L, Beauregard A, Laederach A. Evaluation of the information content of RNA structure mapping data for secondary structure prediction. RNA. 2010;16:1108–1117. doi: 10.1261/rna.1988510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kladwang W, Das R. A mutate-and-map strategy for inferring base pairs in structured nucleic acids: proof of concept on a DNA/RNA helix. Biochemistry. 2010;49:7414–7416. doi: 10.1021/bi101123g. [DOI] [PubMed] [Google Scholar]
- 27.Cho M. Coherent two-dimensional optical spectroscopy. Chem Rev. 2008;108:1331–1418. doi: 10.1021/cr078377b. [DOI] [PubMed] [Google Scholar]
- 28.Pyle AM, Murphy FL, Cech TR. RNA substrate binding site in the catalytic core of the Tetrahymena ribozyme. Nature. 1992;358:123–128. doi: 10.1038/358123a0. [DOI] [PubMed] [Google Scholar]
- 29.Duncan CD, Weeks KM. SHAPE analysis of long-range interactions reveals extensive and thermodynamically preferred misfolding in a fragile group I intron RNA. Biochemistry. 2008;47:8504–8513. doi: 10.1021/bi800207b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kladwang W, Cordero P, Das R. A mutate-and-map strategy accurately infers the base pairs of a 35-nucleotide model RNA. RNA. 2011;17:522–534. doi: 10.1261/rna.2516311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shapiro BA, Yingling YG, Kasprzak W, Bindewald E. Bridging the gap in RNA structure prediction. Curr Opin Struct Biol. 2007;17:157–165. doi: 10.1016/j.sbi.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 32.Lemay JF, Penedo JC, Tremblay R, Lilley DM, Lafontaine DA. Folding of the adenine riboswitch. Chem Biol. 2006;13:857–868. doi: 10.1016/j.chembiol.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 33.Rieder R, Lang K, Graber D, Micura R. Ligand-induced folding of the adenosine deaminase A-riboswitch and implications on riboswitch translational control. Chembiochem. 2007;8:896–902. doi: 10.1002/cbic.200700057. [DOI] [PubMed] [Google Scholar]
- 34.Lemay JF, et al. Comparative study between transcriptionally- and translationally-acting adenine riboswitches reveals key differences in riboswitch regulatory mechanisms. PLoS Genet. 2011;7:e1001278. doi: 10.1371/journal.pgen.1001278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Noeske J, et al. An intermolecular base triple as the basis of ligand specificity and affinity in the guanine- and adenine-sensing riboswitch RNAs. Proc Natl Acad Sci USA. 2005;102:1372–1377. doi: 10.1073/pnas.0406347102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Efron B, Tibshirani RJ. An Introduction to the Bootstrap. Chapman & Hall; 1998. [Google Scholar]
- 37.Wu M, Tinoco I., Jr RNA folding causes secondary structure rearrangement. Proc Natl Acad Sci USA. 1998;95:11555–11560. doi: 10.1073/pnas.95.20.11555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Vicens Q, Gooding AR, Laederach A, Cech TR. Local RNA structural changes induced by crystallization are revealed by SHAPE. RNA. 2007;13:536–548. doi: 10.1261/rna.400207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Leontis NB, Westhof E. The 5S rRNA loop E: chemical probing and phylogenetic data versus crystal structure. RNA. 1998;4:1134–1153. doi: 10.1017/s1355838298980566. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mandal M, et al. A glycine-dependent riboswitch that uses cooperative binding to control gene expression. Science. 2004;306:275–279. doi: 10.1126/science.1100829. [DOI] [PubMed] [Google Scholar]
- 41.Lipfert J, et al. Structural transitions and thermodynamics of a glycine-dependent riboswitch from Vibrio cholerae. J Mol Biol. 2007;365:1393–1406. doi: 10.1016/j.jmb.2006.10.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Kwon M, Strobel SA. Chemical basis of glycine riboswitch cooperativity. RNA. 2008;14:25–34. doi: 10.1261/rna.771608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Butler EB, Xiong Y, Wang J, Strobel SA. Structural basis of cooperative ligand binding by the glycine riboswitch. Chem Biol. 2011;18:293–298. doi: 10.1016/j.chembiol.2011.01.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang L, Serganov A, Patel DJ. Structural insights into ligand recognition by a sensing domain of the cooperative glycine riboswitch. Mol Cell. 2010;40:774–786. doi: 10.1016/j.molcel.2010.11.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Monod J, Wyman J, Changeux JP. On the nature of allosteric transitions: a plausible model. J Mol Biol. 1965;12:88–118. doi: 10.1016/s0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 46.Hajdin CE, Ding F, Dokholyan NV, Weeks KM. On the significance of an RNA tertiary structure prediction. RNA. 2010;16:1340–1349. doi: 10.1261/rna.1837410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tijerina P, Mohr S, Russell R. DMS footprinting of structured RNAs and RNA–protein complexes. Nature Protoc. 2007;2:2608–2623. doi: 10.1038/nprot.2007.380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nikolova EN, et al. Transient Hoogsteen base pairs in canonical duplex DNA. Nature. 2011;470:498–502. doi: 10.1038/nature09775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Korzhnev DM, Religa TL, Banachewicz W, Fersht AR, Kay LE. A transient and low-populated protein-folding intermediate at atomic resolution. Science. 2010;329:1312–1316. doi: 10.1126/science.1191723. [DOI] [PubMed] [Google Scholar]
- 50.Lucks JB, et al. Multiplexed RNA structure characterization with selective 2′-hydroxyl acylation analyzed by primer extension sequencing (SHAPE-Seq) Proc Natl Acad Sci USA. 2011;108:11063–11068. doi: 10.1073/pnas.1106501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Das R, Karanicolas J, Baker D. Atomic accuracy in predicting and designing noncanonical RNA structure. Nature Methods. 2010;7:291–294. doi: 10.1038/nmeth.1433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Yoon S, et al. HiTRACE: high-throughput robust analysis for capillary electrophoresis. Bioinformatics. 2011;27:1798–1805. doi: 10.1093/bioinformatics/btr277. [DOI] [PubMed] [Google Scholar]
- 53.Darty K, Denise A, Ponty Y. VARNA: Interactive drawing and editing of the RNA secondary structure. Bioinformatics. 2009;25:1974–1975. doi: 10.1093/bioinformatics/btp250. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.