Abstract
Developing an HIV-1 vaccine has been hampered by the inability of immunogens to induce broadly neutralizing antibodies (bnAbs) that protect against infection. Previously, we used knockin (KI) mice expressing a prototypical gp41-specific bnAb, 2F5, to demonstrate that immunological tolerance triggered by self-reactivity of the 2F5 H chain, impedes bnAb induction. Here, we generate KI models expressing H chains from two other HIV-1 Abs: 4E10 (another self-/polyreactive, α-gp41 bnAb) and 48d (an α-CD4 inducible, non-polyreactive Ab), and find a similar developmental blockade consistent with central B-cell deletion in 4E10, but not in 48d VH KI mice. Furthermore, in KI strains expressing the complete 2F5 and 4E10 Abs as BCRs, we find that residual splenic B-cells arrest at distinct developmental stages, yet exhibit uniformly low surface Ig densities, elevated basal activation, profoundly muted responses to BCR ligation, and when captured as hybridoma mAb lines, maintain their dual (gp41/lipid) affinities and capacities to neutralize HIV-1, establishing a key role for anergy in suppressing residual 2F5 or 4E10-expressing B-cells. Importantly, serum IgGs from naïve 2F5 and 4E10 KI strains selectively eliminate gp41 and lipid binding, respectively, suggesting B-cells expressing 2F5 or 4E10 as BCRs exhibit specificity for a distinct spectrum of host antigens, including selective interactions by 2F5 BCR+ B-cells (i.e., and not 4E10 BCR+ B-cells) with residues in self-antigen(s) that mimic its gp41 neutralization epitope.
Introduction
Key to the development of an effective HIV-1 vaccine will be the ability to elicit broadly neutralizing antibodies (bnAbs) (1). Experimental evidence supporting this notion comes from the robust protection conferred by bnAbs, either when passively transferred at physiological levels, preceding SHIV challenge in non-human primate (2-5) or more recently, in humanized mice, prior to HIV-1 infection (6). Furthermore, passive infusion of bnAbs significantly mitigates viral rebound in HIV-1 infected subjects for which HAART therapy has been disrupted (7). Unfortunately, bnAbs have not been elicited at significant levels by immunization, and arise only in a minority of HIV-1 infected subjects, typically years after transmission (8, 9). The recent isolation of a number of new bnAbs from chronically-infected subjects has expanded our understanding of vulnerable HIV-1 Env regions as vaccine targets (10).
One important Env vaccine target is the membrane proximal external region (MPER), a conserved, linear, and well-characterized region in gp41 (11) containing several bnAb epitopes, that is critical for HIV-1 fusion with target cells (12). Explanations why bnAbs are so poorly elicited include direct effects of topological constraints in Env that limit epitope accessibility (13-18) and indirect effects such as preferential elicitation of non-neutralizing responses by immunodominant epitopes on non-native Env conformations (19, 20). Additionally, unusual traits of bnAbs i.e., extraordinary somatic mutation, frequencies and HCDR3 lengths may disfavor their generation (9, 21). However, an additional hypothesis has been proposed for the dearth of bnAbs in infected patients and vaccinees: the depletion, inactivation, and/or modification of bnAb-producing B-cells via immunological tolerance (22). This hypothesis arose from the observation that two well studied MPER-specific bnAbs, 2F5 (23) and 4E10 (24), exhibited self-/polyeactivity in vitro (25), a finding which now extends to several other recently isolated bnAb lineages (10, 21).
The tolerizing processes of clonal deletion, anergy, and receptor editing have been extensively studied in mice expressing autoreactive B-cell receptors (BCR) (26-30) and we previously demonstrated that expression of the 2F5 H chain V(D)J rearrangement, either when paired to many endogenous L chains [2F5 VH knock-in (KI) mice], or with the 2F5 L chain [2F5 “complete” KI mice], results in profound deletion of bnAb-expressing immature B-cells in the BM (31, 32). Furthermore, residual 2F5 KI B-cells express reduced levels of IgM on their surface, suggesting their ability to signal through BCR is compromised (33). These results are consistent with the 2F5 H chain being sufficiently autoreactive to trigger profound B-cell tolerance in vivo, and are similar to other mice expressing highly self-/polyreactive BCRs (26, 34, 35).
4E10 and 2F5 have specificity for adjacent, yet distinct, epitopes in the gp41 MPER (36, 37), and in initial autoantigen-binding assays, exhibited a different spectrum of cross-reactivities(25): for example, 4E10 bound HIV-1 membrane lipids such as Cardiolipin (CL) with higher affinity than 2F5, but the latter preferentially cross-reacted with Centromere B. Furthermore, distinct conserved mammalian self-antigens that are avidly bound by the 2F5 and 4E10 mAbs have been identified: for 2F5, the XXX chemokine receptor (CMTM3) and kynureninase (KYNU); for 4E10, splice factor 3B subunit 3 (SF3B3) (38). KYNU and CMTM3 contain the complete (ELDKWA) and core (DKW) neutralizing epitope recognized by the 2F5 bnAb, whereas SF3B3 has no obvious homology to its neutralizing epitope NWFDIT (38). In addition, this study identified an uncommonly high degree of polyreactivity for 4E10, in contrast to a minimal amount for 2F5 (38). Collectively, these findings predict that B-cells carrying 2F5 and 4E10 bnAbs as BCRs should be tolerized in vivo by markedly different selecting agents.
In this study, we generate KI strains expressing the H chains of the 4E10 bnAb, and as a control, the HIV-1 non-neutralizing Ab 48d, and find that only KI mice expressing MPER bnAb H chains trigger a profound early BM developmental blockade, a finding consistent with the self-reactivity of both the 2F5 and 4E10 bnAb H chains being sufficient to trigger clonal B-cell deletion. We also compare KI mice expressing the full 2F5 and 4E10 bnAbs as BCR, and find that while clonal deletion and silencing profoundly suppress B-cells in both strains, their distinct residual B-cell numbers/distributions and serum IgG specificities indicate a distinct spectrum of self-antigens are involved in these processes, including selective cross-reactivity of 2F5 (but not 4E10) with self-antigen(s) that mimic its neutralization epitope .
Materials and Methods
Mice and flow cytometry
Female C57BL/6 RAG-1−/− and C57BL/6 IgHa mouse strains were purchased from The Jackson Laboratory. 4E10 VH+/+ and 48d VH+/+ KI mice were generated based on published methods for engineering the 2F5 VH+/+ KI strain (31), whereas 4E10 V +/+L and complete KI strains were constructed as previously described to generate 2F5 VL+/+ and complete KI strains, respectively (32); site-directed targeting verification and germline transmission of KI alleles are described in the results section and detailed in Figs. 1, and S1. WT IgHb/WT IgHa and 4E10 IgHb/WT IgHa F1 mice were generated by breeding C57BL/6 IgHa congenic mice with 4E10 IgHb and WT IgHb mice, respectively. All strains used in this study were housed in the MSRBII Vivarium at the Duke Human Vaccine Institute in a pathogen-free environment under AAALAC guidelines and all serum sample collection procedures were carried out in accordance with IACUC and the Duke University IBC-approved animal protocols.
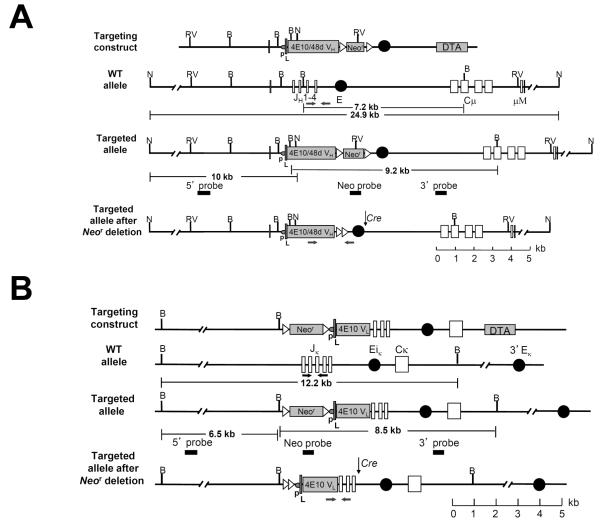
Figure 1. Targeted replacement of the mouse Igh and Igκ loci with the 4E10/48d VH(DH)JH and 4E10 VκJκ rearrangements, respectively.
(A-B) Site-directed strategies used to knock-in VH and VL regions, respectively, showing the Ig targeting constructs, targeted Ig alleles (shown before and after homologous recombination) and the targeted alleles after Cre-mediated neo cassette deletion. Restriction fragment sizes are indicated for both wild-type and targeted loci. B=BamHI. N=NdeI. RV=EcoRV. Genotyping primers are denoted by arrows. A. Genomic structure of the targeted Igh allele, showing the endogenous mouse JH cluster and Cμ region, as well as the 4E10/48d VH expression cassette, comprised of a J558 H10 family promoter (p), the H10 split leader sequence (L), and the rearranged 4E10/48d VH(DH)JH (4E10/48dVH) coding segment. B. Genomic structure of the targeted endogenous mouse Jκ cluster and Cκ region, as well as the 4E10 VL expression cassette, comprised of a VκOx1 promoter (p), the VκOx1 split leader sequence (L), and the rearranged 4E10 VκJκ (4E10VL) coding segment.
Flow cytometric analysis was performed as described previously (31). Briefly, single-cell suspensions from BM, spleen, mesenteric lymph node and peritoneal cavity lavage were isolated from 6-12-week-old naïve 4E10 and 2F5 KI strains and, for comparison, WT (C57BL/6) littermates. 106 cells were suspended in FACS buffer containing 1× PBS (pH 7.2), 3% FBS (Sigma-Aldrich), and 0.01% sodium azide, and B cells were stained with premixed combinations of fluorochrome-labeled mAbs at titration determined optimal concentrations, and total B cells were gated as singlet, live, CD19+, and/or B220+ lymphocytes. All Abs were from BD unless otherwise stated. Primary labeled mAbs used were: Pacific Blue, APC, or Texas Red-conjugated α-B220 (clone RA3-6B2), PE-Cy7 α-CD19, FITC-conjugated α-IgD (clone 11-26), FITC, APC or PE-Cy7-conjugated α-IgM (clone 15F9), PE-conjugated α-CD21, PE-Cy7-labeled α-CD23 (eBiosciences), APC-conjugated α-CD93 (eBiosciences), FITC conjugated α-CD43, PE-conjugated α-BP-1, APC-labeled α-HSA, PE-conjugated α-kappa, and FITC-conjugated α-lambda1-3. Depending on the experiment, either Propidium Iodide (PI) or v-amine live/dead violet dye (Invitrogen) was used to exclude dead cells, and for secondary staining, Texas-Red-conjugated Streptavadin. All FACS analysis was performed using a BD LSRII flow cytometer and data was acquired and analyzed using FACSDiva (BD) and FlowJo (Tree Star) software, respectively.
ELISA and Luminex analysis of serum Abs
Serum samples were collected from naïve 6-12 wk mice, and total serum Ab concentrations of all subclasses were determined by Luminex assay using a MILLIPLEX Mouse Immunoglobulin Isotyping kit (Millipore) and a Bio-Rad Luminex Bead Array Reader. Quantitative measurements of serum IgM and IgG-specific binding to MPER epitope peptides SP62 (containing the 2F5 nominal MPER epitope) and MPR.03 (containing both 2F5 and 4E10 nominal MPER epitopes) were used in 2F5 and 4E10 KI strains, respectively, using 10-point serum dilution curves, based on previously-published methods(31, 32). Plates were coated either directly with peptide (for 2F5-based assays), or with streptavidin followed by MPR.03-biotin (for 4E10-based assays). Binding Abs were detected using AP-conjugated goat anti-mouse μ or γHC-specific reagents (both from Southern Biotech) and plates were developed using AttoPhos Fluorescent Substrate (Promega). Serum Ab endpoint titers, rather than concentrations were used, as we have done previously, in order to more reliably detect low MPER and CL titers in naïve (unimmunized) 2F5 KI strains (32), and were calculated as the reciprocal of the highest serum dilution used where >3 background binding fluorescence were still observed.
Analysis of proximal and distal BCR signaling
Phosphotyrosine protein immunoblotting assays were performed as previously described (39). Briefly, 5×106 splenic B cells were isolated from WT or 4E10 complete KI mice by negative depletion of CD43+ cells using anti-mouse CD43 MACS beads (Miltenyi Biotech), and B cell purity (>95%) was determined by flow cytometry. Purified splenic B cells were incubated at 37°C in DMEM media (Invitrogen) containing 10% FBS (Hyclone) and stimulated with 20μg/ml anti-IgM F(ab’)2 (Southern Biotech) for various times. Whole cell lysates were prepared by lysing cell pellets in lysis buffer containing 1% MP-40, 1mM EDTA, PhosSTOP and complete EDTA-free protease inhibitor (Roche) and insoluble cellular debris was removed by centrifugation. Cell extracts were fractionated by reducing SDS-PAGE gel (Invitrogen) and transferred to PVDF membranes using Xcell II blot module (Invitrogen). Protein phosphotyrosine was detected by antibody clone 4G10 (Millipore) followed by HRP labeled goat α-mouse IgG2b Ab (Jackson ImmunoResearch). The same blot was stripped by Restore Western Blot Stripping Buffer (Thermo Scientific) and re-probed with α-Actin Ab (Sigma-Aldrich) followed by HRP labeled goat anti-mouse IgG Ab (Jackson ImmunoResearch). Blots were developed using the SuperSignal West Pico Substrate (Thermo Scientific Pierce).
Calcium flux assays were performed using fluo-4, following the manufacturer’s instructions (Invitrogen). Briefly, splenic cells were stained with Pacific Blue-labeled α-B220 and APC-labeled α-CD93, washed once with staining buffer (PBS containing 5% FCS, 2mM CaCl2 and 2mM MgCl2) and resuspended in staining buffer at a concentration of 107 cell/ml. Fluo-4 working solution was prepared by mixing Fluo-4 stock solution (1mM) with Plutonic solution (10% V/V) at a ratio 1:1. 1ml splenic cells were loaded with 2μl fluo-4 working solution and incubated in 37°C water bath for 30 minutes followed by washing with staining buffer twice. Cells were incubated at 37°C for 5 minutes before activated by 20, 5 or 1μg/ml anti-IgM F(ab’)2 (Southern Biotech) or 2 μg/ml ionomycin (Sigma) as a positive control, for 3 minutes. Data was acquired on BD LSRII and analyzed using Flowjo software.
For CFSE studies, splenocytes were resuspended in HBSS with 0.1% BSA at 107cell/ml and incubated with 5 μM of carboxyfluorescein diacetate succinimidyl ester (CFSE, Life Technologies) at 37°C for 10 minutes and immediately quenched with 10 vol of RPMI+10% FCS and washed 3 times with RPMI+10% FCS. Cells were then stimulated in vitro with 20, 5 or 1 μg/ml α-IgM F(ab)’2 for 5 days, stained with APC-labeled α-B220, and analyzed using LSRII (BD Biosciences). Data analysis was performed using the FlowJo software (Tree Star) and dead cells (PI+) were excluded from analysis.
BrdU labeling
Mice were injected i.p. with 200μl of 3mg/ml Bromodeoxyuridine (BrdU, BD Pharmingen) in PBS every 12hrs for 4 days. Spleen cells were then harvested and stained with PE-TxRed-labeled α-B220 (BD) and APC-labeled α-CD93 (ebioscience), fixed, permeabilized, and incorporated BrdU was detected with FITC-labeled α-BrdU Ab, using the FITC BrdU Flow Kit (BD Pharmingen) all according to the manufacturer’s protocol. FACS analysis was performed using a BD LSRII flow cytometer and data was acquired and analyzed using FACSDiva (BD) and FlowJo (Tree Star) software, respectively.
Hybridoma generation and analysis
Splenocytes from 4E10 or 2F5 complete KI mice were electrofused with NS0 Bcl-2 myeloma partner cells based on previously described methods (32). Briefly, after electrofusion, cells were seeded into 96-well plates at 1×103 B-cells/well in DMEM medium containing 15% FCS and Hypoxanthine-aminopterin-thymidine media supplement (Sigma-Aldrich). After incubation at 37°C, 8% CO2 for 2 weeks, plates were screened under a microscope for cell growth and culture supernatants from wells with cell colonies were assayed for Ab production, MPER/cardiolipin binding as well as the ability to neutralize HIV-1 virus. Ab production was determined by ELISA to measure total IgM, IgG or IgA using goat anti-mouse Ig (H+L) (Southern Biotech) as capture antibody and horseradish peroxidase (HRP)-conjugated goat anti-mouse IgM, IgG and IgA (Bethyl laboratories) as detection antibody, respectively. SureBlue TMB HRP substrate (KPL) was added for 5 min after which the reaction was stopped by 1N HCL and measured at 450nm. MPER and cardiolipin binding were determined by ELISA using 2F5 nominal MPER epitope peptide SP62 or biotin-labeled 4E10 nominal MPER epitope peptide MPR.03 binding to plated coated streptavidin and cardiolipin as target antigens and a mixture of HRP labeled anti-mouse IgM+IgG+IgA as detection antibodies. HIV-1 neutralization ability was determined by the TZM-bl pseudovirus infectivity assay using bnAbs-sensitive HIV-1 strain MN.3 as previously described (31, 32).
Certain randomly selected clones, including both MPER+ and MPER− clones, were subcloned at least one more time until cell lines were deemed to be monoclonal. For certain subclones, purified mAbs were purified from supernatants, and quantitative ELISAs to measure MPER, NIH-3T3 cytoplasmic/nuclear antigen, and cardiolipin reactivities of purified mAbs were performed, also as previously described (32). Additionally IgV regions from mAb lines were cloned/sequenced based on published methods (32) using either leader peptide-specific forward primers in the 4E10 or 2F5 KI expression cassette or degenerate forward HC or LC-specific forward primers (40), in combination with reverse C region-specific degenerate primers.
Results
Generation and characterization of 4E10 and 48d VH KI mice
Previously, we used knockin (KI) mice expressing the H chain VH(DH)JH rearrangement of a prototypical bnAb, the HIV-1 gp41-specific 2F5 Ab, to demonstrate a profound developmental blockade in the BM, either when paired to many endogenous L chains [2F5 VH KI mice], or with the 2F5 L chain [2F5 VH × VL “complete” KI mice], consistent with 2F5 H chain self-reactivity triggering profound deletion of immature B-cells in the BM, and thus impeding bnAb induction (31, 32). To explore if HC self-reactivity extends to other gp41-specific bnAbs, and to formally exclude that may be a spurious effect of expressing chimeric H chains (i.e. human V+murine C regions) in vivo, we generated KI mice expressing the somatically-mutated VH(D)JH rearrangements from two other well-characterized human HIV-1 mAbs: 4E10 (24), another self-/polyreactive MPER-specific bnAb with an unusually long H chain complementarity determining region (HCDR3), and, for contrast, 48d, a mAb specific for the HIV-1 envelope co-receptor CCR5-binding site (41), with an unremarkable HCDR3 (i.e., of average length and hydrophobicity), and no in vitro polyreactivity (M. Alam, unpublished results). 4E10 and 48d VH KI mice were constructed using the same targeting constructs and strategies previously used to generate 2F5 VH KI mice (31, 32), and in order to mimic their expected locations as primary rearrangement events, involve their site-directed replacement of the murine Igh JH1-4 segments (Fig. 1A), thus allowing all physiologically-relevant H chain diversification processes (including CSR, somatic hypermutation, and VH replacement) to be retained. Several independent 4E10 and 48d VH KI ES clones harboring expected homologous replacements were confirmed by Southern blot (Figs. S1A, B) and homozygous offspring bearing germline transmission of 4E10 and 48d VH(DH)JH rearrangements were identified by PCR (Figs. S1C, D).
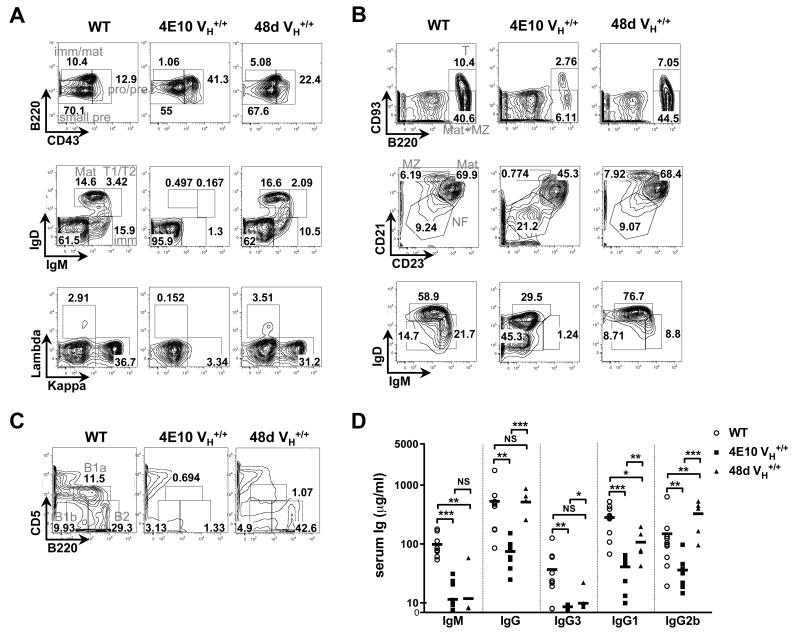
The effects of the targeted 4E10 and 48d VH rearrangements on B-cell development were then evaluated by comparing BM, splenic, and peritoneal B-cell compartments in homozygous 4E10 and 48d VH KI mice to those in C57BL/6 littermates (Fig. 2). Indeed, 4E10 VH KI mice had a major blockade at the small pre-B to immature B transition (Fig. 2A), as well as residual splenic B-cell populations with reduced surface Ig densities (Fig. 2B); phenotypes previously reported in 2F5 VH KI mice (31). In contrast, 48d VH KI mice exhibited BM and splenic B-cell subset frequencies as well as B-cell surface Ig densities comparable to WT (C57BL/6) littermates (Figs. 2A, B), demonstrating that the chimeric 48d H chain functions optimally in supporting B-cell development. Interestingly, naïve 4E10 and 48d VH KI mice both had significantly reduced peritoneal B-1a subsets(Fig. 2C), as well as reduced levels of serum isotypes predominantly associated with natural Ab production by innate B-cells i.e. IgM and IgG3 (Fig. 2D), results consistent with the notion that KI models bearing numerous H chain VDJ specificities, including (but not restricted) to those with self-reactivity, cannot support B-1 B-cell development (42). Nevertheless, the profound blockade in early BM development selectively observed in 4E10 VH KI mice, is consistent with self-reactivity of the 4E10 H chain, as we have previously seen with the 2F5 H chain, being sufficient to trigger central deletion of B-cells expressing it on their surface as BCR.
Figure 2. Analysis of B-cell development and serum Ig subclass levels in C57BL/6 (WT), 4E10 VH+/+ and 48d VH+/+ KI strains.
(A-C). Representative FACS contour histograms (representative of three experiments) comparing BM, splenic, and peritoneal B-cell development in 4E10 VH+/+ and 48d VH+/+ KI mice (expressing the somatically mutated H chains of the polyreactive, MPER-specific bnAb 4E10 and the non-neutralizing HIV-1 mAb 48d, respectively). Also shown as controls are WT (C57BL/6) littermates; all mice were 8-12 wk females. A. Analysis of BM B-cell subsets, either less differentiated subpopulations (upper panels), based on Hardy subfractionation, more differentiated subpopulations (middle panels), as revealed by surface IgM and IgD staining as previously described (32), or based on surface L chain κ and λ1-3 expression (lower panels). Indicated B-cell subsets were pre-gated as singlet, live lymphocytes (upper panels), or as singlet, live, total (CD19+B220+) B-cells (middle and lower panels), respectively. Numbers indicate percentages of B-cells in each subset, with subsets defined as follows: pro/pre=pro-B/large pre-B (fractions A-C’; B220loCD43+), small pre=small pre-B (fraction D; B220loCD43−), imm/mat=mature+immature (fractions E-F; B220hiCD43−); imm=immature (IgMint/hiIgD−), T1/T2=transitional 1+2 (IgMhiIgDlo/int), and mat=mature (IgMintIgDhi). B. Analysis of splenic B-cell subpopulations, as revealed either by surface CD93 and CD23 expression of live lymphocytes (top row), and by surface CD21 and CD23 or IgM and IgD expression of live, total (CD19+B220+) B-cells (middle and lower rows, respectively). Numbers indicate percentages of B-cells in each gate, and B-cell subsets indicated in upper panels are defined as follows: T=transitional (B220+CD93+), Mat+MZ (B220+CD93−), whereas those indicated in the middle row are defined as: NF=newly formed i.e. transitional (CD21−CD23−), MZ=marginal zone (CD23intCD21hi), and Mat=mature follicular B2 (CD23hiCD21int). C. Analysis of peritoneal B-cell subsets as revealed by surface staining of live lymphocytes with B220 and CD5 with numbers in each gate indicated, and B-cell subsets are defined as follows: B1a (B220intCD5int), B1b (B220intCD5−), B2 (B220+CD5−). D. Analysis of total Ig subclass levels in 4E10 VH+/+ and 48d VH+/+ KI mice, or WT (C57BL/6) littermates. Serum samples were collected from 6–12 wk female naïve mice, and serum concentrations for all Ig subclasses were determined by ELISA analysis. Each symbol represents an individual mouse and horizontal lines represent averages for each group. Significance values were determined using a two-tailed student test. NS: P>0.05, *P≤0.05, **P ≤0.01, ***p ≤0.001.
Generation of 4E10 VH × VL KI mice and characterization of 4E10 and 2F5 KI strains
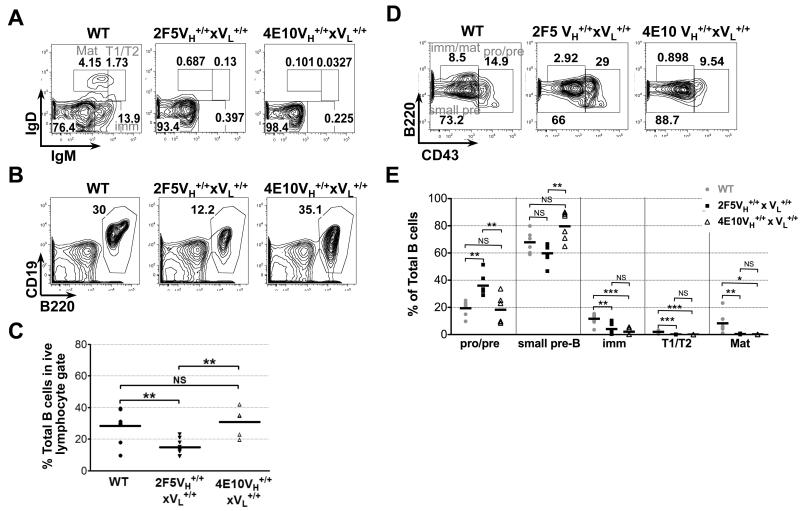
To evaluate the effect of expressing the complete (VH × VL) 4E10 mAb as BCR, and to more thoroughly compare B-cell compartments and serum Ig subclasses between KI mice expressing targeted 4E10 or 2F5 rearrangements in all homozygous strains (i.e., VH, VL, and complete), we constructed 4E10 VL KI mice using the identical targeting strategy and constructs used for generating 2F5 VL KI mice (31, 32). The pre-rearranged 4E10 VkJk rearrangement derived from the original (somatically mutated) 4E10 mAb was targeted into the murine Igκ loci in place of the Jκ1-3 gene segments to mimic its expected location as the primary rearrangement event, thus allowing retention of κLC secondary rearrangements (Fig. 1B). 4E10 VL targeted ES clones and germline-transmitted homozygous mice were confirmed by Southern blot (Fig. S1E) and genomic DNA PCR amplifications (Fig. S1F), respectively. Finally, 4E10 complete (VH+/+ × VL+/+) KI mice were generated by crossbreeding 4E10 homozygous VH and VL KI lines.
Consistent with profound clonal deletion of B-cells expressing either gp41-specific bnAb as a B-cell receptor (BCR), 4E10 and 2F5 complete KI mice, relative to C57BL/6 littermates, had a similar, major blockade at the pre-B to immature B transition, as measured both in terms of B-cell subset frequencies (Fig. 3A) and absolute numbers (Table 1). However, additional, minor developmental differences between 4E10 and 2F5 KI strains were also identified (Fig. 3B-E, table 1). In particular, 2F5 complete KI mice, relative to 4E10 complete KI mice, also had reduced frequencies and absolute numbers of total (CD19+B220+) BM B-cells (Fig. 3B,C, Table 1), as well as an additional, modest blockade, at the pro/large pre-B to small pre-B transition (Figs. 3D,E, Table 1), which we have reported previously (32). Interestingly, 4E10 VH KI mice also exhibited this same, earlier blockade (Fig. S2A; upper panels), which may reflect suboptimal interactions between 4E10 H chains and mouse surrogate L-chains, resulting in poor surface expression andinefficient allelic exclusion of endogenous H chains (Fig. S3), even more than we reported for the 2F5 H chain (31).
Figure 3. Flow cytometric comparison of BM B-cell development in C57BL/6 (WT), and 4E10/2F5 complete (VH+/+ × VL+/+) KI strains.
Shown are FACS contour plot histograms (representative of three experiments) indicating frequencies of either: (A) less differentiated BM B-cell subsets (fractionated/annotated as indicated in Fig. 2A), (B) total (singlet/live/lymphocyte, CD19+B220+) BM B-cells, or (D) more differentiated BM B-cell subsets (fractionated/annotated as in Fig. 2A). Also shown is statistical analysis of (C) total BM B-cell frequencies, or (E) BM B-cell subset frequencies (based on flow cytometric gating shown in Figs. 3A and D, respectively), with each closed circle, closed triangle and open square representing individual 6-12 wk female naïve C57BL/6 (WT), 2F5 complete KI, and 4E10 complete KI mouse, respectively. Horizontal lines represent averages for each group. Significance values were determined using a two-tailed student test. NS: P>0.05, *P≤0.05, **P ≤0.01, ***p ≤0.001.
Table 1. Comparison of B-cell subset absolute numbers in BM and spleen of WT, and 2F5 or 4E10 complete (VH+/+ × VL+/+) KI mice.
| Bone Marrowb | |||||||
|---|---|---|---|---|---|---|---|
|
| |||||||
| Total B-cells | ProB/large pre-B | Small pre-B | Immature B | T1/T2 | Mature | ||
|
|
|||||||
| Mouse | n | B220+CD19+c | B220loCD43+ | B220loCD43− | IgMint/hiIgD− | IgMhiIgDlo/int | IgMintIgDhi |
| WT | 6 | 82.4±16.0a | 15.7±6.8 | 50.0±16.8 | 9.7±2.0 | 1.8±0.6 | 5.1±1.6 |
| 2F5VH+/+×VL+/+ | 6 | 41.7±9.4 | 15.5±4.8 | 24.7±8.4 | 2.5±1.2 | 0.013±0.005 | 0.079±0.021 |
| 4E10VH+/+×VL+/+ | 5 | 68.2±9.5 | 12.5±4.5 | 54.5±7.1 | 1.7±1.0 | 0.016±0.007 | 0.094±0.022 |
|
| |||||||
| Spleen | |||||||
|
|
|||||||
| Total B-cells | NF/T | MZ | Mature | ||||
|
|
|||||||
| Mouse | n | B220+CD19+ | CD21−CD23− | CD23intCD21hi | CD23hiCD21int | ||
|
|
|||||||
| WT | 8 | 106.4±18.0 | 18.1±3.4 | 5.4±1.6 | 70.2±13.3 | ||
| 2F5VH+/+×VL+/+ | 6 | 1.9±0.3 | 0.71±0.20 | 0.073±0.019 | 0.50±0.07 | ||
| 4E10VH+/+×VL+/+ | 5 | 4.4±0.7 | 1.6±0.2 | 0.016±0.008 | 0.28±0.07 | ||
|
|
|||||||
Values are absolute cell numbers (×105±SEM)
Bone marrow cells were taken from both femurs for FACS analysis.
Cells were gated on singlet/live/lymphocytes. B-cell subset annotations are based on those defined in Figs. 2A, B.
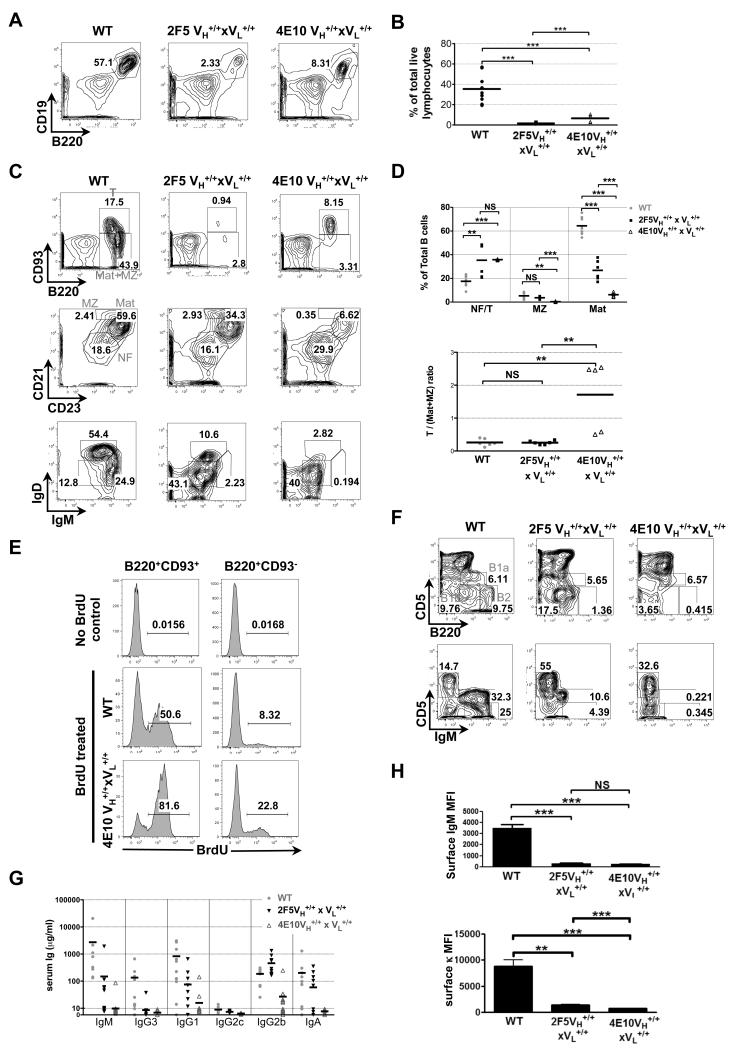
4E10 and 2F5 complete KI strains also had several notable differences in residual B-cell compartments (Figs. 4A-F) and serum Ig subclass distributions (Fig. 4G). First, 2F5 complete KI strains had significantly reduced frequencies and absolute numbers of total (CD19+B220+) splenic B-cells than 4E10 complete KI strains (Fig. 4A,B, Table 1), but residual splenic B-cells in 4E10 complete KI mice preferentially accumulated as phenotypically less differentiated i.e., newly-formed (CD21−CD23−)/transitional (CD93+) B-cell subsets (Fig. 4C,D, Table 1). The transitional nature of residual 4E10 complete KI splenic B-cells was confirmed by demonstrating their rapid in vivo turnover, as measured by incorporation of BrdU by the majority of CD93+ splenic B-cells from continuously-labeled 4E10 complete KI mice (Fig. 4E). In contrast, 2F5 complete KI strains had a more heterogeneous peripheral B-cell subset distribution, with modest accumulation of newly formed/transitional B-cells, but also substantial mature B2 (B220+CD93−) and marginal zone (MZ) (CD23IntCD21High) B-cell subsets (Figs. 4C,D, Table 1). Additionally, although both 2F5 and 4E10 complete KI mice had significantly lower frequencies of B-2 peritoneal B-cells (i.e., in addition to reduced B-1a subsets that were also seen in all VH KI strains, including 48d; Fig. 2C), these subsets were most profound in 4E10 complete KI mice, and also extended to the B1-b subset (Fig. 4F), coinciding with their near-complete elimination of serum IgM and IgG3 levels (Fig. 4G). Finally, 4E10 complete KI strains also had significant reductions in all other serum Ig isotypes (i.e., IgG1, IgG2b, IgA), relative to C57BL/6 littermates (Fig. 4G), consistent with their early blockade in peripheral B2 development; in contrast, 2F5 complete KI mice had less reduced levels of total serum IgG (Fig. S4), and Ig subclass distributions most similar to 2F5 and 48d VH KI mice, including selectively increased IgG2b (Figs. 2D, 4G, S4). Overall, these findings therefore suggest that 2F5 and 4E10-expressing B-cells undergo different degrees of clonal deletion in BM and peritoneal compartments and accumulate at distinct stages of splenic development.
Figure 4. Distribution of splenic and peritoneal B-cell subsets in C57BL/6 (WT) and 4E10/2F5 complete (VH+/+ × VL+/+) KI strains.
A. FACS contour plot histograms (representative of three experiments) showing frequencies of total (singlet/live/lymphocyte, B220+CD19+) B-cells in spleens of 6-12 wk naïve mice. B. Statistical analysis of total splenic B-cell percentages (based on flow cytometric analysis shown in Fig. 4A, with each circle, square and triangle representing an individual 6-12 wk C57BL/6 (WT), 2F5 complete KI, and 4E10 complete KI mouse, respectively. Horizontal lines represent averages for each group. Significance values were determined using a two-tailed student test. NS: P>0.05, *P≤0.05, **P ≤0.01, ***p ≤0.001. C. Representative FACS contour plot histograms showing analysis of splenic B-cell subsets derived from 8-12 wk female naïve mice, fractionated and annotated as indicated in Fig. 2B. D. Statistical analysis of splenic B-cell subsets from individual WT or 2F5/4E10 complete KI mice, represented either as frequencies of newly formed/transitional (CD21−CD23−), MZ (CD23intCD21hi), and Mat (CD23hiCD21int) subsets (upper panel) or transitional (B220+CD93+)/Mat+MZ (B220+CD93−) ratios (lower panel). E. In vivo turnover of the predominant, transitional splenic B-cell subset in 4E10 complete KI mice. 8 wk naïve female naïve 4E10 complete KI or control C57BL/6 (WT) mice (lower and middle panels, respectively) were continuously labeled with BrdU for 4 days, prior to flow cytometric enumeration of the percent of BrdU+ B-cells within singlet/live, lymphocyte-gated transitional (B220+CD93+) and mature/MZ(B220+CD93−) splenic B-cell subsets, as indicated in the shaded histograms. Also shown as negative controls are C57BL/6 (WT) mice without BrdU treatment (top panel). Data is representative of two mice/group. F. Representative FACS contour plot histograms showing analysis of peritoneal B-cell subsets as revealed by surface staining of live lymphocytes with B220 and either CD5 (upper row) or IgM (bottom row), with numbers in each gate indicated, and B-cell subsets indicated in the upper row are defined as for Fig. 2C. G. Analysis of total serum Ig subclass levels in C57BL/6 (WT) mice and 2F5/4E10 complete KI strains. Serum samples were collected from 6-12-wk naive mice, and Ig subclass concentrations were determined by Luminex analysis. Each symbol represents an individual mouse and horizontal lines represent averages for each group. H. Flow cytometric analysis of sIg densities in total (singlet/live/lymphocyte, B220+CD19+) splenic B-cells from 6-12 wk naïve mice, as measured by Median Fluorescence Intensity (MFI) quantifications of surface κL chain (left panel) or surface IgM (right panel) expression.
However, despite the above differences, residual surface Ig+ B-cells from 2F5/4E10 VH or complete KI strains, relative to those in control (C57BL/6 and 48 VH KI) mice, expressed uniformly reduced BCRs densities i.e., lower sIgM and sIgD levels, and reduced BCR densities not just restricted to T3 B-cells, but also include more differentiated (mature+MZ) splenic subsets (Figs. 2B, 4C: lower panels), as well as BM B-cells (Fig. S3) (32). Interestingly, the degree of sIg reductions in total splenic B-cells in all 4E10 and 2F5 KI strains (as quantitated either by surface kappa or IgM MFIs) (Fig. 4H) were greater in complete KI strains than VH KI strains (Fig. S2E), with 4E10 complete KI mice having the greatest reductions (Fig. 4H; lower panel). Since the degree of sIg reduction across these strains also inversely correlates with the differentiation stage at which their residual peripheral B-cells accumulate, it is possible that these quantitative differences could be a manifestation of progressively higher levels of self-reactivity retained by residual B-cells across these KI species, as has been seen in comparison α-DNA KI models having varying degrees of self-antigen affinity (35, 43). Nevertheless, these phenotypic data collectively suggest that anergy is the common mechanism suppressing most residual 4E10 or 2F5 H chain expressing B-cells, regardless of the peripheral developmental stage at which they are arrested.
The majority of residual 2F5 and 4E10-expressing B-cells are functionally anergic
The uniform reduced sIgM and sIgD densities in most splenic B-cells from 2F5/4E10 KI complete KI mice (Figs. 2-4) is unexpected in that it does not resemble the classic anergic phenotype (IgMlo/−IgDhi) originally reported in the HEL transgenic system (44, 45), believed to occur predominantly at the splenic T3 checkpoint (46). However, it is now becoming apparent that anergy is more complex than previously thought, both in terms of the stages it may occur and the phenotypic heterogeneity of functionally anergic B-cells (reviewed in (33)). For example, functionally anergic B-cells have been reported in later splenic B2 differentiation in some autoreactive BCR models (27, 47-53) and B-cells functionally analogous to anergic cells, with low densities of both sIgM and sIgD are observed at varying degrees in α-DNA KI models having differing affinities for self-antigen (43).
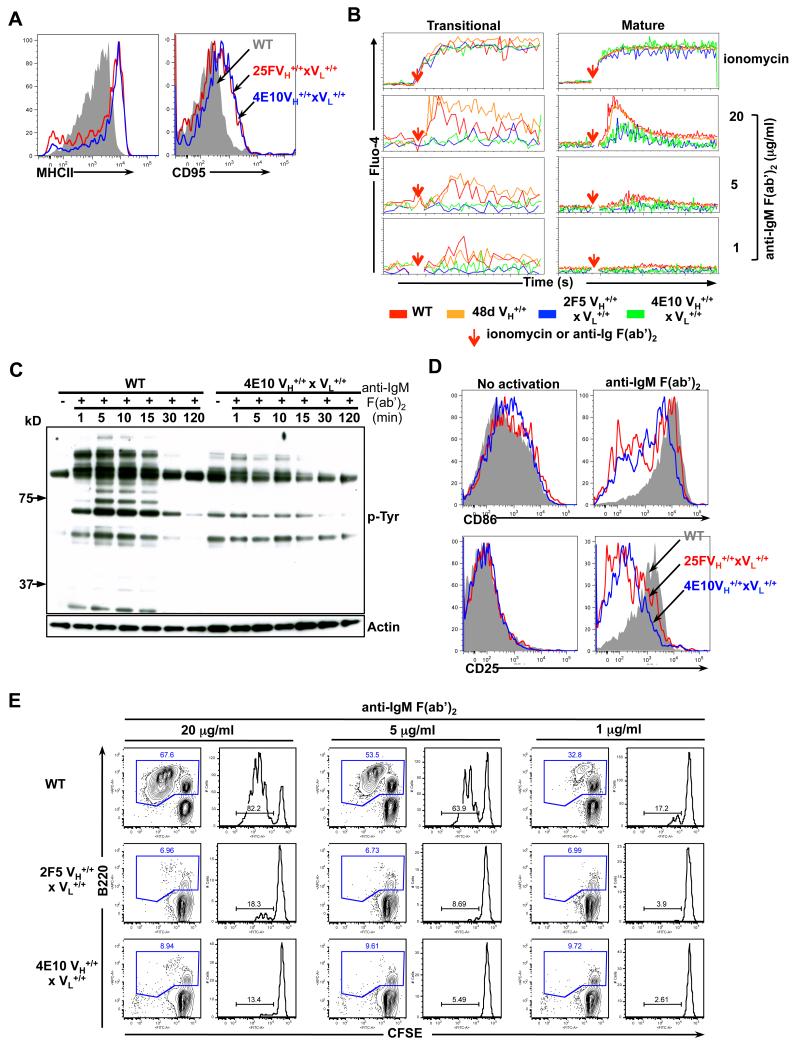
To test if the splenic B-cells expressing these uniformly low sIg densities in our MPER bnAb complete KI models represented bona fide anergic B-cells, we first examined total (B220+CD19+) splenic B-cells from complete KI strains by flow cytometry for basal activation of surface activation markers. We found the total peripheral B-cells in naïve 4E10 and 2F5 complete KI B-cells, relative to WT (C57BL/6) littermate B-cells had uniformly upregulated surface expression of MHC class II and CD95, two phenotypic hallmarks of anergic peripheral B-cells (33) (Fig. 5A), while CD25, CD40, CD69, CD80 and CD86 were unchanged (data not shown). That basal expression of these activation markers was increased in the majority of total B-cells from both KI strains suggests that anergy is not restricted to the T3 transitional subset, since they only make up a minority of splenic B-cell compartments in the 2F5 complete KI strains, which has a predominant B220+CD93− (mature and MZ) B-cell subset. To confirm this uniformly anergic phenotype of MPER bnAb-expressing B-cells functionally, we examined calcium mobilization in response to BCR ligation using fluo-4 flow cytometric staining, which allowed comparison of proximal signaling in splenic transitional (B220+CD93+) and mature B2 (B220+CD93−) subsets of 4E10 and 2F5 complete KI strains, relative to comparable subsets from control (i.e., WT BL/6 and 48d VH KI) mice. Indeed, both transitional and mature B2 B-cell subsets in either 2F5 or 4E10 complete KI mice exhibited significantly muted calcium induction in response to BCR crosslinking (Fig. 5B), demonstrating B-cell anergy, regardless of maturational stage.
Figure 5. Phenotypic analysis of basal activation levels and functional analysis of signaling responses to in vitro BCR cross-linking in MPER bnAb KI splenic B-cells.
A. Flow cytometric analysis of surface MHC class II and CD95 expression in naïve WT and 4E10 or 2F5 complete KI B-cells, represented as grey shadow and blue or red line histograms, respectively. B. Calcium flux analysis of control WT C57BL/6 or 48d VH KI and 2F5 or 4E10 complete KI B-cells (shown in red, orange, blue and green lines, respectively). Pre-stained splenocytes from naive mice were loaded with 1μM Fluo-4, and either baseline levels of calcium, or those in response to either ionomycin (used as a positive control) or anti-IgM F(ab’)2 stimulation were added (denoted by red arrows), were detected by flow cytometry in transitional (B220+CD93+) or mature (B220+CD93−) B-cell subsets. C. Phosphorylated protein analysis of proximal signaling responses to in vitro BCR aggregation in purified WT or 4E10 complete KI splenic B cells, either incubated with medium alone (time=0) or stimulated with 20 μg/ml α-IgM F(ab’)2 for indicated times. Shown are either total phosphotyrosine protein levels, revealed by immunoblotting with the α-phosphotyrosine Ab 4G10 (upper panel) or total protein levels, revealed by re-probing the same blots with an actin-specific Ab (lower panel). (D-E). Distal signaling responses to in vitro BCR aggregation. D. Flow cytometric determination of surface expression of early/intermediate activation markers CD25 and CD86 in total naïve WT or 4E10 and 2F5 complete KI splenic B-cells (grey shadow or blue and red line histograms, respectively) either unstimulated (left panels) or in response to in vitro BCR crosslinking with 20 μg/ml α-IgM F(ab’)2 for 24h (right panels). E. Analysis of WT or 4E10 and 2F5 complete KI splenic B-cell proliferation after BCR aggregation as determined using a flow cytometric CFSE-based cell division assay. Splenocytes from naïve splenic B-cells were stimulated with 20, 5 or 1 μg/ml α-IgM F(ab’)2 with numbers in blue representing total (B220+) B-cell percentages, and those in right panels representing the percentage of dividing cells within the total (B220+) B-cell gate.
Finally, to further confirm the anergic status of MPER bnAb complete KI B-cells functionally, we monitored several other independent proximal and distal signaling responses to BCR ligation in total splenic 4E10 and 2F5 complete KI B-cells. Indeed, protein tyrosine phosphorylation immunoblotting of total naïve 4E10 complete KI splenic B-cells revealed their higher basal levels of tyrosine phosphorylation, relative to those from WT mice, and their failure to induce additional tyrosine phosphorylation upon BCR aggregation by anti-IgM F(ab’)2 (Fig. 5C). Additionally, upon in vitro BCR crosslinking of total splenic naïve B-cells from either WT, 4E10 or 2F5 complete KI splenic naïve B-cells, the majority of total 4E10 and 2F5 complete KI B-cells failed to upregulate basal expression of early/intermediate activation markers CD25 and CD69 (Fig. 5D: lower panels and data not shown) or the late activation marker CD86 (Fig. 5D: upper panels) and exhibited abrogated proliferation (Fig. 5E). As with B-cell from 4E10 complete KI mice, total splenic B-cells from 4E10 VH KI mice, the majority which are mature (B220+CD93−) B-cells, also had uniformly increased basal surface expression of MHC class II and CD95, and in response to BCR ligation, also exhibited muted proliferation, CD25, CD69 and CD86 upregulation, and calcium induction, relative to total splenic 48d VH KI B-cells (data not shown). Collectively, these data confirm that functional silencing is the predominant mechanism controlling most residual 2F5 and 4E10-specific B-cells in vivo.
Characterization of splenic hybridomas from complete 2F5 and 4E10 KI mice
In addition to their low surface Ig levels, it is interesting that 2F5/4E10 VH and complete KI strains exhibit elevated surface LC κ/λ ratios, relative to WT and 48d VH KI mice (Fig. S3F), indicating biased pairing to κL chains by both bnAb H chains. In this regard, it is also intriguing that central deletion triggered by 2F5/4E10 H chain self-reactivity occurs independently of endogenous LC pairing (Figs. 2A, S3A) (32), and that most BM 2F5 complete KI B-cells, when cultured in vitro (under conditions which permit rescue from clonal deletion) undergo extensive κLC editing, but still retain an anergic phenotype (32). This suggests that: i) pairing constraints imparted by 2F5/4E10 H chains prevent λ editing, and that ii) κ-editing only partially removes self-reactivity. We therefore asked if, in vivo, residual splenic 4E10/2F5 KI B-cells, with their uniformly lower BCR expression, attempt L chain editing (or any other V region modifications) prior to being routed into an anergic pathway.
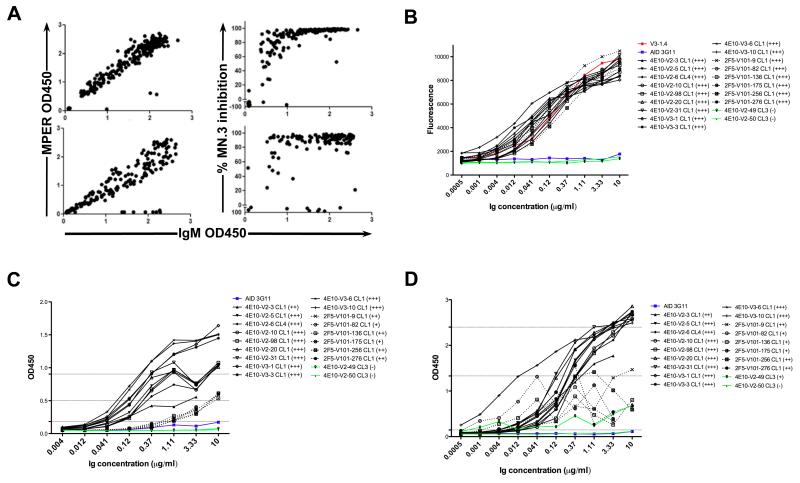
To examine this, hybridomas from naïve 4E10 and 2F5 complete KI splenic B cells were generated and the percentage of clones retaining MPER epitope specificity and neutralization ability were determined. Plates were first screened for cell growth, with 6.7% (320/4800) and 16.8% (510/3040) of wells from 4E10 and 2F5 fusions exhibited growth, respectively, and subcloned lines were screened for Ab secretion, with 150 4E10 and 225 2F5 hybridomas identified, all of the IgM isotype (Table 2). The majority, 95.3% (143/150), of 4E10 Ab-producing hybridoma lines bound the MPR.03 peptide (containing the 4E10 neutralization epitope), and 99.6% (224/225) of Ab-producing 2F5 lines bound the SP62 peptide (containing the 2F5 neutralizing epitope) (Fig. 6A; Table 2). Cardiolipin ELISA assays also revealed that 100% of MPER-reactive hybridomas maintained cardiolipin reactivities from both KI strains (Data not shown). Additionally, all hybridoma culture supernatants that bound the MPER also neutralized HIV MN.3 pseudovirus in standard TZM-bl assays (Fig. 6A).
Table 2. Summary of isotypic distribution and MPER reactivities in primary screen of hybridoma clones derived from 2F5 and 4E10 complete KI spleensa.
| Fusion ID | Mice | Total seeded wells |
Wells with cell growthb |
IgM+c | IgG+ | IgA+ | MPER+d |
|---|---|---|---|---|---|---|---|
| 2F5-V101 | 2F5VH+/+ × VL+/+ | 3040 | 510 | 225 | 0 | 0 | 224 |
| 4E10-V2 | 4E10VH+/+ × VL+/+ | 3200 | 146 | 50 | 0 | 0 | 43 |
| 4E10-V3 | 4E10VH+/+ × VL+/+ | 1600 | 174 | 100 | 0 | 0 | 100 |
All fusions were performed using NS0-bcl2 myeloma fusion partner lines and represent a different individual mouse.
Culture plates were screened under microscope for cell growth.
Ig levels /isotypes of all cloned hybridoma lines were determined by ELISA as described in Materials and Methods. Lines were considered positive if >3× above background OD binding was detected in 1:40 diluted supernatants.
MPER reactivity of 2F5 or 4E10 hybridoma lines was determined by ELISA to their nominal MPER epitopes (as specified by SP62 and MPR.03 peptides, respectively). Lines were considered positive if >3× above background OD binding was detected in 1:2 diluted supernatants.
Figure 6. Antigenic reactivity and neutralization profiles of splenic hybridoma lines isolated from naïve 2F5 or 4E10 complete KI mice.
A. MPER reactivity and HIV-1 neutralization profiles of B-cell hybridoma sub-cloned lines derived from primary screens of fusions using naïve 2F5 or 4E10 complete KI spleens. Data is represented as scatter plots, with MPER nominal epitope reactivities (left panels) or % MN.3 neutralization inhibition scores (right panels) plotted against supernatant IgM concentrations in 2F5 and 4E10 complete KI hybridoma lines (upper and lower panels, respectively). (B-D). Antigenic reactivity profiles of purified mAbs selected from primary screens. Shown are representative ELISA binding curves of purified 4E10 and 2F5 complete KI-derived mAbs (solid and dashed lines, respectively) to their (B) MPER neutralizing epitopes (as specified by MPR.03 peptides), (C) NIH-3T3 cytoplasmic/nuclear components, and (D) cardiolipin, all carried out as previously described (32). V3-1.4 (a 2F5 IgM mAb)(32) or 4E10-V2-6 CL4 (a representative 4E10 IgM mAb identified in this study, bearing 4E10 VH/VL sequences) and AID 3G11 (a non-neutralizing IgM mAb lacking self-polyreactivity) (69) were used as positive and negative controls for binding self-/polyreactive components, respectively. Criteria for positivity were arbitrarily set at saturating concentrations, and relative to control mAbs as follows: (+++) > 80% of (4E10-V2-6 CL4 or V3-1.4) binding, (++) = 50-80% of (4E10-V2-6 CL4 or V3-1.4) binding, and (+) = 25-50% of (4E10-V2-6 CL4 or V3-1.4) binding, but >AID 3G11 binding.
To determine IgH/IgL V region sequences of 4E10 and 2F5 KI strain hybridomas, we selected 51 4E10 KI lines and 30 2F5 KI lines from our primary screens for an additional round of subcloning in order to confirm monoclonality (Table 3). All 30 cloned 2F5 KI hybridoma lines retained their original knocked-in 2F5 VH and VL rearrangements, which had either no, limited, or silent somatic mutations (Table 3). 49/51 of the 4E10 KI hybridoma lines also retained their original 4E10 VH and VL regions/sequences, but interestingly, the two exceptions secreted unusually high amounts of Ab, and both underwent VH replacement of their 4E10 H chains, as well as completely lost their lipid/MPER reactivities and neutralization abilities (Tables 3, 4; Figs. 6B-D). Finally, to fully verify reactivity and neutralization profiles, we also generated purified mAbs from 11 MPER+ representative cloned 4E10 lines, 2 MPER− representative cloned 4E10 lines and for comparison, 6 MPER+ representative cloned 2F5 lines. All purified MPER+ 2F5 and 4E10 mAbs maintained comparable reactivity to NIH-3T3 cell and their respective nominal MPER epitopes (Figs. 6B, C). Furthermore, all 2F5 and 4E10 MPER+ mAbs maintained CL reactivities, but as anticipated, 4E10 KI hybridomas bound CL at generally higher affinities than 2F5 KI hybridomas (Fig. 6D); consistent with differential CL affinities reported for 4E10 and 2F5 bnAbs (25, 54, 55).
Table 3. Individual Ag reactivities and Ig usage properties of hybridomas derived from 2F5 and 4E10 complete KI splenic B cells.
| Reactivity Profilea |
Ig Usage Profilesb |
||||||
|---|---|---|---|---|---|---|---|
| Clone ID No. | MPER | CL | Neutralization Ability |
VH Rearrangeemnt Used |
VH Mutation Type/Location |
VL Rearrangemen t Used |
VL Mutation Type/Location |
|
|
|
||||||
| 4E10-V2-3 CL1 | +++ | N/A | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-5 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-6 CL4 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-10 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-12 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-13 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-20 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-30 CL1 | +++ | N/A | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-31 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-34 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-35 CL4 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-41 CL3 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-47 CL5 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-56 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-60 CL5 | +++ | +++ | +++ | KI 4E10Vh VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-62 CL1-1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-85 CL1 | +++ | N/A | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-98 CL1-1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-100 CL2 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-101 CL2 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-104 CL2 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-105 CL1 | +++ | N/A | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-108 CL8 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-112 CL7 | +++ | N/A | N/A | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-114 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-122 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-123 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-126 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-127 CL4 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-131 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-134 CL3 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-141 CL2-1 | +++ | N/A | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-143 CL3-1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-1 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-3 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-6 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-10 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-16 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-17 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-18 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-19 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-22 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-24 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-27 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-33 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-35 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-36 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-37 CL1 | +++ | +++ | +++ | KI 4E10Vh VDJ | no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V3-40 CL1 | +++ | +++ | +++ | KI 4E10VH VDJ J558.26.116/DFL |
no mutation | KI 4E10VL VDJ | no mutation |
| 4E10-V2-49 CL3 | - | - | - | 16.1/partial 4E10VHc J558.50.143/DSP |
N/A | KI 4E10VL VDJ | no mutation |
| 4E10-V2-50 CL3 | - | - | - | 2.2/ partial 4E10VH |
N/A | KI 4E10VL VDJ | no mutation |
|
| |||||||
| 2F5-V101-1 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-9 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-11 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-14 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-29 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-30 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-33 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-37 CL1 | +++ | ++ | +++ | KI 2F5Vh VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-50 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-74 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-77 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-82 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-103 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-115 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | N/A | N/A |
| 2F5-V101-124 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | N/A | N/A |
| 2F5-V101-125 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-130 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | N/A | N/A |
| 2F5-V101-136 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-143 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | N/A | N/A |
| 2F5-V101-160 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-165 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-175 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-202 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-215 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-247 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | N/A | N/A |
| 2F5-V101-256 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-276 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-282 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-299 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
| 2F5-V101-305 CL1 | +++ | ++ | +++ | KI 2F5VH VDJ | no mutation | KI 2F5VL VDJ | no mutation |
MPER, CL reactivities and neutralization ability were determined as described in Materials and Method. Criteria for positivity were arbitrarily set at saturating concentrations, and relative to control mAbs as follows: (+++) >80% of (4E10-V2-6 CL4 or V3-1.4) binding, (++) = 50-80% of (4E10-V2-6 CL4 or V3-1.4) binding, and (+) = 25-50% of (4E10-V2-6 CL4 or V3- 1.4) binding, but >13H11 (or AID 3G11) binding.
cDNAs from cloned hybridoma lines were amplified using leader-specific and K-specific primers, and PCR products were cloned and sequenced in both orientations as described in the Materials and Methods. KI VH and VL regions were analyzed for mutations using Lasergene software.
Heavy chains that cannot align to 4E10 VH were analyzed using the Ig BLAST algorithm. These 2 clones keep 3′end partial 4E10 VH sequence but replace their 5’end with endogenous VH segments in a region of the 4E10 VH that coinciding with a cryptic Recombination Signal Sequence (RSS) containing a perfect consensus embedded nonamer and heptamer (70).
N/A: not available.
Table 4. Neutralization profiles of purified IgM mAbs selected from cloned hybridoma lines.
| SVAd |
92UG037 |
MN |
SF162 |
6535 |
BG1168 |
ZM14654 |
DU156 |
|
|---|---|---|---|---|---|---|---|---|
| Clone ID | IC50c | IC50 | IC50 | IC50 | IC50 | IC50 | IC50 | IC50 |
| 2F5a | >50 | 0.18 | <0.023 | 2.01 | 9.83 | 2.13 | >50 | >50 |
| 4E10a | >50 | 0.11 | <0.023 | 2.50 | 0.94 | 3.54 | 0.38 | 0.21 |
| V3-1.4a | >50 | 0.09 | <0.023 | 1.92 | 1.56 | 1.25 | 2.79 | >50 |
| AID 3G11b | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 13H11b | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 2F5-V101-282 CL1 | >50 | 0.23 | <0.011 | 3.07 | 1.38 | 2.85 | 2.56 | >25 |
| 2F5-V101-9 CL1 | >25 | 0.05 | 0.01 | 3.15 | 1.26 | 1.58 | 1.83 | >25 |
| 2F5-V101 136 CL1 | >25 | 0.12 | 0.03 | 4.40 | 1.54 | 2.19 | 1.75 | 12.44 |
| 2F5-V101-175 CL1 | >25 | 0.13 | 0.07 | 3.87 | 1.63 | 2.84 | 1.99 | >25 |
| 2F5-V101-256 CL1 | >25 | 0.10 | <0.011 | 3.16 | 1.05 | 1.67 | 1.41 | >25 |
| 4E10-V2-5 CL1 | >50 | 0.98 | 0.04 | 10.31 | 2.46 | 7.81 | 0.57 | 0.29 |
| 4E10-V2-98 CL1 | >50 | 0.65 | 0.07 | 9.50 | 3.90 | 13.41 | 0.57 | 0.28 |
| 4E10-V2-6 CL4 | >50 | 0.69 | 0.15 | 11.87 | 4.03 | 15.71 | 1.58 | 0.49 |
| 4E10-V2-3 CL1 | >25 | 0.43 | 0.21 | 8.88 | 0.46 | 4.05 | 0.12 | 0.09 |
| 4E10-V3-1 CL1 | >25 | 0.31 | 0.19 | 5.37 | 0.23 | 2.23 | 0.10 | 0.03 |
| 4E10-V3-6 CL1 | >25 | 0.40 | 0.10 | 7.19 | 0.50 | 3.48 | 0.10 | 0.05 |
| 4E10-V2-49 CLe | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
| 4E10-V2-50 CLe | >50 | >50 | >50 | >50 | >50 | >50 | >50 | >50 |
Recombinant human 2F5, 4E10 and mouse mAb V3-1.4 were used as positive control antibodies.
Mouse mAbs AID 3G11 and 13H11 were used as negative control.
Shown are 50% neutralization concentrations (IC50) of affinity-purified mAbs required to neutralize each HIV-1 isolate listed in the TZM-bl cell assay.
SVA was used as a negative control virus.
Clones underwent VH replacement.
Finally, 8 purified 4E10 and 5 purified 2F5 mAbs were selected for assay against a panel of viral isolates in TZM-bl neutralization assays. Interestingly, the 2F5 mAbs (all containing unmodified V regions) were found to neutralize with similar potency and breadth as the control (human recombinant IgG) 2F5 Ab, whereas the six 4E10 mAbs with unmodified V regions neutralized with a modestly lower potency than the control (human recombinant IgG) 4E10 Ab (Table 4). One potential explanation for these differences may be related to our previous finding that mouse 2F5 IgM mAbs in unreduced form (containing monomeric/pentameric mixtures) unexpectedly neutralize more potently than reduced (monomeric) mouse 2F5 IgM or IgG (32), thus counteracting the lower potency of 2F5 Abs containing mouse, rather than human C regions (31). In this regard, given 4E10’s neutralization epitope in native HIV-1 viruses may not be as conformationally accessible as 2F5’s (56), 4E10 pentameric IgM mAbs, may thus not be capable of enhancing neutralization potency by whatever mechanism 2F5 pentameric IgM mAbs achieves this, due to structural (size) constraints.
Nevertheless, our above data collectively indicate that a significant fraction of anergic peripheral B-cells from naïve 2F5 and 4E10 KI mice retain their original, functional specificity in vivo, and have not undergone VH or VL modification, prior to (or after) acquiring their anergic phenotypes.
Serum Ab specificities of naïve 4E10 and 2F5 KI strains reveal distinct routes of positive selection
Despite the predominant, uniformly anergic phenotype of residual 2F5 and 4E10 complete KI B-cells, their distinct numbers/peripheral subset distributions, and the recent demonstration that 4E10 and 2F5 bnAbs exhibit markedly different polyreactivity profiles and bind distinct autoantigens in vitro (38) nevertheless predict that distinct positive selection routes are used in vivo by a minority subset of persisting peripheral 2F5 and 4E10 KI B2-cells that can break anergy and spontaneously secrete serum Igs. To test this possibility, we compared serum IgM and IgG Abs from naive 4E10 and 2F5 complete KI mice for their reactivity to two components required for their neutralization abilities: their peptide neutralizing epitopes and membrane lipids, either or both which could also be predicted to be removed due to self-/polyreactivity.
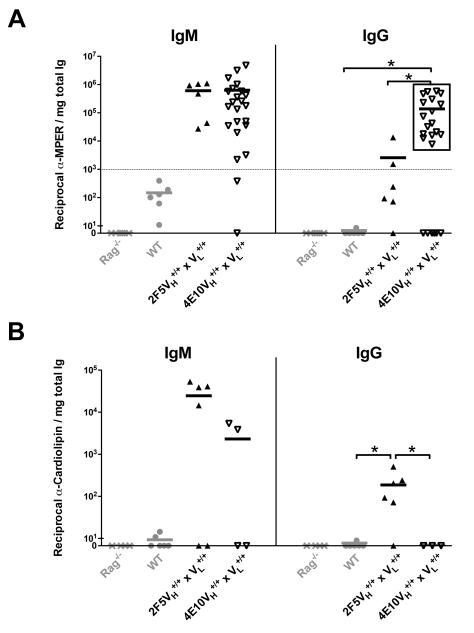
We found that MPER and CL-specific serum IgM levels were comparable to background levels in the WT (C57BL/6) strain (data not shown), but total serum IgM production in both complete KI strains relative to WT mice was profoundly reduced (Fig. 4G), and thus was not a fair comparison. For all strains, we therefore normalized MPER and CL-specific IgM to total IgM levels, and as expected, found that the low amounts of serum IgM secreted in 4E10 and 2F5 complete KI mice retained substantial MPER and CL-specific reactivity (Fig. 7A-B; left panels), consistent with serum IgM originating predominantly from residual self-reactive B-cell pools, either from the profoundly depleted peritoneal fractions, and/or anergic immature/transitional self-reactive B-cells.
Figure 7. MPER epitope and lipid reactivities of serum Abs from naïve 2F5 and 4E10 KI strains.
Shown are reciprocal endpoint titers of serum IgM and IgG-specific binding in 4E10 or 2F5 complete KI strains (grey and black rectangles, respectively) and control WT (C57BL/6) littermates or RAG1-deficient mice to (A) MPER 4E10/2F5 neutralization epitope-containing peptide MPR.03 or to (B) Cardiolipin (CL), with each rectangle representing an individual mouse and horizontal lines representing mean titers for each group. 4E10 complete KI mice having reciprocal α-MPER IgG >1000 were grouped as having “high MPER neutralization epitope reactivity” (framed in black rectangle) and compared with WT as well as 2F5 complete KI mice. For all indicated groups, serum samples were collected from 6-12 wk naïve and subjected to quantitative ELISA based on previous methods (32) and as described in Materials and Methods, with endpoint titers calculated as the reciprocal of the highest serum dilution used in which >3 background binding fluorescence were still observed. Data shown is normalized to the total IgM or IgG levels (mg/ml) shown in Fig 4D and S4A, respectively. Significance values were determined using a two-tailed student test. NS: P>0.05, *P≤0.05, **P ≤0.01, ***p ≤0.001.
In contrast, serum IgG-specific MPER and CL titers in 4E10 and 2F5 KI strains, when normalized to total serum IgG levels, revealed strikingly distinct binding profiles (Fig. 7). In particular, 2F5 KI strains had serum IgG-specific CL titers significantly higher than background (WT/B6) levels, whereas 4E10 KI strains completely lacked CL reactivity (Fig. 7A; right panel), mirroring the higher CL affinities, relative to 2F5, of the original (IgG) human 4E10 mAb (25) and IgM+ 4E10 KI hybridomas in this study (Fig. 6D), and suggesting more stringent elimination of CL reactivity in 4E10-expressing IgG+ mature B2-cells. Conversely, 2F5 complete KI mice exhibited minimal binding to their MPER epitope, whereas a large fraction of 4E10 complete KI mice maintained binding to theirs (Fig. 7B; right panel), suggesting more stringent selection against MPER binding in 2F5-expressing B-cells. Collectively, these data are consistent with distinct self-antigens driving the tolerization of 2F5 and 4E10 BCR-expressing B-cells in vivo, with positive selection of residual 2F5 KI cells involving the elimination of self-reactive residues associated with binding the nominal MPER epitope, whereas that of 4E10 KI cells involving elimination of binding to membrane lipids (and/or other correlating self-components comprising 4E10’s general polyreactivity, but not associated with binding to the 4E10 MPER epitope).
Discussion
This study provides key mechanistic insight into why gp41-targeted bnAbs like 2F5 and 4E10, whose epitopes are both located in the gp41 MPER HR2 region, are difficult to elicit. We demonstrate that site-directed targeting of either 4E10 or 2F5 VH/VL pre-rearrangements results in a common, profound blockade in early BM B-cell ontogeny, coinciding the pre-B to immature B-cell transition, the developmental stage where such bnAbs would normally be expressed by B-cells as surface antigen receptors. These results are similar to other transgenic models expressing autoreactive BCRs with high affinity for self-antigens (26, 34, 35) and are thus consistent with antigen-mediated clonal deletion being the predominant, common mechanism suppressing B-cells expressing gp41 bnAbs as BCR. Moreover, KI mice expressing 2F5 or 4E10 VH rearrangements alone (i.e., paired to multiple endogenous mouse L chains, instead of to those expressing 2F5 or 4E10 VL rearrangements, respectively) also exhibit a profound developmental blockade, suggesting that 2F5/4E10 H chain self-reactivities are sufficient to trigger central deletion, as seen in other KI mice with site-directed targeting of autoreactive H chains containing long and/or positively charged HCDR3 regions like the α-DNA H chain 3H9-76R (35, 57). Furthermore, that we observe an additional, modest blockade in both 2F5 and 4E10 VH KI mice at the pro/large pre-B to small pre-B transition suggests that 2F5/4E10 H chains may also partially impact B-cell development even prior to BCR expression. This finding is also consistent with previous studies demonstrating selection against pre-BCR+ B-cells bearing HCDR3s that are elongated and/or enriched in positively charged/aromatic residues (58-61), and could either be due to inefficient H chain/surrogate L chain pairing and/or pre-BCR-mediated self-reactivity (reviewed in (42, 62-64)). Finally and importantly, the fact that normal BM and splenic B2 B-cell development is supported in KI mice expressing the VH of 48d, a human HIV-1 mAb bearing a HCDR3 of average charge and length (and constructed in identical fashion as those expressing 2F5/4E10 VH regions), argues against the possibility that expression of chimeric 2F5/4E10 Abs (i.e., bearing human V+mouse C regions) non-specifically limits B-cell development in our KI models.
An intriguing question our results raise is whether clonal deletion represents a global mechanism for limiting all B-cells expressing MPER epitope-specific bnAbs. Of relevance to this issue is recent identification of 10E8, a potent MPER-specific bnAb lacking in vitro polyreactivity, but having an extraordinarily high degree of somatic mutations (65). Three explanations could account for the reported differences observed between 4E10/2F5 and 10E8 with respect to in vitro self-/polyreactivity: a) not all MPER-specific bnAb-expressing B-cells may indeed be clonally deleted in vivo, b) bnAbs like 10E8 have no polyreactivity, yet bind specific autoantigen(s) and thus would only be detectable in vitro in protein array scans (and not general Hep-2A or a pre-defined auto-antigen panel screens), akin to the minimally polyreactive bnAb 2F5, which avidly and selectively binds KYNU and CMTM3 (38), and whose physiologically-relevant autoreactivity has been confirmed via the in vivo measurement of negative B-cell selection (31,32), and c) bnAbs like 10E8 may originate from a rare subset of B-cell clones isolated from HIV-1-infected patients which acquired additional somatic mutations at some point during affinity maturation in germinal centers (GC), that eliminated their poly/self-reactivity, yet maintained their neutralization specificity. Future studies involving generation and characterization of KI mice expressing 10E8 will allow these possibilities to be distinguished.
Another key, related question raised by this study is whether precursors in the naïve B-cell repertoire expressing germline (i.e. unmutated common ancestor; UCA) forms of original, (i.e. somatically mutated) bnAbs derived from chronically-infected HIV-1 subjects, like 2F5 and 4E10, undergo central clonal deletion or alternatively, are normally deleted later in peripheral differentiation, during or after the GC reaction. With respect to the 2F5 bnAb lineage specifically, UCA-expressing B-cell precursors are also likely profoundly deleted early in BM development, based on several initial observations. First, it has recently been demonstrated in vitro that the 2F5 UCA binds lipids with higher affinity, exhibits stronger HEp-2A reactivity than the original 2F5 Ab, and depending on the allelic variant, retain MPER epitope reactivity at varying degrees (66). Additionally, the deduced somatic mutations acquired by the original 2F5 H chain do not include HCDR3 residues involved in MPER reactivity (37), lipid reactivity (56), and/or positively charged amino acids believed to be critical for general polyreactivity (57). Importantly, and consistent with this notion, we have recently demonstrated in KI mice expressing the 2F5 UCA VH rearrangement, a similar blockade in BM development (i.e., at the pre-B to immature B-cell stage) relative to those expressing the original 2F5 VH rearrangement (L.Verkoczy, unpublished observations), suggesting HC self-reactivity of mAb 2F5 is already encoded in unmutated B-cell precursors of the 2F5 lineage. On the other hand, the 4E10 UCA is expressed normally as BCR when transfected in vitro in B-cells (67) and in HEp-2A ANA and general autoreactivity assays, exhibits low affinity for various self-antigens (including lipids), and lacks MPER specificity (B.Haynes, unpublished observations), suggesting in vivo, 4E10-expressing B-cells may be deleted only after acquiring somatic mutations during affinity maturation. Generation and direct comparison of KI mice expressing complete (VH × VL) UCAs of 4E10, 2F5, and 10E8 will be critical to address where in development precursors of MPER-specific lineages are normally deleted.
Mechanistically, it is intriguing that we demonstrate anergy in 4E10 and 2F5 (but not control 48d) KI B-cells at not only the T3 developmental checkpoint, but also in splenic mature B2 cells, and furthermore, in 4E10/2F5-expressing B-cells with uniformly low Ig densities i.e., rather than just reduced sIgM levels. Although this distribution/phenotype of functionally anergic cells is atypical, similar observations have been seen in certain other autoreactive Ig transgenic systems (27, 47-53), and has been proposed to represent a general mechanism to lower BCR affinity to autoantigens in B-cells with residual self-reactivity in later B-cell differentiation (35, 43, 46). In the studies we report here in 4E10 and 2F5 KI models, where residual B-cells express extremely low levels of sIgM (and sIgD), we cannot formally exclude two intriguing alternative possibilities we are investigating further: either 2F5/4E10 themselves exhibit cross-reactivities to residues in sIgM or sIgD, or a subset of residual B-cells undergo “H chain locus suicide recombination”, a CSR-mediated regulatory mechanism proposed for irreversibly stopping BCR expression (68). In the context of the 2F5 KI model specifically, where we previously demonstrated that most B-cells expressing 2F5 H chains paired with endogenous mouse LCs (either in 2F5 VH+/+ KI mice, or via L chain editing in cultured 2F5 complete KI BM B-cells) lose MPER binding, yet remain anergic, the general lowering of sIg for the purpose of lowering affinity to self-antigens could represent a last-resort “safeguard” during affinity maturation to temporarily deal with residual self-reactivity of the 2F5 H chain as one that is “uneditable” i.e., with self-reactive residues for which VH replacement is not possible and cannot be eliminated by most (or all) L chain “editors”. Consistent with this notion is the fact that the 2F5 VH sequence lacks a consensus cryptic RSS (32), as well as our recent demonstration in 2F5 KI strains of “affinity reversion” i.e., selection for somatic mutations that eliminate multiple self-reactive HCDR residues (many also associated with binding the 2F5 MPER neutralization epitope), in a subset of persisting, unresponsive mature B2 cells, a process we propose may allow direct rescue of self-reactive bnAb+ B-cells from clonal deletion in GCs (Verkoczy et al., submitted).
4E10 and 2F5 bind neighboring epitopes in the MPER HR2 region (37) and use a similar mode of HIV-1 neutralization, requiring sequential engagement of Env membrane lipids, followed by interaction with their respective MPER epitopes (56). Our comparative studies of 2F5 and 4E10 KI strains here additionally demonstrate that these two bnAbs originate from B-cells profoundly limited in vivo by the same fundamental, overriding immune tolerance mechanisms: deletion and anergy. However, the underlying biology responsible for tolerization of 4E10 and 2F5-expressing B-cells have different features, with general polyreactivity (including, or correlating with, lipid reactivity) and neutralization epitope-associated self-reactivity appearing to play disparate roles, as revealed by their selective losses in serum IgG fractions from these two strains. In particular, that 2F5 complete KI serum IgG and cultured B-cells from sorted mature B2 compartments lose their MPER neutralization epitope specificity (but retain most of their basal lipid reactivity) suggests positive selection of 2F5-expressing B-cells involves eliminating self-antigenic residues associated with its neutralization epitope, whereas the stringent loss of the significant initial lipid reactivity of serum IgG from 4E10 complete KI mice (but most retain 4E10 neutralization epitope specificity) implies that positive-selection of peripheral 4E10 KI B-cells instead occurs via selectively eliminating lipid binding (and/or other correlating reactivities comprising 4E10’s polyreactivity). The distinct residual specificities of 2F5 and 4E10 KI serum IgG for these two neutralization components is intriguing in light of the recent findings of Yang et al. in which KYNU and CMTM3, the conserved mammalian self-antigen targets identified for 2F5, perfectly mimic its complete and core neutralization epitopes, respectively, whereas SF3B3, the primary antigen target of 4E10, has no homology to its epitope (38). Furthermore, in this study, 4E10 was found to have considerable polyreactivity, in addition to its higher lipid affinity, than 2F5 (25, 54).
Collectively, our results therefore provide the first in vivo evidence supporting the notion that 4E10 and 2F5 receptor-expressing B-cells exhibit specificity for markedly distinct host antigens, and suggests this involves 2F5-expressing B-cells (but not 4E10-expressing B-cells) having cross-reactivity for residues in self-antigen(s) that mimic its neutralization epitope. This in turn may have important implications for HIV-1 vaccine research, in that immunization strategies aimed at circumventing tolerance/anergy of bnAbs directed to distinct neutralization MPER epitopes may need to consider designing “minimal” immunogens that mimic components in distinct host antigens. More generally, beyond the field of HIV vaccinology, this study’s identification of KI strains expressing two, related, but distinct anti-viral Igs (i.e., one with high polyreactivity vs. one with selective reactivity for host antigens mimicked by an infectious agent), together, provide an elegant, comparative series of B-cell tolerance models to examine how protective Ab responses are regulated by infectious pathogens which use molecular mimicry for immune evasion.
Supplementary Material
Acknowledgements
We thank the Duke Center for AIDS Research (CFAR) sequencing facility, and the DHVI Flow cytometry and Immune Reconstitution Facilities for expert assistance, as well as Garnett Kelsoe for discussions and critical review.
Abbreviations used
- bnAbs
broadly neutralizing antibodies
- MPER
Membrane-Proximal External Region of gp41
- Env
HIV-1 Envelope
- UCA
Unmutated Common Ancestor
Footnotes
Supported by a CAVD grant from the Bill and Melinda Gates Foundation (to BFH and LKV), the Center For HIV/AIDS Vaccine Immunology NIAID/NIH grant U19AI067854 (to BFH), and NIH grant R01AI087202 (to LKV).
References
- 1.McElrath MJ, Haynes BF. Induction of immunity to human immunodeficiency virus type-1 by vaccination. Immunity. 2010;33:542–554. doi: 10.1016/j.immuni.2010.09.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mascola JR, Stiegler G, VanCott TC, Katinger H, Carpenter CB, Hanson CE, Beary H, Hayes D, Frankel SS, Birx DL, Lewis MG. Protection of macaques against vaginal transmission of a pathogenic HIV-1/SIV chimeric virus by passive infusion of neutralizing antibodies. Nature medicine. 2000;6:207–210. doi: 10.1038/72318. [DOI] [PubMed] [Google Scholar]
- 3.Hessell AJ, Poignard P, Hunter M, Hangartner L, Tehrani DM, Bleeker WK, Parren PW, Marx PA, Burton DR. Effective, low-titer antibody protection against low-dose repeated mucosal SHIV challenge in macaques. Nature medicine. 2009;15:951–954. doi: 10.1038/nm.1974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hessell AJ, Rakasz EG, Poignard P, Hangartner L, Landucci G, Forthal DN, Koff WC, Watkins DI, Burton DR. Broadly neutralizing human anti-HIV antibody 2 G12 is effective in protection against mucosal SHIV challenge even at low serum neutralizing titers. PLoS pathogens. 2009;5:e1000433. doi: 10.1371/journal.ppat.1000433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hessell AJ, Rakasz EG, Tehrani DM, Huber M, Weisgrau KL, Landucci G, Forthal DN, Koff WC, Poignard P, Watkins DI, Burton DR. Broadly neutralizing monoclonal antibodies 2F5 and 4E10 directed against the human immunodeficiency virus type 1 gp41 membrane-proximal external region protect against mucosal challenge by simian-human immunodeficiency virus SHIVBa-L. Journal of virology. 2010;84:1302–1313. doi: 10.1128/JVI.01272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Balazs AB, Chen J, Hong CM, Rao DS, Yang L, Baltimore D. Antibody-based protection against HIV infection by vectored immunoprophylaxis. Nature. 2012;481:81–84. doi: 10.1038/nature10660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Trkola A, Kuster H, Rusert P, Joos B, Fischer M, Leemann C, Manrique A, Huber M, Rehr M, Oxenius A, Weber R, Stiegler G, Vcelar B, Katinger H, Aceto L, Gunthard HF. Delay of HIV-1 rebound after cessation of antiretroviral therapy through passive transfer of human neutralizing antibodies. Nature medicine. 2005;11:615–622. doi: 10.1038/nm1244. [DOI] [PubMed] [Google Scholar]
- 8.McMichael AJ, Borrow P, Tomaras GD, Goonetilleke N, Haynes BF. The immune response during acute HIV-1 infection: clues for vaccine development. Nature reviews. Immunology. 2010;10:11–23. doi: 10.1038/nri2674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haynes BF, Kelsoe G, Harrison SC, Kepler TB. B-cell-lineage immunogen design in vaccine development with HIV-1 as a case study. Nature biotechnology. 2012;30:423–433. doi: 10.1038/nbt.2197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Verkoczy L, Kelsoe G, Moody MA, Haynes BF. Role of immune mechanisms in induction of HIV-1 broadly neutralizing antibodies. Current opinion in immunology. 2011;23:383–390. doi: 10.1016/j.coi.2011.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Haynes BF, Montefiori DC. Aiming to induce broadly reactive neutralizing antibody responses with HIV-1 vaccine candidates. Expert review of vaccines. 2006;5:579–595. doi: 10.1586/14760584.5.4.579. [DOI] [PubMed] [Google Scholar]
- 12.Wyatt R, Sodroski J. The HIV-1 envelope glycoproteins: fusogens, antigens, and immunogens. Science. 1998;280:1884–1888. doi: 10.1126/science.280.5371.1884. [DOI] [PubMed] [Google Scholar]
- 13.Richman DD, Wrin T, Little SJ, Petropoulos CJ. Rapid evolution of the neutralizing antibody response to HIV type 1 infection. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:4144–4149. doi: 10.1073/pnas.0630530100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kwong PD, Doyle ML, Casper DJ, Cicala C, Leavitt SA, Majeed S, Steenbeke TD, Venturi M, Chaiken I, Fung M, Katinger H, Parren PW, Robinson J, Van Ryk D, Wang L, Burton DR, Freire E, Wyatt R, Sodroski J, Hendrickson WA, Arthos J. HIV-1 evades antibody-mediated neutralization through conformational masking of receptor-binding sites. Nature. 2002;420:678–682. doi: 10.1038/nature01188. [DOI] [PubMed] [Google Scholar]
- 15.Wei X, Decker JM, Wang S, Hui H, Kappes JC, Wu X, Salazar-Gonzalez JF, Salazar MG, Kilby JM, Saag MS, Komarova NL, Nowak MA, Hahn BH, Kwong PD, Shaw GM. Antibody neutralization and escape by HIV-1. Nature. 2003;422:307–312. doi: 10.1038/nature01470. [DOI] [PubMed] [Google Scholar]
- 16.Labrijn AF, Poignard P, Raja A, Zwick MB, Delgado K, Franti M, Binley J, Vivona V, Grundner C, Huang CC, Venturi M, Petropoulos CJ, Wrin T, Dimitrov DS, Robinson J, Kwong PD, Wyatt RT, Sodroski J, Burton DR. Access of antibody molecules to the conserved coreceptor binding site on glycoprotein gp120 is sterically restricted on primary human immunodeficiency virus type 1. Journal of virology. 2003;77:10557–10565. doi: 10.1128/JVI.77.19.10557-10565.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Frey G, Peng H, Rits-Volloch S, Morelli M, Cheng Y, Chen B. A fusion-intermediate state of HIV-1 gp41 targeted by broadly neutralizing antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2008;105:3739–3744. doi: 10.1073/pnas.0800255105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Binley JM, Ban YE, Crooks ET, Eggink D, Osawa K, Schief WR, Sanders RW. Role of complex carbohydrates in human immunodeficiency virus type 1 infection and resistance to antibody neutralization. Journal of virology. 2010;84:5637–5655. doi: 10.1128/JVI.00105-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Schief WR, Ban YE, Stamatatos L. Challenges for structure-based HIV vaccine design. Current opinion in HIV and AIDS. 2009;4:431–440. doi: 10.1097/COH.0b013e32832e6184. [DOI] [PubMed] [Google Scholar]
- 20.Stamatatos L, Morris L, Burton DR, Mascola JR. Neutralizing antibodies generated during natural HIV-1 infection: good news for an HIV-1 vaccine? Nature medicine. 2009;15:866–870. doi: 10.1038/nm.1949. [DOI] [PubMed] [Google Scholar]
- 21.Bonsignori M, Alam SM, Liao HX, Verkoczy L, Tomaras GD, Haynes BF, Moody MA. HIV-1 antibodies from infection and vaccination: insights for guiding vaccine design. Trends in microbiology. 2012;20:532–539. doi: 10.1016/j.tim.2012.08.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Haynes BF, Moody MA, Verkoczy L, Kelsoe G, Alam SM. Antibody polyspecificity and neutralization of HIV-1: a hypothesis. Human antibodies. 2005;14:59–67. [PMC free article] [PubMed] [Google Scholar]
- 23.Muster T, Steindl F, Purtscher M, Trkola A, Klima A, Himmler G, Ruker F, Katinger H. A conserved neutralizing epitope on gp41 of human immunodeficiency virus type 1. Journal of virology. 1993;67:6642–6647. doi: 10.1128/jvi.67.11.6642-6647.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Stiegler G, Kunert R, Purtscher M, Wolbank S, Voglauer R, Steindl F, Katinger H. A potent cross-clade neutralizing human monoclonal antibody against a novel epitope on gp41 of human immunodeficiency virus type 1. AIDS research and human retroviruses. 2001;17:1757–1765. doi: 10.1089/08892220152741450. [DOI] [PubMed] [Google Scholar]
- 25.Haynes BF, Fleming J, St Clair EW, Katinger H, Stiegler G, Kunert R, Robinson J, Scearce RM, Plonk K, Staats HF, Ortel TL, Liao HX, Alam SM. Cardiolipin polyspecific autoreactivity in two broadly neutralizing HIV-1 antibodies. Science. 2005;308:1906–1908. doi: 10.1126/science.1111781. [DOI] [PubMed] [Google Scholar]
- 26.Nemazee DA, Burki K. Clonal deletion of B lymphocytes in a transgenic mouse bearing anti-MHC class I antibody genes. Nature. 1989;337:562–566. doi: 10.1038/337562a0. [DOI] [PubMed] [Google Scholar]
- 27.Erikson J, Radic MZ, Camper SA, Hardy RR, Carmack C, Weigert M. Expression of anti-DNA immunoglobulin transgenes in non-autoimmune mice. Nature. 1991;349:331–334. doi: 10.1038/349331a0. [DOI] [PubMed] [Google Scholar]
- 28.Goodnow CC. Transgenic mice and analysis of B-cell tolerance. Annual review of immunology. 1992;10:489–518. doi: 10.1146/annurev.iy.10.040192.002421. [DOI] [PubMed] [Google Scholar]
- 29.Tiegs SL, Russell DM, Nemazee D. Receptor editing in self-reactive bone marrow B cells. The Journal of experimental medicine. 1993;177:1009–1020. doi: 10.1084/jem.177.4.1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gay D, Saunders T, Camper S, Weigert M. Receptor editing: an approach by autoreactive B cells to escape tolerance. The Journal of experimental medicine. 1993;177:999–1008. doi: 10.1084/jem.177.4.999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Verkoczy L, Diaz M, Holl TM, Ouyang YB, Bouton-Verville H, Alam SM, Liao HX, Kelsoe G, Haynes BF. Autoreactivity in an HIV-1 broadly reactive neutralizing antibody variable region heavy chain induces immunologic tolerance. Proceedings of the National Academy of Sciences of the United States of America. 2010;107:181–186. doi: 10.1073/pnas.0912914107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Verkoczy L, Chen Y, Bouton-Verville H, Zhang J, Diaz M, Hutchinson J, Ouyang YB, Alam SM, Holl TM, Hwang KK, Kelsoe G, Haynes BF. Rescue of HIV-1 broad neutralizing antibody-expressing B cells in 2F5 VH × VL knockin mice reveals multiple tolerance controls. J Immunol. 2011;187:3785–3797. doi: 10.4049/jimmunol.1101633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cambier JC, Gauld SB, Merrell KT, Vilen BJ. B-cell anergy: from transgenic models to naturally occurring anergic B cells? Nature reviews. Immunology. 2007;7:633–643. doi: 10.1038/nri2133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hartley SB, Crosbie J, Brink R, Kantor AB, Basten A, Goodnow CC. Elimination from peripheral lymphoid tissues of self-reactive B lymphocytes recognizing membrane-bound antigens. Nature. 1991;353:765–769. doi: 10.1038/353765a0. [DOI] [PubMed] [Google Scholar]
- 35.Chen C, Nagy Z, Radic MZ, Hardy RR, Huszar D, Camper SA, Weigert M. The site and stage of anti-DNA B-cell deletion. Nature. 1995;373:252–255. doi: 10.1038/373252a0. [DOI] [PubMed] [Google Scholar]
- 36.Montero M, van Houten NE, Wang X, Scott JK. The membrane-proximal external region of the human immunodeficiency virus type 1 envelope: dominant site of antibody neutralization and target for vaccine design. Microbiology and molecular biology reviews: MMBR. 2008;72:54–84. doi: 10.1128/MMBR.00020-07. table of contents. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zwick MB, Komori HK, Stanfield RL, Church S, Wang M, Parren PW, Kunert R, Katinger H, Wilson IA, Burton DR. The long third complementarity-determining region of the heavy chain is important in the activity of the broadly neutralizing anti-human immunodeficiency virus type 1 antibody 2F5. Journal of virology. 2004;78:3155–3161. doi: 10.1128/JVI.78.6.3155-3161.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang G, Holl TM, Liu Y, Li Y, Lu X, Nicely NI, Kepler TB, Alam SM, Liao HX, Cain DW, Spicer L, Vandeberg JL, Haynes BF, Kelsoe G. Identification of autoantigens recognized by the 2F5 and 4E10 broadly neutralizing HIV-1 antibodies. The Journal of experimental medicine. 2013;210:241–256. doi: 10.1084/jem.20121977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Verkoczy L, Moody MA, Holl TM, Bouton-Verville H, Scearce RM, Hutchinson J, Alam SM, Kelsoe G, Haynes BF. Functional, non-clonal IgMa-restricted B cell receptor interactions with the HIV-1 envelope gp41 membrane proximal external region. PloS one. 2009;4:e7215. doi: 10.1371/journal.pone.0007215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Tiller T, Busse CE, Wardemann H. Cloning and expression of murine Ig genes from single B cells. Journal of immunological methods. 2009;350:183–193. doi: 10.1016/j.jim.2009.08.009. [DOI] [PubMed] [Google Scholar]
- 41.Thali M, Moore JP, Furman C, Charles M, Ho DD, Robinson J, Sodroski J. Characterization of conserved human immunodeficiency virus type 1 gp120 neutralization epitopes exposed upon gp120-CD4 binding. Journal of virology. 1993;67:3978–3988. doi: 10.1128/jvi.67.7.3978-3988.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hardy RR. B-1 B cell development. J Immunol. 2006;177:2749–2754. doi: 10.4049/jimmunol.177.5.2749. [DOI] [PubMed] [Google Scholar]
- 43.Li Y, Li H, Weigert M. Autoreactive B cells in the marginal zone that express dual receptors. The Journal of experimental medicine. 2002;195:181–188. doi: 10.1084/jem.20011453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Goodnow CC, Crosbie J, Adelstein S, Lavoie TB, Smith-Gill SJ, Brink RA, Pritchard-Briscoe H, Wotherspoon JS, Loblay RH, Raphael K, et al. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 1988;334:676–682. doi: 10.1038/334676a0. [DOI] [PubMed] [Google Scholar]
- 45.Goodnow CC, Brink R, Adams E. Breakdown of self-tolerance in anergic B lymphocytes. Nature. 1991;352:532–536. doi: 10.1038/352532a0. [DOI] [PubMed] [Google Scholar]
- 46.Merrell KT, Benschop RJ, Gauld SB, Aviszus K, Decote-Ricardo D, Wysocki LJ, Cambier JC. Identification of anergic B cells within a wild-type repertoire. Immunity. 2006;25:953–962. doi: 10.1016/j.immuni.2006.10.017. [DOI] [PubMed] [Google Scholar]
- 47.Roark JH, Bui A, Nguyen KA, Mandik L, Erikson J. Persistence of functionally compromised anti-double-stranded DNA B cells in the periphery of non-autoimmune mice. International immunology. 1997;9:1615–1626. doi: 10.1093/intimm/9.11.1615. [DOI] [PubMed] [Google Scholar]
- 48.Nguyen KA, Mandik L, Bui A, Kavaler J, Norvell A, Monroe JG, Roark JH, Erikson J. Characterization of anti-single-stranded DNA B cells in a non-autoimmune background. J Immunol. 1997;159:2633–2644. [PubMed] [Google Scholar]
- 49.Rojas M, Hulbert C, Thomas JW. Anergy and not clonal ignorance determines the fate of B cells that recognize a physiological autoantigen. J Immunol. 2001;166:3194–3200. doi: 10.4049/jimmunol.166.5.3194. [DOI] [PubMed] [Google Scholar]
- 50.Culton DA, O’Conner BP, Conway KL, Diz R, Rutan J, Vilen BJ, Clarke SH. Early preplasma cells define a tolerance checkpoint for autoreactive B cells. J Immunol. 2006;176:790–802. doi: 10.4049/jimmunol.176.2.790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borrero M, Clarke SH. Low-affinity anti-Smith antigen B cells are regulated by anergy as opposed to developmental arrest or differentiation to B-1. J Immunol. 2002;168:13–21. doi: 10.4049/jimmunol.168.1.13. [DOI] [PubMed] [Google Scholar]
- 52.Kilmon MA, Rutan JA, Clarke SH, Vilen BJ. Low-affinity, Smith antigen-specific B cells are tolerized by dendritic cells and macrophages. J Immunol. 2005;175:37–41. doi: 10.4049/jimmunol.175.1.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Acevedo-Suarez CA, Kilkenny DM, Reich MB, Thomas JW. Impaired intracellular calcium mobilization and NFATc1 availability in tolerant anti-insulin B cells. J Immunol. 2006;177:2234–2241. doi: 10.4049/jimmunol.177.4.2234. [DOI] [PubMed] [Google Scholar]
- 54.Sanchez-Martinez S, Lorizate M, Katinger H, Kunert R, Nieva JL. Membrane association and epitope recognition by HIV-1 neutralizing anti-gp41 2F5 and 4E10 antibodies. AIDS research and human retroviruses. 2006;22:998–1006. doi: 10.1089/aid.2006.22.998. [DOI] [PubMed] [Google Scholar]
- 55.Scherer EM, Zwick MB, Teyton L, Burton DR. Difficulties in eliciting broadly neutralizing anti-HIV antibodies are not explained by cardiolipin autoreactivity. AIDS. 2007;21:2131–2139. doi: 10.1097/QAD.0b013e3282a4a632. [DOI] [PubMed] [Google Scholar]
- 56.Alam SM, Morelli M, Dennison SM, Liao HX, Zhang R, Xia SM, Rits-Volloch S, Sun L, Harrison SC, Haynes BF, Chen B. Role of HIV membrane in neutralization by two broadly neutralizing antibodies. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:20234–20239. doi: 10.1073/pnas.0908713106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Li H, Jiang Y, Prak EL, Radic M, Weigert M. Editors and editing of anti-DNA receptors. Immunity. 2001;15:947–957. doi: 10.1016/s1074-7613(01)00251-5. [DOI] [PubMed] [Google Scholar]
- 58.Minegishi Y, Conley ME. Negative selection at the pre-BCR checkpoint elicited by human mu heavy chains with unusual CDR3 regions. Immunity. 2001;14:631–641. doi: 10.1016/s1074-7613(01)00131-5. [DOI] [PubMed] [Google Scholar]
- 59.Ivanov II, Schelonka RL, Zhuang Y, Gartland GL, Zemlin M, Schroeder HW. Development of the expressed Ig CDR-H3 repertoire is marked by focusing of constraints in length, amino acid use, and charge that are first established in early B cell progenitors. J Immunol. 2005;174:7773–7780. doi: 10.4049/jimmunol.174.12.7773. [DOI] [PubMed] [Google Scholar]
- 60.Martin DA, Bradl H, Collins TJ, Roth E, Jack HM, Wu GE. Selection of Ig mu heavy chains by complementarity-determining region 3 length and amino acid composition. J Immunol. 2003;171:4663–4671. doi: 10.4049/jimmunol.171.9.4663. [DOI] [PubMed] [Google Scholar]
- 61.Wang H, Ye J, Arnold LW, McCray SK, Clarke SH. A VH12 transgenic mouse exhibits defects in pre-B cell development and is unable to make IgM+ B cells. J Immunol. 2001;167:1254–1262. doi: 10.4049/jimmunol.167.3.1254. [DOI] [PubMed] [Google Scholar]
- 62.von Boehmer H, Melchers F. Checkpoints in lymphocyte development and autoimmune disease. Nature immunology. 2010;11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- 63.Vettermann C, Jack HM. The pre-B cell receptor: turning autoreactivity into self-defense. Trends in immunology. 2010;31:176–183. doi: 10.1016/j.it.2010.02.004. [DOI] [PubMed] [Google Scholar]
- 64.Herzog S, Jumaa H. Self-recognition and clonal selection: autoreactivity drives the generation of B cells. Current opinion in immunology. 2012;24:166–172. doi: 10.1016/j.coi.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 65.Huang J, Ofek G, Laub L, Louder MK, Doria-Rose NA, Longo ,NS, Imamichi H, Bailer RT, Chakrabarti B, Sharma SK, Alam SM, Wang T, Yang Y, Zhang B, Migueles SA, Wyatt R, Haynes BF, Kwong PD, Mascola JR, Connors M. Broad and potent neutralization of HIV-1 by a gp41-specific human antibody. Nature. 2012;491:406–412. doi: 10.1038/nature11544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Alam SM, Liao HX, Dennison SM, Jaeger F, Parks R, Anasti K, Foulger A, Donathan M, Lucas J, Verkoczy L, Nicely N, Tomaras GD, Kelsoe G, Chen B, Kepler TB, Haynes BF. Differential reactivity of germ line allelic variants of a broadly neutralizing HIV-1 antibody to a gp41 fusion intermediate conformation. Journal of virology. 2011;85:11725–11731. doi: 10.1128/JVI.05680-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ota T, Doyle-Cooper C, Cooper AB, Huber M, Falkowska E, Doores KJ, Hangartner L, Le K, Sok D, Jardine J, Lifson J, Wu X, Mascola JR, Poignard P, Binley JM, Chakrabarti BK, Schief WR, Wyatt RT, Burton DR, Nemazee D. Anti-HIV B Cell Lines as Candidate Vaccine Biosensors. J Immunol. 2012;189:4816–4824. doi: 10.4049/jimmunol.1202165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Peron S, Laffleur B, Denis-Lagache N, Cook-Moreau J, Tinguely A, Delpy L, Denizot Y, Pinaud E, Cogne M. AID-driven deletion causes immunoglobulin heavy chain locus suicide recombination in B cells. Science. 2012;336:931–934. doi: 10.1126/science.1218692. [DOI] [PubMed] [Google Scholar]
- 69.Jiang C, Zhao ML, Scearce RM, Diaz M. Activation-induced deaminase-deficient MRL/lpr mice secrete high levels of protective antibodies against lupus nephritis. Arthritis and rheumatism. 2011;63:1086–1096. doi: 10.1002/art.30230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Zhang Z, Zemlin M, Wang YH, Munfus D, Huye LE, Findley HW, Bridges SL, Roth DB, Burrows PD, Cooper MD. Contribution of Vh gene replacement to the primary B cell repertoire. Immunity. 2003;19:21–31. doi: 10.1016/s1074-7613(03)00170-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.