Abstract
Background
Exclusion from a social group is an effective way to avoid parasite transmission. This type of social removal has also been proposed as a form of collective defense, or social immunity, in eusocial insect groups. If parasitic modification of host behavior is widespread in social insects, the underlying physiological and neuronal mechanisms remain to be investigated. We studied this phenomenon in honey bees parasitized by the mite Varroa destructor or microsporidia Nosema ceranae, which make bees leave the hive precociously. We characterized the chemical, behavioral and neurogenomic changes in parasitized bees, and compared the effects of both parasites.
Results
Analysis of cuticular hydrocarbon (CHC) profiles by gas chromatography coupled with mass spectrophotometry (GC-MS) showed changes in honey bees parasitized by either Nosema ceranae or Varroa destructor after 5 days of infestation. Levels of 10-HDA, an antiseptic important for social immunity, did not change in response to parasitism. Behavioral analysis of N. ceranae- or V. destructor- parasitized bees revealed no significant differences in their behavioral acts or social interactions with nestmates. Digital gene expression (DGE) analysis of parasitized honey bee brains demonstrated that, despite the difference in developmental stage at which the bee is parasitized, Nosema and Varroa-infested bees shared more gene changes with each other than with honey bee brain expression gene sets for forager or nurse castes.
Conclusions
Parasitism by Nosema or Varroa induces changes to both the CHC profiles on the surface of the bee and transcriptomic profiles in the brain, but within the social context of the hive, does not result in observable effects on her behavior or behavior towards her. While parasitized bees are reported to leave the hive as foragers, their brain transcription profiles suggest that their behavior is not driven by the same molecular pathways that induce foraging behavior.
Keywords: Varroa destructor, Nosema ceranae, Social immunity, Transcriptome, Cuticular hydrocarbons
Background
Behavioral defenses are “strikingly effective” mechanisms for combating parasites but are often overlooked by studies addressing the effects of a parasite on the host immune system and related physiological processes [1]. For a behavior to be considered as an anti-parasite defense the parasite must impose a cost on the host and the behavior should limit or eliminate the parasite [2,3]. Parasite avoidance behavior is found across animal taxa, where it manifests as behaviors such as cleaning and avoiding feces or avoiding diseased conspecifics [4]. For example, normally gregarious spiny Caribbean lobsters shun lobsters that carry a lethal virus, even before the diseased lobster is visibly infected [5]. However, not all behaviors are anti-parasite, as parasites can also alter the behavior of their hosts in ways that are ultimately beneficial to the parasite or its offspring [6,7].
Living in groups can further complicate host-parasite interactions since group members can also play a role in the regulation of anti-parasite behavior. Collective defenses against parasites within a social group, called social immunity, are physiological, behavioral or organizational adaptations that prevent the transmission of the parasite [8,9]. A common social defense is the removal of the parasitized individuals from the social group. Indeed, in social insects, parasitized individuals can remove themselves from the group, which corresponds to an altruistic self-removal [10], or nestmates can modify their interaction with those individuals to prevent parasite transmission [6,9]. For example, in honey bees, very different types of stress, including exposure to the mite Varroa[11,12], the microsporidia Nosema ceranae[13,14], or immune challenge [15] have been shown to induce precocious foraging or forager-like physiological and behavioral characteristics. Leaving the colony to perform outside activities, like foraging, limits contact in the hive, especially with castes of great importance (e.g. queen, brood), and thus, the spread of parasites into the colony [9]. However, despite parasite modification of host behavior being widespread in animals, the underlying physiological and neuronal mechanisms are poorly understood, especially in social insects.
In order to better understand this phenomenon, we analyzed how two different parasites, an ectoparasite (Varroa destructor) and endoparasite (Nosema ceranae), affect the physiology and the brain neurogenomic state of honey bees. The Varroa destructor mite infects the larval cell immediately before capping where it feeds on the developing pupae and completes its reproductive cycle. It weakens the honey bee by feeding on its hemolymph and transmitting viruses, such as deformed wing virus (DWV), which are correlated with its effect on honey bee survivorship [16-18]. Nosema species are obligate, intracellular spore-forming fungal parasites that infect honey bee adults from emergence by spreading through the hive, most likely through the activities of cleaning and trophallaxis [19]. Once a worker has ingested Nosema spores, the spores develop in the intestine of the bee, where the germinated microsporidian infects the epithelial cell layer of the midgut and consumes the energy of the cell [20,21]. However, despite Varroa and Nosema infections varying greatly in their pathologies, they affect honey bee behavior and learning abilities in similar ways (see Table 1). For example, both Varroa-infested and Nosema-infected bees showed impaired orientation abilities at the hive entrance [22,23].
Table 1.
Comparative pathologies of Varroa destructor and Nosema ceranae
| Varroa destructor | Nosema ceranae | |
|---|---|---|
| Type of parasite |
ectoparasite |
endoparasite |
| Mode of action |
Sucks hemolymph/transmits viruses |
Attacks epithelium of the gut |
| Mode of transmission |
Enters brood cell before operculation; phoresis-carried between cells by adult bees (males and females) |
Oral transmission between workers, matrices of the hive |
| Stage(s) attacked |
Nymph/adult |
Adult |
| Physiological effects |
Reduced weight, metabolism, vitellogenin titers, and proportion of normal hemocytes; increased ecdysteroid titers [24,25] |
Increase oxidative stress; degeneration of gut epithelium [26]; reduced vitellogenin level [27]; induces pheromone (ethyl oleate) production [28] |
| Lifespan decrease [12] |
Lifespan decrease |
|
| Behavioral effects |
Impaired orientation [22] |
Impaired orientation [29] |
| |
Accelerated maturation [11,12] |
Accelerated maturation [13,14] |
| Faster habituation and lower response probability to odor stimulus but no change in sucrose response [30] | Increased sucrose response based on PER test [31] |
Since there is a robust association between brain gene expression in the honey bee and its behavioral state [29,32,33], we measured the brain transcriptional changes induced by V. destructor and N. ceranae and determined whether they induce similar brain host responses. We also characterized the cuticular hydrocarbon (CHC) profiles of honey bees parasitized by V. destructor or N. ceranae and recorded nestmate interactions in observation hives in order to detect nestmate aggression toward parasitized bees. Indeed, challenging the immune system of bees with lipopolysaccharides or other non-living immune stimulants changed the CHC profiles of the bees, involved in social recognition, which lead to modified and aggressive conspecific contacts in a laboratory-based nestmate recognition assay [34,35]. Bees infected with the virus DWV, that showed changes in their CHC profiles, were also ejected from the hive at higher rates than healthy bees, notably from healthy hives [36]. Finally, we determined whether parasitism can affect the level of production of 10-HDA that contributes to social immunity of the colony. This compound, produced in the mandibular glands of bees, displays antiseptic properties in the royal jelly [37].
Results
Experiment 1: Chemical analysis of Nosema ceranae- or Varroa destructor-parasitized bees
Cuticular hydrocarbon profiles
Whether parasites induced changes in the cuticular hydrocarbon profiles (alkanes, alkenes, alkynes and methylalkanes) was tested in 11 to 12 bees per colony at days 5 and 10 post-emergence. The hydrocarbons were extracted from control and parasitized bees and analyzed and identified using gas chromatography (GC) followed by mass spectrometry (MS).
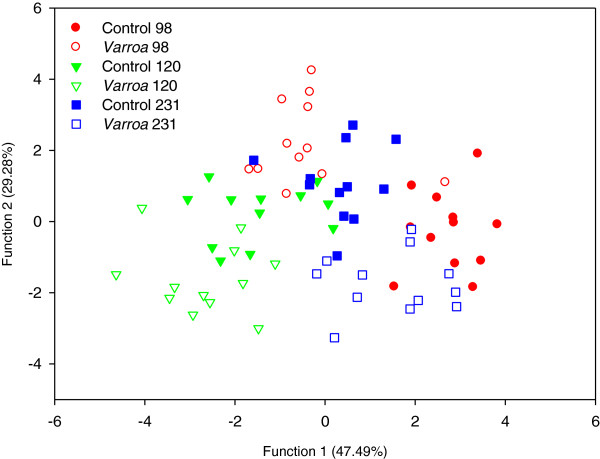
We did not find new compounds in parasitized bees (Nosema and Varroa) as compared to control groups. Comparisons of the relative proportions of peaks corresponding to specific compounds did not reveal overwhelming differences between the Nosema-infected and control groups (Additional file 1: Table S1). However, the comparison of the overall chemical profiles, via discriminant analysis, of Nosema-infected bees and their control counterparts at days 5 and 10 showed highly significant differences for each colony (Colony 98: Wilks’ λ = 0.015, F39,92 = 7.39, p < 0.0001; Colony 120: Wilks’ λ = 0.011, F39,.95 = 8.73, p < 0.0001; Colony 231: Wilks’ λ = 0.037, F27,102 = 7.92, p < 0.0001; Figure 1). For each colony of origin, the squared Mahalanobis distance between Nosema-infected and control bees at 5 days old was not significant after Bonferoni correction (Table 2). Conversely, Mahalanobis chemical distances for older bees (10-day-old) were all statistically significant between infected and control bees (Table 2). The cuticular hyrocarbon profiles changed also with aging in both control and parasitized bees (Figure 1, Table 2). Cuticular profiles of worker bees were quite specific as it was possible to correctly assign between 72-100% of all workers from each age and infected groups (Additional file 2: Table S2).
Figure 1.
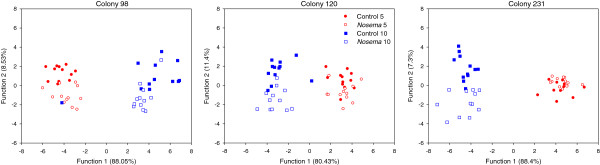
Nosema-infected bees developed different cuticular hydrocarbons profiles. Discriminant analysis based on the cuticular hydrocarbons profiles of Nosema-infected and control bees at day 5 and 10. The analysis was repeated on bees from 3 different colonies (N = 11–12 bees/colony and treatment). Young (5 days) parasitized and control bees did not display different chemical profiles, but 5 days later both profiles were distinct (see Table 2).
Table 2.
Pair-wise squared Mahalanobis distances between the chemical profile of Nosema-infected and control bees at day 5 and 10
| Colony # | Treatment | Nosema5 | Control10 | Nosema10 |
|---|---|---|---|---|
| Colony 98 |
Control5 |
7.2 |
68.62 |
47.92 |
|
P = 0.023 |
P < 0.001 |
P < 0.001 |
||
| |
Nosema5 |
|
72.85 |
58.03 |
|
P < 0.001 |
P < 0.001 |
|||
| |
Control10 |
|
|
9.55 |
| |
P = 0.0055 |
|||
| Colony 120 |
Control5 |
6.54 |
77.64 |
74 |
|
P = 0.034 |
P < 0.001 |
P < 0.001 |
||
| |
Nosema5 |
|
77.93 |
77.45 |
|
P < 0.001 |
P < 0.001 |
|||
| |
Control10 |
|
|
9.59 |
|
P = 0.0035 | ||||
| Colony 231 |
Control5 |
4.28 |
31.22 |
34.92 |
|
P = 0.044 |
P < 0.001 |
P < 0.001 |
||
| |
Nosema5 |
|
31.89 |
34.09 |
|
P < 0.001 |
P < 0.001 |
|||
| |
Control10 |
|
|
5.99 |
| P = 0.0057 | ||||
P-values that are significant at the 5% probability after Bonferroni's correction (P < 0.0083) are indicated in bold. In the treatment column, 5 and 10 indicate the age of the bees. The experiment was repeated on bees from 3 different colonies.
For Varroa-parasitized bees, the cuticular hydrocarbon profiles were compared with control bees on day 5 only (see Methods). The discriminant analysis also showed significant differences overall between parasitized and non-parasitized bees (Wilks’ λ = 0.027, F75,253 = 3.79, p < 0.0001, Figure 2). Mahalanobis chemical distances between Varroa-infested and control bees were highly significant and the colony of origin also had an important influence on the cuticular hydrocarbon profiles (Table 3). Between 75 and 100% of the individuals were correctly classified according to their chemical profiles (Additional file 2: Table S3).
Figure 2.
Varroa-infested bees developed different cuticular hydrocarbons profiles. Discriminant analysis based on the cuticular hydrocarbons profiles of Varroa-infested and control bees. The analysis was repeated on bees from 3 different colonies (N = 12 bees/colony and treatment).
Table 3.
Pair-wise squared Mahalanobis distances between the chemical profile of Varroa-parasitized and control bees
| Treatment | Varroa98 | Control120 | Varroa120 | Control231 | Varroa231 |
|---|---|---|---|---|---|
| Control98 |
17.46 |
18.02 |
28.31 |
9.61 |
6.2 |
|
P < 0.001 |
P < 0.001 |
P < 0.001 |
P = 0.0015 |
P = 0.038 |
|
|
Varroa98 |
|
12.38 |
18.19 |
6.4 |
17.07 |
|
P = 0.0012 |
P < 0.001 |
P = 0.032 |
P < 0.001 |
||
| Control120 |
|
|
9.09 |
8.34 |
17.7 |
| |
P = 0.0025 |
P = 0.005 |
P < 0.001 |
||
|
Varroa120 |
|
|
|
16.68 |
17.8 |
| |
|
P < 0.001 |
P < 0.001 |
||
| Control231 |
|
|
|
|
9.72 |
| P = 0.0014 |
P-values that are significant at the 5% probability after Bonferroni's correction (P < 0.0083) are indicated in bold. The numbers 98, 120 and 231 indicate the colony origin of the bees.
The relative proportion of each hydrocarbon affected by Nosema and Varroa is listed in (Additional file 1: Table S1). Some of them were significantly affected by the parasites and age but there was no consistent effect of age and parasite on the relative proportions of each compound.
10-HDA levels
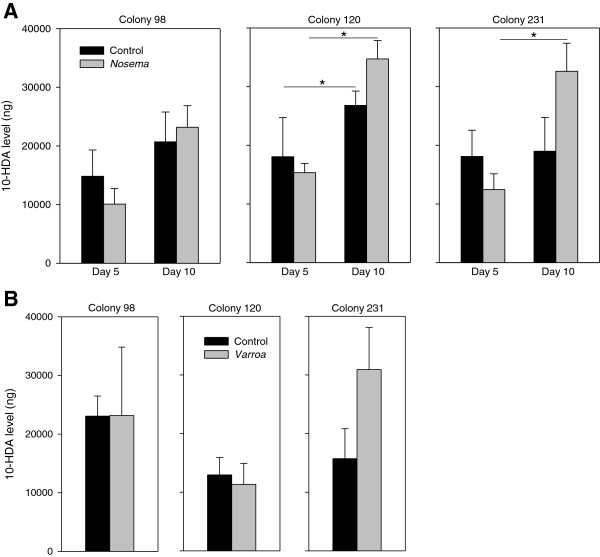
We compared 10-HDA levels in the heads of Nosema- and Varroa-parasitized bees to control bees from the same colony (Figure 3A and B). For the comparison of Nosema-parasitized and control bees, there was no significant difference in 10-HDA levels, but for two colonies 10-HDA showed a significant increase with age (Figure 3A). Levels of 10-HDA did not differ between Varroa-parasitized and control bees (Figure 3B).
Figure 3.
Parasitism did not affect the levels of 10-HDA levels in (A) Nosema- or (B) Varroa-parasitized bees. The 10-HDA levels did not differ between Nosema-infected and control bees at day 5 and 10 but increase with age in colony 120 and 231 (except for control) (Kruskall-Wallis tests: Colony 98: H = 4.53, P = 0.2; Colony 120: H = 19.32, P < 0.001; Colony 231: H = 8.57, P = 0.035; * denotes significant differences after Conover-Iman post-hoc tests, P < 0.05). Varroa did not modify the 10-HDA levels (Mann–Whitney tests: Colony 98: P = 0.38, Colony 120: P = 0.68; Colony 231: P = 0.058).
Experiment 2: Behavioral analysis of Nosema ceranae- or Varroa destructor-parasitized bees
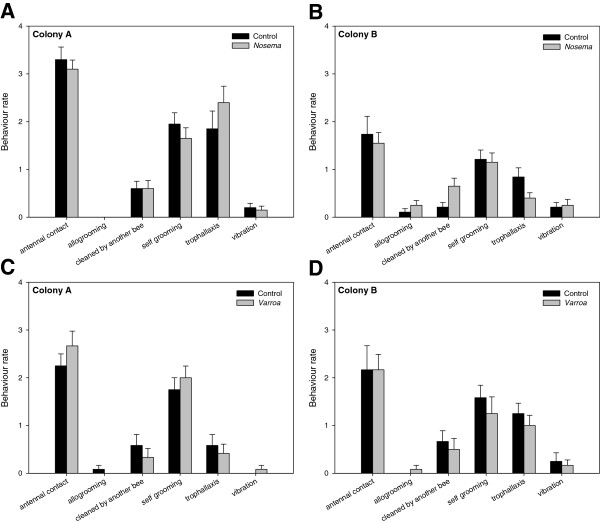
In two four-frame observation hives, we quantified a suite of social behaviors (antennal contact, allogrooming, self-grooming, cleaned by another bee, trophallaxis and vibration) undertaken or received by Nosema-parasitized (N = 40) and control bees (N = 39). No significant differences were observed between parasitized and healthy bees in the rate of behavioral acts or social interactions (Figure 4A and B). Similarly, Varroa-parasitized bees did not display different behavior and were not treated differently by nestmates as compared to control individuals (N = 24 for each group) (Figure 4C and D). In addition, we did not see any agonistic behavior toward parasitized bees during the observation periods (600 min for Nosema-parasitized and 585 min for control bees; 360 min for both Varroa-parasitized and control bees).
Figure 4.
Parasitism did not induce different behavioral treatment by colony nestmates. The rate of behavioral acts or interactions did not differ between Nosema-infected and control bees (A and B), and Varroa-infested and control bees (C and D) (Mann–Whitney tests, P > 0.05 for each behavior). The experiment was performed on two colonies (19–20 bees/state in the Nosema experiment and 12 bees/state in the Varroa experiment).
Experiment 3: Brain transcriptomics of Nosema ceranae- or Varroa destructor- parasitized bees
The brain transcriptome modifications induced by parasitism were determined in bees originating from two different colonies using digital gene expression (DGE) analysis. Two DGE-tag libraries were generated for each experimental group, control, Nosema-infected and Varroa-infested. For each library more than 100,000,000 tags were produced that were then narrowed down to around 9,300 unique gene hits for the honey bee (Additional file 3: Table S4). These distinct tags and their genomic count are available from NCBI Gene Expression Omnibus (GEO) database with the accession number: GSE41109.
The number of genes whose expression was affected in the bee brain by Nosema- and Varroa-parasitism was markedly different. In Varroa-infested bees 455 genes changed overall (225 up- and 230 downregulated) while in Nosema-infected bees only 57 genes responded differentially (12 up- and 45 downregulated) at an adjusted P-value < 0.05 (Additional file 4: Table S5).
Gene Ontology analysis revealed that gene expression changes in Varroa-infested brains (represented by 265 fly orthologs) were most significantly overrepresented in the metallopeptidase functional group with changes in both directions (Table 4). Varroa-infested bees showed decreased expression of glutamate and GABA receptor-related genes (GB15851, GB14954, GB13604, GB15167, GB18621), and the dopamine receptor, Amdop1, and overexpression of ascorbate/aldarate metabolism genes (GB17015, GB16747, GB14956, GB15000), which include dopamine and serotonin metabolism [38-40]. Conversely, both glutamate decarboyxlase 1, which synthesizes GABA neurotransmitter, and the GABA neurotransmitter transporter 1B were overexpressed in Varroa-parasitized brains (Table 4). Nosema-infected brains, for which only 24 genes had Flybase orthologs, showed no significant patterns in the classification of functional groups. Several genes that are involved in immune and antioxidant activity, defensin-1, peroxidase, esterase A2, glucose oxidase, flavin-containing monooxygenase, were upregulated.
Table 4.
Functional analysis of genes regulated by Varroa parasitism
| Biological process/ Molecular Function | P-value | # upregulated genes | # downregulated genes |
|---|---|---|---|
| Metallopeptidase activity |
0.00193 |
5 |
4 |
| Ascorbate and aldarate metabolism |
0.0106 |
4 |
0 |
| GPCR, family 3, C-terminal |
0.0126 |
0 |
3 |
| Metalloendopeptidase activity |
0.0127 |
2 |
4 |
| Nucleophile |
0.0150 |
2 |
3 |
| Carbohydrate binding |
0.0170 |
2 |
5 |
| Integral/intrinsic to plasma membrane |
0.0243 |
4 |
6 |
| Glutamate receptor activity |
0.0320 |
0 |
4 |
| Pattern/ Polysaccharide binding |
0.0331 |
1 |
4 |
| Starch and sucrose metabolism |
0.0338 |
3 |
1 |
| Peptidase activity, acting on L-amino acid peptides |
0.0379 |
10 |
5 |
| Hydrolase |
0.0431 |
20 |
16 |
| Developmental growth |
0.0471 |
3 |
3 |
| Metalloprotease | 0.0482 | 2 | 2 |
Gene ontology biological process and molecular function categories that were overrepresented in the brain transcriptomic profile are shown (P < 0.05).
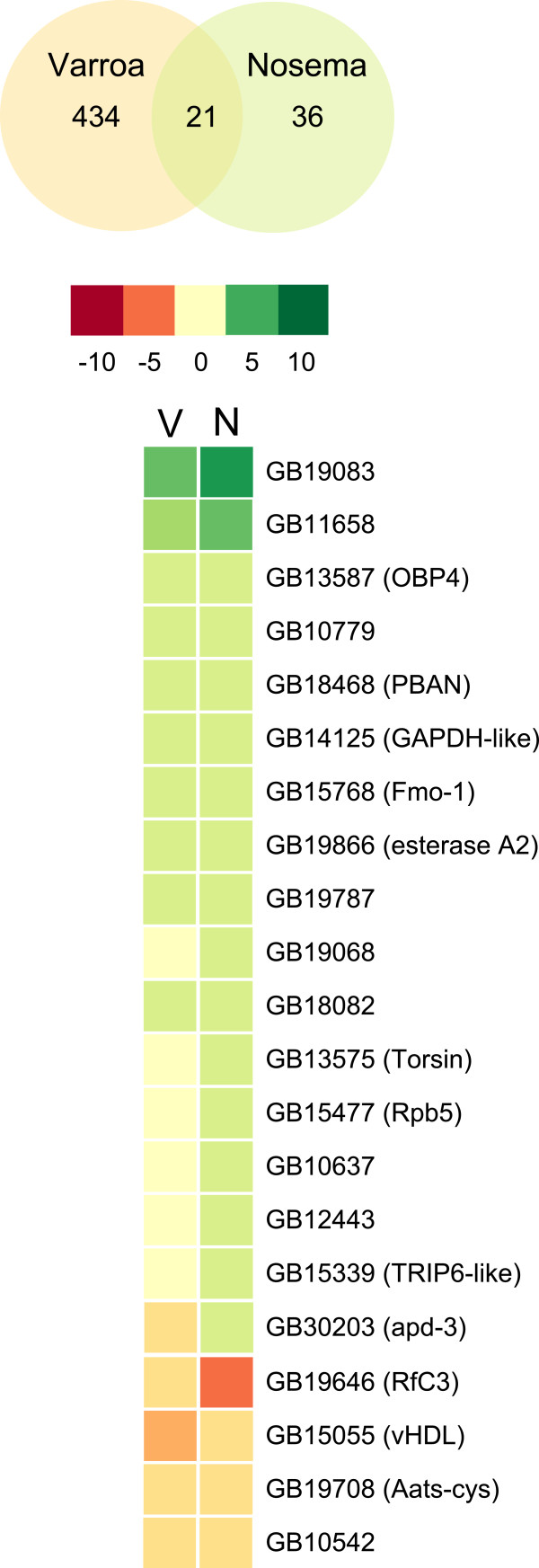
Despite the difference in overall number of genes affected by both parasites as compared to controls and the 245 genes that changed between Varroa and Nosema parasitism (Additional file 4: Table S5), Nosema- and Varroa- parasitized bees shared more gene changes with each other (21 genes, Figure 5) than expected by chance (5.6 times more genes). In addition, Nosema parasitism caused a brain gene expression profile that was similar to the profile of bees parasitized by Varroa; except for apidermin 3 (GB30203), genes that were up- and downregulated by Nosema were also up- and downregulated by Varroa, giving a significant pattern (χ2 = 11.049, P < 0.001, Figure 5). We also tested gene expression overlap with brain gene expression data from nurse/forager, i.e. genes known to be upregulated in nurse brains as compared to forager brains and the other way round [41]. We found respectively 8 and 34 genes affected by Nosema and Varroa parasites to overlap with the nurse/forager sets (Additional file 4: Table S5) but neither of these gene sets was significantly similar to nurse and forager bees (χ2 = 0.08, P = 0.78 and χ2 = 0.58, P = 0.45 for Nosema and Varroa, respectively).
Figure 5.
Expression levels show similar directionality for 20 brain genes commonly affected by Nosema and Varroa parasites. For the total number of genes changed by parasitism by Varroa (N = 455 genes) or Nosema (N = 57 genes), a statistically significant number of genes occur in both lists (N = 21) with 20 genes expressed in the same direction (Exact hypergeometric probability test: P < 0.0001). Color scale for the heatmap (red to green) indicates log2 transcription ratios where red color indicates underexpression of the gene in the parasitized bee and green color indicates overexpression. For each gene, the accession number and annotation are indicated.
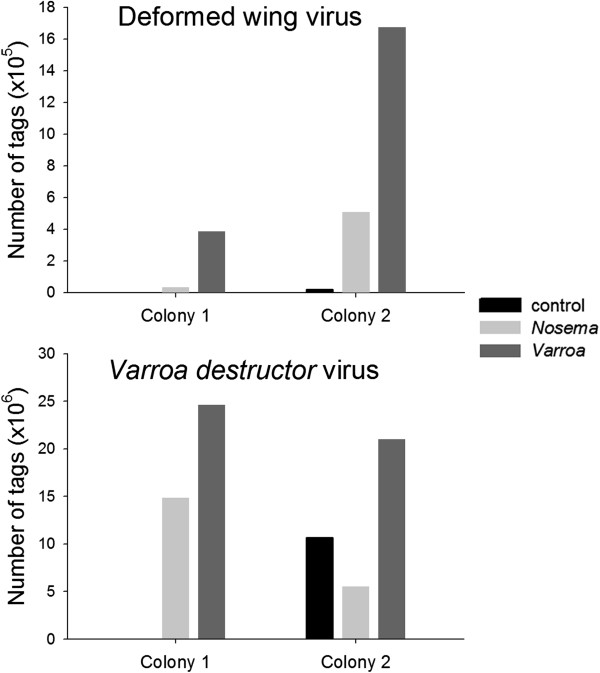
Using the DGE-tag libraries, we determined whether parasites affected the viral landscape in the bee brain. We looked for presence and abundance of 9 viruses: Chronic bee paralysis virus RNA 1, Chronic bee paralysis virus RNA 2, Sacbrood virus, Deformed wing virus, Black queen cell virus, Acute bee paralysis virus, Kashmir bee virus, Varroa destructor virus 1 and Israel acute paralysis virus. Only two viruses were identified in the bee brains, Deformed wing virus (DWV) and Varroa destructor virus (Figure 6). Varroa-infested bees had the highest levels of DWV compared to control bees and higher levels of Deformed wing virus than Nosema-infected bees (Figure 6, Additional file 5). Nosema-infected bees also had higher levels of DWV than control bees, at the limit of statistical significance. Varroa destructor virus levels were not statistically different across control, Varroa- and Nosema- parasitized bees.
Figure 6.
Deformed wing virus titers increased in the honey bee brain with parasitism. Viral titers are expressed as the total number of tags in a sample for each colony individually (Colony 1 and Colony 2). Deformed wing virus (DWV) levels were significantly higher in Varroa-infested bees compared to Nosema- and control bees (Generalized Linear Model with Cox-Reid method for estimating dispersion; Varroa vs. control: P < 0.05, Varroa vs. Nosema:P < 0.01), while increased DWV levels in Nosema-infested bees were marginally significant (P < 0.10). Varroa destructor virus (VDV) titers did not differ between parasitized and control bees (For P-values see Additional file 5: Table S6.)
Discussion
In this study, we demonstrated that two parasites, Varroa destructor and Nosema ceranae, with distinct, differing pathologies both modified the physiology and transcriptomic profiles in the brain of their honey bee host. Parasitized honey bees exhibited changes in their CHC profiles but showed no differences in behaviors between parasitized and healthy bees. In addition we observed no significant aggressive behavior towards parasitized bees, nor any change in social interactions.
A previous study found that bees parasitized by Varroa exhibit a modified CHC at their emergence [42]. Our data shows that this change is lasting. However, our behavioral results differ from recent studies that observed increased general social interactions or aggressive behaviors towards immune-challenged bees, but differences in the experimental design of our study may account for those differences. In a series of studies, increased social and aggressive behaviors towards immune-suppressed workers were observed a few hours after treatment in assays that were conducted in the laboratory [34,35]. Our study employed natural conditions of a four-frame observation hive using bees that had been parasitized in the days prior to the experiment, in order to understand if nestmates respond to parasitized bees within the context of general activity of the hive. Moreover we chose to focus on bees that were Varroa-parasitized but asymptomatic for DWV. In contrast, a second study found that DWV-infected bees that exhibited deformed wing symptoms were detected and removed from the hive by nestmates [36]. Thus our results do not contradict previous studies but reflect the subtle nature of parasitism by Varroa or Nosema that, when resulting in precocious departure from the hive, is more likely due to altruistic self-removal, acting as a mechanism of social immunity. Indeed, it may be less costly for the colony that sick or parasitized bees leave the colony of their own accord, rather than recruiting nestmates to exclude those bees via aggressive behaviors. In that case, bees might distinguish sick bees based on different CHC profiles but not discriminate them, except in the case of extremely sick bees that cannot leave on their own, such as bees exhibiting deformed wing symptoms. We also found a change of the chemical profile with age which is consistent with previous studies (nurse vs forager, see [43]). In honey bee hives, older bees segregate themselves from young bees by olfactory discrimination of cuticular hydrocarbons as they correspond to different age groups [44]. Indeed, older bees both emit and respond to a more complex bouquet of cuticular hydrocarbons than younger bees [43,44]. Since Nosema and Varroa-parasitized bees age faster, it is possible that they exhibited a CHC profile of old bees. In addition, the CHC profile is shaped by the genotype, nutrition, environment and physiological state [45,46]. Therefore, it is possible that their nestmates did not respond to the parasitized bees because their chemical profiles could not be distinguished from the chemical profile of others bees of different ages, physiological status and genotypes. These results highlight the importance of testing for biological effects within the hive when trying to draw conclusions about honey bee behavior.
If parasitism by Varroa or Nosema induces precocious foraging, one would expect the parasitized bees to show physiological changes similar to the transition to a forager bee. Levels of 10-HDA increased with the age of the bee confirming the study of Plettner et al.[47], but did not change in response to Nosema or Varroa parasitism. Thus the production of antiseptic compounds, like 10-HDA, in the food is age- or task-dependent but not regulated by the presence of parasites. However, further investigation of different type of pathogens or parasites, would give more insight as to whether antiseptic production can vary according to the infection level of the hive.
Our results also demonstrate that parasites alter the brain of the honey bee host, whether they were parasitized at the pupal (Varroa) or the adult stage (Nosema). In addition, twenty genes, nearly one half of those detected in Nosema-infested bee brains, show a shared expression pattern between Varroa and Nosema-infested bees. Their functions are diverse but several genes stand out as interesting for their possible roles in oxidative stress, neural function and foraging behavior. Flavin-containing monooxygenase FMO GS-OX-like 3-like (FMO3) and torsin-like protein (torp4a) are overexpressed and replication factor C subunit 5-like (RfC5) is downregulated in Varroa-infested and Nosema-infected bees. FMO3 is part of the FMO family known to react to xenobiotic stress in other organisms [48]. In Drosophila, the Torp4a ortholog (dtorsin) is involved in dopamine metabolism and locomotion [49] and the RfC5 ortholog (RfC3) plays a role in neurogenesis [50], indicating that both parasites can modify brain function. The honey bee gene, Pheromone biosynthesis-activating neuropeptide (PBAN) is also over-expressed in both Varroa-infested and Nosema-infected bees compared to controls. In honey bees, PBAN neuropeptide levels are significantly higher in nectar foragers than pollen foragers [51]. In Lepidoptera, PBAN is linked to regulation of sex pheromone production [52], where pheromone production is JH-dependent, and JH primes the pheromone gland in adult females to respond to PBAN[53]. JH is also responsible for “priming” the foraging behavior in honey bees, though no link has been established between PBAN and JH in honey bees.
The presence of viruses may also account for similarities in gene expression between Nosema and Varroa-parasitized bees. We found increased levels of DWV in the brains of both types of parasitized bees and therefore cannot exclude that the observed changes are actually caused by an increase in DWV titer. DWV is a positive-strand RNA picorna-like virus that has been detected and can actively replicate in the heads [54] and brains of honey bees, specifically the mushroom bodies, visual and antennal lobe neuropils [55]. The virus is closely associated with Varroa infestation [56] and thus it was not unusual that it occurred in higher levels in Varroa-infested brains. On the other hand, we did not expect to find an increase in DWV levels in N. ceranae-infected bees, as a negative correlation between these two pathogens in the midgut of the bee was recently reported [57], which suggests that N. ceranae and DWV may compete for resources in the degenerated midgut, but not in the brain, where Nosema is not found.
Despite the statistically significant number of shared genes that change, Varroa and Nosema-infested brains demonstrate different patterns of expression that may reflect the different pathologies of the two parasites. Bees that were parasitized by Varroa as developing pupae exhibit more gene changes compared to controls than bees that were inoculated with Nosema ceranae as one-day old adults. This apparent disparity in gene expression changes may be due to long-lasting brain developmental changes induced during pupal development that persist in adult bees. The genes affected by N. ceranae infection could not be sorted by functional group analysis but several immune-related and antioxidant genes, including defensin-1, peroxidase, esterase A2, glucose oxidase, were upregulated indicating that the blood–brain barrier in honey bees, although not well studied, may be compromised by a parasitic attack. Genes involved in the oxidative response to stress were also upregulated in the guts of N. ceranae-infected honey bees [58], suggesting a systemic response throughout the honey bee in response to the microsporidian.
The impact of Varroa on the brain transcriptome suggests a decrease in learning and memory that may result from parasitism during development. This brain response would explain the actual learning impairment and losses of foragers induced by Varroa[23,59]. Based on functional group analysis, Varroa-infested bees show decreased expression of glutamate and GABA receptor-related genes, the dopamine receptor, Amdop1, and overexpression of ascorbate/aldarate metabolism genes. Inhibition or suppression of glutamate receptors disrupts memory formation in honey bees [26,30,60]. GABA receptors are present throughout the mushroom bodies [61], a region important for learning and memory, especially in foragers [62]. GABAnergic interneurons also form part of the olfactory conditioned learning pathway [63]. Yet the simultaneous increase in the expression of glutamate decarboxylase and GABA neurotransmitter transporter with a decrease in GABA receptor targets signals either compensatory mechanisms at work or a disruption in GABAnergic network. Analysis of the neuroanatomical changes in the Varroa-parasitized brain could resolve whether decreased expression of GABA and glutamate receptors leads to a reduction in their numbers. The dopamine receptor Amdop1 is higher in newly born cells in the mushroom body than older cells [64] and in Drosophila, it is required for aversive and appetitive learning [65]. Finally, the cAMP pathway and its targets in the mushroom bodies are important mechanisms for learning in bees [66]. Several genes linked to the cAMP and calcium signaling cascades are downregulated in Varroa brains: Adenylate cyclase type 10-like, Ryanodine receptor[67] and voltage-dependent calcium channel subunit (GB10696).
Compared to the transcriptomic changes in the honey bee brain that accompany the switch from nurse to forager, we found relatively few genes that changed in response to Nosema ceranae or Varroa destructor infestation. Brain expression profiles of Varroa and Nosema- parasitized bees bear a greater resemblance to each other than to the reported profiles of typical foragers or nurses. Thus, their early departures from the hive may not be induced by mechanisms of normal behavioral development, but perhaps by an alternative mechanism that results in altruistic self-removal. Indeed, certain genes that are typically upregulated in foraging bee brains (Inos, Kr-h1) are downregulated in Varroa-infested honey bee brains [29,41,68,69]. Foraging activity is an especially demanding activity for learning and memory in the honey bee [63], but parasitized bees seems to have deficiencies at this level (see above). Therefore, altogether these results suggest that Varroa and Nosema-parasitized bees seem to not be true foragers, much like CO2-treated bees that left the hive, but also disappeared, at higher rates than control bees [10]. However, this will require confirmation in a more natural context (colony level).
Neither of the parasites, Varroa destructor nor Nosema ceranae, attacks the honey bee brain directly, yet we observed transcriptional changes in the brains of honey bees in response to parasitism. Thus, these changes, that are most likely triggered by a reaction in another tissue (e.g. midgut, fat bodies, hemolymph), highlight a link between the immune system, the brain, and perhaps, behavior in the honey bee. While parasitized bees are reported to behave like foragers, by leaving the hive, their brain transcription profiles suggest that their behavior is not driven by the same molecular pathways that induce foraging behavior. Whether the transcriptional changes observed are due to host immune response, parasite protein release or viruses that propagate in the brain is not known. LPS-challenged bees also behave more like foragers than same-aged bees [15], even without parasitic or viral challenges, but proteomic analysis of parasitized insects, grasshopper (by a nematode) and tsetse fly (by Trypansoma brunei) detected changes in the host brain [70,71] and proteins released by the parasite that may affect host behavior [70].
Conclusion
Stress response in the honey bee to parasitism by Varroa destructor or Nosema ceranae shows similarities in their features; both parasites induce changes in CHC profiles and similar transcriptional profiles in the brain. That these parasitized bees are not attacked by their nestmates suggests that they leave the hive voluntarily, perhaps propelled by gene expression changes in the brain, showing altruistic behavior as predicted by Rueppel et al.[10]. This social removal may be a general and conserved response to parasitism, given that it was observed with extremely different types of parasites: a mite (ectoparasite) [11,12] and a single cell microsporidian (endoparasite) [13,14]. As to what these bees do once they have left the hive still needs to be examined. Are they normal foragers but with shorter life spans, less efficient foragers due to learning and memory deficiencies or do they leave the hive and wander aimlessly in the landscape? Emerging tracking technology will allow us to answer these questions and determine the role of the parasitized honey bee within the colony. In addition, such studies that incorporate behavioral, genomic and physiological components will help us to better understand current worldwide declines of honey bee populations that are often characterized by an unusual loss of adult bees from the colony [72].
Methods
Bees and parasites
This experiment was performed using hives of a hybrid of Apis mellifera mellifera and A.m. ligustica located at the Institut National de la Recherche Agronomique in Avignon, France. Nosema-treated bees were individually fed 2 uL of 50% sugar solution with a mixture of freshly extracted spores of Nosema ceranae at a concentration of 50,000 spores/uL. The presence of N. ceranae was confirmed by PCR analysis [73]. Guts were dissected at the end of the experiments and no spores were found in the control bees (data not shown). Varroa-parasitized bees were obtained following a similar procedure described in [74]. Colonies that were not treated with miticide were used and the queen was caged to stop egg-laying so that Varroa mites had no cells to parasitize. Meanwhile, the queen from a different colony was transferred into a queen-excluder for 7 days with an empty frame. Then the frame containing new brood (young larvae) was transferred into the colony that had a caged queen and sealed brood. The new brood on this frame was then uniformly parasitized by Varroa mites. Three weeks after the queen laid eggs in the frame enclosed in the queen-excluder, the frame was removed from the colony and newly emerging adults were picked from capped cells in order to verify whether the cell was infested with Varroa. Varroa-parasitized bees with deformed wings were discarded due to their extremely short lifespan.
Experiment 1: Chemical analysis of Nosema ceranae- or Varroa destructor-parasitized bees
Experiments on Nosema and Varroa were performed separately but following the same procedure. In each experiment bees from three colonies were used. One day after their emergence, parasitized and non-parasitized (control) bees (N = 40–60 bees/state/colony) were color painted (Lackstift, Motip, Netherlands) on the thorax according to their state and colony origin and then all transferred into a host colony that was Varroa-treated and Nosema-free. At this age bees are easily accepted by the colony since they are lacking the recognition cues, including cuticular hydrocarbons [75]. The hydrocarbon profiles, which are genetically and environmentally acquired, change progressively with age [75,76]. After 5 and 10 days in the Nosema experiment, and after 5 days in the Varroa experiment, bees were collected and stored at −80°C for later chemical analysis. Varroa-parasitized bees were not collected at day 10, because they were more difficult to find in the hive at that age and in sufficient number for later analysis (shorter lifespan or had left the hive).
Cuticular hydrocarbon profiles
Hydrocarbons were extracted by individually immersing bees for 5 minutes at room temperature in 1,900 μL of isohexane and 100 μL of eicosane (C20) at 25 ng/μL as an internal standard. Each sample was concentrated under a stream of nitrogen to a volume of 10 μL, of which 1 μL was injected into a fast gas chromatograph (GC) (Shimadzu 2014, Japan) equipped with a split-splitless inlet, a flame ionization detector, and a capillary column Equity 5 (15 m x 0.10 mm, 0.10 μm film thickness). Samples were injected in split mode and hydrogen was used as a carrier gas with a column flow of 0.55 ml/min. The oven temperature was held at 70°C for 30 sec., increased from 70°C to 150°C at 40°C/min., from 150°C to 320°C at 10°C/min., and held at 320°C for 10 min. Peaks from the paint were identified and automatically removed by comparing the profile of non-marked bees to paint-marked bees and paint diluted in isohexane.
The structure of cuticular compounds present in the profiles was determined by performing gas chromatography coupled with mass spectrometry (GC-MS). Two μL of sample was injected into a GC-MS Thermo Scientific Trace GC Ultra ISQ equipped with a split-splitless inlet, an ISQ electron impact ion source, and a Thermo TR-5 column (20 m x 0.10 mm, 0.10 μm film thickness). The column flow was 0.4 ml/min. and the oven temperature was held at 50°C for 43 sec., increased from 50°C to 150°C at 20°C/min., from 150°C to 300°C at 10°C/min., and held at 300°C for 10 min.
For statistical analysis of the chemical profiles, only peaks that were reproducibly quantifiable in all samples were used. Each peak area was standardized according to Reyment [77]. To determine whether parasitized and control bees could be distinguished on the basis of their cuticular profiles and assess the profile similarity, a stepwise discriminant analysis was performed with Statistica 8.0. (StatSoft® Inc.). In addition, Mahalanobis distances between all pairwise groups were calculated as estimates for the chemical distances between each group. P-values were adjusted for multiple comparisons using Bonferroni’s correction. The effect of parasitism on the relative proportion of each compound was determined by using Mann–Whitney U tests.
10-HDA levels
Chemical compounds were extracted by crushing individual heads in 200 μL of methanol and 100 μl of decanoic acid (250 ng/μl; internal standard) for 2 min. 30 sec. at 50 Hz with a Mini-Mill Pulverisette 23 (Fritsch, France). The solution was centrifuged at 4,000 g for 40 min. Twenty μL of the supernatant were collected, concentrated under nitrogen stream and then derivatized with 5 μL of bistrimethylsilyltrifluoroacetamide. The solution was agitated and left at room temperature for 40 min. The derivatized sample was then diluted in 10 μL of isohexane and 1 μL of this solution was injected into the GC (Shimadzu 2014, Japan). The samples were injected in split mode. Hydrogen was used as carrier gas. Oven temperature was set at 100°C, then increased to 200°C at 40°C/min. and to 250°C at 10°C/min. and held at 250°C for 2 min. Identification and quantification of 10-HDA were based on retention times of synthetic compounds (Cayman Chemical, France). The confirmation of 10-HDA compound was done by mass spectrometer (Thermo Scientific Trace GC Ultra ISQ). Nosema and Varroa effects on 10-HDA synthesis were determined by using Kruskall-Wallis and Mann–Whitney U tests, respectively.
Experiment 2: Behavioral analysis of Nosema ceranae- or Varroa destructor-parasitized bees
To determine whether parasitized bees are treated differently than healthy, control bees, we recorded social interactions between focal bees and nestmates in two four-frame observation hives. Nosema- (70 bees/state/observation hive) and Varroa-parasitized bees (30 bees/state/observation hive) were obtained as previously described, tag-numbered (Opalith Plättchen, Friedrich Wienold, Germany) and introduced into the observation hives. The two experiments were carried out separately. We performed focal sampling behavioral observation on randomly-picked bees for 15 min (19–20 bees/state/observation hive for the Nosema experiment and 12 bees/state/observation hive for the Varroa experiment). The regular behaviors recorded during the observation period were: antennal contacts, allo-grooming, cleaned by another bee, self-grooming, trophallaxis and being vibrated (vibration signal, see [78]). The agonistic behaviors were: mandibular openings, bites, and stinging. We then determined whether the rate of each behavior performed during the observation period differed between parasitized and non- parasitized bees using Mann–Whitney U tests.
Experiment 3: Brain transcriptomics of Nosema ceranae- or Varroa destructor -parasitized bees
Three treatment groups were used: control bees that had no presence of Varroa in their brood cells, Varroa-infested bees that had Varroa in their cells, and Nosema-infected bees that received the Nosema sugar solution. Each treatment group was obtained as previously described (see Bees and parasites) and then each group composed of one day old bees (N = 20 bees/treatment group) were housed in different plastic cages (10.5 × 7.5 × 11.5 cm). Keeping the bees in cages allowed us to remove the potential effect of hive environment and capture only the effect of the parasite on brain gene expression. The experiment was repeated on 2 different colonies.
Brain dissection
After 10 days in cages, bees were sacrificed by flash freezing in liquid nitrogen. Heads of the bees were separated from the body and the cuticle scratched with surgical tweezers before storing in RNAlater®-ICE solution (Life Technologies) at −20°C for 16–18 hours, according to manufacturer’s instructions. Brains were dissected on ice under a dissection microscope to remove all traces of the optic lobe and then stored at −80°C.
RNA isolation
For each cage, 3 pools of 3 bee brains were homogenized in 500 μL of TRIzol Reagent (Life Technologies), phase-separated with chloroform/Trizol and the aqueous phase removed for precipitation with 70% ethanol. The resulting aqueous-ethanol solution was loaded onto an RNeasy mini spin column of the Qiagen RNeasy Mini Kit (Qiagen). RNA isolation was performed according to the manufacturer’s instructions, starting with washing with Buffer RW1. Genomic DNA was removed from samples using an RNA-free DNase set (Qiagen). RNA was quantified by spectrophotometry using the Nanodrop 1000 (Thermo Scientific). Then, RNA isolated from the 3 pools was equally combined.
Digital gene expression
For each treatment, two brain pools (one per colony) were analyzed. Sample preparation was performed using the DGE DpnII Sample preparation kit (Illumina) (ref.FC-102-1007) according to the manufacturer's instructions. Briefly, 2 μg of total RNA was incubated with magnetic oligo(dT) beads. Non poly-adenylated RNA was removed by several washes. Reverse transcription was performed on captured RNAs followed by the synthesis of the second strand of cDNA. Captured double stranded DNA was digested using DpnII. A ligation was performed with Illumina's GEX DpnII adapter 1 followed by a digestion using MmeI resulting in the release of tags. Those tags were ligated using Illumina's GEX Adapter 2, amplified by PCR (15 cycles) and purified on acrylamide gel. Libraries were validated using an Agilent DNA1000 BioAnalyzer chip, denatured using 0.1 N NaOH, diluted to 8 pM and sequenced on a Hiseq2000 using a Sequence by Synthesis technique.
Analysis and mapping of DGE tags
Image analyses and base-calling were conducted using the HiSeq Control Software (HCS 1.4.5.0) and RTA component (RTA 1.12.4.0). Extraction of 16 bp tags (reads were trimmed for adaptor sequence) and tag counting were performed using home-made Perl script.Sequences were first aligned (using the Illumina's sequencing analysis software, CASAVA 1.8) to transcripts of the Apis mellifera genome version 4.5 downloaded from NCBI. Only those 16 bp tags that were perfect matches were retained. Those tags that could not be aligned to transcripts were re-aligned to the complete Apis mellifera genome (version 4.5).
Mapping was also performed on sequences of honey bee virus genomes (Chronic bee paralysis virus RNA 1: GenBank EU122229, Chronic bee paralysis virus RNA 2: EU122230, Sacbrood virus: AF092924, Deformed wing virus: AJ489744, Black queen cell virus: AF183905, Acute bee paralysis virus: AF150629, Kashmir bee virus: AY275710, Varroa destructor virus 1: AY251269 and Israel acute paralysis virus: EF219380).
The package DESeq from Bioconductor was used to conduct the analysis [79]. Genes (i.e. tags that matched a transcript) and tags that were only aligned to the genome were analyzed separately. Tags that occur less than one time in a million, in two or more samples, were filtered from the analysis. DESeq estimates variance-mean dependence in count data from DGE-tag libraries and tests for differential expression based on a model using the negative binomial distribution. Genes were considered to be differentially expressed between two treatments at an adjusted P-value < 0.05. The P-value was adjusted for multiple testing with the Benjamini-Hochberg procedure, which controls for false discovery rate.
Analysis of gene expression profiles
Genes that overlapped between gene lists were identified by creating Venn diagrams using GeneVenn [80]. Exact hypergeometric probability test was performed to test the statistical significance of the overlap between two gene lists [81]. DAVID 6.7 [82,83] was used to determine the enriched functional groups, based on GO terms, within the complete list of expressed genes containing those genes with Flybase orthologs.
We also determined whether Nosema and Varroa modified the brain gene expression profile in a manner consistent with some previously characterized behavioral phenotypes or in a completely different way. To explore this idea, we compared the parasite effects to nurse/forager profiles that were obtained with microarray analysis [41], using Chi-square tests with Yates correction.
Competing interests
The authors have declared no competing interests.
Authors’ contributions
YLC, CMM and CA conceived of the study and its experimental design. CMM, DC and CA conducted the experimental protocol with the honey bees, ED and DB performed the chemical analysis, ED and CA performed the behavioral observations, CMM performed brain dissections and RNA isolation, HP and JPD carried out the sequencing and statistical analysis of sequencing data, CMM and CA analyzed and interpreted the data, and CMM, CA and YLC drafted manuscript. All authors read and approved the final manuscript.
Supplementary Material
Relative proportion of each CHC compound in control and Nosema or Varroa parasitized bees. Changes in proportion induced by parasitism were determined with Mann–Whitney U tests (P < 0.05 are in bold).
Percentage of correct assignments of Nosema-infected and control bees based on their cuticular hydrocarbons profiles. In the treatment column, 5 and 10 indicate the age of the bees. The last four columns show the number of bees classified in the different group treatment by the discriminant analysis. The experiment was repeated on bees from 3 different colonies. Table S3 Percentage of correct assignments of Varroa-infested and control bees based on their cuticular hydrocarbons profiles. The last six columns show the number of bees classified in the different group treatment by the discriminant analysis. The numbers 98, 120 and 231 indicate the colony origin of the bees.
Summary of DGE sequencing results. The analysis was performed on two colonies.
Lists of genes affected in the bee brain by Nosema or Varroa parasitism. Corresponding honey bee gene, Drosophila ortholog and genes also up- or downregulated in the brain of nurses and foragers are shown.
Effects of Nosema and Varroa parasites on the prevalance of Deformed Wing Virus and Varroa Destructor Virus levels in the bee brain. Comparisons of viral titers (A) Nosema vs Control, (B) Varroa vs Control and (C) Varroa vs Nosema using Cox-Reid method for estimating dispersion and Generalized Linear Model (GLM) for statistical test.
Contributor Information
Cynthia M McDonnell, Email: cynthia.mcdonnell@avignon.inra.fr.
Cédric Alaux, Email: cedric.alaux@avignon.inra.fr.
Hugues Parrinello, Email: Hugues.Parrinello@mgx.cnrs.fr.
Jean-Pierre Desvignes, Email: jean-pierre.desvignes@univ-amu.fr.
Didier Crauser, Email: crauser@avignon.inra.fr.
Emma Durbesson, Email: emma.durbesson@live.fr.
Dominique Beslay, Email: dominique.beslay@avignon.inra.fr.
Yves Le Conte, Email: leconte@avignon.inra.fr.
Acknowledgements
We thank Stéphanie Rialle for performing the statistical analysis of the viral titer data. This work was funded by a BEEDOC grant (FP7, RTD REG/E.4 (2009)D/561221) to YLC.
References
- Moore J. Parasites and the Behavior of Animals. Oxford: Oxford University Press; 2002. [Google Scholar]
- Hart BL. Behavioral adaptations to pathogens and parasites–5 strategies. Neurosci Biobehav Rev. 1990;14:273–294. doi: 10.1016/S0149-7634(05)80038-7. [DOI] [PubMed] [Google Scholar]
- Hart BL. Behavioral adaptations to parasites—an ethological approach. J Parasitol. 1992;78:256–265. doi: 10.2307/3283472. [DOI] [PubMed] [Google Scholar]
- Curtis V, de Barra M, Aunger R. Disgust as an adaptive system for disease avoidance behavior. Phil Trans R Soc B. 2011;366:389–401. doi: 10.1098/rstb.2010.0117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behringer DC, Butler MJ IV, Shields JD. Avoidance of disease in social lobsters. Nature. 2006;441:421. doi: 10.1038/441421a. [DOI] [PubMed] [Google Scholar]
- Libersat F, Delago A, Gal R. Manipulation of host behavior by parasitic insects and insect parasites. Annu Rev Entomol. 2009;54:189–207. doi: 10.1146/annurev.ento.54.110807.090556. [DOI] [PubMed] [Google Scholar]
- Klein SL. Parasite manipulation of the proximate mechanisms that mediate social behavior in vertebrates. Physiol Behavi. 2003;79:441–449. doi: 10.1016/S0031-9384(03)00163-X. [DOI] [PubMed] [Google Scholar]
- Wilson-Rich N, Spivak M, Fefferman NH, Starks PT. Genetic, individual, and group facilitation of disease resistance in insect societies. Annu Rev Entomol. 2009;54:405–423. doi: 10.1146/annurev.ento.53.103106.093301. [DOI] [PubMed] [Google Scholar]
- Cremer S, Armitage SA, Schmid-Hempel P. Social immunity. Curr Biol. 2007;17:R693–702. doi: 10.1016/j.cub.2007.06.008. [DOI] [PubMed] [Google Scholar]
- Rueppell O, Hayworth MK, Ross NP. Altruistic self-removal of health-compromised honey bee workers from their hive. J Evol Biol. 2010;23:1538–1546. doi: 10.1111/j.1420-9101.2010.02022.x. [DOI] [PubMed] [Google Scholar]
- Downey DL, Higo TT, Winston ML. Single and dual parasitic mite infestations on the honey bee Apis mellifera L. Insect Soc. 2000;47:171–176. doi: 10.1007/PL00001697. [DOI] [Google Scholar]
- Janmaat AF, Winston ML. The influence of pollen storage area and Varroa jacobsoni Oudemans parasitism on temporal caste structure in honey bees (Apis mellifera L.) Insect Soc. 2000;47:177–182. doi: 10.1007/PL00001698. [DOI] [Google Scholar]
- Dussaubat C, Maisonnasse A, Crauser D, Beslay D, Costagliola G, Soubeyrand S, Kretzchmar A, Le Conte Y. Flight behavior and pheromone changes associated to Nosema ceranae infection of honeybee workers (Apis mellifera) in field conditions. J Invert Pathol. 2013;113:42–51. doi: 10.1016/j.jip.2013.01.002. [DOI] [PubMed] [Google Scholar]
- Goblirsch M, Huang ZY, Spivak M. Physiological and behavioral changes in honey bees (Apis mellifera) induced by Nosema ceranae infection. PLoS One. 2013;8:e58165. doi: 10.1371/journal.pone.0058165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaux C, Kemper N, Krezschmar A, Le Conte Y. Brain, physiological and behavioral modulation induced by immune stimulation in honeybees (Apis mellifera): A potential mediator of social immunity? Brain Behav Immun. 2012;26:1057–1060. doi: 10.1016/j.bbi.2012.04.004. [DOI] [PubMed] [Google Scholar]
- Le Conte Y, Ellis M, Ritter W. Varroa mites and honey bee health: can Varroa explain part of the colony losses? Apidologie. 2010;41:353–363. doi: 10.1051/apido/2010017. [DOI] [Google Scholar]
- Rosenkranz R, Aumeier P, Ziegelmann B. Biology and control of Varroa destructor. J Invert Pathol. 2010;103:S96–S119. doi: 10.1016/j.jip.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Yang X, Cox-Foster D. Effects of parasitization by Varroa destructor on survivorship and physiological traits of Apis mellifera in correlation with viral incidence and microbial challenge. Parasitol. 2007;134:405–412. doi: 10.1017/S0031182006000710. [DOI] [PubMed] [Google Scholar]
- Higes M, Martín-Hernández R, Meana A. Nosema ceranae in Europe: an emergent type C nosemosis. Apidologie. 2010;41:375–392. doi: 10.1051/apido/2010019. [DOI] [Google Scholar]
- Fries I, Feng F, da Silva A, Slemenda SB, Pieniazek NJ. Nosema ceranae n. sp. (Micropora, Nosematidae) morphological and molecular characterization of a microsporidian parasite of the Asian honey bee Apis cerana (Hymenoptera, Apidae) Europ J Protistol. 1996;32:356–365. doi: 10.1016/S0932-4739(96)80059-9. [DOI] [Google Scholar]
- Williams BAP. Unique physiology of host-parasite interactions in microsporidia infections. Cell Microbiol. 2009;11:1551–1560. doi: 10.1111/j.1462-5822.2009.01362.x. [DOI] [PubMed] [Google Scholar]
- Kralj J, Fuchs S. Nosema sp. influences flight behavior of infected honey bee (Apis mellifera) foragers. Apidologie. 2010;41:21–28. doi: 10.1051/apido/2009046. [DOI] [Google Scholar]
- Kralj J, Fuchs S. Parasitic Varroa destructor mites influence flight duration and homing abiligy of infested Apis mellifera foragers. Apidologie. 2006;37:577–587. doi: 10.1051/apido:2006040. [DOI] [Google Scholar]
- Amdam GV, Hartfelder K, Norberg K, Hagen A, Omholt SW. Altered physiology in worker honey bees (Hymenoptera: Apidae) infested with the mite Varroa destructor (Acari: Varroidae): a factor in colony loss during overwintering? J Econ Entomol. 2004;97:741–747. doi: 10.1603/0022-0493(2004)097[0741:APIWHB]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- Bowen-Walker PL, Gunn A. The effect of the ectoparasitic mite, Varroa destructor on adult worker honeybee (Apis mellifera) emergence weights, water, protein, carbohydrate, and lipid levels. Entomol Exp Appl. 2001;101:207–217. doi: 10.1046/j.1570-7458.2001.00905.x. [DOI] [Google Scholar]
- Kucharski R, Mitri C, Grau Y, Maleszka R. Characterization of a metabotropic glutamate receptor in the honeybee (Apis mellifera): implications for memory formation. Invertebr Neurosci. 2007;7:99–108. doi: 10.1007/s10158-007-0045-3. [DOI] [PubMed] [Google Scholar]
- Antunez K, Martin-Hernandez R, Prieto L, Meana A, Zunino P, Higes M. Immune suppression in the honey bee (Apis mellifera) following infection by Nosema ceranae (Microsporidia) Environ Microbiol. 2009;11:2284–2290. doi: 10.1111/j.1462-2920.2009.01953.x. [DOI] [PubMed] [Google Scholar]
- Dussaubat C, Maisonnasse A, Alaux C, Tchamitchan S, Brunet JL, Plettner E, Belzunces LP, Le Conte Y. Nosema spp. infection alters pheromone production in honey bee (Apis mellifera) J Chem Ecol. 2010;36:522–525. doi: 10.1007/s10886-010-9786-2. [DOI] [PubMed] [Google Scholar]
- Whitfield CW, Cziko AM, Robinson GE. Gene expression profiles in the brain predict behavior in individual honey bees. Science. 2003;302:296–299. doi: 10.1126/science.1086807. [DOI] [PubMed] [Google Scholar]
- Mußig L, Richlitzki A, Rößler R, Eisenhardt D, Menzel R, Leboulle G. Acute disruption of the NMDA receptor subunit NR1 in the honeybee brain selectively impairs memory formation. J Neurosci. 2010;30:7817–7825. doi: 10.1523/JNEUROSCI.5543-09.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mayack C, Naug D. Energetic stress in the honeybee Apis mellifera from Nosema ceranae infection. J Invert Pathol. 2009;100:185–188. doi: 10.1016/j.jip.2008.12.001. [DOI] [PubMed] [Google Scholar]
- Ament SA, Blatti CA, Alaux C, Wheeler MM, Toth AL, Le Conte Y, Hunt GJ, Guzman-Novoa E, DeGrandi-Hoffman G, Uribe-Rubio JL, Amdam GV, Page RE Jr, Rodriguez-Zas SL, Robinson GE, Sinha S. New meta-analysis tools reveal common transcriptional regulatory basis for multiple determinants of behavior. P Natl Acad Sci USA. 2012;109:E1801–E1810. doi: 10.1073/pnas.1205283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zayed A, Robinson GE. Understanding the relationship between gene expression and social behavior: lessons from the honey bee. Annu Rev Genet. 2012;46:589–613. doi: 10.1146/annurev-genet-110711-155517. [DOI] [PubMed] [Google Scholar]
- Richard F-J, Aubert A, Grozinger CM. Modulation of social interactions by immune stimulation in honey bee, Apis mellifera, workers. BMC Biol. 2008;6:50–62. doi: 10.1186/1741-7007-6-50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard F-J, Holt HL, Grozinger CM. Effects of immunostimulation on social behavior, chemical communication and genome-wide gene expression in honey bee workers (Apis mellifera) BMC Genom. 2012;13:558. doi: 10.1186/1471-2164-13-558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baracchi D, Fadda A, Turillazzi S. Evidence for antiseptic behavior towards sick adult bees in honey bee colonies. J Insect Physiol. in press. [DOI] [PubMed]
- Blum MS, Novak AF, Taber S III. 10-hydroxy-Δ2-decenoic acid, an antibiotic found in royal jelly. Science. 1959;130:452–453. doi: 10.1126/science.130.3373.452. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy S, Duan SX, von Moltke LL, Greenblatt DJ, Court MH. Validation of serotonin (5-hydroxtryptamine) as an in vitro substrate probe for human UDP-glucuronosyltransferase (UGT) 1A6. Drug Metab Dispos. 2003;31:133–139. doi: 10.1124/dmd.31.1.133. [DOI] [PubMed] [Google Scholar]
- Krishnaswamy S, Hao Q, von Moltke LL, Greenblatt DJ, Court MH. Evaluation of 5-hydroxytryptophol and other endogenous serotonin (5-hydroxytryptamine) analogs as substrates for UDP-glucuronosyltransferase 1A6. Drug Metab Dispos. 2004;32:862–869. doi: 10.1124/dmd.32.8.862. [DOI] [PubMed] [Google Scholar]
- Itäaho K, Court M, Uutela P, Kostiainen R, Radominska-Pandya A, Finel M. Dopamine is a low-affinity and high-specificity substrate for the human UDP-glucuronosyltransferase 1A10. Drug Metab Dispos. 2009;37:768–775. doi: 10.1124/dmd.108.025692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaux C, Le Conte Y, Adams HA, Rodriguez-Zas S, Grozinger CM, Sinha S, Robinson GE. Regulation of brain gene expression in honey bees by brood pheromone. Genes Brain Behav. 2009;8:309–319. doi: 10.1111/j.1601-183X.2009.00480.x. [DOI] [PubMed] [Google Scholar]
- Salvy M, Martin C, Bagneres AG, Provost E, Roux M, Le Conte Y, Clement JL. Modifications of the cuticular hydrocarbon profile of Apis mellifera worker bees in the presence of the ectoparasitic mite Varroa jacobsoni in brood cells. Parasitol. 2001;122:145–159. doi: 10.1017/s0031182001007181. [DOI] [PubMed] [Google Scholar]
- Kather R, Drijfhout FP, Martin SJ. Task group differences in cuticular lipids in the honey bee Apis mellifera. J Chem Ecol. 2011;37:205–212. doi: 10.1007/s10886-011-9909-4. [DOI] [PubMed] [Google Scholar]
- Scholl J, Naug D. Olfactory discrimination of age-specific hydrocarbons generates behavioral segregation in a honeybee colony. Behav Ecol Sociobiol. 2011;65:1967–1973. doi: 10.1007/s00265-011-1206-2. [DOI] [Google Scholar]
- Howard RW, Blomquist GJ. Ecological, behavioral and biochemical aspects of insect hydrocarbons. Annu Rev Entomol. 2005;50:371–393. doi: 10.1146/annurev.ento.50.071803.130359. [DOI] [PubMed] [Google Scholar]
- Blomquist GJ, Bagneres AG. Insect hydrocarbons: biology, biochemistry and chemical ecology. Cambridge: Cambridge University Press; 2010. [Google Scholar]
- Plettner E, Sutherland GRJ, Slessor KN, Winston ML. Why not be a queen? Regioselectivity in mandibular secretions of honeybee castes. J Chem Ecol. 1995;21:1017–1029. doi: 10.1007/BF02033805. [DOI] [PubMed] [Google Scholar]
- Phillips IR, Shephard EA. Flavin-containing monooxygenases: mutations, disease and drug response. Trends Pharmacol Sci. 2008;29:294–301. doi: 10.1016/j.tips.2008.03.004. [DOI] [PubMed] [Google Scholar]
- Wakabayashi-Ito N, Doherty OM, Moriyama H, Breakefield XO, Gusella JF, O'Donnell JM, Ito N. dtorsin, the Drosophila ortholog of the early-onset dystonia TOR1A (DYT1), plays a novel role in dopamine metabolism. PLoS One. 2011;6:e26183. doi: 10.1371/journal.pone.0026183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumüller RA, Richter C, Fischer A, Novatchkova M, Neumuller KG, Knoblich JA. Genome-wide analysis of self-renewal in Drosophila neural stem cells by transgenic RNAi. Cell Stem Cell. 2011;8:580–593. doi: 10.1016/j.stem.2011.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brockmann A, Annangudi SP, Richmond TA, Ament SA, Xie F, Southey BR, Rodriguez-Zas SR, Robinson GE, Sweedler JV. Quantitative petidomics reveal brain peptide signatures of behavior. Proc Natl Acad Sci USA. 2009;106:2383–2388. doi: 10.1073/pnas.0813021106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rafaeli A. Pheromone biosynthesis activating neuropeptide (PBAN): Regulatory role and mode of action. Gen Comp Endocrinol. 2009;162:69–78. doi: 10.1016/j.ygcen.2008.04.004. [DOI] [PubMed] [Google Scholar]
- Fan Y, Rafaeli A, Gileadi C, Kubli E, Applebaum SW. Drosophila melanogaster sex peptide stimulates juvenile hormone synthesis and depresses sex pheromone production in Helicoverpa armigera. J Insect Physiol. 1999;45:127–133. doi: 10.1016/S0022-1910(98)00106-1. [DOI] [PubMed] [Google Scholar]
- Zioni N, Soroker V, Chejanovsky N. Replication of Varroa destructor virus 1 (VDV-1) and a Varroa destructor virus 1-deformed wing virus recombinant (VDV-1-DWV) in the head of the honey bee. Virol. 2011;417:106–112. doi: 10.1016/j.virol.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Shah KS, Evans EC, Pizzorno MC. Localization of deformed wing virus (DWV) in the brains of the honeybee, Apis mellifera Linnaeus. Virol J. 2009;6:182–188. doi: 10.1186/1743-422X-6-182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genersch E, Aubert M. Emerging and re-emerging viruses of the honey bee (Apis mellifera L.) Vet Res. 2010;41:54–73. doi: 10.1051/vetres/2010027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Costa C, Tanner G, Lodesani M, Maistrello L, Neumann P. Negative correlation between Nosema ceranae spore loads and deformed wing virus infection levels in adult honey bee workers. J Invert Pathol. 2011;108:224–225. doi: 10.1016/j.jip.2011.08.012. [DOI] [PubMed] [Google Scholar]
- Dussaubat C, Brunet JL, Higes M, Colbourned JK, Lopez J, Choi JH, Martin-Hernandez R, Botias C, Cousin M, McDonnell C, Bonnet M, Belzunces LP, Moritz RFA, Le Conte Y, Alaux C. Gut pathology and responses to the microsporidium Nosema ceranae in the honey bee Apis mellifera. PLoS One. 2012;7:e37017. doi: 10.1371/journal.pone.0037017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kralj J, Brockmann A, Fuchs S, Tautz J. The parasitic mite Varroa destructor affects non-associative learning in honey bee foragers, Apis mellifera L. J Comp Physiol A. 2007;193:363–370. doi: 10.1007/s00359-006-0192-8. [DOI] [PubMed] [Google Scholar]
- Si A, Helliwell P, Maleszka R. Effects of NMDA receptor antagonists on olfactory learning and memory in the honeybee (Apis mellifera) Pharmacol Biochem Behav. 2004;77:191–197. doi: 10.1016/j.pbb.2003.09.023. [DOI] [PubMed] [Google Scholar]
- Fahrbach SE. Structure of the mushroom bodies of the insect brain. Annu Rev Entomol. 2006;51:209–232. doi: 10.1146/annurev.ento.51.110104.150954. [DOI] [PubMed] [Google Scholar]
- Farris SM, Robinson GE, Fahrbach SE. Experience and age-related outgrowth of intrinsic neurons in the mushroom bodies of the adult worker honeybee. J Neurosci. 2001;21:6395–6404. doi: 10.1523/JNEUROSCI.21-16-06395.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hammer M, Menzel R. Learning and memory in the honeybee. J Neurosci. 1995;15:1617–1630. doi: 10.1523/JNEUROSCI.15-03-01617.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurshan PT, Hamilton IS, Mustard JA, Mercer AR. Developmental changes in expression patterns of two dopamine receptor genes in mushroom bodies of the honeybee, Apis mellifera. J Comp Neurol. 2003;466:91–103. doi: 10.1002/cne.10864. [DOI] [PubMed] [Google Scholar]
- Kim YC, Lee HG, Han KA. D1 dopamine receptor dDA1 is required in the mushroom body neurons for aversive and appetitive learning in Drosophila. J Neurosci. 2007;27:7640–7647. doi: 10.1523/JNEUROSCI.1167-07.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Menzel R, Müller U. Learning and memory in honeybees: from behavior to neural substrates. Annu Rev Neurosci. 1996;19:379–404. doi: 10.1146/annurev.ne.19.030196.002115. [DOI] [PubMed] [Google Scholar]
- Shakiryanova D, Zettel GM, Gu T, Hewes RS, Levitan ES. Synaptic neuropeptide release induced by octopamine without Ca2+ entry into the nerve terminal. Proc Natl Acad Sci USA. 2011;108:4477–4481. doi: 10.1073/pnas.1017837108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fussnecker BL, Grozinger CM. Dissecting the role of Kr-h1 brain gene expression in foraging behavior in honey bees (Apis mellifera) Insect Mol Biol. 2008;17:515–522. doi: 10.1111/j.1365-2583.2008.00819.x. [DOI] [PubMed] [Google Scholar]
- Lutz CC, Rodriguez-Zas SL, Fahrbach SE, Robinson GE. Transcriptional response to foraging experience in the honey bee mushroom bodies. Dev Neurobiol. 2012;72:153–166. doi: 10.1002/dneu.20929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biron DG, Marché L, Ponton F, Loxdale HD, Galéotti N, Renault L, Joly C, Thomas F. Behavioural manipulation in a grasshopper harbouring hairworm: a proteomics approach. Proc R Soc B. 2005;272:2117–2126. doi: 10.1098/rspb.2005.3213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefevre T, Thomas F, Rave S, Patre D, Renault L, Le Bourligu L, Cuny G, Biron DG. Trypanosoma brucei brucei induces alteration in the head proteome of the tsetse fly vector Glossina palpalis gambiensis. Insect Mol Biol. 2007;16:651–660. doi: 10.1111/j.1365-2583.2007.00761.x. [DOI] [PubMed] [Google Scholar]
- Van Engelsdorp D, Meixner MD. A historical review of managed honey bee populations in Europe and the United States and the factors that may affect them. J Invert Pathol. 2010;103:S80–S95. doi: 10.1016/j.jip.2009.06.011. [DOI] [PubMed] [Google Scholar]
- Alaux C, Brunet JL, Dussaubat C, Mondet F, Tchamitchan S, Cousin M, Brillard J, Baldy A, Belzunces LP, Le Conte Y. Interactions between Nosema microspores and a neonicotinoid weaken honeybees (Apis mellifera) Environ Microbiol. 2010;12:774–782. doi: 10.1111/j.1462-2920.2009.02123.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alaux C, Dantec C, Parrinello H, Le Conte Y. Nutrigenomics in honey bee: digital gene expression analysis of pollen’s nutritive effects on healthy and varroa-parasitized bees. BMC Genomics. 2011;12:496. doi: 10.1186/1471-2164-12-496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Breed MD, Perry S, Biostad LB. Testing the blank slate hypothesis: why honey bee colonies accept young bees. Insect Soc. 2004;51:12–16. doi: 10.1007/s00040-003-0698-9. [DOI] [Google Scholar]
- Page RE, Metcalf RA, Metcalf RL, Erickson EH, Lampman RL. Extractable hydrocarbons and kin recognition in honeybee. J Chem Ecol. 1991;17:745–756. doi: 10.1007/BF00994197. [DOI] [PubMed] [Google Scholar]
- Reyment RA. Compositional data analysis. Terra Rev. 1989;1:29–34. [Google Scholar]
- Schneider SS, Lewis LA. The vibration signal, modulatory communication and the organization of labor in honey bees, Apis mellifera. Apidologie. 2004;35:117–131. doi: 10.1051/apido:2004006. [DOI] [Google Scholar]
- Anders S, Huber W. Differential expression analysis for sequence count data. Genome Biol. 2010;11:R106. doi: 10.1186/gb-2010-11-10-r106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Priooznia M, Nagarajan V, Deng Y. GeneVenn – a web application for comparing gene lists using Venn diagrams. Bioinformation. 2007;1:420–422. doi: 10.6026/97320630001420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SK, Lund J, Kiraly M, Duke K, Jiang M, Stuart JM, Eizinger A, Wylie BN, Davidson GS. A gene expression may for Caenorhabditis elegans. Science. 2001;293:2087–2092. doi: 10.1126/science.1061603. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Systematic and integrative analysis of large gene lists using DAVID Bioinformatics Resources. Nature Protoc. 2009;4:44–57. doi: 10.1038/nprot.2008.211. [DOI] [PubMed] [Google Scholar]
- Huang DW, Sherman BT, Lempicki RA. Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists. Nucleic Acids Res. 2009;37:1–13. doi: 10.1093/nar/gkn923. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Relative proportion of each CHC compound in control and Nosema or Varroa parasitized bees. Changes in proportion induced by parasitism were determined with Mann–Whitney U tests (P < 0.05 are in bold).
Percentage of correct assignments of Nosema-infected and control bees based on their cuticular hydrocarbons profiles. In the treatment column, 5 and 10 indicate the age of the bees. The last four columns show the number of bees classified in the different group treatment by the discriminant analysis. The experiment was repeated on bees from 3 different colonies. Table S3 Percentage of correct assignments of Varroa-infested and control bees based on their cuticular hydrocarbons profiles. The last six columns show the number of bees classified in the different group treatment by the discriminant analysis. The numbers 98, 120 and 231 indicate the colony origin of the bees.
Summary of DGE sequencing results. The analysis was performed on two colonies.
Lists of genes affected in the bee brain by Nosema or Varroa parasitism. Corresponding honey bee gene, Drosophila ortholog and genes also up- or downregulated in the brain of nurses and foragers are shown.
Effects of Nosema and Varroa parasites on the prevalance of Deformed Wing Virus and Varroa Destructor Virus levels in the bee brain. Comparisons of viral titers (A) Nosema vs Control, (B) Varroa vs Control and (C) Varroa vs Nosema using Cox-Reid method for estimating dispersion and Generalized Linear Model (GLM) for statistical test.