Abstract
Tubulin is subject to a reversible post-translational modification involving polyglutamylation and deglutamylation of glutamate residues in its C-terminal tail. This process plays key roles in regulating the function of microtubule associated proteins, neuronal development, and metastatic progression. This study describes the synthesis and testing of three phosphinic acid-based inhibitors that have been designed to inhibit both the glutamylating and deglutamylating enzymes. The compounds were tested against the polyglutamylase TTLL7 using tail peptides as substrates (100 µM) and the most potent inhibitor displayed an IC50 value of 150 µM. The incorporation of these compounds into tubulin C-terminal tail peptides may lead to more potent TTLL inhibitors.
The cytoskeleton of the eukaryotic cell plays key roles in determining cellular organization, shape, motility, and life cycle and is therefore absolutely essential for cellular survival. The major structural components of the cytoskeleton are microtubules, which are hollow cylindrical polymers formed by the self-assembly of α- and β-tubulin monomers.1 In the assembled microtubule, the C-termini, or tails, of the tubulin subunits are projected outward into solution and contribute to defining the surface of the filament. These tails are subject to reversible post-translational modifications (PTM's) such as detyrosination, polyglutamylation, and polyglycylation.2,3 It is becoming increasingly clear that these modifications affect both microtubule dynamics and interactions with microtubule associated proteins (MAPS) in cells, and therefore serve as control elements in a variety of biological processes.
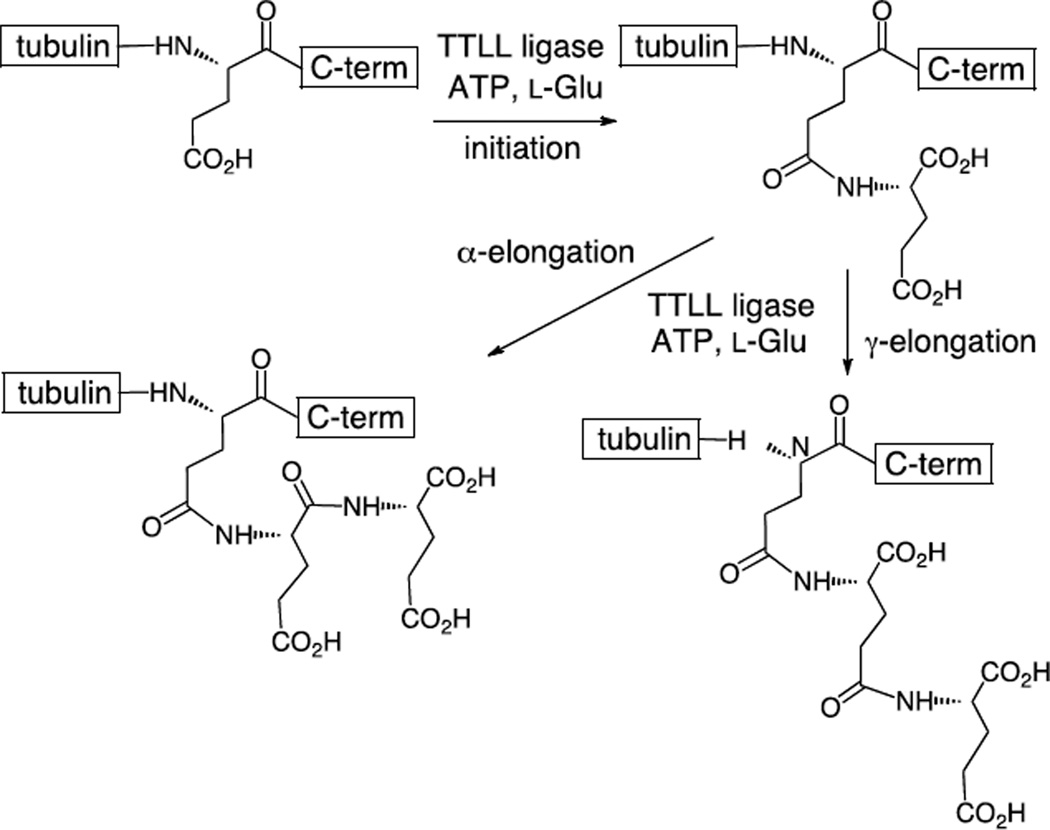
Tubulin polyglutamylation occurs at the C-termini of both α- and β-tubulin.4–7 This typically involves the addition of one to six additional glutamate residues, and the overall extent of tubulin polyglutamylation increases during development.8–11 The first glutamate is added to the side chain of a main chain glutamate to form an isopeptide bond in a process called initiation (Figure 1). Subsequent glutamate residues could conceivably be added to either the α-carboxylate or the γ-carboxylate in elongation steps. HPLC analyses using synthetic peptides have indicated that α-elongations predominantly occur during human brain tubulin polyglutamylation.8,10,11 These PTM's are catalyzed by a series of ATP-dependent amino acid ligases that are members of the "tubulin-tyrosine ligase-like" (TTLL) family of enzymes.6 These enzymes belong to the ATP-grasp family of ligases that include the prototypical member D-alanine-D-alanine ligase as well as tubulin-tyrosine ligase (TTL).12,13 Of the thirteen known TTLL enzymes in the human genome, ten have been implicated as glutamylases.2 In vitro studies using recombinant enzyme have only been performed on one of these, TTLL7, and it has been reported that this enzyme is capable of catalyzing both initiation and elongation.14 As mentioned previously, this PTM is reversible and the enzymes that remove the glutamate residues from tubulin have recently been identified as members of the soluble cytosolic carboxypeptidase (CCP) family.15,16 Four CCP members have been implicated as tubulin deglutamylases; however, in vitro activity has not yet been demonstrated for most of them.
Figure 1.
The initiation and elongation steps of tubulin polyglutamylation catalyzed by the TTLL enzymes.
Polyglutamylation has been shown to regulate the activity of the microtubule associated molecular motors kinesin and dynein.3,17,18 Not surprisingly, polyglutamylating enzymes are crucial for normal neuronal development.19,5 Tubulin polyglutamylation has also been implicated in positively regulating the activity of the microtubule severing enzyme spastin,20 a protein that is mutated in more than 40% of patients diagnosed with hereditary spastic paraplegias.21 Loss of spastin function has been implicated in defects in mitosis,22 late stage cytokinesis events,23 as well as dendritic arborization.24 Moreover, it has been found that prostate and pancreatic cancer cells display higher levels of polyglutamylation than normal cells.25,26 In particular, a recent study showed that TTLL4 is highly expressed in pancreatic cancer cells and knockdown of TTLL4 attenuated their growth,25 supporting the idea of using TTLL enzymes as therapeutic targets for small molecule inhibitors. Furthermore, hyperglutamylation has been linked to neurodegeneration in mouse models and inhibition of the TTLL1 polyglutamylase reversed this neurodegenerative phenotype.15 Thus, potent inhibitors of the tubulin polyglutamylation cycle could play key roles in understanding the structure and function of these enzymes and could serve as lead compounds in the development of therapies based on interfering with tubulin PTM levels.
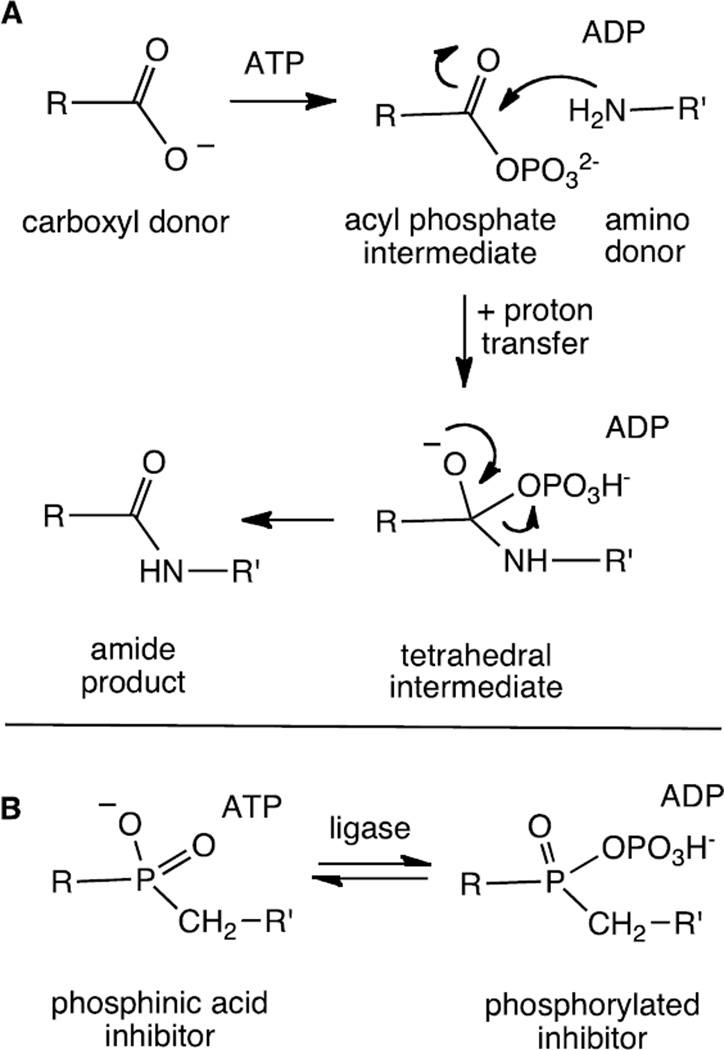
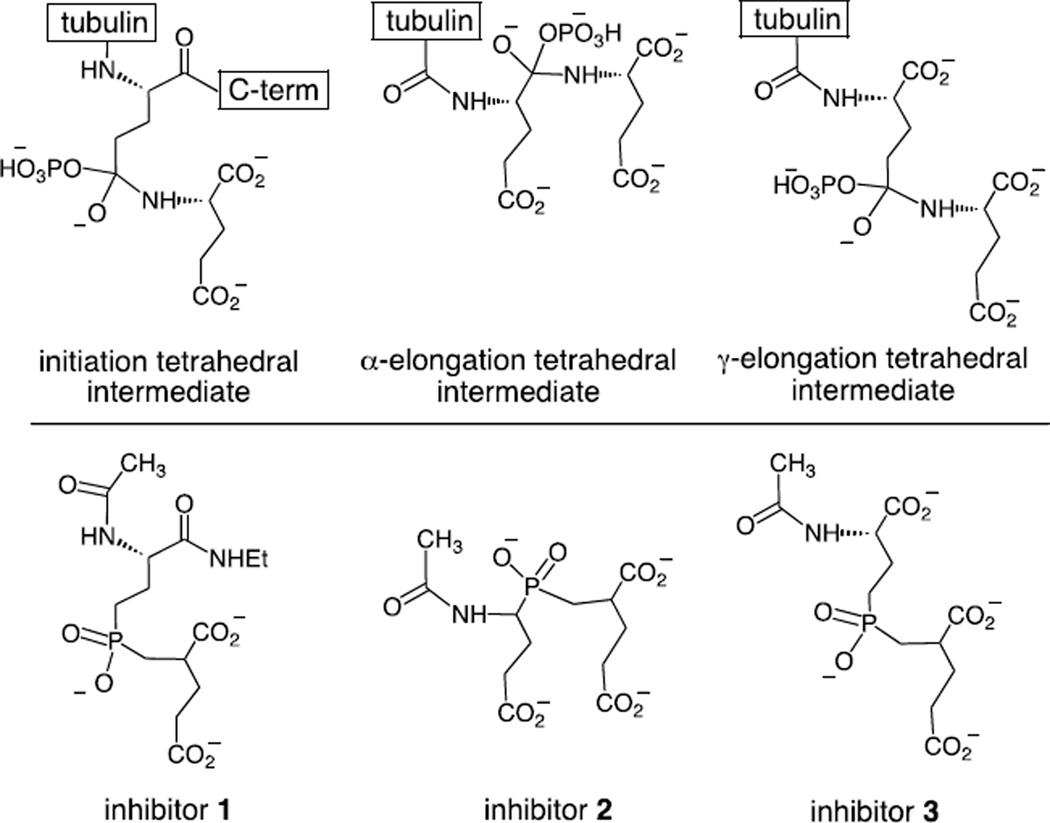
Phosphinic acids are known to serve as effective inhibitors of both ATP-dependent ligases and carboxypeptidases.27–38 The tetrahedral geometry and negative charge serves as an excellent mimic of the tetrahedral intermediate formed in the ligase reaction (Figure 2). In some instances, the bound phosphinic acid may be phosphorylated to generate a tightly bound phosphorylated inhibitor.13,33–35 The potency of these inhibitors is often quite impressive and KI values in the low nanomolar range have been reported for both the ligases and the carboxypeptidases.27,28,32,36,37 In order to target the tubulin glutamylation enzymes, three phosphinic acid inhibitors were designed (Figure 3). Inhibitor 1 is expected to mimic the tetrahedral intermediate formed in the initiation step and inhibitors 2 and 3 are expected to mimic the intermediates formed in the α- and γ-elongation steps, respectively. These compounds are expected to serve as inhibitors of the corresponding deglutamylation enzymes as well. In this study we describe the synthesis of inhibitors 1–3 and their inhibition of the TTLL7 reaction using a tubulin tail peptide as substrate.
Figure 2.
A. The mechanism employed by the ATP-dependent amino acid ligases. B. The structure of phosphinic acid-based inhibitors and the potential phosphorylation of the inhibitor within the ligase active site.
Figure 3.
The structure of the potential intermediates involved in tubulin polyglutamylation and the corresponding inhibitors targeted against them.
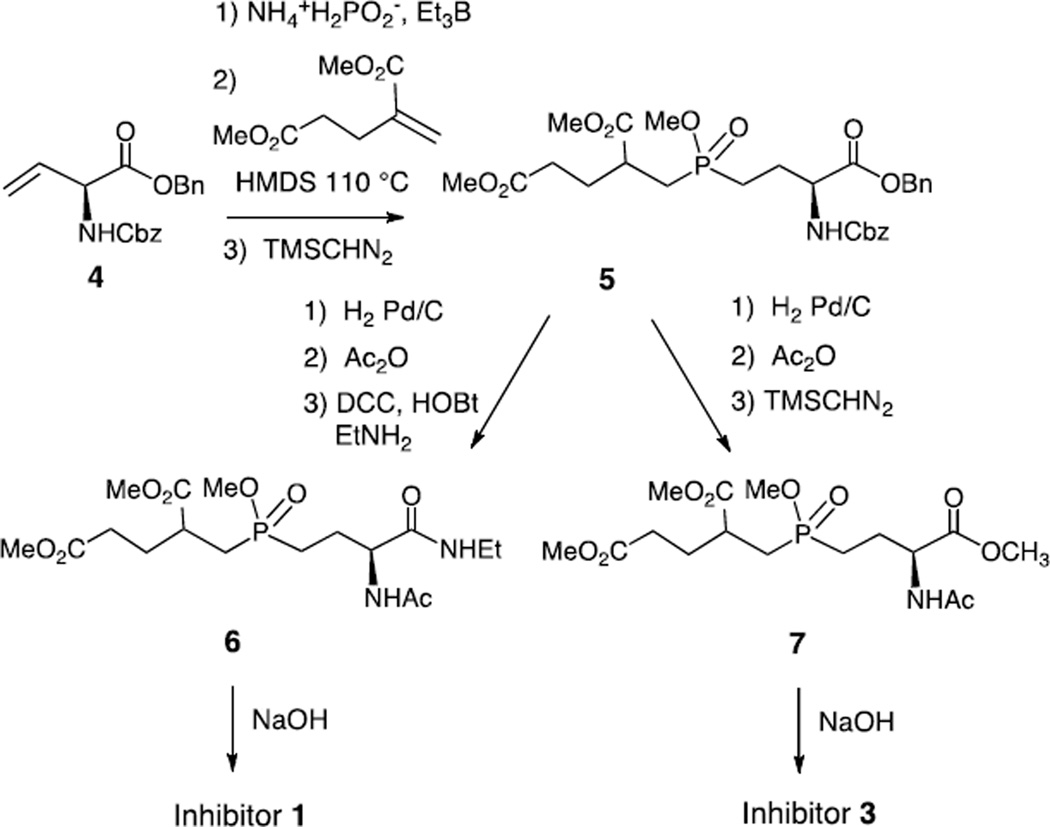
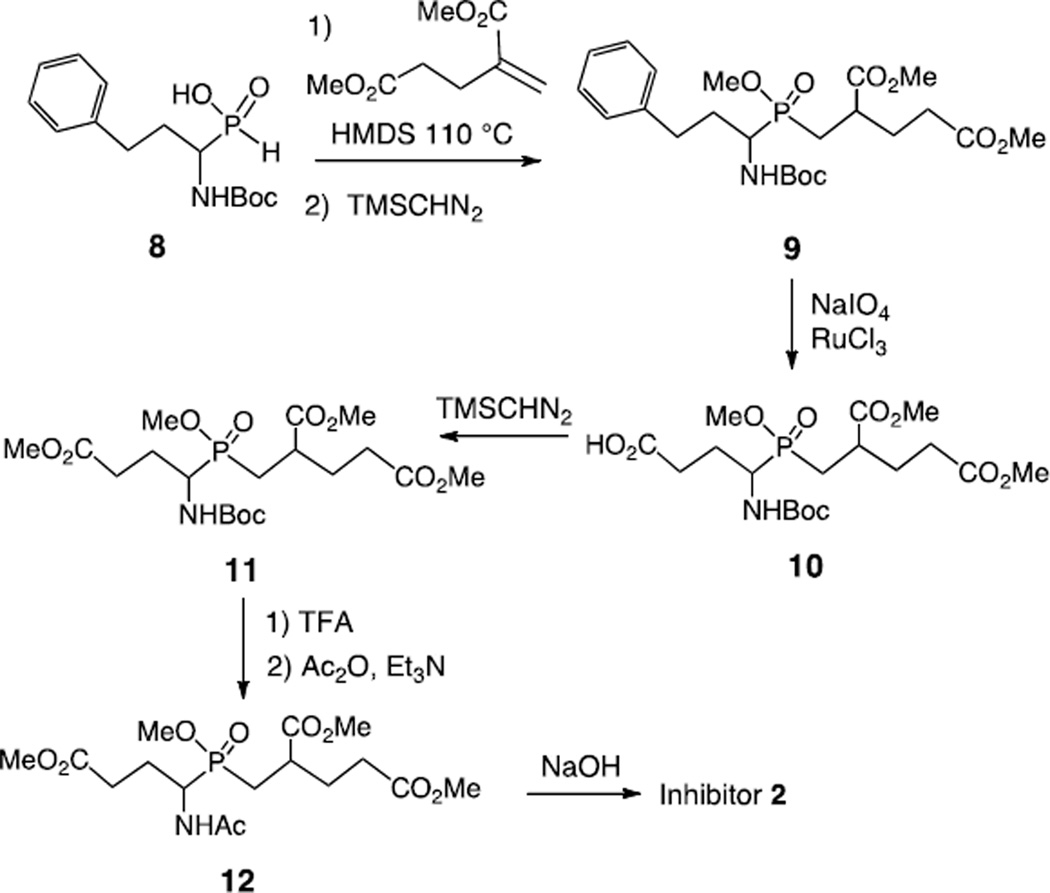
Inhibitors 1 and 3 resemble compounds previously prepared for the inhibition of folylpoly-γ-glutamate handling-enzymes and were synthesized in an analogous fashion.39,27 The synthesis of inhibitor 1 began with the treatment of the known alkene (S)-4 with ammonium hypophosphite and triethylborane to give the mono-alkylphosphinic acid (Scheme 1).39,40 This acid was silylated and added to dimethyl 2-methylenepentanedioate to generate the di-alkylphosphinic acid.41,42 This was immediately methylated to give compound 5 as a mixture of four diastereomers. The Cbz and benzyl groups were removed by hydrogenolysis and the resulting amino acid was acetylated and coupled to ethylamine to give compound 6. Demethylation was performed with NaOH to give inhibitor 1 as a mixture of two diastereomers. The precursor to inhibitor 3, compound 7, was also prepared by the hydrogenolysis of compound 5 followed by acetylation and methylation. Global demethylation using NaOH produced inhibitor 3 as a mixture of two diastereomers. In order to prepare inhibitor 2, a strategy reported by Georgiadis et al. was employed in which a phenyl group is used as a carboxyl synthon (Scheme 2).41 The known compound 8 was silylated and added to dimethyl 2-methylenepentanedioate to generate the di-alkylphosphinic acid. This was immediately methylated to give compound 9 as a mixture of eight diastereomers. Oxidation of the phenyl ring was performed using RuCl3 and NaIO4 to give the carboxylic acid 10 which was methylated to give compound 11. The Boc group was removed with TFA and the free amine was acetylated with acetic anhydride to give compound 12. Demethylation of the methyl esters by treatment with NaOH gave inhibitor 2 as a mixture of four diastereomers. With all three inhibitors, no attempts were made to separate the diastereomers and the mixtures were used directly in inhibition studies.
Scheme 1.
Scheme 2.
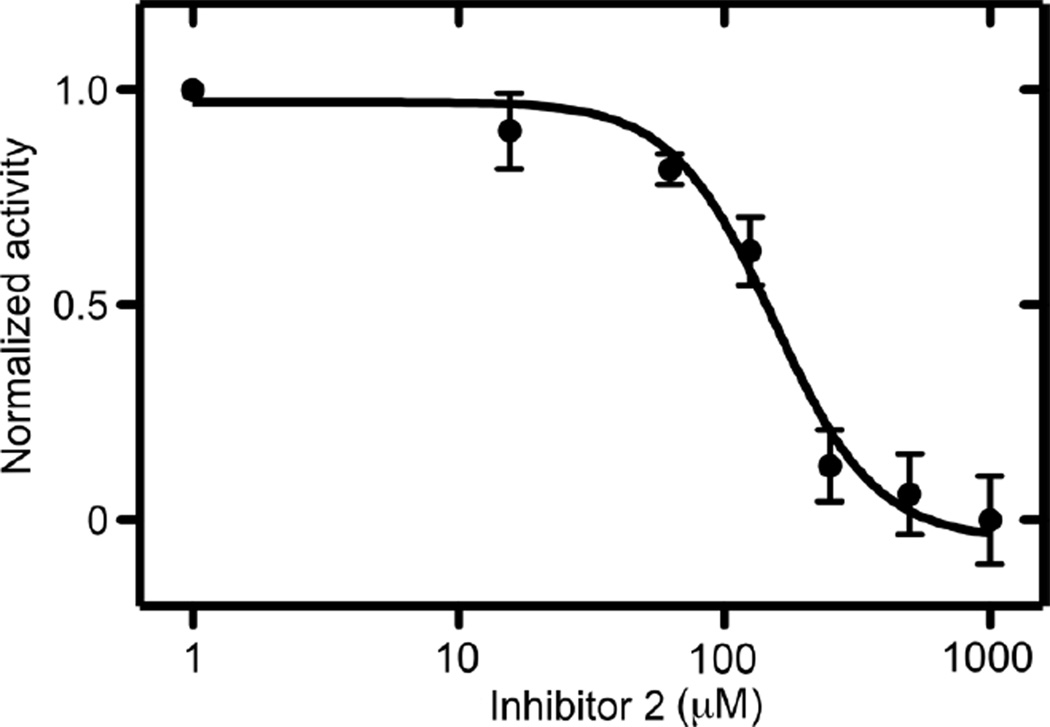
The activity of the enzyme was monitored using a synthetic N-acetlyated βIVb-tubulin C-terminal peptide (Ac-DATAEEEGEFEEEAEEEVA) as substrate (see supplementary data for details). The peptide (100 µM) was incubated with recombinant mouse TTLL7 in the presence of glutamate, ATP, Mg2+, and varying amounts of inhibitors 1–3. Control reactions lacked ATP and glutamate. Under these conditions, the predominant reaction product consisted of monoglutamylated peptide with small amounts (<20%) of di-glutamylated peptide (see supplementary material Figure S1). The extent of reaction was determined using ESI TOF LC-MS by comparing the decrease in intensity of the signal of the unmodified peptide to that of the corresponding control reaction. The most potent inhibition was observed with compound 2 which showed an IC50 value of 150 µM (Figure 4). Compounds 1 and 3 were weaker inhibitors and only showed inhibition at mM concentrations. All three compounds were also tested for their ability to inhibit the reaction catalyzed by the related enzyme, tubulin tyrosine ligase (TTL) but failed to show any inhibition at mM concentrations.12 This indicates that the inhibition by inhibitor 2 is specific for the glutamylase TTLL7 and does not interfere with the activity of the closely related family member tubulin tyrosine ligase.
Figure 4.
A plot of normalized activity versus the concentration of inhibitor 2 (log plot) for the inhibition of tubulin C-terminal peptide (100 µM) glutamylation by TTLL7.
In summary, we have prepared a series of phosphinic acid pseudo-dipeptides designed to inhibit the enzymes of tubulin polyglutamylation/deglutamylation. Of these compounds, inhibitor 2 was shown to inhibit the activity of mouse TTLL7 with an IC50 of 150 µM, whereas inhibitors 1 and 3 were only active in the millimolar range. Inhibitor 2 was designed to mimic an α-elongation step of polyglutamylation. While TTLL7 has been reported to be capable of catalyzing both initiation and elongation, it is not yet known whether the elongation steps involve α- or γ-additions.14 It is likely that the stronger binding of inhibitor 2 reflects the ability of TTLL7 to catalyze α-elongations. This is consistent with reports that α-elongation linkages have been observed in tubulin isolated from mouse brain.8,10,11 Alternatively, inhibitor 2 may simply be acting as an initiation inhibitor and the additional negative charge (as compared to the designed initiation inhibitor 1) favors binding to an electropositive peptide-binding groove in the enzyme (vide infra). The modest IC50 value observed with these inhibitors likely reflects the fact that the true substrate is an intact tubulin protein within a microtubule, as opposed to a dipeptide.9 It is clear from this work, and that of others, that peptides containing 15 or more residues corresponding to the C-terminus of tubulin can act as substrates for some polyglutamylases including TTLL7; however, the minimal substrate length is not known for each isozyme.43 In the case of the related enzyme tubulin tyrosine ligase (TTL), studies on substrate specificity with C-terminal peptides have been performed.44 This ligase adds a single tyrosine to the C-terminus of de-tyrosinated α-tubulin and it has been shown that the activity drops off dramatically with peptides containing less than 10 residues. An inspection of the structure of TTL explains this phenomenon as an elongated stripe of conserved positively charged residues connects the active site to the N-terminal domain.12 These residues interact with the highly negatively charged tubulin tail and contribute significantly to peptide substrate binding. This is also thought to be the case with the TTLL enzymes and can help to explain why dipeptide-based inhibitors are not able to access much of the binding affinity. Similar observations have been made with phosphinic acid inhibitors that target the amino acid ligases of peptidoglycan biosynthesis.28,31 The D-glutamic acid-adding enzyme MurD adds a D-glutamate residue onto the alanine of UDP-N-acetylmuramic acid-L-Ala (UDP-MurNAc-L-Ala). When the phosphinic acid dipeptide corresponding to L-Ala-D-Glu was tested as an inhibitor, an IC50 of > 1 mM was observed. However, when the same phosphinic acid was attached to UDP either via a spacer or via a MurNAc residue, the IC50 values dropped to 0.7 µM and less than 1.0 nm, respectively. Thus, the potency of phosphinic acid-based ligase inhibitors can be dramatically affected by the inclusion of "remote" functional groups into the structures. In future studies, we will embed the phosphinic acids within tubulin C-terminal peptides.45 These compounds are anticipated to be potent inhibitors of both the TTLL glutamylases and the CCP deglutamylases.
Supplementary Material
Acknowledgements
We thank Duck-Yeon Lee from the Biochemistry Core Facility of the National Heart, Lung and Blood Institute (NHLBI) for his suggestions and assistance with experiments. This research was supported by the Natural Sciences and Engineering Research Council of Canada (NSERC) (MET) and the intramural program of the National Institutes of Health (NIH) (AR-M).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Supplementary data
Supplementary data including the full experimental and spectral details associated with this article can be found, in the online version, at doi:
References
- 1.Wade RH. Mol. Biotech. 2009;43:177–191. doi: 10.1007/s12033-009-9193-5. [DOI] [PubMed] [Google Scholar]
- 2.Garnham CP, Roll-Mecak A. Cytoskeleton. 2012;69:442–463. doi: 10.1002/cm.21027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Janke C, Bulinski JC. Nat. Rev. Mol. Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 4.van Dijk J, Rogowski K, Miro J, Lacroix B, Edde B, Janke C. Mol. Cell. 2007;26:437–448. doi: 10.1016/j.molcel.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 5.Ikegami K, Mukai M, Tsuchida J, Heier RL, MacGregor GR, Setou M. J. Biol. Chem. 2006;281:30707–30716. doi: 10.1074/jbc.M603984200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janke C, Rogowski K, Wloga D, Regnard C, Kajava AV, Strub JM, Temaruk N, van Dijk J, Boucher D, Dorsselaer AV, Suryavanshi S, Gaertig J, Edde B. Science. 2005;308:1758–1762. doi: 10.1126/science.1113010. [DOI] [PubMed] [Google Scholar]
- 7.Edde B, Rossier J, Le Caer JP, Desbruyeres E, Gros F, Denoulet P. Science. 1990;247:83–85. doi: 10.1126/science.1967194. [DOI] [PubMed] [Google Scholar]
- 8.Redeker V. Meth. Cell Biol. 2010;95:77–103. doi: 10.1016/S0091-679X(10)95006-1. [DOI] [PubMed] [Google Scholar]
- 9.Regnard C, Audebert S, Desbruyeres E, Denoulet P, Edde B. Biochemistry. 1998;37:8395–8404. doi: 10.1021/bi9804131. [DOI] [PubMed] [Google Scholar]
- 10.Redeker V, Rusconi F, Mary J, Prome D, Rossier J. J. Neurochem. 1996;67:2104–2114. doi: 10.1046/j.1471-4159.1996.67052104.x. [DOI] [PubMed] [Google Scholar]
- 11.Redeker V, Le Caer JP, Rossier J, Prome JC. J. Biol. Chem. 1991;266:23461–23466. [PubMed] [Google Scholar]
- 12.Szyk A, Deaconescu AM, Piszczek G, Roll-Mecak A. Nat. Struct. Mol. Biol. 2011;18:1250–1259. doi: 10.1038/nsmb.2148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fan C, Moews PC, Walsh CT, Knox JR. Science. 1994;266:439–443. doi: 10.1126/science.7939684. [DOI] [PubMed] [Google Scholar]
- 14.Mukai M, Ikegami K, Sugiura Y, Takeshita K, Nakagawa A, Setou M. Biochemistry. 2009;48:1084–1093. doi: 10.1021/bi802047y. [DOI] [PubMed] [Google Scholar]
- 15.Rogowski K, van Dijk J, Magiera MM, Bosc C, Deloulme JC, Bosson A, Peris L, Gold ND, Lacroix B, Grau MB, Bec N, Larroque C, Desagher S, Holzer M, Andrieux A, Moutin MJ, Janke C. Cell. 2010;143:564–578. doi: 10.1016/j.cell.2010.10.014. [DOI] [PubMed] [Google Scholar]
- 16.Kimura Y, Kurabe N, Ikegami K, Tsutsumi K, Konishi Y, Kaplan OI, Kunitomo H, Y I, Blacque OE, Setou M. J. Biol. Chem. 2010;285:22936–22941. doi: 10.1074/jbc.C110.128280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Suryavanshi S, Edde B, Fox LA, Guerrero S, Hard R, Hennessey T, Kabi A, Malison D, Pennock D, Sale WS, Wloga D, Gaertig J. Curr. Biol. 2010;20:435–440. doi: 10.1016/j.cub.2009.12.062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kubo T, Tanagisawa H, Tagi T, Hirono M, Kamiya R. Curr. Biol. 2010;20:441–445. doi: 10.1016/j.cub.2009.12.058. [DOI] [PubMed] [Google Scholar]
- 19.Ikegami K, Heier RL, Taruishi M, Takagi H, Mukai M, Shimma S, Tairo S, Hatanaka K, Morone N, Yao I, Campbell PK, Yuasa S, Janke C, MacGregor GR, Setou M. Proc. Nat. Acad. Sci. USA. 2007;104:3213–3218. doi: 10.1073/pnas.0611547104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lacroix B, van Dijk J, Gold ND, Guizetti J, Aldrian-Herrada G, Rogowski K, Gerlich DW, Janke C. J. Cell Biol. 2010;189:945–954. doi: 10.1083/jcb.201001024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Roll-Mecak A, McNally FJ. Curr. Opin. Cell Biol. 2010;22:96–103. doi: 10.1016/j.ceb.2009.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang D, Rogers GC, Buster DW, Sharp DJ. J. Cell Biol. 2007;177:231–242. doi: 10.1083/jcb.200612011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Guizetti J, Schermelleh L, Mantler J, Maar S, Poser I, Leonhardt H, Muller-Reichert T, Gerlich DW. Science. 2011;331:1616–1620. doi: 10.1126/science.1201847. [DOI] [PubMed] [Google Scholar]
- 24.Jinushi-Nakao S, Arvind R, Amikura R, Kinameri E, Liu AW, Moore AW, Walton A. Neuron. 2007;56:963–978. doi: 10.1016/j.neuron.2007.10.031. [DOI] [PubMed] [Google Scholar]
- 25.Kashiwaya K, Nakagawa H, Hosokawa M, Mochizuki Y, Ueda K, Piao L, Chung S, Hamamoto R, Eguchi H, Ohigashi H, Ishikawa O, Janke C, Shinomura Y, Nakamura Y. Cancer Res. 2010;70:4024–4032. doi: 10.1158/0008-5472.CAN-09-4444. [DOI] [PubMed] [Google Scholar]
- 26.Soucek K, Kamaid A, Phung AD, Kubala L, Bulinski JC, Harper RW, Eiserich JP. Prostate. 2006;66:954–965. doi: 10.1002/pros.20416. [DOI] [PubMed] [Google Scholar]
- 27.Valiaeva N, Bartley D, Kuono T, Coward JK. J. Org. Chem. 2001;66:5146–5154. doi: 10.1021/jo010283t. [DOI] [PubMed] [Google Scholar]
- 28.Gegnas LD, Waddell ST, Chabin RN, Reddy S, Wong KK. Bioorg. Med. Chem. Lett. 1998;8:1643–1648. doi: 10.1016/s0960-894x(98)00285-6. [DOI] [PubMed] [Google Scholar]
- 29.Miller DJ, Hammond SM, Anderluzzi D, Bugg TDH. J. Chem. Soc. Perkin Trans. I. 1998:131–142. [Google Scholar]
- 30.Zeng B, Wong KK, Pompliano DL, Reddy S, Tanner ME. J. Org. Chem. 1998;63:10081–10086. [Google Scholar]
- 31.Tanner ME, Vaganay S, van Heijenoort J, Blanot D. J. Org. Chem. 1996;61:1756–1760. doi: 10.1021/jo951780a. [DOI] [PubMed] [Google Scholar]
- 32.Ellsworth BA, Tom NJ, Bartlett PA. Chem. Biol. 1996;3:37–44. doi: 10.1016/s1074-5521(96)90082-4. [DOI] [PubMed] [Google Scholar]
- 33.Hiratake J, Kato H, Oda J. J. Am. Chem. Soc. 1994;116:12059–12060. [Google Scholar]
- 34.Abell LM, Villafranca JJ. Biochemistry. 1991;30:6135–6141. doi: 10.1021/bi00239a008. [DOI] [PubMed] [Google Scholar]
- 35.Duncan K, Walsh CT. Biochemistry. 1988;27:3709–3714. doi: 10.1021/bi00410a028. [DOI] [PubMed] [Google Scholar]
- 36.Barinka C, Hlouchova K, Rovenska M, Majer P, Dauter M, Hin N, Ko YS, Tsukamoto T, Slusher BS, Konvalinka J, Lubkowski J. J. Mol. Biol. 2008;376:1438–1450. doi: 10.1016/j.jmb.2007.12.066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Suzuki K, Muto Y, Fushihara K, Kanemoto Ki, Iida H, Sato E, Kikuchi C, Matsushima T, Kato E, Nomoto M, Yoshioka S, Hidemi I. J. Pharmacol. Exp. Ther. 2004;309:607–615. doi: 10.1124/jpet.103.062729. [DOI] [PubMed] [Google Scholar]
- 38.Dive V, Georgiadis D, Matziari M, Makaritis A, Beau F, Cuniasse P, Yiotakis A. Cell. Mol. Life Sci. 2004;61:2010–2019. doi: 10.1007/s00018-004-4050-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Feng Y, Coward JK. J. Med. Chem. 2006;49:770–788. doi: 10.1021/jm050871p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Liu S, Ben RN. Org. Lett. 2005;7:2385–2388. doi: 10.1021/ol050677x. [DOI] [PubMed] [Google Scholar]
- 41.Georgiadis D, Matziari M, Vassilou S, Dive V, Yiotakis A. Tetrahedron. 1999;55:14635–14648. [Google Scholar]
- 42.Thottathil JK, Ryono DE, Przybyla CA, Moniot JL, Neubeck R. Tetrahedron Lett. 1984;25:4741–4744. [Google Scholar]
- 43.Westermann S, Plessmann U, Weber K. FEBS Lett. 1999;459:90–94. doi: 10.1016/s0014-5793(99)01227-2. [DOI] [PubMed] [Google Scholar]
- 44.Rudiger M, Wehland J, Weber K. Eur. J. Biochem. 1994;220:309–320. doi: 10.1111/j.1432-1033.1994.tb18627.x. [DOI] [PubMed] [Google Scholar]
- 45.Yiotakis A, Vassilou S, Jiracek J, Dive V. J. Org. Chem. 1996;61:6601–6605. doi: 10.1021/jo9603439. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.