Abstract
The objective of this study was to characterize the temporal variability of fluoroquinolone resistance mechanisms among Escherichia coli colonizing the gastrointestinal tract of hospitalized patients. Patients with new fluoroquinolone-resistant E. coli (FQREC) colonization were followed with serial fecal sampling until discharge or death. Genetic mechanism(s) of resistance for all FQREC isolates were characterized, including mutations in gyrA and parC and efflux pump overexpression. Of 451 subjects, 73 (16.2%) became newly colonized with FQREC. There was significant variability in regard to temporal changes in resistance mechanisms and levofloxacin MICs among isolates from individual patients. Compared to patients with transient colonization, patients with persistent colonization were more likely to have a urinary catheter (P=0.04), diarrhea (P=0.04), and a longer duration of hospitalization (22 and 9.0 mean days, respectively; P=0.01) prior to sampling. Our data demonstrate the significant variability of resistance mechanisms in colonizing E. coli isolates among hospitalized patients.
1. Introduction
The rapid increase in the prevalence of fluoroquinolone-resistant Escherichia coli (FQREC) in recent years is of significant public health concern (Lautenbach et al., 2004). The major mechanisms leading to FQ resistance in E. coli include 1) mutations in the genes encoding the drug targets DNA gyrase and topoisomerase IV, most commonly in the gyrA and parC genes in the quinolone resistance-determining region (QRDR), and 2) overproduction of the AcrAB-TolC drug efflux pump (Jacoby, 2005; Li and Nikaido, 2009).
In vitro studies characterizing the emergence of FQ resistance in E. coli have demonstrated that selection of resistance occurs in a stepwise fashion, with increasing numbers of mutations leading to correspondingly higher FQ minimum inhibitory concentrations (MICs) (Kern et al., 2000; Chang et al., 2007; Singh et al., 2012). In the clinical setting, studies have also suggested that MICs to FQs in E. coli are typically higher in organisms with a greater number of resistance mutations (e.g., in target enzymes or genes mediating efflux) (Komp Lindgren et al., 2003; Lautenbach et al., 2006a; Morgan-Linnell et al., 2009; Moon et al., 2010). However, these studies have focused on isolates derived from clinical infections, whereas FQ resistance likely arises at the level of gastrointestinal tract colonization (Richard et al., 2001; Donskey, 2006).
Characterizing the stepwise accumulation of resistance mutations in colonizing E. coli isolates from individual patients is critical to enhanced understanding of the development of FQ resistance in the clinical setting, including informing potential strategies targeting specific resistance mechanisms to limit the emergence of FQREC. Therefore, we conducted this study to characterize the temporal changes in FQ resistance and resistance mutations among adult inpatients with new FQREC gastrointestinal tract colonization. In addition, we compared characteristics of patients who demonstrated transient FQREC colonization (i.e., FQREC colonization demonstrated on only one occasion) versus those with persistent colonization (i.e., multiple FQREC isolates over time).
2. Materials and methods
2.1. Study design and setting
This prospective cohort study was conducted at two hospitals in the University of Pennsylvania Health System (UPHS) in Philadelphia: the Hospital of the University of Pennsylvania (HUP), a 725-bed academic tertiary care medical center, and Penn Presbyterian Medical Center (PPMC), a 344-bed urban community hospital. As previously described (Lautenbach et al., 2006a; Lautenbach et al., 2009), three annual fecal surveillance surveys were performed hospital-wide at the two hospitals during the study years 2002, 2003, and 2004. For the present study, target units were selected from the two hospitals based on high prevalence rates of FQREC characterized by the three surveys (two units at PPMC and four units at HUP). The selected units included general medicine, oncology, rehabilitation, and intensive care units.
Subsequently, each unit was surveyed for a 3-month time period, with all patients admitted to the target units eligible for inclusion in the present study cohort. On the first day of a unit survey, all patients hospitalized on the unit at 8:00 AM were identified and approached, with subsequent enrollment in the study if informed consent was obtained. For those patients who agreed to participate, fecal samples via a perirectal swab were obtained and submitted to the HUP Clinical Microbiology Laboratory for processing. Patients were followed longitudinally and continued to have fecal samples submitted every 48 to 72 hours (depending on patient availability) until the time of hospital discharge or death. New patients admitted to the unit during the survey period were also eligible to be enrolled in the study. Any patient transferred to another unit of the hospital continued to be followed until the time of hospital discharge or death. At the end of the three months, all patients currently undergoing surveillance continued to be followed until the time of hospital discharge or death. However, no new patients were enrolled during the third month of the survey to allow for complete follow up of all patients already enrolled. Each patient was included as a subject only once, with only the first episode of eligibility included. The study was approved by the institutional review board of the University of Pennsylvania.
2.2 Data collection
Data were abstracted from the Pennsylvania Integrated Clinical and Administrative Research Database (PICARD) (Barton et al., 2005; Lee et al., 2009), which includes demographic, laboratory, pharmacy, and billing information. Information for all patients was collected on the following: baseline demographics, year of the surveillance culture, hospital of admission, transfer from another institution or nursing home, admissions to UPHS in the 30 days prior to sampling, service location at the time of sampling (i.e., medical versus surgical), and number of hospital days prior to sampling. The presence of the following comorbid conditions was documented at the time of the sampling: diabetes mellitus, malignancy, renal insufficiency (creatinine 2.0 mg/dL or the requirement of dialysis), HIV infection, solid organ or hematopoietic stem cell transplant, neutropenia (absolute neutrophil count <500/mm3), significant cardiovascular disease (e.g., severe congestive heart failure), significant respiratory disease (e.g., severe chronic obstructive pulmonary disease, chronic bronchitis), and any surgical procedure performed in the 30 days prior to sampling. Data on the presence of a urinary catheter, central venous catheter, or diarrhea prior to the initial surveillance culture were collected for all patients. Furthermore, data on antimicrobial therapy, chemotherapy, and steroids or other immunosuppressive agents administered during the 30 days prior to fecal sampling was ascertained.
2.3. Microbiological methods
Detection of E. coli from fecal samples was performed as described previously (Lautenbach et al., 2006a; Lautenbach et al., 2009). Given the multi-step nature of development of FQ resistance in a given isolate, organisms with MICs in the susceptible but elevated range (e.g., with early single mutations) may be critical in explaining the emergence and dissemination of FQ resistance (Gales et al., 2000; Kern et al., 2000; Chang et al., 2007; Singh et al., 2012). As such, for the present study, low-level FQ resistance (i.e., reduced FQ susceptibility) and high-level FQ resistance were defined as a levofloxacin MIC 0.25 g/mL but <8 g/mL and 8 g/mL, respectively. The QRDR of gyrA and parC were amplified and sequenced as previously described (Lautenbach et al., 2006a; Lautenbach et al., 2009). Overexpression of AcrAB was measured indirectly by the organic solvent tolerance assay as previously validated (White et al., 1997; Wang et al., 2001). Two sets of primers were used to detect the plasmid-encoded fluoroquinolone resistance gene qnr as previously described (Lautenbach et al., 2006a).The genetic relatedness of E. coli isolates was determined by molecular typing using pulsed field gel electrophoresis (PFGE) (Lautenbach et al., 2006a), with all results analyzed using the Fingerprinting II Informatix Software v 3.0 (Bio-Rad Laboratories, Inc., Hercules, CA) and interpreted according to established criteria (Goering and Tenover, 1997).
2.4. Statistical analysis
The incidence of new FQREC colonization during the study period was calculated, with three stages of FQREC colonization identified, as follows: 1) no FQREC colonization (levofloxacin MIC <0.25 μg/mL); 2) low-level FQREC colonization (levofloxacin MIC 0.25 μg/mL but <8.0 μg/mL); and 3) high-level FQREC colonization (levofloxacin MIC ≥8 μg/mL). For each patient with new FQREC colonization, resistance mechanisms (e.g., accumulation of mutations) of the isolates were described. For any patient with more than one FQREC isolate identified over time, all FQREC isolates were similarly characterized. Genetic mechanism(s) of resistance for all FQREC isolates were characterized by focusing specifically on mutations in gyrA and parC, as well as the presence of OST.
Characteristics of patients with colonization with one FQREC isolate (i.e., transient colonization) versus multiple FQREC isolates (i.e., persistent colonization) during the sampling period were compared, including demographic variables, comorbid conditions, and antimicrobial use in the 30 days prior to initial sampling. Continuous variables were compared using the Student’s t-test or Wilcoxon rank-sum test and categorical variables were compared using the χ2 or Fisher’s exact test. Bivariable analyses were then performed to determine the association between patient characteristics and colonization with more than one FQREC isolate during the sampling period. All statistical calculations were performed using commercially available software (STATA v11.0; StataCorp LP, College Station, Texas).
3. Results
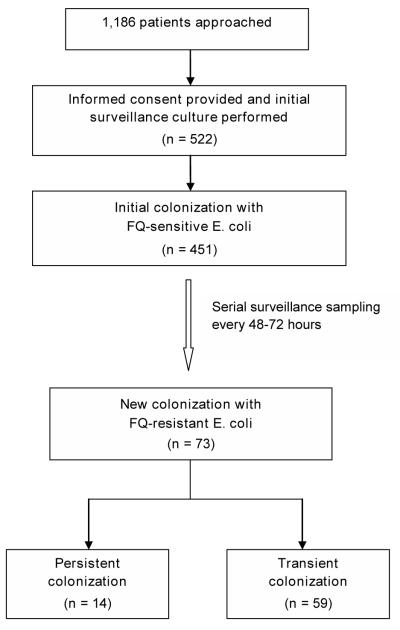
During the study period, a total of 1,186 hospitalized patients were approached for enrollment (Figure 1). Of these, 522 (44.0%) provided informed consent and had an initial fecal swab obtained. Notably, there were no significant differences with regard to mean age, race and ethnicity, year of enrollment, and hospital of admission (i.e., HUP versus PPMC) when comparing patients who did and did not enroll in the study.
Figure 1.
Study flow diagram.
Of the 522 patients who had an initial sample obtained, 429 (82.2%) were hospitalized at HUP while 93 (17.8%) were hospitalized at PPMC. Subsequently, 516 patients had fecal specimens that revealed E. coli, of which 451 (87.4%) were FQ-susceptible. These 451 patients who were initially colonized with FQ-susceptible E. coli represented the primary study cohort. These subjects underwent serial surveillance sampling during hospitalization with 73 (16.2%) having a subsequent culture positive for FQREC. Among these unique 73 patients, there were a total of 98 E. coli isolates with FQ resistance during the sampling period, as follows: 53 (54.1%) isolates with low-level resistance (levofloxacin MIC 0.25 μg/mL but <8.0 μg/mL) and 45 (45.9%) isolates with high-level resistance (levofloxacin MIC ≥8 μg/mL). Molecular characteristics of these FQREC isolates are shown in Table 1. The median number of gyrA mutations for E. coli isolates with high-level resistance versus low-level resistance was 2.0 and 1.0, respectively (P<0.001). The median number of parC mutations for E. coli isolates with high-level resistance versus low-level resistance was 1.0 and 0.0, respectively (P<0.001). The qnr gene was not detected in any study isolate. Finally, isolates with high-level resistance were more likely to be OST-positive compared to isolates with low-level resistance (P<0.001).
Table 1.
Mechanisms of Resistance in Colonizing Escherichia coli Isolates Among Hospitalized Patients
| Isolatesa | MICb |
gyrA mutations n (%) |
No.gyrA mutations Median (IQR) |
parC mutations n (%) |
No.parC mutations Median (IQR) |
OST positive n (%) |
Total mechanisms of resistance n (%) |
|---|---|---|---|---|---|---|---|
| Resistance | 0:12 (13.2%) | ||||||
| (n=92) | 0.75 | 72 (78.3%) | 1 (1, 2) | 37 (40.2%) | 0 (0, 1) | 35 (38.4%) | 1:38 (41.8%) |
| 2: 24 (26.3%) | |||||||
| 3: 17 (18.7%) | |||||||
|
| |||||||
| High-level | 0:0 (0.0%) | ||||||
| resistancec | 32 | 40 (93.0%) | 2 (2, 2) | 34 (79.1%) | 1 (1, 1) | 25 (58.1%) | 1: 8 (19.0%) |
| (n=43) | 2: 17 (40.5%) | ||||||
| 3: 17 (40.5%) | |||||||
|
| |||||||
| Low-level | 0: 12 (24.5%) | ||||||
| resistanced | 0.38 | 32 (65.3%) | 1 (0, 1) | 3 (6.1%) | 0 (0, 0) | 10 (20.4%) | 1: 30 (61.2%) |
| (n=49) | 2: 7 (14.3%) | ||||||
| 3: 0 (0.0%) | |||||||
MIC = minimum inhibitory concentration; OST = organic solvent tolerance; IQR = interquartile range.
6 isolates without information on mutations.
Median levofloxacin MIC by Etest.
Levofloxacin MIC 8 g/mL.
Levofloxacin MIC 0.25 g/mL but <8 g/mL.
A total of 14 (19.2%) of the 73 patients with new colonization with FQREC had >1 FQREC culture during the sampling time period. Among these 14 unique patients, there were 39 isolates with FQ resistance (low-level and high-level). Temporal changes in levofloxacin MIC values, resistance mechanisms, as well as clonal relationships among the 14 patients with >1 FQREC isolate during sampling are shown in Table 2. Thirty-seven out of 39 isolates were successfully characterized by PFGE. Comparison of the 37 isolates as a group demonstrated no evidence of clustering during the study period (i.e., indication of an outbreak). Subsequently, the variability in PFGE patterns within sample sets for each patient was assessed, with five patients demonstrating clonally related isolates (C, K, M, Q, V; Table 2).
Table 2.
Temporal Changes in Escherichia coli Isolates During Hospitalization
| Subject no. | No. isolate(s) |
Days from initial sample |
MICa (μg/mL) |
gyrA mutation(s) |
parC mutation(s) |
OST | PFGE pattern(s)b |
|---|---|---|---|---|---|---|---|
| 1 | 3 Susc. | 0-5 | |||||
| Resistant | 7 | 0.75 | Ser83Leu | Neg | Neg | A | |
| 9 Susc. | 9-41 | ||||||
| Resistant | 44 | 0.75 | Neg | Neg | Neg | B | |
| 3 Susc. | 46-52 | ||||||
|
| |||||||
| 2 | 8 Susc. | 0-19 | |||||
| Resistant | 21 | 0.25 | Ser83Leu | Neg | Pos | C | |
| 4 Susc. | 24-31 | ||||||
| Resistant | 33 | 0.75 | Ser83Leu | Neg | Pos | D | |
| 4 Susc. | 35-42 | ||||||
| Resistant | 45 | 0.25 | Asp87Tyr | Neg | Pos | C | |
| 5 Susc. | 47-56 | ||||||
| Resistant | 59 | 32 | Ser83Leu;Asp87Gly | Ser80Ile | Pos | E | |
|
| |||||||
| 3 | 4 Susc. | 0-10 | |||||
| Resistant | 12 | 0.38 | Ser83Leu | Neg | Neg | F | |
| Resistant | 14 | 0.25 | Asp87Tyr | Neg | Neg | G | |
| 6 Susc. | 17-28 | ||||||
|
| |||||||
| 4 | 6 Susc. | 0-14 | |||||
| Resistant | 17 | ≥ 32 | Ser83Leu; Asp87Tyr | Ser80Arg; Glu84Val | Neg | H | |
| 7 Susc. | 19-33 | ||||||
| Resistant | 35 | ≥ 32 | Ser83Leu; Asp87Gly | Ser80Ile; Glu84Gly | Pos | I | |
| 7 Susc. | 38-52 | ||||||
| Resistant | 54 | 0.25 | Asp87Tyr | Ser80Ile | Neg | J | |
|
| |||||||
| 5 | 6 Susc. | 0-12 | |||||
| Resistant | 14 | ≥ 32 | Ser83Leu; Asp87Tyr | Ser80Arg; Glu84Val | Pos | K | |
| 1 Susc. | 17 | ||||||
| Resistant | 19 | ≥ 32 | Ser83Leu,Asp87Asn | Neg | Pos | K | |
| 4 Susc. | 21-31 | ||||||
| Resistant | 33 | 0.5 | Ser83Leu | Neg | Neg | K | |
| 1 Susc. | 35 | ||||||
|
| |||||||
| 6 | 2 Susc. | 0-2 | |||||
| Resistant | 4 | ≥ 32 | Ser83Leu; Asp87Gly | Neg | Pos | L | |
| Resistant | 7 | 0.25 | Asp87Tyr | Neg | Pos | M | |
| Resistant | 9 | 0.25 | Asp87Tyr | Neg | N/A | M | |
| 8 Susc. | 11-28 | ||||||
|
| |||||||
| 7 | 3 Susc. | 0-4 | |||||
| Resistant | 7 | 1 | Neg | Neg | Neg | N | |
| 2 Susc. | 9-11 | ||||||
| Resistant | 14 | ≥ 32 | Ser83Leu; Asp87Asn | Ser80Ile | Neg | O | |
| Resistant | 16 | ≥ 32 | Ser83Leu; Asp87Asn | Ser80Ile; Presentc | Pos | P | |
| Resistant | 18 | ≥ 32 | Ser83Leu,Asp87Tyr | Ser80Ile | Neg | N/A | |
|
| |||||||
| 8 | 1 Susc. | 0 | |||||
| Resistant | 2 | ≥ 32 | Ser83Leu; Asp87Asn | Ser80Ile | Pos | Q | |
| 4 Susc. | 4-11 | ||||||
| Resistant | 14 | 0.125 | N/A | N/A | N/A | R | |
| Resistant | 16 | ≥ 32 | Ser83Leu,Asp87Asn | Presentc | Pos | Q | |
| Resistant | 18 | ≥ 32 | Ser83Leu; Asp87Asn | Ser80Ile | Pos | Q | |
|
| |||||||
| 9 | 1 Susc. | 0 | |||||
| Resistant | 2 | ≥ 32 | Ser83Leu,Asp87Asn | Ser80Ile | Neg | S | |
| 1 Susc. | 5 | ||||||
| Resistant | 9 | 0.75 | Ser83Leu | Neg | Neg | T | |
| 3 Susc. | 12-16 | ||||||
|
| |||||||
| 10 | 14 Susc. | 0-41 | |||||
| Resistant | 43 | 0.19 | Neg | Neg | Pos | U | |
| 1 Susc. | 46 | ||||||
| Resistant | 53 | ≥ 32 | Ser83Leu; Asp87Asn | Neg | Neg | V | |
| Resistant | 55 | ≥ 32 | Ser83Leu; Asp87Asn | Neg | Neg | V | |
| 2 Susc. | 60-63 | ||||||
|
| |||||||
| 11 | 1 Susc. | 0 | |||||
| Resistant | 4 | 8 | Ser83Leu; Asp87Asn | Ser80Ile | Neg | W | |
| 14 Susc. | 6-42 | ||||||
| Resistant | 46 | ≥ 32 | Ser83Leu; Asp87Asn | Ser80Ile; Glu84Val | Neg | X | |
|
| |||||||
| 12 | 2 Susc. | 0-3 | |||||
| Resistant | 6 | 0.38 | Neg | Neg | Neg | Y | |
| 4 Susc. | 8-17 | ||||||
| Resistant | 20 | 0.5 | Neg | Neg | Neg | Z | |
| Susc. | 22 | ||||||
| Resistant | 24 | ≥ 32 | Neg | Ser80Ile | Neg | N/A | |
|
| |||||||
| 13 | 2 Susc. | 0-2 | |||||
| Resistant | 5 | 0.25 | 1gyrA | AA | |||
| Resistant | 7 | 0.25 | 1gyrA | BB | |||
|
| |||||||
| 14 | 3 Susc. | 0-5 | |||||
| Resistant | 7 | 0.5 | Ser83Leu | Neg | Neg | CC | |
| 4 Susc. | 17-21 | ||||||
| Resistant | 26 | 0.38 | Ser83Leu; Asp87Asn | Neg | Pos | DD | |
| 1 | Susc. 28 | ||||||
Susc. = susceptible; MIC = minimum inhibitory concentration; OST = organic solvent tolerance; PFGE = pulsed field gel electrophoresis; Neg = negative; Pos = positive; N/A = not available.
Levofloxacin MIC by Etest.
Isolates with the same designated letter are considered to be clonally related.
A single parC mutation was present, but unable to be further characterized.
Characteristics of patients with >1 FQREC isolate during sampling are compared to those with only one FQREC isolate in Table 3. Notably, patients with persistent as opposed to transient colonization with FQREC during the sampling period were more likely to have received cefepime (50.0% and 17.0%, respectively; P=0.02) and vancomycin (50.0% and 20.3%, respectively; P=0.04) in the 30 days prior to sampling. These patients were also more likely to have had a urinary catheter (78.6% and 44.1%, respectively; P=0.04) and diarrhea (28.6% and 6.8%, respectively; P=0.04) present prior to the initial culture.
Table 3.
Characteristics of Hospitalized Patients with Transient vs Persistent Fluoroquinolone-Resistant E. coli (FQREC) Gastrointestinal Colonization
| Variable | Single FQREC isolate (n=59)a |
Multiple FQREC isolates (n=14)a |
P value |
|---|---|---|---|
| Age, mean years (SD) | 63.5 (18.5) | 61.5 (13.3) | 0.62 |
|
| |||
| Female sex | 31 (52.5) | 6 (42.9) | 0.56 |
|
| |||
| Non-white race | 26 (44.1) | 6 (42.9) | >0.99 |
|
| |||
| Duration of hospitalization prior to culture date, mean days (SD) |
9.0 (13.8) | 22 (33.5) | 0.01 |
|
| |||
| PPMC admission | 15 (25.4) | 2 (14.3) | 0.50 |
|
| |||
| Year of cultureb | 36 (61.0) | 5 (35.7) | 0.13 |
|
| |||
| Admitted from a nursing home | 5 (8.5) | 0 (0.0) | 0.58 |
|
| |||
| Transferred from another hospital | 12 (20.3) | 2 (14.3) | >0.99 |
|
| |||
| UPHS admit ≤30 days prior to culture date |
22 (37.3) | 5 (35.7) | >0.99 |
|
| |||
| Surgical service | 9 (15.3) | 3 (21.4) | 0.69 |
|
| |||
| Urinary catheter | 26 (44.1) | 11 (78.6) | 0.04 |
|
| |||
| Mechanical ventilation | 13 (22.0) | 5 (35.7) | 0.31 |
|
| |||
| Central venous catheter | 28 (50.0) | 11 (78.6) | 0.07 |
|
| |||
| Diarrhea present | 4 (6.8) | 4 (28.6) | 0.04 |
|
| |||
| Diabetes mellitus | 15 (25.4) | 1 (7.1) | 0.17 |
|
| |||
| Neutropenia | 0 (0.0) | 0 (0.0) | …. |
|
| |||
| Cirrhosis | 3 (5.1) | 1 (7.1) | >0.99 |
|
| |||
| HIV | 1 (1.7) | 1 (7.1) | 0.35 |
|
| |||
| Malignancy | 13 (22.0) | 5 (35.7) | 0.31 |
|
| |||
| Transplant | 4 (7.1) | 2 (15.4) | 0.32 |
|
| |||
| Renal insufficiency | 13 (22.0) | 0 (0.0) | 0.06 |
|
| |||
| Surgical procedure ≤30 days prior to culture date |
15 (25.4) | 6 (42.9) | 0.21 |
|
| |||
| Receipt of immunosuppression ≤30 days prior to culture date |
9 (15.3) | 1 (7.1) | 0.68 |
|
| |||
| Receipt of antimicrobial therapy ≤30 days prior to culture datec |
|||
|
| |||
| Levofloxacin | 11 (18.6) | 0 (0.0) | 0.11 |
|
| |||
| Gentamicin | 1 (1.7) | 2 (14.3) | 0.09 |
|
| |||
| Cefepime | 10 (17.0) | 7 (50.0) | 0.02 |
|
| |||
| Flagyl | 8 (13.6) | 5 (35.7) | 0.11 |
|
| |||
| Vancomycin | 12 (20.3) | 7 (50.0) | 0.04 |
|
| |||
| Piperacillin-tazobactam | 0 (0.0) | 1 (7.1) | 0.19 |
|
| |||
| Any antibiotic | 25 (42.4) | 10 (71.4) | 0.07 |
SD = standard deviation; PPMC = Penn Presbyterian Medical Center; UPHS = University of Pennsylvania Health System.
Data are presented as numbers (percentages), except where noted.
Reference year 2002.
Only antimicrobial agents with P<0.20 are shown.
4. Discussion
In this 3-year study, we found that 73 (16.2%) patients became newly colonized with FQREC during hospitalization. Of these 73 patients, 14 (19.2%) had >1 FQREC isolate on serial surveillance cultures. Notably, there was significant variability in regard to temporal changes in both resistance mechanisms, as well as levofloxacin MICs, among isolates from individual patients. There was little evidence for persistent colonization with the same FQREC clone among patients with >1 FQREC isolate on serial cultures.
The present study, to our knowledge, is the first to characterize longitudinal changes in resistance mechanisms in FQREC isolates from the same patient during hospitalization. Specifically, we found that the majority of patients who were newly colonized with FQREC had only one resistant isolate during serial surveillance performed during hospitalization. In addition, all of these patients had surveillance cultures that were positive for FQ-susceptible E. coli. It is possible that these 59 patients demonstrated resolution of FQREC colonization, or that they were predominantly colonized with FQ-susceptible strains which dominated on subsequent samplings such that the previous FQREC isolate could not be identified.
In contrast, 14 patients who developed new FQREC colonization had more than one resistant isolate during the sampling period. Compared to patients who had only one FQREC isolate during surveillance, these patients were more likely to have had a urinary catheter or diarrhea, as well as a longer duration of hospitalization, prior to the sampling date. All of these factors, notably instrumentation (e.g., indwelling catheters) may have increased the risk of developing new and persistent gastrointestinal and/or urinary tract colonization with FQREC. Furthermore, patients with more than one FQREC isolate were more likely to have received cefepime and/or vancomycin in the 30 days prior to sampling. It is likely that receipt of these antimicrobial agents reflected greater severity of illness overall in hospitalized patients with new FQREC colonization.
Interestingly, only 4 of these patients (subjects #2, 7, 10, and 12) had isolates that progressed longitudinally from no FQREC colonization to low-level FQREC and ultimately to high-level FQREC. The progression in FQ resistance evidenced in our study usually involved acquisition of an additional gyrA mutation and/or a parC mutation as opposed to changes in efflux pump overexpression. However, as the low-level and high-level resistant isolates in these 4 individual patients were not clonally related by PFGE, it is unlikely that the temporal increase in levofloxacin resistance was due to colonization with E. coli strains with high mutation rates (e.g., allowing for relatively rapid development of full FQ resistance).
Rather than de novo mutations accounting for the observed increase in the number of resistance mechanisms, it is likely that patients were colonized with more than one FQREC strain during hospitalization. Indeed, a previous study performed at the same institution (Lautenbach et al., 2006b) demonstrated that several subjects who were recently discharged from the hospital were colonized with more than one FQREC strain during the surveillance period. Along these lines, FQ-susceptible strains were detected between that of FQREC isolates for the majority of the 14 patients in the present study. Furthermore, some patients had more than one E. coli strain with high-level FQ resistance as determined by differences in resistance mechanisms and lack of clonality (i.e., subjects #4, #7, and #11). These findings suggest that patients may be colonized with multiple E. coli strains with varying levels of FQ resistance, and further studies are needed to identify determinants of subsequent infection with FQREC as opposed to FQ-susceptible strains in multiply-colonized patients.
Finally, 5 (35.7%) of the 14 patients had a clonally-related strain detected on more than one sample during serial surveillance (subjects #2, 5, #6, #8, #10) This may have reflected variability in which colonies were sampled during processing. However, the results suggest that duration of colonization and/or predominance of a particular FQREC clone may significantly vary in an individual patient, and future studies will need to evaluate potential risk factors, including specific resistance mechanisms, for persistence of a particular colonizing FQREC strain in hospitalized patients.
There are potential limitations of our study. Selection bias is a potential concern; however, although only ~45% of eligible subjects were enrolled, participants and nonparticipants were similar in regard to demographic characteristics. Sampling variability may have limited the detection of all colonizing FQREC isolates from a single patient. Finally, the present study was conducted in a single healthcare system, and these results may not be generalizable to other institutions.
In conclusion, the results of our study highlight the significant variability in resistance mechanisms in colonizing E. coli isolates among hospitalized patients. The emergence and persistence of FQ resistance is complex, and future studies are need to evaluate selection pressure for specific resistance mechanisms during hospitalization, as well as risk factors for infection with FQREC strains in multiply-colonized patients.
Acknowledgments
Funding. This work was supported by a Public Health Service grant of the Centers for Disease Control and Prevention (RS1/CCR320627-01) to E.L. This study was also supported, in part, by the National Institutes of Health [K24 AI080942 to E.L.], a Commonwealth Universal Research Enhancement Program grant from the Pennsylvania State Department of Health (to E.L.), and the Centers for Disease Control and Prevention Epicenters Program (U54-CK000163 to E.L.).
Role of the funding agency. The funding agencies had no role in the design and conduct of the study; collection, management, analysis, and interpretation of the data; or preparation, review, or approval of the manuscript.
Footnotes
Potential conflicts of interest. E.L. has received research grant support from AstraZeneca and Cubist. All other authors: no conflicts reported.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Barton TD, Fishman NO, Weiner MG, LaRosa LA, Lautenbach E. High rate of coadministration of di- or tri-valent cation-containing compounds with oral fluoroquinolones: risk factors and potential implications. Infect Control Hosp Epidemiol. 2005;26:93–9. doi: 10.1086/502493. [DOI] [PubMed] [Google Scholar]
- Chang TM, Lu PL, Li HH, Chang CY, Chen TC, Chang LL. Characterization of fluoroquinolone resistance mechanisms and their correlation with the degree of resistance to clinically used fluoroquinolones among Escherichia coli isolates. J Chemother. 2007;19:488–94. doi: 10.1179/joc.2007.19.5.488. [DOI] [PubMed] [Google Scholar]
- Donskey CJ. Antibiotic regimens and intestinal colonization with antibiotic-resistant gram-negative bacilli. Clin Infect Dis. 2006;43(Suppl 2):62–9. doi: 10.1086/504481. [DOI] [PubMed] [Google Scholar]
- Gales AC, Gordon KA, Wilke WW, Pfaller MA, Jones RN. Occurrence of single-point gyrA mutations among ciprofloxacin-susceptible Escherichia coli isolates causing urinary tract infections in Latin America. Diagn Microbiol Infect Dis. 2000;36:61–4. doi: 10.1016/s0732-8893(99)00121-2. [DOI] [PubMed] [Google Scholar]
- Goering RV, Tenover FC. Epidemiological interpretation of chromosomal macro-restriction fragment patterns analyzed by pulsed-field gel electrophoresis. J Clin Microbiol. 1997;35:2432–3. doi: 10.1128/jcm.35.9.2432-2433.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacoby GA. Mechanisms of resistance to quinolones. Clin Infect Dis. 2005;41(Suppl 2):S120–6. doi: 10.1086/428052. [DOI] [PubMed] [Google Scholar]
- Kern WV, Oethinger M, Jellen-Ritter AS, Levy SB. Non-target gene mutations in the development of fluoroquinolone resistance in Escherichia coli. Antimicrob Agents Chemother. 2000;44:814–20. doi: 10.1128/aac.44.4.814-820.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komp Lindgren P, Karlsson A, Hughes D. Mutation rate and evolution of fluoroquinolone resistance in Escherichia coli isolates from patients with urinary tract infections. Antimicrob Agents Chemother. 2003;47:3222–32. doi: 10.1128/AAC.47.10.3222-3232.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenbach E, Strom BL, Nachamkin I, et al. Longitudinal trends in fluoroquinolone resistance among Enterobacteriaceae isolates from inpatients and outpatients, 1989-2000: differences in the emergence and epidemiology of resistance across organisms. Clin Infect Dis. 2004;38:655–62. doi: 10.1086/381549. [DOI] [PubMed] [Google Scholar]
- Lautenbach E, Metlay JP, Bilker WB, Edelstein PH, Fishman NO. Association between fluoroquinolone resistance and mortality in Escherichia coli and Klebsiella pneumoniae infections: the role of inadequate empirical antimicrobial therapy. Clin Infect Dis. 2005;41:923–9. doi: 10.1086/432940. [DOI] [PubMed] [Google Scholar]
- Lautenbach E, Fishman NO, Metlay JP, et al. Phenotypic and genotypic characterization of fecal Escherichia coli isolates with decreased susceptibility to fluoroquinolones: results from a large hospital-based surveillance initiative. J Infect Dis. 2006a;194:79–85. doi: 10.1086/503046. [DOI] [PubMed] [Google Scholar]
- Lautenbach E, Tolomeo P, Mao X, et al. Duration of outpatient fecal colonization due to Escherichia coli Isolates with decreased susceptibility to fluoroquinolones: longitudinal study of patients recently discharged from the hospital. Antimicrob Agents Chemother. 2006b;50:3939–43. doi: 10.1128/AAC.00503-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lautenbach E, Metlay JP, Weiner MG, et al. Gastrointestinal tract colonization with fluoroquinolone-resistant Escherichia coli in hospitalized patients: changes over time in risk factors for resistance. Infect Control Hosp Epidemiol. 2009;30:18–24. doi: 10.1086/592703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee I, et al. Risk factors for fluconazole-resistant Candida glabrata bloodstream infections. Arch Intern Med. 2009;169:379–83. doi: 10.1001/archinte.169.4.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li XZ, Nikaido H. Efflux-mediated drug resistance in bacteria: an update. Drugs. 2009;69:1555–623. doi: 10.2165/11317030-000000000-00000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moon DC, Seol SY, Gurung M, et al. Emergence of a new mutation and its accumulation in the topoisomerase IV gene confers high levels of resistance to fluoroquinolones in Escherichia coli isolates. Int J Antimicrob Agents. 2010;35:76–9. doi: 10.1016/j.ijantimicag.2009.08.003. [DOI] [PubMed] [Google Scholar]
- Morgan-Linnell SK, Becnel Boyd L, Steffen D, Zechiedrich L. Mechanisms accounting for fluoroquinolone resistance in Escherichia coli clinical isolates. Antimicrob Agents Chemother. 2009;53:235–41. doi: 10.1128/AAC.00665-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Richard P, Delangle MH, Raffi F, Espaze E, Richet H. Impact of fluoroquinolone administration on the emergence of fluoroquinolone-resistant gram-negative bacilli from gastrointestinal flora. Clin Infect Dis. 2001;32:162–6. doi: 10.1086/317551. [DOI] [PubMed] [Google Scholar]
- Singh R, Swick MC, Ledesma KR, et al. Temporal interplay between efflux pumps and target mutations in development of antibiotic resistance in Escherichia coli. Antimicrob Agents Chemother. 2012;56:1680–5. doi: 10.1128/AAC.05693-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang H, Dzink-Fox JL, Chen M, Levy SB. Genetic characterization of highly fluoroquinolone-resistant clinical Escherichia coli strains from China: role of acrR mutations. Antimicrob Agents Chemother. 2001;45:1515–21. doi: 10.1128/AAC.45.5.1515-1521.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White DG, Goldman JD, Demple B, Levy SB. Role of the acrAB locus in organic solvent tolerance mediated by expression of marA, soxS, or robA in Escherichia coli. J Bacteriol. 1997;179:6122–6. doi: 10.1128/jb.179.19.6122-6126.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]