Abstract
Background
Heavy alcohol consumption increases risk of developing squamous cell carcinoma of the head and neck (SCCHN). Alcohol metabolism to cytotoxic and mutagenic intermediates acetaldehyde and reactive oxygen species is critical for alcohol-drinking-associated carcinogenesis. We hypothesized that polymorphisms in alcohol metabolism-related and antioxidant genes influence SCCHN survival.
Methods
Interview and genotyping data (64 polymorphisms in 12 genes) were obtained from 1227 white and African-American cases from the Carolina Head and Neck Cancer Epidemiology study, a population-based case–control study of SCCHN conducted in North Carolina from 2002 to 2006. Vital status, date and cause of death through 2009 were obtained from the National Death Index. Kaplan–Meier log-rank tests and adjusted hazard ratios were calculated to identify alleles associated with survival.
Results
Most tested SNPs were not associated with survival, with the exception of the minor alleles of rs3813865 and rs8192772 in CYP2E1. These were associated with poorer cancer-specific survival (HRrs3813865, 95%CI = 2.00, 1.33–3.01; HRrs8192772, 95%CI = 1.62, 1.17–2.23). Hazard ratios for 8 additional SNPs in CYP2E1, GPx2, SOD1, and SOD2, though not statistically significant, were suggestive of differences in allele hazards for all-cause and/or cancer death. No consistent associations with survival were found for SNPs in ADH1B, ADH1C, ADH4, ADH7, ALDH2, GPx2, GPx4, and CAT.
Conclusions
We identified some polymorphisms in alcohol and oxidative stress metabolism genes that influence survival in subjects with SCCHN. Previously unreported associations of SNPs in CYP2E1 warrant further investigation.
Keywords: Head and neck neoplasms/epidemiology, Genes, Survival, Alcohol drinking/metabolism, Oxidative stress
1. Introduction
Head and neck cancers typically include cancers of the oral cavity, pharynx, and larynx. There were estimated to be 53,640 new cases and 11,520 deaths from oral cavity, pharyngeal, and laryngeal cancers in the U.S. in 2013 [1]. Globally in 2008, oral cavity tumors were among the top 10 incident cancers in men world-wide, and among the top 10 fatal cancers in men in developing countries [2]. Five-year relative survival for laryngeal, oral cavity, and pharyngeal cancer patients averages about 80% for localized cases, 50% for regional cases, and 33% for metastatic cases, with somewhat lower survival for laryngeal compared to oral cavity and pharyngeal cancers [3]. Age-adjusted mortality rates for African-American men with oral cavity and pharynx cancers are double those of men of other races, and disparities are especially pronounced for men with laryngeal cancer (4.7 deaths per 100,000 population in African-Americans compared to 2.1 in whites, 1.9 in Native Americans and Hispanics, and 0.7 in Asian/Pacific Islanders) [4].
It is estimated that 75% of new cases of SCCHN in the United States are caused by tobacco use, especially cigarette smoking, and/or drinking of alcoholic beverages [5]. In the past few decades the proportional incidence of oropharyngeal tumors associated with carcinogenic human papillomavirus sub-types has risen; such tumors appear to have better prognosis than non-HPV oropharyngeal tumors [6]. Multiple studies have reported associations between SCCHN incidence and polymorphisms in alcohol metabolism genes, especially ADH1B, ADH1C, ADH4, ADH7, ALDH2, and CYP2E1 [7–16]. The primary biological mechanism responsible for this effect is hypothesized to be high levels of cytotoxic and mutagenic acetaldehyde, the metabolic intermediate between ethanol and acetate. Acetaldehyde, when associated with consumption of alcoholic beverages, is classified as a known human carcinogen by the International Agency for Research on Cancer [17]. Also, alcohol metabolism through CYP2E1, probably mediated by alcohol-induced stabilization of the CYP2E1 protein [18], is known to result in production of increased levels of reactive oxygen species (ROS) [19]. DNA damage by acetaldehyde or ROS has been suggested as one of the key mechanisms of alcohol drinking-associated carcinogenesis, and variation in associated pathway genes may modify progression and survival. Therefore, it is also of interest to know whether polymorphisms in genes encoding enzymes protective against oxidative stress (SOD, GPx, CAT) are associated with altered survival in SCCHN patients.
In addition to its role in alcohol metabolism, CYP2E1, as a member of the cytochrome P450 oxidative system, is involved in metabolism of xenobiotics and drugs [20], including the platinum-containing chemotherapeutic agents used to treat many head and neck cancers. Although CYP2E1 enzymatically inactivates some substrates, it has also been shown to bioactivate many compounds that are possibly carcinogenic; for example, the tobacco carcinogen N-nitrosamines, 4-(methylnitrosamino)-1-(3-pyridyl)-1-buta-none [21].
While many studies have examined genetic associations with SCCHN incidence, there have been few reports of the effect of gene polymorphisms in alcohol metabolism and oxidative stress-related genes on survival of subjects with SCCHN. For example, only two studies investigated whether selected ADH1C and CYP2E1 polymorphisms influence prognosis, and they reported evidence of an association with advanced clinical stages or higher recurrence [11,22]. Further, no studies have examined the influence on post-diagnosis survival of genetic variation in the oxidative stress pathways.
We examined the effect on survival of 64 SNPs in ADH1B, ADH1C, ADH4, ADH7, ALDH2, and CYP2E1 alcohol metabolism genes, and in CAT, GPx1, GPx2, GPx4, SOD1, and SOD2 antioxidant genes, using exposure, genetic, clinical, and outcome data from cases included in a large North Carolina (NC) population-based case–control study of SCCHN that included both white and African-American subjects.
2. Materials and methods
2.1. Study population
Cases for this analysis were obtained from the Carolina Head and Neck Cancer Epidemiology study (CHANCE), a population-based case–control study [23,24].
All cases of squamous cell carcinoma of the oral cavity, pharynx, and larynx diagnosed in 46 NC counties between January 1, 2002 and February 28, 2006 were eligible for enrollment. Rapid identification of cases was conducted by the NC Central Cancer Registry [24]. CHANCE cases included ICD-O-3 topography codes C0.00–C14.8, and C32.0–C32.9, excluding salivary gland tumors (C07.9, C08.0–C08.9), nasopharynx (C11.0–C11.9), nasal cavity (C30.0), and nasal sinuses (C31.0–C31.9). ICD-O-3 morphology codes included were 8010/3, 8051/3, 8083/3, 8071/3, 8072/3, 8073/3, 8074/3, and 8076/3. Benign tumors, carcinomas in situ, papillary carcinomas, and adenoid carcinomas were excluded. This analysis further excluded 21 lip cancers (C00.3–C00.9, C14.2), 26 cases of “other” race, and 115 without genotyping data, producing a study analysis group of 1227 cases, of which 922 were white/European-American and 305 were black/African-American.
Case tumors were classified into anatomic sub-site according to the following 5 ICD-O categories that are also used by the International Head and Neck Cancer Epidemiology (INHANCE) Consortium [25]: (1) oral cavity: C02.0–C02.3, C03.0, C03.1, C03.9, C04.0, C04.1, C04.8, C04.9, C05.0, C06.0–C06.2, C06.8, and C06.9; (2) oropharynx: C01.9, C02.4, C05.1, C05.2, C09.0, C09.1, C09.8, C09.9, C10.0–C10.4, C10.8, and C10.9; (3) oral cavity–oropharynx–hypopharynx NOS: C02.8, C02.9, C05.8, C05.9, C14.0, C14.2, and C14.8; (4) hypopharynx: C12.9, C13.0–C13.2, C13.8, and C13.9; and (5) larynx: C32.0–C32.3, and C32.8–C32.9.
Written informed consent was obtained from all subjects. The study was approved by the Biomedical Institutional Review Board at the University of North Carolina at Chapel Hill.
2.2. Outcome assessment
We determined whether death had occurred in study participants by December 31, 2009, and, if so, the date and cause, through linkage with the United States National Death Index (NDI). The NDI is a national file of identified death record information compiled from computer files submitted by State vital statistics offices [26]; of all US sources of national death data, NDI has demonstrated the highest sensitivity and is the only national source with a coded cause of death field suitable for research purposes [27]. CHANCE collected multiple NDI matching data: social security number (SSN), date of birth (DOB), sex, race, state of residence, and name. Therefore there was a high proportion (>75%) of perfect/very close to perfect matches on SSN, DOB, and sex. The remaining near matches were further confirmed by examining the United States Social Security Death Index, which is created from Social Security Administration (SSA) records of persons with social security numbers whose deaths were reported to the SSA, usually by mortuaries, attorneys, or family members. A very small number were further confirmed by obituary search of newspaper websites. Partial matches on name, but having in common only a few SSN digits and parts of the DOB, that could not be confirmed by SSDI or obituary were excluded as non-matches. If the initial three digits of the first-listed cause of death code were C01–C06, C09, C10, C12–C14, or C32, the cause of death was classified as head and neck cancer.
2.3. SNP selection and genotyping
Blood samples were obtained at the time of questionnaire administration by nurse-interviewers trained in phlebotomy. If the subject was not willing or able to consent to the blood draw, they were asked to contribute a buccal cell sample via mouthrinse.
For this analysis, 75 SNPs (69 tag SNPs and 6 candidate SNPs found in prior studies to be associated with aerodigestive cancer incidence, breast cancer survival, alcohol dependence, or interaction with genes in the proposed carcinogenic pathway for vinyl chloride) were selected in 12 genes that are part of two metabolic pathways: ADH1B, ADH1C, ADH4, ADH7, ALDH2, and CYP2E1 in the alcohol metabolism pathway in the upper aerodigestive tract; and CAT, SOD1, SOD2, GPx1, GPx2, and GPx4 in the oxidative stress metabolic pathway. Tag SNPs, chosen to represent the genetic variation within each of the 12 candidate genes (gene and 2000 bp upstream and downstream) were selected using the Genome Variation Server [28], using SNPs that were polymorphic in either CEU or YRI HapMap Release 2 (unrelated only), and with the following parameters: allele frequency cutoff 10%, 0.8 R2 threshold minimum for variations to belong to the same cluster, 85% minimum data coverage for tag SNPs, 70% minimal data coverage for a variation to be potentially clustered with others.
To control for potential population stratification, we selected 157 Ancestry Informative Markers (AIMs) to maximize (1) the difference in allele frequencies (delta) between European and African populations in the HapMap data (CEU versus YRI), and (2) the Fisher’s information criterion (FIC). AIMs were prioritized based on having the highest delta and FIC values in the following order: 90% European/10% African, 10% European/90% African, and 50% European/50% African. This allowed AIMs to represent the entire expected ancestral distribution of the study population. Individual estimates of percentage African ancestry were calculated from 145 successfully genotyped AIMs using maximum likelihood estimation (MLE) methods previously described [29–31]. AIMs were chosen to differentiate only between African and European ancestry, so we derived individual ancestry estimates for the two groups that summed to 1.0.
Genotyping was done by the Mammalian Genotyping Core Facility, University of North Carolina at Chapel Hill, using the Illumina GoldenGate genotyping assay with Sentrix Array matrix and 96-well standard microtiter plates. Technicians were blinded to case/control status of samples, and samples from both cases and controls were tested on each plate, along with DNA controls and random sample duplicates. Assay intensity data and genotype cluster images for all SNPs were individually reviewed to identify those with inadequate signal intensity or indistinguishable genotype clusters. Blind duplicates of 109 samples were genotyped to verify reliability of genotype calls. Tests of Hardy–Weinberg equilibrium (HWE) were conducted in controls stratified by race, and those SNPs for which the p-value was <0.001 were judged to be out of HWE.
DNA samples from 1266 CHANCE SCCHN cases of white or African-American race were genotyped. Of these, 38 failed to genotype, and 1 was gender discrepant, leaving 1227 cases with genotyping data. Of the 75 tag and candidate SNPs for which genotyping was attempted, 8 tag SNPs and 1 candidate SNP were excluded from analyses due to inadequate signal intensity or inability to distinguish genotype clusters, and 2 tag SNPs were excluded due to genotypes being out of HWE at the p < 0.001 level, leaving 64 tag and candidate SNPs available for analyses. Genotype calls for blind duplicates were concordant for all SNPs. All 1227 cases had less than 4% missing SNP results.
2.4. Covariate measurement
Trained nurse-interviewers conducted an in-person interview with each subject. For this analysis only self-reported, non-proxy data were included. Questions were asked about demographics (age, sex, race-ethnicity, education, income, health insurance), cigarette smoking, drinking of alcoholic beverages, diet, oral health, medical history, family history of cancer, and textual information on co-morbid conditions.
We made an initial selection of covariates based on a priori knowledge regarding their potential association with survival. Covariates obtained from the interview included age, sex, race; duration of cigarette smoking; ever-use of non-cigarette tobacco products, ever exposure to environmental tobacco smoke (ETS) at work, ever exposure to ETS at home; whether the subject was covered by health insurance on diagnosis date, had a routine dental visit in the past 10 years, ever had a loose permanent tooth due to disease, ever used mouthwash; family history of SCCHN; household poverty as defined by federal guidelines for both income and number of persons supported, highest attained education level; and presence of selected co-morbid diseases: liver disease including hepatitis, cardiovascular and/or lung disease, renal disease and/or diabetes, and other cancers. Alcohol consumption, although measured, was not included in analyses because it may be an intermediate in the causal pathway between some of the selected SNPs and survival.
Clinical information about the tumor and its treatment were abstracted from the subjects’ medical record. Pathology reports were reviewed by the study pathologist (WF) and a head and neck cancer surgeon (MW). Tumor and treatment characteristics obtained included TNM and summary stages, angiolymphatic invasion status, and cancer treatment (surgery, radiation, and/or chemotherapy). These clinical-pathologic factors were chosen a priori as potential confounders.
2.5. Statistical analysis
For all-cause death, months of survival were calculated by subtracting the date of diagnosis from the date of death, and subjects who were still alive on 12/31/2009 were censored on that date. For head-and-neck-cancer-specific death (hereafter referred to as cancer death or cancer-specific death), months of survival were calculated by subtracting the date of diagnosis from the date of cancer death, while subjects who died of another cause were censored as of the date of death; subjects who were still alive on 12/31/2009 were censored on that date. Kaplan–Meier survival plots, and hazard ratios for the independent effects of SNPs from Cox proportional hazards regression models, were implemented in SAS® 9.2.
Kaplan–Meier plots and log-rank tests for all-cause and cancer-specific survival were constructed for the two alleles of each SNP. Kaplan–Meier plots and log-rank tests were also constructed for all-cause and cancer specific survival for the two races, for all head-and-neck cancers combined as well as separately for each anatomic sub-site.
A dominant genetic model (at least one minor allele versus referent of no minor alleles) was implemented because, for many of the SNPs, the number of subjects homozygous for the minor allele was too small to permit estimation of precise effect measure estimates.
Potential covariates were selected using a step-wise backwards elimination from a full model (which contained demographic, socioeconomic status (SES), tumor, clinical, co-morbid diseases, oral health, and tobacco exposure variables, as listed in Table 2) and comparison of each reduced model to the full model. There was no collinearity between any of the explanatory variables except possibly between race and ancestry. If eliminating a set of covariates did not change the ln(HR) for any SNP by a difference of at least 0.10, those covariates were eliminated from subsequent models. The final Cox models for all-cause and cancer-specific death included the demographic, SES, tumor, clinical, co-morbid diseases, oral health, and tobacco exposure variables listed in Table 3 footnotes. With self-reported race included in the model the ancestry variable was not important to all-cause or cancer-specific death for the genetic variants studied, so for parsimony, and because ancestry and race showed evidence of collinearity, ancestry was not included in final models.
Table 2.
Descriptives and univariate hazard ratios for non-SNP variables – one explanatory variable per model (complete case analysis).
| Death from all causes (n = 545) |
Death from head and neck cancer (n =227) |
|||||||
|---|---|---|---|---|---|---|---|---|
| Potential covariate (type) | Covariate values | # (%) of total cases |
# Died/ # survived |
HR (95% CI)a | Unadjusted p-value (bold if <0.05) |
# Died/ # survived |
HR (95% CI)a | Unadjusted p-value (bold if <0.05) |
| Demographics | ||||||||
| Age category (ordinal) | 20–49 years | 239 (19.5%) | 87/152 | 1.10 (1.05–1.15) | 2.1E-05 | 42/197 | 1.08 (1.00–1.15) | 0.043 |
| 50–54 years | 189 (15.4%) | 79/110 | 30/159 | |||||
| 55–59 years | 207 (16.9%) | 82/125 | 35/172 | |||||
| 60–64 years | 205 (16.7%) | 87/118 | 37/168 | |||||
| 65–69 years | 168 (l3.7%) | 90/78 | 33/135 | |||||
| 70–74 years | 135 (11.0%) | 71/64 | 27/108 | |||||
| 75–80 years | 84 (6.8%) | 49/35 | 23/61 | |||||
| Sex (categorical) | Female | 289 (23.6%) | 126/163 | 1.00 (ref) | 58/231 | 1.00 (ref) | ||
| Male | 938 (76.4%) | 419/519 | 1.04 (0.85–1.27) | 0.678 | 169/769 | 0.91 (0.68–1.23) | 0.551 | |
| Race (categorical) | White/European-American | 922 (75.1%) | 383/539 | 1.00 (ref) | 169/753 | 1.00 (ref) | ||
| Black/African-American | 305 (24.9%) | 162/143 | 1.42 (1.18–1.71) | 1.8E-04 | 58/247 | 1.14 (0.84–1.53) | 0.403 | |
| Proportion African ancestry (continuous) (mean % in those who died) | 1227 | 0.27/0.21 | 1.64 (1.28–2.10) | 1.0E-04 | 0.24/0.24 | 1.19 (0.79–1.79) | 0.401 | |
| Tumor/treatment characteristics | ||||||||
| Anatomic subsite (categorical) | Oral cavity (referent) | 172 (14.0%) | 91/81 | 1.00 (ref) | 38/134 | 1.00 (ref) | ||
| Hypopharynx | 55 (4.5%) | 39/16 | 1.66 (1.14–2.41) | 0.009 | 15/40 | 1.46 (0.80–2.65) | 0.215 | |
| Larynx | 443 (36.1%) | 197/246 | 0.76 (0.59–0.97) | 0.027 | 78/365 | 0.72 (0.49–1.07) | 0.103 | |
| Oral cavity–oropharynx– | 224 (18.3%) | 90/134 | 0.73 (0.54–0.97) | 0.033 | 42/182 | 0.83 (0.53–1.28) | 0.393 | |
| hypopharynx NOS | ||||||||
| Oropharynx | 333 (27.1%) | 128/205 | 0.65 (0.50–0.86) | 0.002 | 54/279 | 0.67 (0.44–1.02) | 0.062 | |
| T stage (ordinal) | T1, T1a, T1b | 392 (31.9%) | 125/267 | 1.38 (1.28–1.49) | 1.9E-16 | 34/358 | 1.62 (1.43-1.82) | 2.2E-15 |
| T2 | 409 (33.3%) | 177/232 | 74/335 | |||||
| T3 | 218 (17.8%) | 119/99 | 55/163 | |||||
| T4, T4a, T4b | 208 (17.0%) | 124/84 | 64/144 | |||||
| N stage (ordinal) | N0 | 657 (53.5%) | 261/396 | 1.26 (1.16–1.37) | 5.2E-08 | 90/567 | 1.47 (1.30–1.68) | 2.6E-09 |
| N1 | 156 (12.7%) | 67/89 | 33/123 | |||||
| N2, N2a, N2b, N2c | 366 (29.8%) | 184/182 | 83/283 | |||||
| N3 | 48 (3.9%) | 33/15 | 21/27 | |||||
| M stage (categorical) | M0 | 1219 (99.3%) | 538/681 | 1.00 (ref) | 222/997 | 1.00 (ref) | ||
| M1 | 8 (0.7%) | 7/1 | 6.95 (3.28–14.70) | 4.0E-07 | 5/3 | 10.19 (4.18–24.88) | 3.4E-07 | |
| Margin status (categorical) | Negative | 396 (32.3%) | 170/226 | 1.00 (ref) | 65/331 | 1.00 (ref) | ||
| Positive | 691 (56.3%) | 317/374 | 1.12 (0.93–1.35) | 0.233 | 137/554 | 1.25 (0.93–1.68) | 0.141 | |
| Missing | 140 (11.4%) | 58/82 | 25/115 | |||||
| Angiolymphatic invasion (Categorical) | Absent | 954 (77.8%) | 412/542 | 1.00 (ref) | 171/783 | 1.00 (ref) | ||
| Present | 107 (8.7%) | 58/49 | 1.48 (1.12–1.94) | 0.006 | 26/81 | 1.54 (1.02–2.33) | 0.039 | |
| Missing | 166 (13.5%) | 75/91 | 30/136 | |||||
| Surgical treatment (categorical) | No | 537 (43.8%) | 268/269 | 1.00 (ref) | 119/418 | 1.00 (ref) | ||
| Yes | 690 (56.2%) | 277/413 | 0.72 (0.61–0.85) | 1.2E-04 | 108/582 | 0.64 (0.50–0.84) | 0.001 | |
| Radiation treatment (categorical) | No | 282 (23.0%) | 105/177 | 1.00 (ref) | 37/245 | 1.00 (ref) | ||
| Yes | 945 (77.0%) | 440/505 | 1.36 (1.10–1.68) | 0.005 | 190/755 | 1.63 (1.15–2.32) | 0.007 | |
| Chemotherapy treatment (categorical) | No | 754 (61.5%) | 326/428 | 1.00 (ref) | 130/624 | 1.00 (ref) | ||
| Yesb | 473 (38.5%) | 219/254 | 1.15 (0.97–1.36) | 0.111 | 97/376 | 1.26 (0.97–1.63) | 0.090 | |
| Comorbid diseases | ||||||||
| Comorbid liver disease and/or hepatitis (categorical) | No | 1113 (90.7%) | 476/637 | 1.00 (ref) | 198/915 | 1.00 (ref) | ||
| Yes | 105 (8.6%) | 63/42 | 1.62 (1.24–2.10) | 3.4E-04 | 25/80 | 1.47 (0.97–2.24) | 0.067 | |
| Missing | 9 (0.7%) | 6/3 | 4/5 | |||||
| Comorbid cardiovascular and/or lung disease (categorical) | No | 887 (72.3%) | 357/530 | 1.00 (ref) | 153/734 | 1.00 (ref) | ||
| Yes | 336 (27.4%) | 186/150 | 1.47 (1.24–1.76) | 1.8E-05 | 72/264 | 1.31 (0.99–1.73) | 0.061 | |
| Missing | 4 (0.3%) | 2/2 | 2/2 | |||||
| Comorbid renal disease and/or diabetes (categorical) | No | 1053 (85.8%) | 451/602 | 1.00 (ref) | 196/857 | 1.00 (ref) | ||
| Yes | 167 (13.6%) | 88/79 | 1.32 (1.05–1.66) | 0.018 | 28/139 | 0.97 (0.65–1.44) | 0.873 | |
| Missing | 7 (0.6%) | 6/1 | 3/4 | |||||
| Comorbid disease: other cancer (categorical) | No | 1019 (83.0%) | 446/573 | 1.00 (ref) | 186/833 | 1.00 (ref) | ||
| Yes | 204 (16.6%) | 96/108 | 1.08 (0.87–1.35) | 0.477 | 38/166 | 1.02 (0.72–1.44) | 0.932 | |
| Missing | 4 (0.3%) | 3/1 | 3/1 | |||||
| # Comorbid conditions (continuous) (mean # in those who died) | 1227 | 1.03/0.72 | 1.24 (1.14–1.34) | 1.2E-07 | 0.95/0.84 | 1.13 (1.00–1.29) | 0.051 | |
| Other potential confounders | ||||||||
| Smoking duration, rounded to nearest whole year (continuous) | 1227 | 36.9/27.9 | 1.02 (1.02–1.03) | 3.5E-16 | 36.4/30.9 | 1.02 (1.01–1.03) | 4.2E-06 | |
| (mean # years in those who died) | ||||||||
| Alcoholic beverages, lifetime consumption of ethanol in ml, | Never-drinkers | 117 (9.5%) | 38/79 | 1.00 (ref) | 20/97 | 1.00 (ref) | ||
| never-users and tertiles of drinking | >0–133,294 | 210 (17.1%) | 61/149 | 0.84 (0.56–1.27) | 0.413 | 28/182 | 0.75 (0.42–1.33) | 0.324 |
| 133,294–757,550 | 318 (25.9%) | 129/189 | 1.24 (0.86–1.78) | 0.251 | 50/268 | 0.90 (0.53–1.51) | 0.683 | |
| 757,550+ | 505 (41.2%) | 277/228 | 1.94 (1.38–2.73) | 1.0E-04 | 113/392 | 1.45 (0.90–2.33) | 0.126 | |
| Missing | 77 (6.3%) | 40/37 | 16/61 | |||||
| Federal poverty guideline (categorical) | At or above | 816 (66.5%) | 307/509 | 1.00 (ref) | 135/681 | 1.00 (ref) | ||
| Below | 356 (29.0%) | 208/148 | 1.87 (1.56–2.23) | 4.1E-12 | 78/278 | 1.53 (1.16–2.03) | 0.003 | |
| Missing | 55 (4.5%) | 30/25 | 14/41 | |||||
| Routine dental visit in last 10 years (categorical) | Yes | 781 (63.7%) | 278/503 | 1.00 (ref) | 127/664 | 1.00 (ref) | ||
| No | 438 (35.7%) | 262/176 | 2.01 (1.70–2.38) | 5.1E-16 | 97/341 | 1.59 (1.22–2.07) | 0.001 | |
| Missing | 8 (0.7%) | 5/3 | 3/5 | |||||
| Environmental tobacco smoke exposure at home (categorical) |
No | 399 (32.5%) | 148/251 | 1.00 (ref) | 62/337 | 1.00 (ref) | ||
| Yes | 827 (67.4%) | 396/431 | 1.36 (1.12–1.64) | 0.001 | 164/663 | 1.33 (0.99–1.78) | 0.056 | |
| Missing | 1 (0.1%) | 1/0 | 1/0 | |||||
| Environmental tobacco smoke | No | 316 (25.8%) | 153/163 | 1.00 (ref) | 64/252 | 1.00 (ref) | ||
| exposure at work (categorical) | Yes | 909 (74.1%) | 391/518 | 0.85 (0.70–1.02) | 0.078 | 162/747 | 0.84 (0.63–1.13) | 0.246 |
| Missing | 2 (0.2%) | 1/1 | 1/1 | |||||
| Ever used non-cigarette tobacco product (categorical) | No | 754 (61.5%) | 329/425 | 1.00 (ref) | 136/618 | 1.00 (ref) | ||
| Yes | 473 (38.5%) | 216/257 | 1.07 (0.90–1.26) | 0.469 | 91/382 | 1.09 (0.83–1.42) | 0.545 | |
| Had health insurance on reference date (categorical) | Yes | 1068 (87.0%) | 459/609 | 1.00 (ref) | 188/880 | 1.00 (ref) | ||
| No | 154 (12.6%) | 82/72 | 1.43 (1.13–1.81) | 0.003 | 37/117 | 1.54 (1.08–2.19) | 0.016 | |
| Missing | 5 (0.4%) | 4/1 | 2/3 | |||||
| Highest education level attained (categorical) | High school or more | 828 (67.5%) | 325/503 | 1.00 (ref) | 140/688 | 1.00 (ref) | ||
| Less than high school | 399 (32.5%) | 220/179 | 1.58 1.33–1.87) | 1.7E-07 | 87/312 | 1.42 (1.09–1.86) | 0.010 | |
| Ever had loose permanent tooth due to disease (categorical) | No | 765 (62.3%) | 308/457 | 1.00 (ref) | 146/619 | 1.00 (ref) | ||
| Yes | 455 (37.1%) | 232/223 | 1.35(1.14–1.60) | 0.001 | 78/377 | 0.94 (0.71-1.23) | 0.639 | |
| Missing | 7 (0.6%) | 5/2 | 3/4 | |||||
| Ever regularly used mouthwash (categorical) | No | 502 (40.9%) | 216/286 | 1.00 (ref) | 88/414 | 1.00 (ref) | ||
| Yes | 719 (58.6%) | 324/395 | 1.07 (0.90–1.27) | 0.469 | 137/582 | 1.11 (0.85–1.45) | 0.462 | |
| Missing | 6 (0.5%) | 5/1 | 2/4 | |||||
| Family history of H and N cancer in 1st degree relatives (categorical) | No | 1206 (98.3%) | 535/671 | 1.00 (ref) | 222/984 | 1.00 (ref) | ||
| Yes | 21 (1.7%) | 10/11 | 1.11 (0.59–2.08) | 0.742 | 5/16 | 1.31 (0.54–3.18) | 0.551 | |
For categorical variables, HRs are in comparison to referent category. For continuous and ordinal variables, HRs are per unit of measurement and are cumulative (e.g. HR per year of age, HR per each additional comorbid disease, HR per each higher stage category).
Of 473 cases who received chemotherapy, 464 (98%) received at least one platinum-containing agent.
Table 3.
SNP hazard ratios, dominant genetic model (adjusted p-values < 0.05 are highlighted).
| DEATH FROM ALL CAUSES |
DEATH FROM HEAD AND NECK CANCER |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Model:SNP unadjusted Complete case analysis n=1227 for mostc SNPs |
Model: SNP & covariatesa Complete case analysis n=1050for mostc SNPs |
Model: SNP & covariatesa summary from 25 imputations n=1227 for all SNPs |
Model: SNP unadjusted Complete case analysis n=1227 for mostc SNPs |
Model: SNP & covariatesb Complete case analysis n=1050 for mostc SNPs |
Model: SNP & covariatesb summ ary f rom 25 impu tation s n=1227 for all SNPs |
||||||||||
| Gene | SNP name | major/ minor allele |
allele frequency (any minor/only major/missing) |
HRd (95% CI) | Bonferroni- corrected p- value (64 tests) (highlighted if <0 05) |
HRd (95% CI) | Bonferroni- correctedp- value (64 tests) (highlighted if <0 05) |
HRd (95% CI) | Bonferroni- corrected p- value (64 tests) (highlighted if <0 05) |
HRd (95% CI) | Bonferroni- corrected p- value (64 tests) (highlighted if <0 05) |
HRd (95% CI) | Bonferroni- corrected p- value (64 tests) (highlighted if <0 05) |
HRd (95% CI) | Bonferroni- corrected p- value (64 tests) (highlighted if <0 05) |
|
|
|
|
|
|
|
|
|||||||||
|
ALCOHOL METABOLISM GENES
|
|||||||||||||||
| ADH1B | rs12507573 | C/A | 855/372/0 | 0.95 (0.80-1.14) | 1 | 1.11 (0.91-1.36) | 1 | 1.05 (0.87-1.27) | 1 | 1.05 (0.79–1.39) | 1 | 1.07 (0.77-1.49) | 1 | 1.12 (0.83-1.50) | 1 |
| rs1042026 | A/G | 512/715/0 | 0.92 (0.78-1.10) | 1 | 1.02 (0.84-1.23) | 1 | 1.01 (0.85-1.20) | 1 | 1.05 (0.81-1.37) | 1 | 1.10 (0.81-1.51) | 1 | 1.04 (0.79-1.36) | 1 | |
| rs7673353 | C/T | 99/1128/0 | 1.20 (0.89-1.62) | 1 | 0.90 (0.65-1.25) | 1 | 0.95 (0.70-1.30) | 1 | 1.14 (0.71-1.82) | 1 | 1.11 (0.61-2.03) | 1 | 1.02 (0.59-1.78) | 1 | |
| rs17028834 | T/C | 86/1141/0 | 1.38 (1.03-1.87) | 1 | 1.03 (0.74-1.44) | 1 | 1.11 (0.82-1.51) | 1 | 1.25 (0.77-2.03) | 1 | 1.25 (0.68-2.27) | 1 | 1.30 (0.74-2.27) | 1 | |
| rs1693457 | T/C | 421/805/1 | 0.91 (0.76-1.08) | 1 | 0.87 (0.71-1.06) | 1 | 0.89 (0.75-1.07) | 1 | 0.89 (0.67-1.17) | 1 | 0.94 (0.68-1.30) | 1 | 0.90 (0.68-1.20) | 1 | |
| rs1229984 | G/A | 31/1196/0 | 0.59 (0.30-1.13) | 1 | 0.65 (0.27-1.57) | 1 | 0.76 (0.39-1.48) | 1 | 0.78 (0.32-1.90) | 1 | 0.70 (0.17-2.84) | 1 | 0.95 (0.38-2.33) | 1 | |
| rs1159918 | G/T | 825/402/0 | 1.08 (0.90-1.29) | 1 | 0.91 (0.75-1.11) | 1 | 0.99 (0.82-1.19) | 1 | 0.97 (0.74-1.28) | 1 | 0.93 (0.67-1.30) | 1 | 0.97 (0.72-1.30) | 1 | |
| rs1229982 | G/T | 527/700/0 | 1.12 (0.95-1.33) | 1 | 0.95 (0.78-1.14) | 1 | 1.06 (0.89-1.25) | 1 | 0.94 (0.72-1.23) | 1 | 0.89 (0.64-1.23) | 1 | 0.96 (0.72-1.27) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| ADH1C | rs2298753 | T/C | 221/1006/0 | 0.82 (0.65-1.02) | 1 | 0.87 (0.68-1.12) | 1 | 0.85 (0.68-1.07) | 1 | 0.89 (0.63-1.26) | 1 | 0.99 (0.67-1.46) | 1 | 0.91 (0.64-1.29) | 1 |
| rs1614972 | C/T | 687/540/0 | 0.93 (0.79-1.10) | 1 | 0.84 (0.70-1.01) | 1 | 0.88 (0.74-1.04) | 1 | 0.84 (0.65-1.09) | 1 | 0.82 (0.61-1.11) | 1 | 0.84 (0.64-1.09) | 1 | |
| rs1391088 | C/A | 178/1049/0 | 0.93 (0.73-1.18) | 1 | 0.86 (0.65-1.13) | 1 | 0.95 (0.74-1.22) | 1 | 0.94 (0.65-1.37) | 1 | 0.82 (0.52-1.30) | 1 | 1.01 (0.68-1.49) | 1 | |
| rs1693482 | C/T | 695/530/2 | 0.90 (0.76-1.07) | 1 | 1.07 (0.89-1.29) | 1 | 1.03 (0.86-1.22) | 1 | 0.95 (0.73-1.24) | 1 | 1.06 (0.78-1.45) | 1 | 1.06 (0.80-1.40) | 1 | |
| rs1631460 | C/G | 704/522/1 | 0.89 (0.75-1.05) | 1 | 1.06 (0.88-1.28) | 1 | 1.02 (0.86-1.21) | 1 | 0.95 (0.73-1.23) | 1 | 1.06 (0.77-1.45) | 1 | 1.06 (0.80-1.39) | 1 | |
| rs11936869 | C/G | 614/613/0 | 0.95 (0.81-1.13) | 1 | 0.90 (0.75-1.08) | 1 | 0.93 (0.78-1.10) | 1 | 0.82 (0.63-1.07) | 1 | 0.85 (0.63-1.16) | 1 | 0.87 (0.66-1.14) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| ADH4 | rs29001227 | A/T | 105/1122/0 | 1.33(1.01-1.76) | 1 | 1.09 (0.80-1.48) | 1 | 1.12 (0.84-1.49) | 1 | 1.36 (0.89-2.08) | 1 | 1.62 (0.92-2.87) | 1 | 1.64 (0.97-2.75) | 1 |
| rs1126672 | C/T | 505/722/0 | 0.93 (0.79-1.11) | 1 | 1.00 (0.83-1.21) | 1 | 1.02 (0.86-1.22) | 1 | 0.82 (0.62-1.07) | 1 | 0.74 (0.53-1.02) | 1 | 0.81 (0.61-1.07) | 1 | |
| rs4699710 | T/C | 585/642/0 | 1.01 (0.85-1.19) | 1 | 1.07 (0.89-1.30) | 1 | 1.09 (0.92-1.30) | 1 | 0.86 (0.66-1.12) | 1 | 0.79 (0.58-1.07) | 1 | 0.88 (0.67-1.15) | 1 | |
| rs10017466 | T/C | 669/557/1 | 1.07 (0.91-1.27) | 1 | 1.08 (0.89-1.30) | 1 | 1.11 (0.93-1.32) | 1 | 0.97 (0.74-1.25) | 1 | 0.91 (0.68-1.24) | 1 | 0.99 (0.76-1.30) | 1 | |
| rs1800759 | C/A | 870/357/0 | 1.12 (0.93-1.35) | 1 | 1.03 (0.83-1.26) | 1 | 1.04 (0.86-1.26) | 1 | 0.97 (0.73-1.29) | 1 | 0.91 (0.65-1.27) | 1 | 0.91 (0.68-1.23) | 1 | |
| rs1800761 | G/A | 474/753/0 | 1.02 (0.86-1.22) | 1 | 1.01 (0.83-1.22) | 1 | 1.02 (0.86-1.22) | 1 | 1.00 (0.76-1.30) | 1 | 1.00 (0.73-1.37) | 1 | 1.05 (0.79-1.39) | 1 | |
| rs3762894 | T/C | 385/842/0 | 0.97 (0.81-1.17) | 1 | 0.96 (0.79-1.17) | 1 | 0.98 (0.82-1.18) | 1 | 0.97 (0.73-1.29) | 1 | 0.96 (0.69-1.33) | 1 | 1.01 (0.76-1.35) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| ADH7 | rs284787 | C/T | 484/743/0 | 0.96 (0.81-1.14) | 1 | 1.03 (0.85-1.25) | 1 | 1.06 (0.89-1.26) | 1 | 1.15 (0.88-1.49) | 1 | 1.23 (0.90-1.66) | 1 | 1.32 (1.00-1.72) | 1 |
| rs894369 | C/G | 391/836/0 | 1.15 (0.97-1.37) | 1 | 1.10 (0.91-1.33) | 1 | 1.08 (0.90-1.29) | 1 | 1.06 (0.80-1.39) | 1 | 0.96 (0.70-1.32) | 1 | 0.98 (0.74-1.30) | 1 | |
| rs17588403 | T/A | 388/839/0 | 0.97 (0.81-1.16) | 1 | 1.06 (0.87-1.30) | 1 | 1.02 (0.85-1.22) | 1 | 0.81 (0.61-1.09) | 1 | 0.84 (0.60-1.17) | 1 | 0.79 (0.59-1.07) | 1 | |
| rs1154454 | T/C | 469/758/0 | 1.12 (0.94-1.33) | 1 | 0.99 (0.82-1.20) | 1 | 0.99 (0.83-1.18) | 1 | 1.06 (0.82-1.39) | 1 | 0.97 (0.71-1.34) | 1 | 0.99 (0.74-1.31) | 1 | |
| rs1154456 | T/C | 648/579/0 | 0.86 (0.73-1.02) | 1 | 1.02 (0.84-1.22) | 1 | 0.95 (0.80-1.13) | 1 | 1.06 (0.82-1.38) | 1 | 1.11 (0.81-1.51) | 1 | 1.11 (0.84-1.46) | 1 | |
| rs1154460 | G/A | 895/329/3 | 0.87 (0.72-1.05) | 1 | 0.87 (0.71-1.06) | 1 | 0.83 (0.69-1.00) | 1 | 0.97 (0.72-1.30) | 1 | 0.90 (0.65-1.25) | 1 | 0.95 (0.71-1.27) | 1 | |
| rs971074 | G/A | 281/946/0 | 0.98 (0.80-1.20) | 1 | 0.86 (0.68-1.07) | 1 | 0.90 (0.73-1.10) | 1 | 0.93 (0.68-1.27) | 1 | 0.86 (0.59-1.25) | 1 | 0.88 (0.63-1.23) | 1 | |
| rs1573496 | C/G | 166/1060/1 | 0.82 (0.64-1.07) | 1 | 0.80 (0.60-1.06) | 1 | 0.81 (0.63-1.05) | 1 | 0.85 (0.58-1.27) | 1 | 0.73 (0.45-1.17) | 1 | 0.79 (0.52-1.19) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| ALDH2 | rs4767939 | A/G | 529/697/1 | 1.12 (0.94-1.32) | 1 | 0.96 (0.79-1.15) | 1 | 1.01 (0.85-1.20) | 1 | 1.07 (0.83-1.40) | 1 | 0.97 (0.72-1.32) | 1 | 1.01 (0.77-1.33) | 1 |
| rs2238151 | T/C | 858/365/4 | 1.18 (0.98-1.43) | 1 | 0.93 (0.75-1.15) | 1 | 0.98 (0.81-1.19) | 1 | 1.16 (0.87-1.55) | 1 | 1.00 (0.70-1.43) | 1 | 1.09 (0.79-1.49) | 1 | |
| rs7312055 | G/A | 144/1083/0 | 1.60 (1.26-2.02) | 0.006 | 1.13(0.87-1.48) | 1 | 1.18 (0.93-1.51) | 1 | 1.29 (0.87-1.91) | 1 | 1.63 (0.92-2.89) | 1 | 1.36 (0.81-2.27) | 1 | |
| rs2158029 | G/A | 57/1170/0 | 1.51 (1.06-2.13) | 1 | 1.07 (0.74-1.57) | 1 | 1.11 (0.78-1.57) | 1 | 0.69 (0.33-1.47) | 1 | 0.62 (0.27-1.46) | 1 | 0.55 (0.25-1.20) | 1 | |
| rs16941667 | C/T | 239/988/0 | 1.05 (0.85-1.29) | 1 | 0.94 (0.75-1.19) | 1 | 0.99 (0.80-1.22) | 1 | 1.25 (0.92-1.70) | 1 | 1.13(0.79-1.61) | 1 | 1.20 (0.87-1.64) | 1 | |
| rs16941669 | T/G | 240/987/0 | 0.87 (0.70-1.08) | 1 | 0.91 (0.72-1.16) | 1 | 0.84 (0.68-1.05) | 1 | 0.91 (0.65-1.28) | 1 | 0.87 (0.59-1.29) | 1 | 0.88 (0.62-1.23) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| CYP2E1 | rs3813865 | G/C | 125/1102/0 | 1.49 (1.16-1.92) | 0.116 | 1.38 (1.05-1.82) | 1 | 1.43(1.11-1.85) | 0.391 | 1.64 (1.13-2.39) | 0.605 | 2.20 (1.40-3.45) | 0.04 | 2.00 (1.33-3.01) | 0.059 |
| rs3813867 | G/C | 83/1143/1 | 0.80 (0.55-1.14) | 1 | 0.70 (0.48-1.03) | 1 | 0.68 (0.47-0.97) | 1 | 0.89 (0.52-1.53) | 1 | 0.67 (0.36-1.25) | 1 | 0.78 (0.45-1.36) | 1 | |
| rs8192772 | T/C | 200/1026/1 | 1.39 (1.12-1.72) | 0.148 | 1.42 (1.13-1.79) | 0.158 | 1.37 (1.11-1.70) | 0.248 | 1.53(1.12-2.10) | 0.539 | 1.82 (1.28-2.58) | 0.055 | 1.62 (1.17-2.23) | 0.231 | |
| rs915908 | G/A | 250/977/0 | 0.71 (0.56-0.89) | 0.184 | 0.82 (0.64-1.06) | 1 | 0.75 (0.60-0.95) | 0.98 | 0.84 (0.60-1.18) | 1 | 0.90 (0.61-1.34) | 1 | 0.85 (0.60-1.20) | 1 | |
| rs915909 | C/T | 59/1167/1 | 1.30 (0.91-1.86) | 1 | 0.96 (0.65-1.41) | 1 | 0.90 (0.63-1.30) | 1 | 1.14 (0.64-2.04) | 1 | 1.09 (0.55-2.17) | 1 | 0.97 (0.51-1.84) | 1 | |
| rs7092584 | C/T | 263/962/2 | 1.26 (1.04-1.54) | 1 | 1.24 (1.00-1.53) | 1 | 1.22 (1.00-1.48) | 1 | 1.31 (0.97-1.77) | 1 | 1.31 (0.94-1.83) | 1 | 1.31 (0.97-1.77) | 1 | |
| rs743535 | C/T | 249/969/9 | 1.30 (1.06-1.58) | 0.677 | 1.20 (0.97-1.49) | 1 | 1.19 (0.97-1.45) | 1 | 1.47 (1.09-1.99) | 0.694 | 1.37 (0.97-1.93) | 1 | 1.46 (1.08-1.99) | 0.957 | |
| rs2249695 | C/T | 603/623/1 | 1.33(1.13-1.58) | 0.053 | 1.12 (0.92-1.35) | 1 | 1.10 (0.92-1.31) | 1 | 1.37 (1.05-1.78) | 1 | 1.43(1.02-2.00) | 1 | 1.33 (0.98-1.78) | 1 | |
| rs28969387 | A/T | 25/1202 /0 | 1.68(1.02-2.77) | 1 | 0.92(0.51-1.66) | 1 | 1.14(0.69-1.90) | 1 | 2.03(1.00-4.11) | 1 | 0.79(0.28-2.24) | 1 | 1.40(0.66-2.99) | 1 | |
| rs11101812 | T/C | 26/1198 /3 | 0.72(0.37-1.39) | 1 | 0.53(0.26-1.07) | 1 | 0.45(0.23-0.87) | 1 | 0.79(0.29-2.12) | 1 | 0.60(0.18-1.97) | 1 | 0.62(0.22-1.74) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
|
OXIDATIVE STRESS GENES
|
|
|
|
|
|
|
|||||||||
| CAT | rs1049982 | C/T | 703/521/3 | 1.08 (0.91-1.28) | 1 | 1.06 (0.88-1.27) | 1 | 1.06 (0.89-1.26) | 1 | 1.03 (0.79-1.33) | 1 | 1.09 (0.81-1.48) | 1 | 1.06 (0.81-1.39) | 1 |
|
|
|
|
|
|
|
|
|||||||||
| GPx1 | rs8179172 | T/A | 51/1176/0 | 1.51 (1.03-2.21) | 1 | 1.12 (0.74-1.72) | 1 | 1.13 (0.76-1.67) | 1 | 0.86 (0.41-1.83) | 1 | 0.52 (0.20-1.33) | 1 | 0.67 (0.30-1.50) | 1 |
| rs1800668 | C/T | 588/638/1 | 0.93 (0.78-1.10) | 1 | 0.97 (0.81-1.16) | 1 | 0.96 (0.81-1.14) | 1 | 0.87 (0.67-1.12) | 1 | 0.79 (0.58-1.08) | 1 | 0.85 (0.65-1.12) | 1 | |
| rs3811699 | A/G | 614/613/0 | 0.96 (0.82-1.14) | 1 | 0.94 (0.78-1.13) | 1 | 0.95 (0.80-1.13) | 1 | 0.89 (0.69-1.16) | 1 | 0.78 (0.58-1.06) | 1 | 0.86 (0.66-1.12) | 1 | |
| rs3448 | C/T | 578/649/0 | 0.90 (0.76-1.07) | 1 | 0.91 (0.75-1.09) | 1 | 0.87 (0.73-1.03) | 1 | 0.85 (0.65-1.10) | 1 | 0.93 (0.69-1.26) | 1 | 0.81 (0.62-1.05) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| GPx2 | rs11623705 | G/T | 227/1000/0 | 0.90 (0.72-1.12) | 1 | 0.91 (0.71-1.17) | 1 | 0.93 (0.74-1.16) | 1 | 1.37 (1.01-1.86) | 1 | 1.41 (0.97-2.04) | 1 | 1.39 (1.01-1.91) | 1 |
| rs2412065 | G/C | 529/698/0 | 1.18 (1.00-1.40) | 1 | 1.02 (0.85-1.23) | 1 | 1.03 (0.87-1.23) | 1 | 1.33 (1.03-1.73) | 1 | 1.25 (0.92-1.70) | 1 | 1.26 (0.96-1.66) | 1 | |
| rs2737844 | C/T | 728/494/5 | 1.24 (1.04-1.47) | 1 | 1.07 (0.88-1.30) | 1 | 1.01 (0.85-1.21) | 1 | 1.22 (0.93-1.60) | 1 | 1.22 (0.87-1.71) | 1 | 1.15 (0.85-1.54) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| GPx4 | rs757229 | G/C | 925/301/1 | 1.03 (0.85-1.26) | 1 | 0.99 (0.80-1.23) | 1 | 1.01 (0.82-1.23) | 1 | 0.95 (0.70-1.27) | 1 | 1.10 (0.78-1.56) | 1 | 0.94 (0.69-1.27) | 1 |
|
|
|
|
|
|
|
|
|||||||||
| SOD1 | rs11910115 | A/C | 62/1165/0 | 1.25 (0.87-1.78) | 1 | 1.15 (0.79-1.68) | 1 | 1.08 (0.75-1.56) | 1 | 1.00 (0.54-1.83) | 1 | 1.25 (0.63-2.51) | 1 | 1.10 (0.57-2.15) | 1 |
| rs4998557 | G/A | 395/831/1 | 1.24 (1.04-1.47) | 1 | 1.07 (0.88-1.30) | 1 | 1.09 (0.91-1.30) | 1 | 1.15 (0.87-1.51) | 1 | 1.08 (0.77-1.50) | 1 | 1.05 (0.78-1.41) | 1 | |
| rs10432782 | T/G | 350/876/1 | 1.25 (1.04-1.49) | 1 | 1.13 (0.93-1.38) | 1 | 1.15 (0.96-1.38) | 1 | 1.29 (0.98-1.70) | 1 | 1.24 (0.90-1.71) | 1 | 1.22 (0.92-1.62) | 1 | |
| rs2070424 | A/G | 222/1005/0 | 1.32 (1.07-1.62) | 0.517 | 1.12 (0.89-1.40) | 1 | 1.17 (0.95-1.44) | 1 | 1.41 (1.03-1.92) | 1 | 1.15 (0.80-1.66) | 1 | 1.30 (0.94-1.80) | 1 | |
| rs1041740 | C/T | 543/684/0 | 0.91 (0.77-1.08) | 1 | 0.96 (0.80-1.16) | 1 | 0.97 (0.82-1.15) | 1 | 0.94 (0.72-1.22) | 1 | 0.88 (0.64-1.20) | 1 | 0.89 (0.68-1.18) | 1 | |
|
|
|
|
|
|
|
|
|||||||||
| SOD2 | rs4342445 | G/A | 473/754/0 | 0.92 (0.77-1.09) | 1 | 1.05 (0.87-1.27) | 1 | 1.01 (0.85-1.21) | 1 | 0.78 (0.59-1.03) | 1 | 0.88 (0.64-1.21) | 1 | 0.87 (0.66-1.16) | 1 |
| rs2842980 | A/T | 510/717/0 | 1.22 (1.03-1.44) | 1 | 1.07 (0.89-1.29) | 1 | 1.16 (0.97-1.37) | 1 | 1.49 (1.15-1.94) | 0.161 | 1.43 (1.05-1.94) | 1 | 1.50 (1.15-1.97) | 0.202 | |
| rs8031 | T/A | 838/389/0 | 0.99 (0.83-1.18) | 1 | 1.05 (0.86-1.29) | 1 | 1.02 (0.85-1.22) | 1 | 1.19 (0.89-1.58) | 1 | 1.12 (0.80-1.56) | 1 | 1.12 (0.83-1.51) | 1 | |
| rs5746134 | C/T | 100/1127/0 | 1.33 (1.00-1.77) | 1 | 0.95 (0.69-1.32) | 1 | 0.99 (0.73-1.33) | 1 | 1.39 (0.90-2.14) | 1 | 1.23 (0.67-2.26) | 1 | 1.36 (0.80-2.32) | 1 | |
| rs2758331 | C/A | 803/424/0 | 0.95 (0.80-1.13) | 1 | 1.09 (0.90-1.33) | 1 | 1.03 (0.86-1.23) | 1 | 1.06 (0.80-1.40) | 1 | 1.10 (0.79-1.52) | 1 | 1.05 (0.78-1.41) | 1 | |
Cox models used to calculate hazard ratios for all-cause death included SNP coded for dominant model, anatomical site, T stage, N stage, M stage, angiolymphatic invasion status, treatment (surgery, chemotherapy), presence of comorbid cardiovascular or lung disease, duration of smoking cigarettes (years), reception of at least one routine dental visit in past 10 years.
Cox models used to calculate hazard ratios for cancer-specific death included SNP coded for dominant model and all covariates listed above for all-cause death models, plus the following additional variables: radiation treatment, presence of comorbid liver disease or hepatitis, sex, race, age, federal poverty level, ever exposure to environmental tobacco smoke at work, ever loss of a tooth due to disease.
These analyses included only those cases who had complete information on the SNP and any covariates in the model. Two-thirds of SNPs (43 of 64) were successfully genotyped for all cases, and 20% of SNPs (n = 13) were missing in only one subject.
HRs represent hazard of any minor allele, compared to hazard of homozygous major allele.
For head and neck cancer, clinicians consider local-regional-distant stage classification to be inadequate for predicting survival; for example, advanced nodal (N) stage is a worse prognosticator than advanced locus disease [32,33]. Therefore, recent studies of survival in head and neck cancer patients most often control for cancer stage in Cox regression models by including separate variables representing tumor (T), node (N), and metastasis (M) stages. In CHANCE, comparison of two sets of hazard ratios and confidence intervals from Cox regression models that included summary stage, versus models that included separate variables for T, N, and M stages, revealed similar estimates from both methods. Therefore we controlled for stage using three separate variables.
In order to include available information from all cases, and because the data were not missing in a monotone pattern, we used the Markov Chain Monte Carlo method implemented in SAS® 9.2 PROC MI and information about non-missing values from all variables in full models (i.e. all variables in Table 2) to impute missing values for variables included in final models. This method imputes missing values for binary variables (such as angiolymphatic invasion) as continuous values from a normal distribution, which we did not round to 0 or 1 because the continuous values are less biased. Hazard ratios reported from those analyses are summaries of HRs from Cox regressions using 25 imputed datasets, calculated using SAS® 9.2 PROC MIANALYZE.
To adjust for Type 1 error due to multiple statistical tests for 64 SNPs, we compared results using three methods of correction: Bonferroni, False Discovery Rate, and False Positive Report Probability. Because results and conclusions were essentially the same from all three models, the simplest and most commonly-used method – Bonferroni – was used to correct p-values.
3. Results
Mean and median follow-up times from diagnosis date through 12/31/2009 were 71.2 and 70.6 months, respectively. As of December 31, 2009, 545 of 1227 cases (44%) had died, of which 227 (42%) died of head and neck cancer. Mean survival in all subjects with SCCHN was 52.5 months (Table 1). Mean/median survival for those with hypopharynx tumors was lower (39/30 months) than for those with other tumor sites (49–55/53–58 months). For subjects who died during the follow-up period, mean/median survival for all-cause death was 30.2/25.1 months, and for cancer-specific death, 24.0/19.5 months.
Table 1.
Survivala within study population.
| Survival (months) |
|||||
|---|---|---|---|---|---|
| n | Mean | 25th percentile | 50th percentile | 75th percentile | |
| All subjects | 1227 | 52.5 | 29.2 | 56.3 | 73.7 |
| Oral cavity | 172 | 49.0 | 23.2 | 52.6 | 71.2 |
| Oropharynx | 333 | 54.7 | 33.7 | 58.0 | 76.4 |
| Hypopharynx | 55 | 39.0 | 16.3 | 30.0 | 65.3 |
| Oral cavity–oropharynx–hypopharynx NOS | 224 | 51.4 | 22.8 | 57.1 | 74.5 |
| Larynx | 443 | 54.3 | 36.1 | 56.9 | 74.3 |
| Subjects who died of any cause | 545 | 30.2 | 13.8 | 25.1 | 42.9 |
| Subjects who died of head and neck cancer | 227 | 24.0 | 12.5 | 19.5 | 32.0 |
Vital status as of 31/12/2009, as determined from NDI.
Descriptive analyses. Table 2 presents the frequency distributions and hazard ratios for the initial set of covariates within the study population.
For both all-cause and cancer-specific survival, more subjects with oropharynx cancer survived compared to those with oral cavity and hypopharynx tumors (Table 2). Improved survival was also associated with: lower tumor and node stages, absence of angiolymphatic invasion, surgical cancer treatment, and shorter duration of smoking. In addition, having survived was also associated with being younger, richer, more educated, having health insurance, and having had at least one routine dental visit in the last 10 years. African-American race was associated with all-cause death but not cancer-specific death in univariate analysis.
SNP hazard ratios and survival plots. Backwards elimination of potential covariates, from 64 full models each containing one SNP (of the 64 studied) and all covariates listed in Table 2, produced a Cox regression model for all-cause death containing the following covariates: SNP being analyzed (one per model); anatomic subsite; T, N, and M stage; angiolymphatic invasion status; treatment (surgery, chemotherapy); co-morbid cardiovascular or lung disease; duration of cigarette smoking; and routine dental care. The final Cox regression model for cancer death contained all those variables, plus radiation treatment, co-morbid liver disease, sex, race, age category, federal poverty group, exposure to ETS at work, and tooth mobility due to disease.
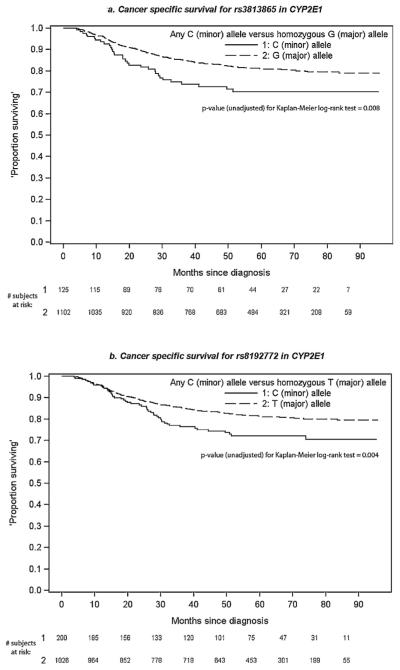
In unadjusted analyses, most SNPs were not associated with significantly increased hazard of all-cause or cancer death (Table 3), and results were generally consistent between models. However, two SNPs in CYP2E1–rs3813865 and rs8192772 – were associated with increased hazard of cancer death after adjustment for confounders in complete-case analysis and Bonferroni correction of p-values, with a similar pattern noted after adjustment for confounders in multiple imputation analysis (Table 3). Carrying the ‘C’ minor allele of rs3813865 was associated with double the hazard of head and neck cancer death compared to the ‘G’ allele (plog-rank = 0.008). Similarly, carrying the ‘C’ minor allele of rs8192772 was associated with about 60% increased cancer death hazard compared to the ‘T’ allele (plog-rank = 0.004). A non-statistically significant association with higher hazard of cancer death was noted in unadjusted Cox regression models (Table 3) and in the p-values (0.008, 0.004) for Kaplan–Meier survival curves for these SNPs (Fig. 1). A non-significant increase in hazard of all-cause death was also noted for these SNPs, due to the association with cancer death (data not shown).
Fig. 1.
Cancer-specific survival for two SNPs in CYP2E1 (Kaplan–Meier survival curves).
Most SNPs had similar survival effects in both African-Americans and whites, though there were suggestive differences for some SNPs (Table 4). Low allele frequencies in one race or the other precluded precise evaluation of statistical differences. For example, carrying the CYP2E1 rs3813865 minor ‘C’ allele, present in 26% of African-Americans and 5% of whites, conferred an increased HR (95% CI) in all subjects of 2.0 (1.3–3.0), which was 2.6 (1.5–4.9) in African-Americans, and 1.4 (0.7–2.7) in whites. Carrying the minor ‘C’ allele of rs8192772, present in 22% of African-Americans and 14% of whites, conferred similar increased risks in blacks and whites, with a HR (95% CI) of 1.6 (1.2–2.2) in all subjects, 1.9 (1.0–3.6) in African-Americans, and 1.5 (1.0–2.2) in whites.
Table 4.
Hazard differences between alleles of selected SNPs.a
| All-cause death (all subjects; n =1227) |
Head and neck cancer death (all subjects; n = 1227) |
H and N cancer death (African-Americans only; n = 305) |
H and N cancer death (Whites only; n = 922) |
||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
p-Values from Kaplan– -Meier log-rank testsd |
p-Values from Kaplan– Meier log-rank testsd |
||||||||||||
| Gene | SNP | Major/ minor allele |
# Cases homozygous for major allele (of 1227) |
Adjusted HR (95% CI)b |
Unadjusted | Bonferroni- corrected (64 tests) |
Adjusted HR (95% CI)c |
Unadjusted | Bonferroni- corrected (64 tests) |
# Cases homozygous for major allele (of 305) |
Adjusted HR (95% CI)c |
# Cases homozygous for major allele (of 922) |
Adjusted HR (95% CI)c |
| CYP2E1 | rs3813865 | G/C | 1102 | 1.43 (1.11–1.85) | 0.002 | 0.108 | 2.00 (1.33–3.01) | 0.009 | 0.560 | 225 | 2.64 (1.48–4.89) | 877 | 1.36 (0.68–2.72) |
| rs8192772 | T/C | 1026 | 1.37 (1.11–1.70) | 0.002 | 0.144 | 1.62 (1.17–2.23) | 0.008 | 0.509 | 237 | 1.88 (0.98–3.62) | 789 | 1.48 (1.00–2.19) | |
| rs7092584 | C/T | 962 | 1.22 (1.00–1.48) | 0.019 | 1.000 | 1.31 (0.97–1.77) | 0.076 | 1.000 | 219 | 1.13 (0.60–2.13) | 743 | 1.42 (0.99–2.02) | |
| rs743535 | C/T | 969 | 1.19 (0.97–1.45) | 0.010 | 0.666 | 1.46 (1.08–1.99) | 0.010 | 0.670 | 212 | 1.37 (0.76–2.45) | 757 | 1.56 (1.08–2.24) | |
| rs2249695 | C/T | 748 | 1.10 (0.92–1.31) | 0.001 | 0.050 | 1.33 (0.98–1.78) | 0.019 | 1.000 | 153 | 1.37 (0.47–3.97) | 595 | 1.27 (0.93–1.74) | |
| GPx2 | rs11623705 | G/T | 1000 | 0.93 (0.74–1.16) | 0.352 | 1.000 | 1.39 (1.01–1.91) | 0.043 | 1.000 | 290 | 0.99 (0.31–3.15) | 710 | 1.35 (0.96–1.90) |
| rs2412065 | G/C | 698 | 1.03 (0.87–1.23) | 0.051 | 1.000 | 1.26 (0.96–1.66) | 0.029 | 1.000 | 103 | 1.50 (0.78–2.88) | 595 | 1.13 (0.82–1.54) | |
| SOD1 | rs10432782 | T/G | 876 | 1.15 (0.96–1.38) | 0.016 | 1.000 | 1.22 (0.92–1.62) | 0.071 | 1.000 | 172 | 1.21 (0.69–2.11) | 704 | 1.33 (0.95–1.86) |
| rs2070424 | A/G | 1005 | 1.17 (0.95–1.44) | 0.008 | 0.507 | 1.30 (0.94–1.80) | 0.029 | 1.000 | 216 | 1.37 (0.81–2.76) | 789 | 1.46 (0.99–2.15) | |
| SOD2 | rs2842980 | A/T | 717 | 1.16 (0.97–1.37) | 0.021 | 1.000 | 1.50 (1.15–1.97) | 0.002 | 0.151 | 136 | 2.01 (1.07–3.76) | 581 | 1.38 (1.00–1.89) |
SNPs were selected for presentation in this table if the confidence interval for the adjusted HR and the Kaplan–Meier log-rank p-value both appeared to depart from the null, for either all-cause or cancer-specific death, or both.
Cox models used to calculate hazard ratios for all-cause death included SNP coded for dominant model (any minor allele versus homozygous major allele), anatomical site, T stage, N stage, M stage, angiolymphatic invasion status, treatment (surgery, chemotherapy), presence of comorbid cardiovascular or lung disease, duration of smoking cigarettes (years), reception of at least one routine dental visit in past 10 years. 25 datasets were imputed, and results were summarized as a single HR.
Cox models used to calculate hazard ratios for cancer-specific death included SNP coded for dominant model (any minor allele versus homozygous major allele) and all covariates listed above for all-cause death models, plus the following additional variables: radiation treatment, presence of comorbid liver disease or hepatitis, sex, race, age, federal poverty level, ever exposure to environmental tobacco smoke at work, ever loss of a tooth due to disease. 25 datasets were imputed, and results were summarized as a single HR.
Kaplan–Meier log-rank test for differences between the survival curves for the two alleles of each SNP.
Three additional SNPs in CYP2E1 (rs7092584, rs743535, rs2249695), and five SNPs in the oxidative stress pathway (rs11623705 and rs2412065 in GPx2, rs10432782 and rs2070424 in SOD1, and rs2842980 in SOD2) displayed suggestive differences in allele hazards for either or both all-cause and cancer death from Kaplan–Meier and log-rank analysis (Table 4).
Examination of hazard ratios stratified by summary stage (Table 5) indicate that carriers of the ‘C’ minor allele of rs3813865 in CYP2E1 experienced higher hazard of death compared to those homozygous for the major allele, for tumors diagnosed at stage II/III and stage IV. In contrast, higher hazard of death was limited to patients with Stage II/III tumors (not Stage IV) carrying the minor allele of rs8192772 in CYP2E1, rs7092584 in CYP2E1, rs11623705 in GPx2, rs2070424 in SOD1, or rs2842980 in SOD2.
Table 5.
Selected SNP hazard ratios of death from head and neck cancer, stratified by summary AJCC stagea (models and covariates as in Table 5).
| Stage I (n = 279) |
Stage II/III (n=431) |
Stage IV (n = 517) |
||||||
|---|---|---|---|---|---|---|---|---|
| Gene | SNP | Major/minor alleles |
# Cases: major/ any minor |
HR (95% CI)b | # Cases: major/ Any minor |
HR (95% CI)b | # Cases: major/ Any minor |
HR (95% CI)b |
| CYP2E1 | rs3813865 | G/C | 251/28 | – | 382/49 | 2.80(1.45–5.44) | 469/48 | 2.18 (1.23–3.88) |
| rs8192772 | T/C | 240/39 | 1.47 (0.38–5.69) | 358/73 | 1.89 (1.10–3.26) | 428/88 | 1.40 (0.90–2.18) | |
| rs7092584 | C/T | 227/52 | 0.95 (0.25–3.67) | 337/93 | 1.77 (1.06–2.96) | 398/118 | 0.97 (0.64–1.47) | |
| rs743535 | C/T | 227/51 | 1.70 (0.50–5.80) | 340/87 | 1.52 (0.88–2.60) | 402/111 | 1.29 (0.86–1.94) | |
| rs2249695 | C/T | 156/123 | 0.73 (0.22–2.37) | 217/213 | 1.48 (0.89–2.46) | 250/267 | 1.25 (0.84–1.88) | |
| GPx2 | rs11623705 | G/T | 229/50 | 1.49 (0.43–5.10) | 348/83 | 1.78 (1.04–3.05) | 423/94 | 1.31 (0.84–2.06) |
| rs2412065 | G/C | 163/116 | 1.25 (0.41–3.83) | 245/186 | 1.32 (0.82–2.13) | 290/227 | 1.32 (0.91–1.92) | |
| SOD1 | rs10432782 | T/G | 211/68 | 1.05 (0.31–3.53) | 297/133 | 1.54 (0.95–2.47) | 368/149 | 0.98 (0.66–1.46) |
| rs2070424 | A/G | 240/39 | 0.35 (0.04–2.85) | 342/89 | 1.81 (1.08–3.04) | 423/94 | 1.10 (0.69–1.76) | |
| SOD2 | rs2842980 | A/T | 174/105 | 1.67 (0.59–4.72) | 245/186 | 1.77 (1.11–2.83) | 298/219 | 1.40 (0.97–2.03) |
SNPs were selected for presentation in this table if the confidence interval for the adjusted HR and the Kaplan–Meier log-rank p-value both appeared to depart from the null, for either all-cause or cancer-specific death, or both.
Cox models used to calculate hazard ratios for head and neck cancer-specific death included SNP coded for dominant model (any minor allele versus homozygous major allele), age, sex, race, anatomical site, T stage, N stage, M stage, angiolymphatic invasion status, surgical treatment, chemotherapy treatment, radiation treatment, presence of comorbid cardiovascular or lung disease, presence of comorbid liver disease or hepatitis, duration of smoking cigarettes (years), ever exposure to environmental tobacco smoke at work, reception of at least one routine dental visit in past 10 years, ever loss of a tooth due to disease, federal poverty level. 25 datasets were imputed, and results were summarized as a single HR.
Table 6 presents hazard ratios for each genotype and chemotherapy combination for the 11 SNPs in Tables 4 and 5, using a common referent group for each SNP consisting of those homozygous for the major allele who did not receive chemotherapy. Among those who did not receive chemotherapy, the minor alleles of four CYP2E1 SNPs (rs3813865, rs8192772, rs7092584, rs743535) were associated with greater hazard of both all-cause and cancer death, and two SNPs in GPx2 (rs11623705, rs2412065) were associated with cancer death (not all-cause). In those who received chemotherapy, carriers of the minor allele had HR point estimates similar to those homozygous for the major allele (i.e. both were less than 1.0) but they were not statistically different.
Table 6.
Analysis of interaction between selected SNPsa and platinum chemotherapy treatment.
| All-cause death |
Cancer death |
||||||||
|---|---|---|---|---|---|---|---|---|---|
| Did not receive platinum chemotherapy |
Received platinum chemotherapy |
Did not receive platinum chemotherapy |
Received platinum chemotherapy |
||||||
| Gene | SNP | # Died/# survived/ missing |
Adjusted HR (95% CI)b |
# Died/# survived/ missing |
Adjusted HR (95% CI)b |
# Died/# survived/ missing |
Adjusted HR (95% CI)c |
# Died/# survived/ missing |
Adjusted HR (95% CI)c |
| CYP2E1 | rs3813865 | ||||||||
| GG | 295/398 | 1.00 (ref) | 180/229 | 0.67 (0.51–0.87) | 114/579 | 1.00 (ref) | 81/328 | 0.55 (0.35–0.86) | |
| GC + CC | 37/33 | 1.40 (0.97–2.02) | 33/22 | 0.92 (0.61–1.40) | 18/52 | 2.34 (1.30–4.23) | 14/41 | 1.13 (0.58–2.20) | |
| rs8192772 | |||||||||
| TT | 270/378 | 1.00 (ref) | 167/211 | 0.67 (0.50–0.88) | 101/547 | 1.00 (ref) | 77/301 | 0.64 (0.40–1.01) | |
| TC + CC | 61/53 | 1.50 (1.11–2.01) | 46/40 | 0.88 (0.61–1.29) | 31/83 | 2.34 (1.50–3.66) | 18/68 | 0.80 (0.43–1.50) | |
| rs7092584 | |||||||||
| CC | 256/352 | 1.00 (ref) | 155/199 | 0.66 (0.49–0.87) | 96/512 | 1.00 (ref) | 73/281 | 0.61 (0.38–0.97) | |
| CT + TT | 74/79 | 1.22 (0.92–1.60) | 58/52 | 0.83 (0.58–1.18) | 35/118 | 1.44 (0.93–2.24) | 22/88 | 0.70 (0.39–1.26) | |
| rs743535 | |||||||||
| CC | 251/356 | 1.00 (ref) | 164/198 | 0.70 (0.53–0.93) | 94/513 | 1.00 (ref) | 73/289 | 0.61 (0.39–0.98) | |
| CT + TT | 79/72 | 1.34 (1.02–1.77) | 47/51 | 0.72 (0.49–1.05) | 37/114 | 1.62 (1.04–2.52) | 21/77 | 0.66 (0.35–1.23) | |
| rs2249695 | |||||||||
| CC | 161/239 | 1.00 (ref) | 85/138 | 0.58 (0.41–0.82) | 60/340 | 1.00 (ref) | 41/182 | 0.50 (0.28–0.88) | |
| CT + TT | 171/192 | 1.01 (0.79–1.28) | 128/112 | 0.77 (0.56–1.06) | 72/291 | 1.30 (0.84–2.00) | 54/186 | 0.80 (0.47–1.36) | |
| GPx2 | rs11623705 | ||||||||
| GG | 268/347 | 1.00 (ref) | 185/200 | 0.70 (0.53–0.91) | 96/519 | 1.00 (ref) | 77/308 | 0.62 (0.40–0.97) | |
| GT + TT | 64/84 | 0.99 (0.73–1.34) | 28/51 | 0.54 (0.34–0.85) | 36/112 | 1.63 (1.02–2.61) | 18/61 | 0.70 (0.36–1.37) | |
| rs2412065 | |||||||||
| GG | 172/252 | 1.00 (ref) | 122/152 | 0.69 (0.51–0.93) | 59/365 | 1.00 (ref) | 56/218 | 0.72 (0.43–1.20) | |
| GC + CC | 160/179 | 1.03 (0.82–1.31) | 91/99 | 0.68 (0.49–0.95) | 73/266 | 1.55 (1.03–2.33) | 39/151 | 0.68 (0.39–1.19) | |
| SOD1 | rs10432782 | ||||||||
| TT | 227/318 | 1.00 (ref) | 144/187 | 0.69 (0.52–0.92) | 89/456 | 1.00 (ref) | 63/268 | 0.53 (0.33–0.86) | |
| TG + GG | 105/113 | 1.19 (0.92–1.53) | 69/63 | 0.74 (0.53–1.04) | 43/175 | 1.16 (0.76–1.78) | 32/100 | 0.72 (0.42–1.24) | |
| rs2070424 | |||||||||
| AA | 267/365 | 1.00 (ref) | 161/212 | 0.66 (0.50–0.86) | 105/527 | 1.00 (ref) | 70/303 | 0.52 (0.33–0.82) | |
| AG + GG | 65/66 | 1.08 (0.80–1.46) | 52/39 | 0.77 (0.54–1.11) | 27/104 | 0.96 (0.57–1.63) | 25/66 | 0.73 (0.41–1.29) | |
| SOD2 | rs2842980 | ||||||||
| AA | 178/266 | 1.00 (ref) | 121/152 | 0.67 (0.49–0.90) | 67/377 | 1.00 (ref) | 46/227 | 0.49 (0.29–0.82) | |
| AT + TT | 154/165 | 1.06 (0.84–1.34) | 92/99 | 0.73 (0.53–1.01) | 65/254 | 1.23 (0.83–1.84) | 49/142 | 0.85 (0.50–1.44) | |
SNPs were selected for presentation in this table if the confidence interval for the adjusted HR and the Kaplan–Meier log-rank p-value both appeared to depart from the null, for either all-cause or cancer-specific death, or both.
Cox models used to calculate hazard ratios for all-cause death included SNP coded for dominant model (any minor allele versus homozygous major allele), anatomical site, T stage, N stage, M stage, angiolymphatic invasion status, treatment (surgery), presence of comorbid cardiovascular or lung disease, duration of smoking cigarettes (years), reception of at least one routine dental visit in past 10 years. 25 datasets were imputed, and results were summarized as a single HR.
Cox models used to calculate hazard ratios for cancer-specific death included SNP coded for dominant model (any minor allele versus homozygous major allele) and all covariates listed above for all-cause death models, plus the following additional variables: radiation treatment, presence of comorbid liver disease or hepatitis, sex, race, age, federal poverty level, ever exposure to environmental tobacco smoke at work, ever loss of a tooth due to disease. 25 datasets were imputed, and results were summarized as a single HR.
To investigate the effect of including alcohol consumption and percent African ancestry variables to the final reduced models, hazard ratios were re-calculated with those variables included. Effect estimates changed very little, if at all (data not shown).
4. Discussion
In this study we evaluated the association of SNPs in genes in alcohol metabolism and oxidative stress pathways with survival after SCCHN diagnosis. Overall, we found that most SNPs were not associated with survival. However, we found that the minor alleles of two SNPs in CYP2E1 were significantly associated with increased hazard for death from head and neck cancer, especially in Stage II/III tumors. Also, carrying the minor allele of these SNPs was associated with worse survival only in those who did not receive chemotherapy. In contrast, having received chemotherapy was associated with improved survival regardless of genotype, although improvement was not statistically significant in carriers of the minor allele.
CYP genes are highly conserved, yet some polymorphisms have been found to be associated with cancer risk and survival. For example, RsaI/rs2031920 and a 96-bp insertion polymorphism in CYP2E1 have been associated with incidence of colorectal and esophageal cancer [21]. It is thought that these effects are produced by CYP2E1 inducibility rather than enzyme function, primarily by a mechanism that stabilizes the protein against degradation [34]. One study suggested that CYP2E1 polymorphisms play a role in the metabolism of drugs used to treat non-Hodgkin lymphoma, with one allele of rs2070673 associated with longer survival [35], and another reported a significant association between CYP2E1 rs2031920 wild-type ‘C’ allele and improved survival from non-small cell lung cancer [36].
Our findings provide evidence that two CYP2E1 gene polymorphisms are associated with survival from head and neck cancer; carriers of the minor allele of these SNPs experienced worse survival than those homozygous for the major allele. This effect appeared only in those who did not receive chemotherapy, which if true, implies that these SNPs do not affect CYP2E1 metabolism of, or induction by, platinum-containing chemotherapeutic agents. The effect did not appear to differ between those who received radiation therapy and those who did not (data not shown). Similar to other studies reporting that apparently non-functional CYP2E1 SNPs influence cancer risk, the two SNPs we identified with the highest survival associations are also in typically non-coding regions of the genome – rs3813865 in the 5′ region near the gene, and rs8192772 in intron 2. Interestingly, some studies have suggested that upstream 5′ mutations in CYP2E1 may affect gene expression and inducibility by ethanol [37]. If these SNPs are not causal, they may instead be in linkage disequilibrium with a causal polymorphism.
Although one previous study reported an association of CYP2E1 rs2031920 with advanced T stage SCCHN [11], ours is the first study to report the association of SNPs in CYP2E1 with survival after SCCHN diagnosis. Effects were observed in both unadjusted models and those adjusted for covariates that are independently associated with survival, suggesting that the association between SNP and survival is independent of other factors. Our results are compatible with the hypothesis that genetic predisposition affects cancer-specific survival after diagnosis, an effect which appears to be at least partly ameliorated by platinum-containing chemotherapeutic agents. Although results for a few other SNPs in other genes in the oxidative stress pathway suggested possible survival differences between genotypes, none were statistically significant, and no SNPs in other alcohol metabolism genes (ADH1B, ADH1C, ADH4, ADH7, ALDH2) were associated with either cancer-specific or overall survival.
CHANCE is one of the largest population-based studies of head and neck cancer ever conducted in the United States, and its population is drawn from a racially diverse region, resulting in the largest number of African-Americans ever enrolled in a molecular epidemiologic study of head and neck cancer. We were also able to adjust for population stratification by using ancestry informative markers. Unfortunately, the number of African-American participants (n = 305) was too small to provide sufficient power to adequately assess racial differences in SNP associations with survival. We studied mostly tag SNPs in order to study polymorphism associations with cancer across entire genes. We used a minor allele frequency threshold of 10% or greater, so we were unable to examine rare variants. Outcome assessment using high-quality identifying variables to link with the gold standard NDI to obtain vital status and cause of death is a strength of our study; any residual errors in determining vital status would tend to drive observed associations nearer the null. While CHANCE obtained detailed information on well-known risk factors such as smoking and drinking, and on many minor risk factors, it did not assess human papillomavirus (HPV) infection. The association of HPV with our SNPs would likely be minimal. If survival effects due to SNP polymorphisms are more pronounced in subjects with rapidly-fatal disease. It is possible that study subjects excluded from analysis because they died soon after diagnosis and before providing a biological sample (n = 52) could have biased our results toward the null. However, we found no evidence of SNP associations with T, N, or summary stage in our data. Although it is possible that our results could be due to chance, the effects persist even after Bonferroni adjustment for multiple comparisons, so we do not feel this is a likely explanation of our findings. In addition, we did not have any data on locoregional recurrence or distant metastasis.
In conclusion, our study found multiple SNPs in CYP2E1 to be statistically associated with cancer-specific survival, but only in those who did not receive chemotherapy. Future genetic factors and survival analyses of head and neck cancer should confirm the association with these SNPs and refine the identification of potentially causal loci in larger and diverse study populations. Such research would improve understanding of differential treatment effects on head and neck tumor progression and metastasis.
Acknowledgments
This work was supported in part by the National Cancer Institute (R01-CA90731; 2T32 CA009330-26) and the National Institute of Environmental Health Sciences (P30ES10126).
Footnotes
Conflict of interest statement The authors have no financial or personal conflicts of interest to report.
References
- [1].American Cancer Society . Cancer facts & figures 2013. American Cancer Society, Inc.; Atlanta: 2013. [Google Scholar]
- [2].American Cancer Society . Global cancer facts & figures. 2nd ed American Cancer Society; Atlanta: 2011. [Google Scholar]
- [3].Howlader N, Noone AM, Krapcho M, Neyman N, Aminou R, Waldron W, et al. SEER cancer statistics review, 1975–2008. National Cancer Institute; Bethesda, MD: 2008. [Google Scholar]
- [4].National Center for Health Statistics . US Mortality Files. Centers for Disease Control and Prevention; 2002–2006. [Google Scholar]
- [5].Blot WJ, McLaughlin JK, Winn DM, Austin DF, Greenberg RS, Preston-Martin S, et al. Smoking and drinking in relation to oral and pharyngeal cancer. Cancer Res. 1988;48(11):3282–7. Epub 01/06/1988. [PubMed] [Google Scholar]
- [6].Ang KK, Harris J, Wheeler R, Weber R, Rosenthal DI, Nguyen-Tan PF, et al. Human papillomavirus and survival of patients with oropharyngeal cancer. N Engl J Med. 2010;363(1):24–35. doi: 10.1056/NEJMoa0912217. Epub 10/06/2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Tai J, Yang M, Ni X, Yu D, Fang J, Tan W, et al. Genetic polymorphisms in cytochrome P450 genes are associated with an increased risk of squamous cell carcinoma of the larynx and hypopharynx in a Chinese population. Cancer Genet Cytogenet. 2010;196(1):76–82. doi: 10.1016/j.cancergencyto.2009.08.015. Epub 08/12/2009. [DOI] [PubMed] [Google Scholar]
- [8].Garcia SM, Curioni OA, de Carvalho MB, Gattas GJ. Polymorphisms in alcohol metabolizing genes and the risk of head and neck cancer in a Brazilian population. Alcohol Alcohol. 2010;45(1):6–12. doi: 10.1093/alcalc/agp078. Epub 06/11/2009. [DOI] [PubMed] [Google Scholar]
- [9].Ruwali M, Khan AJ, Shah PP, Singh AP, Pant MC, Parmar D. Cytochrome P450 2E1 and head and neck cancer: interaction with genetic and environmental risk factors. Environ Mol Mutagen. 2009;50(6):473–82. doi: 10.1002/em.20488. Epub 01/04/2009. [DOI] [PubMed] [Google Scholar]
- [10].Oze I, Matsuo K, Suzuki T, Kawase T, Watanabe M, Hiraki A, et al. Impact of multiple alcohol dehydrogenase gene polymorphisms on risk of upper aerodigestive tract cancers in a Japanese population. Cancer Epidemiol Biomarkers Prev. 2009;18(11):3097–102. doi: 10.1158/1055-9965.EPI-09-0499. Epub 29/10/2009. [DOI] [PubMed] [Google Scholar]
- [11].Olivieri EH, da Silva SD, Mendonca FF, Urata YN, Vidal DO, Faria Mde A, et al. CYP1A2*1C, CYP2E1*5B, and GSTM1 polymorphisms are predictors of risk and poor outcome in head and neck squamous cell carcinoma patients. Oral Oncol. 2009;45(9):e73–9. doi: 10.1016/j.oraloncology.2009.03.004. Epub 16/05/2009. [DOI] [PubMed] [Google Scholar]
- [12].Canova C, Hashibe M, Simonato L, Nelis M, Metspalu A, Lagiou P, et al. Genetic associations of 115 polymorphisms with cancers of the upper aerodigestive tract across 10 European countries: the ARCAGE project. Cancer Res. 2009;69(7):2956–65. doi: 10.1158/0008-5472.CAN-08-2604. Epub 03/04/2009. [DOI] [PubMed] [Google Scholar]
- [13].Soya SS, Vinod T, Reddy KS, Gopalakrishnan S, Adithan C. CYP2E1 polymorphisms and gene–environment interactions in the risk of upper aerodigestive tract cancers among Indians. Pharmacogenomics. 2008;9(5):551–60. doi: 10.2217/14622416.9.5.551. Epub 10/05/2008. [DOI] [PubMed] [Google Scholar]
- [14].Hashibe M, McKay JD, Curado MP, Oliveira JC, Koifman S, Koifman R, et al. Multiple ADH genes are associated with upper aerodigestive cancers. Nat Genet. 2008;40(6):707–9. doi: 10.1038/ng.151. Epub 27/05/2008. [DOI] [PubMed] [Google Scholar]
- [15].Buch SC, Nazar-Stewart V, Weissfeld JL, Romkes M. Case–control study of oral and oropharyngeal cancer in whites and genetic variation in eight metabolic enzymes. Head Neck J Sci Spec. 2008;30(9):1139–47. doi: 10.1002/hed.20867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Boccia S, Cadoni G, Sayed-Tabatabaei FA, Volante M, Arzani D, De Lauretis A, et al. CYP1A1, CYP2E1, GSTM1, GSTT1, EPHX1 exons 3 and 4, and NAT2 polymorphisms, smoking, consumption of alcohol and fruit and vegetables and risk of head and neck cancer. J Cancer Res Clin Oncol. 2008;134(1):93–100. doi: 10.1007/s00432-007-0254-5. Epub 06/07/2007. [DOI] [PubMed] [Google Scholar]
- [17].Secretan B, Straif K, Baan R, Grosse Y, El Ghissassi F, Bouvard V, et al. A review of human carcinogens – part E: tobacco, areca nut, alcohol, coal smoke, and salted fish. Lancet Oncol. 2009;10(11):1033–4. doi: 10.1016/s1470-2045(09)70326-2. Epub 06/11/2009. [DOI] [PubMed] [Google Scholar]
- [18].Koop DR. Alcohol metabolism’s damaging effects on the cell: a focus on reactive oxygen generation by the enzyme cytochrome P450 2E1. Alcohol Res Health. 2006;29(4):274–80. Epub 28/08/2007. [PMC free article] [PubMed] [Google Scholar]
- [19].Lu Y, Cederbaum AI. CYP2E1 and oxidative liver injury by alcohol. Free Radic Biol Med. 2008;44(5):723–38. doi: 10.1016/j.freeradbiomed.2007.11.004. Epub 15/12/2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Lieber CS. Cytochrome P-4502E1: its physiological and pathological role. Physiol Rev. 1997;77(2):517–44. doi: 10.1152/physrev.1997.77.2.517. Epub 01/04/1997. [DOI] [PubMed] [Google Scholar]
- [21].Trafalis DT, Panteli ES, Grivas A, Tsigris C, Karamanakos PN. CYP2E1 and risk of chemically mediated cancers. Expert Opin Drug Metab Toxicol. 2010;6(3):307–19. doi: 10.1517/17425250903540238. Epub 16/01/2010. [DOI] [PubMed] [Google Scholar]
- [22].Wang D, Ritchie JM, Smith EM, Zhang Z, Turek LP, Haugen TH. Alcohol dehydrogenase 3 and risk of squamous cell carcinomas of the head and neck. Cancer Epidemiol Biomarkers Prev. 2005;14(3):626–32. doi: 10.1158/1055-9965.EPI-04-0343. Epub 16/03/2005. [DOI] [PubMed] [Google Scholar]
- [23].Divaris K, Olshan AF, Smith J, Bell ME, Weissler MC, Funkhouser WK, et al. Oral health and risk for head and neck squamous cell carcinoma: the Carolina Head and Neck Cancer Study. Cancer Causes Control. 2010;21(4):567–75. doi: 10.1007/s10552-009-9486-9. Epub 06/01/2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Hakenewerth AM, Millikan RC, Rusyn I, Herring AH, North KE, Barnholtz-Sloan JS, et al. Joint effects of alcohol consumption and polymorphisms in alcohol and oxidative stress metabolism genes on risk of head and neck cancer. Cancer Epidemiol Biomarkers Prev. 2011;20(11):2438–49. doi: 10.1158/1055-9965.EPI-11-0649. Epub 24/09/2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Hashibe M, Brennan P, Benhamou S, Castellsague X, Chen C, Curado MP, et al. Alcohol drinking in never users of tobacco, cigarette smoking in never drinkers, and the risk of head and neck cancer: pooled analysis in the International Head and Neck Cancer Epidemiology Consortium. J Natl Cancer Inst. 2007;99(10):777–89. doi: 10.1093/jnci/djk179. Epub 17/05/2007. [DOI] [PubMed] [Google Scholar]
- [26].Division of Vital Statistics. National Center for Health Statistics. Centers for Disease Control and Prevention National Death Index. 2002–2009.
- [27].Cowper DC, Kubal JD, Maynard C, Hynes DM. A primer and comparative review of major US mortality databases. Ann Epidemiol. 2002;12(7):462–8. doi: 10.1016/s1047-2797(01)00285-x. Epub 16/10/2002. [DOI] [PubMed] [Google Scholar]
- [28].NHLBI Program for Genomic Applications [database on the Internet] SeattleSNPs. Seattle, WA: Available from: http://pga.gs.washington.edu. [cited 22.02.08–25.04.08] [Google Scholar]
- [29].Barnholtz-Sloan JS, Chakraborty R, Sellers TA, Schwartz AG. Examining population stratification via individual ancestry estimates versus self-reported race. Cancer Epidemiol Biomarkers Prev. 2005;14(6):1545–51. doi: 10.1158/1055-9965.EPI-04-0832. Epub 09/06/2005. [DOI] [PubMed] [Google Scholar]
- [30].Barnholtz-Sloan JS, McEvoy B, Shriver MD, Rebbeck TR. Ancestry estimation and correction for population stratification in molecular epidemiologic association studies. Cancer Epidemiol Biomarkers Prev. 2008;17(3):471–7. doi: 10.1158/1055-9965.EPI-07-0491. Epub 20/03/2008. [DOI] [PubMed] [Google Scholar]
- [31].Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, et al. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31(8):1417–23. doi: 10.1093/carcin/bgq128. Epub 18/06/2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Brandwein-Gensler M, Smith RV. Prognostic indicators in head and neck oncology including the new 7th edition of the AJCC staging system. Head Neck Pathol. 2010;4(1):53–61. doi: 10.1007/s12105-010-0161-y. Epub 20/03/2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Takes RP, Rinaldo A, Silver CE, Piccirillo JF, Haigentz M, Jr, Suarez C, et al. Future of the TNM classification and staging system in head and neck cancer. Head Neck. 2010;32(12):1693–711. doi: 10.1002/hed.21361. Epub 02/03/2010. [DOI] [PubMed] [Google Scholar]
- [34].Roberts BJ, Song BJ, Soh Y, Park SS, Shoaf SE. Ethanol induces CYP2E1 by protein stabilization. Role of ubiquitin conjugation in the rapid degradation of CYP2E1. J Biol Chem. 1995;270(50):29632–35. doi: 10.1074/jbc.270.50.29632. Epub 15/12/1995. [DOI] [PubMed] [Google Scholar]
- [35].Han X, Zheng T, Foss FM, Lan Q, Holford TR, Rothman N, et al. Genetic polymorphisms in the metabolic pathway and non-Hodgkin lymphoma survival. Am J Hematol. 2010;85(1):51–6. doi: 10.1002/ajh.21580. Epub 24/12/2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Haque AK, Au W, Cajas-Salazar N, Khan S, Ginzel AW, Jones DV, et al. CYP2E1 polymorphism, cigarette smoking, p53 expression, and survival in non-small cell lung cancer: a long term follow-up study. Appl Immunohistochem Mol Morphol. 2004;12(4):315–22. doi: 10.1097/00129039-200412000-00005. Epub 13/11/2004. [DOI] [PubMed] [Google Scholar]
- [37].Neafsey P, Ginsberg G, Hattis D, Johns DO, Guyton KZ, Sonawane B. Genetic polymorphism in CYP2E1: population distribution of CYP2E1 activity. J Toxicol Environ Health B Crit Rev. 2009;12(5–6):362–88. doi: 10.1080/10937400903158359. Epub 26/02/2010. [DOI] [PubMed] [Google Scholar]