Summary
In studies of protein complexes for which high-resolution structural data are unavailable, it is often still possible to determine both nearest-neighbor relationships between subunits and atomic-resolution details of these interactions. The eukaryotic 26S proteasome, a ~2.5 MDa protein complex with at least 33 different subunits, is a prime example. Important information about quaternary organization and assembly of proteasomes has been gained using a combination of sequence alignments with related proteins of known tertiary structure, molecular modeling, and disulfide engineering to allow oxidative crosslinking between predicted polypeptide neighbors. Here we provide detailed protocols for engineered cysteine crosslinking of yeast proteasome subunits in whole cell extracts, in active 26S proteasome complexes first isolated by native polyacrylamide gel electrophoresis, and in subcomplexes that function as potential assembly intermediates.
Keywords: disulfide engineering; proteasome, site-directed mutagenesis, nondenaturing polyacrylamide gel electrophoresis (native PAGE), substrate overlay, Saccharomyces cerevisiae, yeast, immunoblotting
1. Introduction
Our understanding of large protein complexes and their assembly has been greatly aided by high-resolution structural studies. Frequently, however, such information is not available, especially for complexes containing many different subunits or exhibiting a high degree of structural or conformational plasticity. The eukaryotic 26S proteasome provides an illuminating example of incomplete atomic-resolution data that could be bridged by biochemical and genetic methods that exploit structural information from simpler but related complexes of known structure (1) (2) (3) (4) (5).
The 26S proteasome consists of a cylindrical 20S proteasome core particle (CP) with a 19S regulatory particle (RP) bound to one or both ends of the CP cylinder (6) (7) (8) (9). For the CP, fourteen different subunits generally comprise the complex, with two identical heteroheptameric β-subunit rings that create a central proteolytic chamber sandwiched by a pair of heteroheptameric α-subunit rings (10) (11). In the yeast Saccharomyces cerevisiaeall but one of the 14 different CP subunits is essential for cell viability. The exception is α3, which can be replaced by a second copy of the α4 subunit (3). A CP assembly chaperone, Pba3-Pba4, ensures that α4 does not take the α3 position in the α ring under most conditions (4). However, cells bearing these alternative CP forms appear to have a growth advantage in certain environments, particularly in the presence of heavy metals.
The ability of α4 to take the α3 position and the regulation of this alternative ring assembly by Pba3-Pba4 were demonstrated most convincingly by engineered disulfide crosslinking studies (3) (4). All the α subunits have sequence and structural homology, and high-resolution crystal structures are available for the yeast CP with the usual heteroheptameric α ring (11). This allowed structural modeling of α4 in the α3 position and the selection of residues that could be substituted with cysteines for induced disulfide bond formation between subunits (3).
For the RP, no atomic-resolution structures are available. A key component of the RP is a heterohexameric ring of AAA+ ATPase subunits that directly contacts the α ring of the CP (9). Discrepant data for the arrangement of these six subunits made it difficult to decide with any confidence what the correct order was or even if it was a uniquely ordered ring (12). Fortunately, crystal structures recently became available for domains derived from related homohexameric ATPases from prokaryotes (13) (14). The outer domain (subcomplex I) of Methanocaldococcus jannaschii proteasome-associated nucleotidase (PAN) could be purified as a stable hexamer (13). Each protomer of subcomplex I has an N-terminal helix followed by an oligosaccharide/oligonucleotide-binding (OB) domain. The helices from neighboring subunits form coiled coils, resulting in a trimer-of-dimers arrangement. Despite poor sequence conservation, accurate alignment of the amino-acid sequence of subcomplex I from M. jannaschii with the N-terminal segments of the eukaryotic proteasomal ATPases is possible. Using the archaeal structure as a guide, these alignments have enabled the selection of residues for cysteine substitution by site-directed mutagenesis and disulfide crosslinking of neighboring yeast proteasomal ATPases (5).
Here we describe detailed methods for engineered disulfide-crosslinking analysis of CP subunits and RP ATPase subunits. These include methods for analyzing crosslinking in yeast whole-cell extracts (Section 3.1), in immunoprecipitated proteasome assembly intermediates that are of low relative abundance (Section 3.2), and in nondenaturing gel-separated 26S proteasome complexes (Section 3.3). Sequence alignment methods, searching for residues suitable for cysteine substitution, and site-directed DNA mutagenesis are not covered here.
2. Materials
2.1 Disulfide crosslinking in yeast whole-cell extracts
Zymolyase Buffer (ZB): 1.2 M sorbitol, 50 mM Tris-HCl, pH 8.0, 0.5 mM MgCl2 (store at room temperature).
Lysis Buffer 1 (LB1: 50 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 1 mM Na-EDTA, 0.1% Triton-X100, (store at room temperature). Add Roche protease inhibitor cocktail (PIC; from pellet) per manufacturer instructions before use.
Lysis Buffer 2 (LB2): 50 mM HEPES-NaOH pH 7.5, 150 mM NaCl, 5 mM MgCl2, (store at room temperature). Add Roche protease inhibitor cocktail (PIC; from pellet) per manufacturer instructions before use.
10X Stop Solution (SS): 10 mM sodium iodoacetate, 50 mM NEM. Stock solutions: 1. 2 M sodium iodoacetate: FW = 207.9. Dissolve 0.416 g/ml (store at −20°C); 500 mM N-ethylmaleimide (NEM): FW125.13. Dissolve 0.063 g/ml in 50% ethanol (store at –20°C).
- Recipe for 3X SDS sample buffer (nonreducing) (for 1 ml; store at –20°C):
0.18 ml 1M Tris-HCl, pH 6.8 0.30 ml 100% glycerol 0.225 ml 20% SDS 0.15 ml 2 M sodium iodoacetate 0.10 ml H2O 45 µl 0.5% (w/v) bromophenol blue solution
2.2 Disulfide crosslinking of native gel-separated complexes
- Recipe for Cell Extraction Buffer (CEB) for native gel separations:
Stock 1 ml 50 mM Tris-HCl (pH 7.4) 1 M 50 µl 5 mM MgCl2 1 M 5 µl 10% glycerol 75% 133 µl 5 mM ATP 1 M 5 µl (avoid repeated freeze/thawing) dH2O 807 µl Make up 1M ATP stock in 50 mM Tris-HCl (pH 7.4); aliquot and freeze at −80°C. 5x native loading buffer: CEB containing 3 µg/ml xylene cyanol
- Recipe for native gel-electrophoresis buffer:
Stock 500 ml 90 mM Tris-borate, pH 8.3 0.9 M 50 ml 5 mM MgCl2 1 M 2.5 ml 1 mM ATP 1 M 500 µl (see CEB recipe) Note: Can be made without ATP and stored for at least 6 mo. at 4˚C. - Recipe for nondenaturing resolving gel (4%)
Stock 10ml (1 gel) 20 ml (2 gels) 4% acrylamide 40% 974 µl 1.95 ml 0.1% bis-acrylamide 2% 519 µl 1.04 ml 5 ml MgCl2 1 M 50 µl 100 µl 1 mM ATP 1 M 10 µl 20 µl (see CEB recipe) 90 mM Tris-borate, pH 8.3 0.9 M 1 ml 2 ml 2.5% Sucrose 25% 1 ml 2 ml Deionized H2O 7.34 ml 1.47 ml APS 10% 100 µl 200 µl TEMED 10 µl 20 µl (see Note 1) - Recipe for nondenaturing stacking gel
Stock 5ml (1 gel) 10 ml (2 gels) 3% acrylamide 40% 300 µl 600 µl 0.3% bis-acrylamide 2% 1.5 ml 3 ml 5 mM MgCl2 1 M 25 µl 50 µl 1 mM ATP 1 M 5 µl 10 µl (see CEB recipe) 50 mM Tris-HCl, pH 6.8 1M 250 µl 500 µl 2.5% sucrose 25% 500 µl 1 ml dH2O 2.4 ml 4.7 ml APS 10% 50 µl 100 µl TEMED 5 µl 10 µl -
Recipe for Suc-LLVY-AMC substrate overlay buffer (OB
Stock 10 ml 50 mM Tris-HCl (pH 7.5) 1 M 500 µl 5 mM MgCl2 1 M 50 µl 10% glycerol 75% 1.33 ml 1 mM ATP 1 M 10 µl (see CEB recipe) 1 mM Suc-LLVY-AMC 10 mM 100 µl {0.04% SDS 20% 20 µl (only for detection of 20S activity)} (10 mM Suc-LLVY-AMC (Sigma, S6510-10MG). Add 1.3 ml of DMSO to the vial and dissolve powder. Make 100 µl aliquots. Store at -80°C. This can be added to OB before or after the native gel has been immersed.)
2.3 Native PAGE–nonreducing SDS PAGE crosslinking analysis of active 26S proteasomes
- Recipe for crosslinking buffer
Stock 100 ml 50 mM HEPES, pH 7.5 1 M 5 ml 150 mM NaCl 4 M 3.75 ml 1 mM MgCl2 1 M 100 µl 10% glycerol 75% 13.3 ml 0.2 mM CuCl2 0.2 M 100 µl Note: the same buffer without CuCl2 should also be prepared. - Recipe for gel slice packing solution
Stock 50 ml dH2O 45.6 ml Tris-HCl, pH 6.8 1.5M 4.17 ml SDS 20% 250 µl agarose LE powder 0.35 g bromophenol blue trace
2.4 Equipment
Standard equipment is used for native and SDS-PAGE and immunoblotting. For the two-dimensional native gel separation followed by nonreducing SDS-PAGE, the following equipment was utilized:
Mini-PROTEAN®3 Electrophoresis Cell (BIO-RAD)
Spacer plate: 1.5 mm integrated spacers
Comb: 10-well comb
Ultraviolet Products MC Chromato-VUE® Transilluminator Model C-62
3. Methods
The distance between the sulfur atoms in a protein disulfide bond is slightly over 2 Å. Site-specific engineering of disulfide bonds between subunits therefore requires a way to predict sites of intersubunit contact with high precision. If an atomic resolution structure of a related protein complex is available, sequence alignment and molecular modeling will allow reasonable guesses for residues to replace with cysteines. Because of the stringent constraints on cysteine side-chain distance and orientation, multiple pairs of substituted residues may need to be tested. It is not generally necessary to mutate cysteine residues naturally present in the polypeptides being tested, although oxidation of these residues (or other oxidation-sensitive amino acids) could potentially alter the structure of the protein during the crosslinking time course, which can also change the migration of the heterodimer on a nonreducing SDS gel. In principle, one also need not eliminate the chromosomally expressed wild-type subunits under investigation, although competition with the cysteine-substituted versions will reduce the crosslinking signal.
A variety of controls are essential for valid interpretation of the disulfide crosslinking results. The crosslinked species should only be present when both of the tested subunits have the engineered cysteine replacements (Fig. 1). The crosslinked subunit dimer should be sensitive to addition of reducing agent prior to nonreducing SDS-gel electrophoresis. To distinguish between a unique pairing of two paralogous subunits versus promiscuous pairings, cysteine mutations at the aligned positions of other subunits in the ring should also be tested for disulfide bond formation with the subunit of interest.
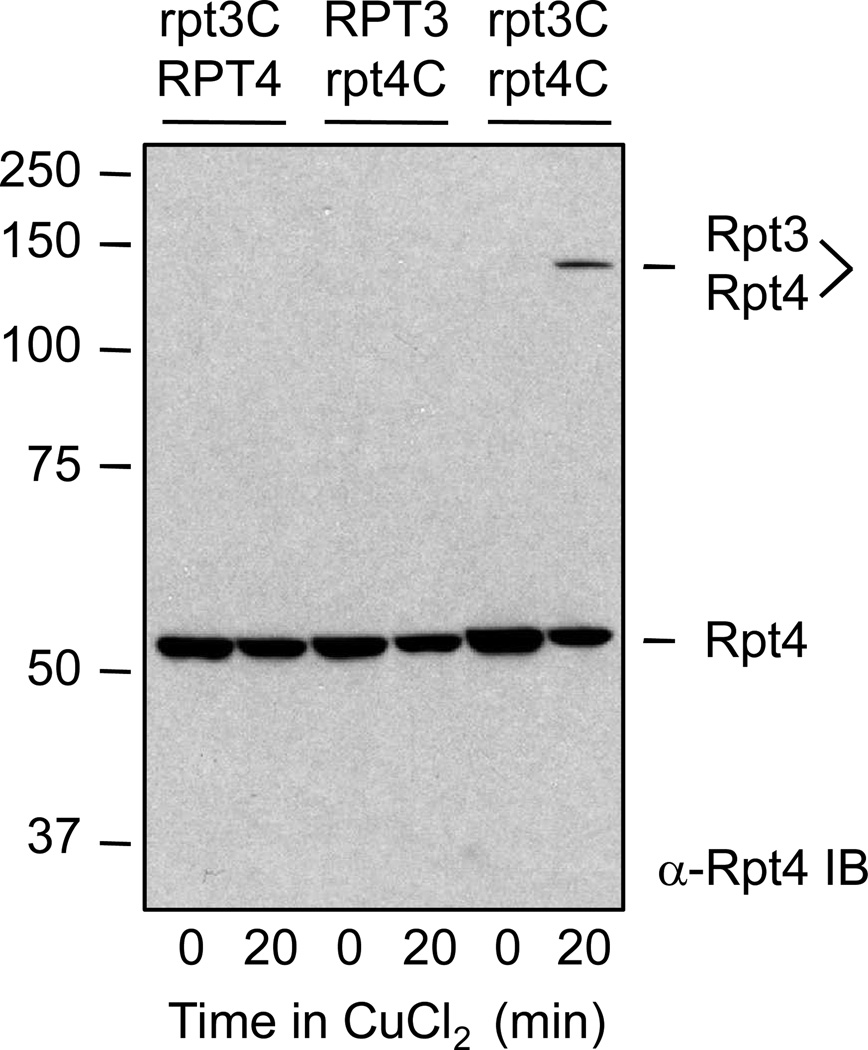
Fig 1.
An example of engineered disulfide crosslinking between two predicted neighboring subunits in the ATPase ring of the S. cerevisiae proteasomal RP. Amino acids were chosen for substitution with cysteine based on primary sequence alignment with the archaeal proteasomal ATPase PAN. The crystal structure of PAN, in turn, suggested residues at the interface between neighboring subunit OB domains that, if replaced with cysteines, could potentially be linked by a disulfide bond. The yeast strains from which whole-cell extracts were made had either an rpt3-I96C or rpt4-R126C mutation or both. Only when both cysteine substitutions were present was a 0.2 mM CuCl2-induced disulfide crosslink observed between the Rpt3 and Rpt4 subunits (last lane). Modified from Ref. 5 (with permission from Cell Press).
3.1 Disulfide crosslinking in yeast whole-cell extracts
3.1.1 Digest cell wall and lyse resulting spheroplasts
Yeast cells are grown under standard conditions. On the previous evening, inoculate desired yeast strains into small cultures (typically 10 ml in 15 ml culture tubes) in rich medium (YPD) or minimal dropout medium if selection for a plasmid(s) with an auxotrophic marker is necessary. In the morning, dilute the overnight cultures to 10 ml with fresh medium to achieve an initial optical density at 600 nm (OD600) of 0.1–0.3. Grow cells for 5–8 h at 30°C with aeration until cells are in mid-logarithmic growth (OD600= 0.8–1.2).
Collect cells by centrifuging in a Sorvall RT6000B tabletop centrifuge or equivalent for 5 min at 1,500×g at room temperature. Resuspend cells in 1 ml of sterile water and transfer to a 1.5 ml Eppendorf tube and spin 20 sec at top speed in a tabletop microcentrifuge to pellet cells.
Resuspend cells in 0.1 ml Zymolyase Buffer (ZB)+30 mM DTT. Leave at room temperature for 15 min. Pellet the cells in a microcentrifuge with a 30 sec spin and discard the supernate. (see Note 2)
Resuspend the cell pellet in 0.1 ml ZB + 4 µl 15 mg/ml Zymolyase 100T (1.5 mg in 0.1 ml TE for stock suspension; 0.6 mg/ml final concentration). Incubate at 30°C for 30 min, keeping cells on a roller or equivalent to keep cells in suspension.
Gently pellet spheroplasts in a microcentrifuge by centrifuging at 1,200×g for 5 min at room temperature. Wash once with 0.5 ml ZB (resuspend by breaking up pellet with disposable pipette tip). Pellet spheroplasts by centrifuging at 1,200×g for 5 min.
Add 100–125 µl ice-cold Lysis Buffer (LB1 or LB2). Stir up pellet with pipette tip to partially resuspend cells. (see Note 3)
Lyse spheroplasts by vortexing three times for 30 sec at top speed and leave on ice for 1 min in between. Pellet cell debris and unbroken cells with a 15,000×g centrifugation for 15 min at 4°C.
Save supernate in a fresh Eppendorf tube on ice. A second centrifugation for 5–10 min can be done if additional clarification of the extract is needed.
3.1.2 CuCl2–induced disulfide crosslinking time course
Save 20 µl of crude lysate for time zero and add to a tube on ice containing 2 µl of 10X Stop Solution (SS).
Add 2 µl of 10 mM CuCl2 to 80 µl of remaining lysate for a final concentration of 0.25 mM. This concentration can be varied depending on the efficiency of crosslinking (usually between 0.2 and 0.5 mM). Leave at room temperature. (see Note 4)
At the desired times, e.g., 15, 30, 60 min, remove 20 µl of lysate and add to 2 µl SS on ice. When the efficiency of crosslinking is established, a single timepoint for oxidative crosslinking will often suffice.
Store samples at –20°C or –80°C or proceed to SDS-PAGE separation.
3.1.3 Nonreducing SDS-PAGE and immunoblot analysis
Thaw samples. For a reduced-sample control, add 1 µl of 1M DTT to the crosslinked sample and leave at RT for 5–10 min.
Add 10 µl 3xSDS sample buffer (no DTT). Place in a boiling water bath for 3 min. Note that the sample with added DTT will turn yellow-green and may have some precipitate, but it will run fine during SDS-PAGE. This control sample should be loaded on the gel last or separated from the nonreduced samples to avoid diffusion of DTT into the nonreduced sample lanes.
Load 10–15 µl of each sample on a PA gel (for 20S proteasome subunit crosslinking, a 10% PA gel is typically used; for RP ATPase crosslinking, an 8% PA gel is appropriate). Electrophorese the samples until the dye front is at the bottom of the gel.
Electroblot proteins from the gel to a PVDF (Millipore) membrane. Crosslinked proteins often run slower than expected from their predicted masses (see Fig. 1), and transfer tends to be less efficient than for the uncrosslinked monomers. Using an Idea Scientific Genie blotting apparatus, transfer for 1–2 h at 20–25V (constant voltage) at 4°C is usually satisfactory.
Process membrane for Western immunoblot analysis using standard methods. An example of an immunoblot demonstrating formation of an engineered disulfide bond between two proteasomal ATPases is shown in Fig. 1.
3.2 Disulfide crosslinking of immuno-purified proteasomal complexes
For protein complexes that are present at low levels, such as many proteasome assembly intermediates, it is sometimes necessary to concentrate the complexes from larger cultures prior to crosslinking. This may require a means to separate the specific complex of interest from related species. For analysis of proteasome assembly intermediates, for example, specific assembly chaperones are only associated at early stages of assembly; thus, an affinity purification based on binding to one of these factors can be used to separate the desired species from later intermediates or mature proteasomes. The following protocol has been used successfully with a 3xFLAG-tagged RP assembly chaperone, Nas2, which dissociates relatively early in RP assembly (5). The purification starts with ~300 OD600 equivalents of yeast cells.
3.2.1 Digest cell wall and lyse spheroplasts
Overnight yeast cultures are grown as in Section 3.1.1, but are diluted into 200 ml of fresh medium the following morning. Cultures are grown until they reach an OD600 of ~2 and harvested by centrifugation for 5 min at 4,200×g (4°C) in a floor centrifuge (Sorvall RC-5B or similar).
Resuspend cells in 50 ml of ice-cold water, transfer to a 50 ml conical tube and centrifuge for 5 min at 1,500 ×g (4°C) in a tabletop centrifuge (Sorvall RT6000B or similar). Cell pellets can be stored at 4°C overnight.
Add 1 ml of ZB containing freshly added 30 mM DTT and resuspend cells. Leave at room temperature for 15–20 min.
Divide the cell suspension between two 1.5 ml Eppendorf tubes and centrifuge for 30 sec in a microcentrifuge.
Resuspend cells in each tube in 1 ml ZB to which 0.12 ml of 5 mg/ml Zymolyase 100T had been freshly added (0.6 mg/ml final concentration). Divide cells from the original two tubes between three new 1.5 ml Eppendorf tubes (roughly 0.75 ml each) and rotate at 30°C for 1 h.
Centrifuge spheroplasts in a microcentrifuge for 5 min at 1,200×g at room temperature, add 1 ml ZB, and repeat centrifugation step. Spheroplast pellets can be saved at this point at 4°C. (see Note 5)
3.2.2 Immunoaffinity purification of complexes
Suspend spheroplasts in each tube in 1 ml of ice-cold LB2 to which 2 mM ATP and protease inhibitors were freshly added. Use a sterile applicator stick or pipette tip to stir up the cells.
Lyse spheroplasts with three 30 sec vortexing steps, leaving the tubes on ice for 1 min in between.
Pellet cell debris with a 15 min centrifugation at 15,000×g at 4°C.
Carefully remove the supernates. Combine supernates if the levels of the desired protein complex are expected to be low.
For each 1 ml of lysate, add 50 µl of a 50% slurry of anti-FLAG M2 agarose beads (Sigma). Rotate for 2 h at 4°C.
Pellet the beads by centrifugation for 5 min at 1,500×g at 4°C and wash with 1 ml LB2 (containing ATP and protease inhibitors). Save the original supernates. Repeat the washing step two additional times, and carefully pull off as much of the liquid as possible after the final centrifugation.
To the beads, add 50 µl of a 200 µg/ml 3xFLAG peptide solution in LB2 (containing ATP and protease inhibitors). Rotate end-over-end for 30 min at 4°C.
After pelleting the beads, carefully remove 50 µl of eluate.
3.2.3 CuCl2-induced disulfide crosslinking of purified complexes
Add 1 µl of 10 mM CuCl2 to each eluate (0.2 mM final). Do the same for 50 µl of the supernate of step 6 in section 3.2.2 (0.5% of input).
Incubate at room temperature for 20–30 min.
Add 15 µl of 3X loading buffer (nonreducing) and heat in boiling water bath for 3 min. Samples can be stored at −20°C or −80°C or can be used immediately.
SDS-PAGE and immunoblotting are carried out as in Section 3.1.
3.3 Native PAGE–nonreducing SDS PAGE crosslinking analysis of active 26S proteasomes
Detailed protocols are provided for native PAGE using a cell lysis protocol involving liquid nitrogen freezing of cell pellets and grinding them to a fine powder. This lysis method largely follows that of Verma et al. (15). The native PAGE protocol is modified from previous procedures (16) (17) (18). The disulfide crosslinking methodology for intact 26S proteasome analysis was developed in two earlier studies (4) (5).
3.3.1 Preparation of cell extract by freezing cells in liquid N2 and grinding into powder
Harvest cells by centrifugation from a 1-liter yeast culture grown to OD600=2.
Resuspend the cells in 25–50 ml of ice-cold water and transfer to a 50 ml conical tube. Centrifuge cells and discard the supernate. The cell pellet can be stored at this point in the tube by flash-freezing in liquid nitrogen and storing at −80°C (pellet is fine for at least 2–3 mo.).
Pour liquid nitrogen into a 12 cm-wide porcelain mortar until fully chilled and filled partly with N2. Using a spatula, scoop the cell paste from the pellet obtained in step 2 into the liquid nitrogen in the mortar. (see Note 6).
Using a liquid nitrogen-cooled pestle, grind the frozen cell paste to a fine powder. Pour fresh liquid nitrogen into the mortar every 2 min to keep the cell powder frozen. It takes 20–30 min to obtain a fine powder. The consistency should be similar to flour. (see Note 7)
Scoop the cell powder using a liquid nitrogen-cooled spatula into liquid nitrogen-cooled 2-ml screw-cap plastic cryotubes (tubes can be filled to the top). Powder can be stored at -80°C for many months.
3.3.2 Extract preparation from pulverized frozen cells
Transfer approximately 0.1 ml of cell powder by tapping from the liquid nitrogen-chilled screw-capped tube into a liquid nitrogen-chilled 1.5 ml Eppendorf tube that had then been placed on ice. Transfer powder immediately after chilling the tubes to avoid thawing during transfer.
Add one volume (0.1 ml) of cell extraction buffer (CEB) to the powder. Vortex on high to mix and speed thawing. Incubate the suspension on ice for 10 min with occasional vortexing to improve protein yield.
Centrifuge the suspension at 15,000×g at 4°C for 10 min in a microcentrifuge and transfer the supernate to a fresh 1.5 ml tube on ice.
Calculate the protein concentration of the extract from a small aliquot (BIO RAD Bradford assay or similar). The concentration is typically 5–20 mg/ml.
Add 5x native loading buffer to remainder of extract.
3.3.3 Native polyacrylamide gel electrophoresis (PAGE) and substrate overlay assay
Set up the native PA gel (1.5-mm thick) as described in Section 2.1 (see Note 8).
Load equal amounts of protein in each lane. 25–50 µg of total protein per lane is typical.
Run the gel at 100 V at 4°C. It takes about 2.5 h for the dye to reach the bottom of the mini-gel.
Carefully transfer the gel from the glass plates in which it was sandwiched to a plastic container containing Overlay Buffer (OB) containing Suc-LLVY-AMC substrate. The stacking gel should first be removed using a razor blade. (see Note 9).
Incubate at 30°C for 30 min with occasional manual rocking.
Photograph the gel on a UV Transilluminator to visualize the active 26S proteasome species.
3.3.4 Disulfide crosslinking and second dimension non-reducing SDS gel separation
Place the gel into a glass dish containing 50 ml of crosslinking buffer without CuCl2. Incubate for 5 min at room temperature with occasional swirling.
Remove the buffer completely. Add 50 ml of crosslinking buffer containing 0.2 mM CuCl2 and incubate at room temperature for 30 min with occasional swirling.
Discard the crosslinking solution and add 50 ml of crosslinking buffer that lacks CuCl2.
Add 5 ml of 10X Stop Solution (SS) to the buffer and incubate for 5 min.
Discard the buffer and add another 50 ml of crosslinking buffer without CuCl2.
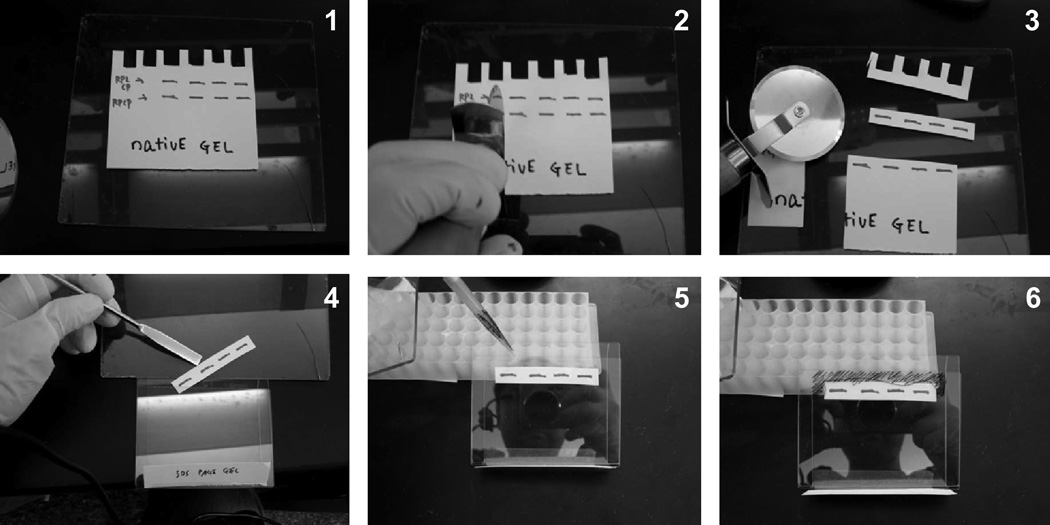
Transfer the gel to a glass plate. Using a UV transilluminator, remove the section of the gel containing the active 26S proteasomes (doubly capped or RP2CP form) using a pizza cutter (see Fig. 2). Note that the AMC signal generated from Suc-LLYV-AMC cleavage gets weaker as a result of the crosslinking.
Using a spatula, place the excised gel slice on top of a nonreducing SDS gel (1.5 mm thick). Carefully pour in the molten gel slice-packing solution around the gel slice and allow the packing solution to solidify (Fig. 2).
Subsequent steps of gel electrophoresis, protein electrotransfer to PVDF, and immunodetection follow standard procedures.
Fig 2.
A simulation of native gel excision with a pizza cutter and transfer of the gel piece to the top of a second-dimension nonreducing SDS gel. All steps would normally be done on a UV transilluminator except the step of adding of molten packing solution (5, 6).
Acknowledgements
We thank Robb Tomko and Mary Kunjappu for critical reading of the manuscript. Work from our laboratory that led to the development of these crosslinking methods was supported by NIH grants GM046904 and GM083050.
Footnotes
If no stacking gel is used, the sucrose should be omitted. Because the acrylamide content is very low (usually 4%), these gels are prone to slow leaks when poured. It is helpful to first seal the bottom where the plates meet the rubber pad with some molten agarose. Plates also need to be thoroughly cleaned and dried. Clean first with distilled water and then with 95% ethanol to assure complete drying. When cleaning the plates, do not use paper towels that might leave fibers, especially if fluorescent substrate overlay assays will be performed: small fibers and particles left by these materials can fluoresce under the UV light and may distort the migration of bands.
Other nondenaturing cell lysis procedures can also be used. An alternative protocol that uses mortar-and-pestle grinding of liquid nitrogen-frozen cells is provided in Section 3.3. Note that spheroplasts, having lost their cell wall, are relatively fragile and should not be vortexed at high speed or pipetted up and down too vigorously.
LB1 was designed for crosslinking with 20S proteasomes, which are very stable structures and not sensitive to ATP. LB2 is used for crosslinking 19S RP subunits, which may be sensitive to ATP and might also be sensitive to the detergent used in LB1. Although no ATP is present in LB2, it may be useful to add it (or a nonhydrolyzable derivative) for some experiments.
Other oxidizing agents that catalyze disulfide bond formation can also be used. An oxidant that has also been used successfully for engineered disulfide crosslinking of proteasome subunits is aqueous iodine used at 0.5 mM (3). A 25 mM stock solution (F.W. 253.8; Sigma) dissolved in 100% ethanol and stored at room temperature in the dark is recommended.
Because large numbers of cells have been concentrated into a small number of tubes, these cell pellets are large. A sterile toothpick or pipette tip can be used to facilitate their resuspension.
When generating cell powder, it is very important to pre-cool with liquid nitrogen everything that will come in contact with the cells or powder in order to prevent them from sticking to the equipment. It is not necessary to keep everything submerged in liquid nitrogen as you work, but it is important to re-cool items periodically either by submersing in liquid nitrogen or in the case of the mortar, filling it part-way with liquid nitrogen.
An alternative way to freeze the cells is to submerge the 50 ml Falcon tube with the frozen cell pellet into liquid nitrogen in a Dewar flask. Once it is cold, wrap the tube in a paper towel, and bang it against the benchtop to fracture the tube. Pick the frozen cell pieces out of the plastic and drop into the chilled mortar.
When grinding the cells, pressing harder does not result in a finer powder, and in fact, pressing too hard can reduce its quality. Allow the weight of the pestle do the grinding, and instead concentrate on a high number of rotations per minute with the pestle. It is useful to press harder onto the pellet for the first full minute or two of grinding to reduce the particle size more rapidly. After that, a gentler stroke with high-speed circular motions should be used.
If the sample has a volume of less than 20 µl, a stacking gel might not be required. If the sample volume is large or the sample is a purified protein, a stacking gel generally gives sharper bands.
The native gel is soft, fragile, and sticky. After carefully prying off one glass plate and removing the stacking gel with a razor blade, an OB-wetted plastic plate separator tool can be used to score the edges of the gel touching the plate spacers (to which the gel tends to stick) and then to fold the gel onto itself (top over bottom) and gently nudge into the buffer. A stream of OB from a squirt bottle can also be used to ease the gel from the plate into the buffer.
References
- 1.Chen P, Hochstrasser M. Autocatalytic subunit processing couples active site formation in the 20S proteasome to completion of assembly. Cell. 1996;86:961–972. doi: 10.1016/s0092-8674(00)80171-3. [DOI] [PubMed] [Google Scholar]
- 2.Arendt CS, Hochstrasser M. Identification of the yeast 20S proteasome catalytic centers and subunit interactions required for active-site formation. Proc. Natl. Acad. Sci. U.S.A. 1997;94:7156–7161. doi: 10.1073/pnas.94.14.7156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Velichutina I, Connerly PL, Arendt CS, et al. Plasticity in eucaryotic 20S proteasome ring assembly revealed by a subunit deletion in yeast. Embo J. 2004;23:500–510. doi: 10.1038/sj.emboj.7600059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kusmierczyk AR, Kunjappu MJ, Funakoshi M, Hochstrasser M. A multimeric assembly factor controls the formation of alternative 20S proteasomes. Nat Struct Mol Biol. 2008;15:237–244. doi: 10.1038/nsmb.1389. [DOI] [PubMed] [Google Scholar]
- 5.Tomko RJ, Jr, Funakoshi M, Schneider K, et al. Heterohexameric ring arrangement of the eukaryotic proteasomal ATPases: implications for proteasome structure and assembly. Mol Cell. 2010;38:393–403. doi: 10.1016/j.molcel.2010.02.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hochstrasser M. Ubiquitin-dependent protein degradation. Annual Review of Genetics. 1996;30:405–439. doi: 10.1146/annurev.genet.30.1.405. [DOI] [PubMed] [Google Scholar]
- 7.Glickman MH, Ciechanover A. The ubiquitin-proteasome proteolytic pathway: destruction for the sake of construction. Physiological Reviews. 2002;82:373–428. doi: 10.1152/physrev.00027.2001. [DOI] [PubMed] [Google Scholar]
- 8.Pickart CM, Cohen RE. Proteasomes and their kin: proteases in the machine age. Nat Rev Mol Cell Biol. 2004;5:177–187. doi: 10.1038/nrm1336. [DOI] [PubMed] [Google Scholar]
- 9.Marques AJ, Palanimurugan R, Matias AC, et al. Catalytic mechanism and assembly of the proteasome. Chem Rev. 2009;109:1509–1536. doi: 10.1021/cr8004857. [DOI] [PubMed] [Google Scholar]
- 10.Chen P, Hochstrasser M. Biogenesis, structure, and function of the yeast 20S proteasome. EMBO J. 1995;14:2620–2630. doi: 10.1002/j.1460-2075.1995.tb07260.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Groll M, Ditzel L, Löwe J, et al. Structure of 20S proteasome from yeast at 2.4 Å resolution. Nature. 1997;386:463–471. doi: 10.1038/386463a0. [DOI] [PubMed] [Google Scholar]
- 12.Fu H, Reis N, Lee Y, et al. Subunit interaction maps for the regulatory particle of the 26S proteasome and the COP9 signalosome. EMBO Journal. 2001;20:7096–7107. doi: 10.1093/emboj/20.24.7096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zhang F, Hu M, Tian G, et al. Structural insights into the regulatory particle of the proteasome from Methanocaldococcus jannaschii. Mol Cell. 2009;34:473–484. doi: 10.1016/j.molcel.2009.04.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Djuranovic S, Hartmann MD, Habeck M, et al. Structure and activity of the N-terminal substrate recognition domains in proteasomal ATPases. Mol Cell. 2009;34:580–590. doi: 10.1016/j.molcel.2009.04.030. [DOI] [PubMed] [Google Scholar]
- 15.Verma R, Chen S, Feldman R, et al. Proteasomal proteomics: identification of nucleotide-sensitive proteasome-interacting proteins by mass spectrometric analysis of affinity-purified proteasomes. Mol Biol Cell. 2000;11:3425–3439. doi: 10.1091/mbc.11.10.3425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hough R, Pratt G, Rechsteiner M. Purification of two high molecular weight proteases in rabbit reticulocyte lysate. J. Biol. Chem. 1987;262:8303–8313. [PubMed] [Google Scholar]
- 17.Glickman MH, Rubin DM, Fried VA, Finley D. The regulatory particle of the Saccharomyces cerevisiae proteasome. Mol. Cell. Biol. 1998;18:3149–3162. doi: 10.1128/mcb.18.6.3149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Elsasser S, Schmidt M, Finley D. Characterization of the proteasome using native gel electrophoresis. Methods Enzymol. 2005;398:353–363. doi: 10.1016/S0076-6879(05)98029-4. [DOI] [PubMed] [Google Scholar]