Abstract
Programmed cell death 4 (Pdcd4) is a novel tumor suppressor, whose expression is frequently down-regulated in several types of cancers. In the present study, we demonstrated that Pdcd4 knockdown up-regulates MAP kinase kinase kinase kinase 1 (MAP4K1) expression and increases phosphorylation of c-Jun. Over-expression of c-Myc in HEK293 cells increases the levels of MAP4K1, MAP4K1 promoter activity, and phospho-c-Jun. Mutation analysis showed that the c-Myc binding site at −536 bp (relative to the initiation ATG) of map4k1 promoter responds to c-Myc regulation. In addition, chromatin immunoprecipitation demonstrated that c-Myc directly binds to map4k1 promoter at this site. Down-regulation of c-Myc reverses MAP4K1 expression and AP-1 activation in Pdcd4 knockdown cells. Moreover, over-expression of dominant negative Tcf4 decreases expression of c-Myc and MAP4K1, JNK activation, and AP-1 dependent transcription. Thus, activation of β-catenin/Tcf dependent transcription in Pdcd4 knockdown cells up-regulates MAP4K1 expression and AP-1 activity via c-Myc. The study presented here further reveals in detail the mechanism of how Pdcd4 inhibits tumor cell invasion and provides a functional connection between β-catenin/Tcf and AP-1 dependent transcription.
Keywords: Pdcd4, MAP4K1, JNK signaling pathway, AP-1, c-Myc
1. Introduction
Programmed cell death 4 (Pdcd4) is a novel tumor suppressor that is frequently down-regulated in several types of cancers. The pdcd4 gene was first identified as a differentially expressed mRNA when cells were treated with apoptosis inducers [1]. Over-expression of Pdcd4 has been shown to induce apoptosis in breast MDA-MB-231 and hepatocellular carcinoma HCC cells [2,3]. In consistence with the induction of cell death, depletion of Pdcd4 promoted cell proliferation [4] and over-expression of Pdcd4 inhibited proliferation [5–7]. However, over-expression of Pdcd4 in human HEK293 and chicken DT40 cells had no effects on apoptosis or cell proliferation [8,9]. In addition, Eto et al. [10] showed that loss of Pdcd4 induced apoptosis in HeLa and C2C12 cells. Thus, the role of Pdcd4 in programmed cell death remains unclear.
Despite the action in programmed cell death, the inhibitory role of Pdcd4 in tumorigenesis has been clearly demonstrated in vitro and in vivo. Over-expression of pdcd4 cDNA inhibits 12-O-tetradecanoylphorbol-13-acetate (TPA)-induced transformation and tumor phenotype in mouse JB6 cells [11,12]. Pdcd4 transgenic mice that overexpress Pdcd4 in the epidermis show significant reductions in 7,12-dimethylbenz(a)anthracene (DMBA)/TPA induced skin papilloma formation and carcinoma incidence [13]. Conversely, knockout of Pdcd4 expression in mice increases DMBA/TPA induced skin papilloma formation and carcinoma occurrence [14]. In addition to inhibiting the tumor promotion stage, Pdcd4 has also been demonstrated to be involved in tumor progression stage. Overexpression of Pdcd4 inhibits colon, breast, and ovarian tumor cell invasion [5,15–18], while knockdown of Pdcd4 promotes tumor cell invasion [17,19,20]. Inhibition of tumor cell invasion by Pdcd4 is attributed, at least in part, by suppressing activator protein-1 (AP-1) dependent transcription that is through inhibiting the transactivation of c-Jun or c-Fos [8,12]. Pdcd4 blocks c-Jun activation by inhibiting the expression of mitogen-activated protein kinase kinase kinase kinase 1 (MAP4K1) (also known as hematopoietic progenitor kinase 1), a kinase upstream of Jun N-terminal kinase (JNK) [18]. MAP4K1 is a mammalian STE-20-like protein serine/threonine kinase which regulates the JNK signaling pathway [21]. MAP4K1 activates JNK through the signaling pathway MAP4K1 →TAKl →MKK4→JNK [22] and does not affect other MAPK signaling pathways, including the ERK and p38 signaling pathways [23]. MAP4K1 is involved in the stress response, proliferation, and apoptosis of hematopoietic cells; however, the expression regulation and functions of MAP4K1 outside of the hematopoietic cells is poorly understood. We previously reported that over-expression of Pdcd4 suppressed MAP4K1 expression, with consequent inhibition of c-Jun activation and AP-1-dependent transcription [18]. In addition, ectopic expression of map4k1 cDNA enhanced c-Jun phosphorylation and activated AP-1 dependent transcription [18], suggesting that MAP4K1 plays a crucial role in c-Jun activation. However, how Pdcd4 regulates the expression of MAP4K1 remains unknown.
Recently, we demonstrated that knockdown of Pdcd4 expression in colon tumor GEO and HT29 cells led to a fibroblast-like morphological change and promoted invasion [19]. In addition, Pdcd4 knockdown resulted in down-regulation of E-cadherin expression, accumulation of β -catenin into the nuclei, and activation of β-catenin/Tcf and AP-1 dependent transcription [19]. Promoting tumor cell invasion by Pdcd4 knockdown was contributed at least in part by c-Myc elevation since knockdown of c-Myc inhibited invasion induced by Pdcd4 knockdown [20]. c-Myc, a proto-oncogene encoding transcription factor, frequently up-regulated protein in all types of human cancers, whose expression is highly correlated with high-grade premalignancy and invasive tumors [24]. c-Myc is an essential protein for embryogenesis and is involved in cell migration, invasion, and metastasis [24,25]. Being a transcription factor, c-Myc is able to function as transcription activator or repressor depending on the recruiting factors [26]. For example, c-Myc can stimulate cyclin D2 and cyclin-dependent kinase 4 expression as repressed p27KIP1 expression for promoting cell cycle progression [24]. In addition, c-Myc also globally influences chromatin structure and affects genetic program [27],
In this study, we provide mechanistic insights of how c-Myc, a target of β-catenin/Tcf dependent transcription, regulates the expression of MAP4K1 and activation of AP-1 dependent transcription.
2. Materials and methods
2.1. Tissue culture
The colon GEO (a gift from Dr. Douglas Boyd, MD Anderson Cancer Center, Houston, TX, USA) and HT29 cells (American Type Culture Collection, ATCC, Manassas, VA) were grown in McCoy's medium containing 10% FBS, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. HEK293 cells were purchased from ATCC and were grown in DMEM medium containing 10% FBS, 2 mM l-glutamine, and 100 U/ml penicillin-streptomycin. Cells were incubated at 37 °C in a humidified atmosphere of 5% CO2 in air.
2.2. Western blot analysis
Aliquots containing 20 to 40 µg of protein were separated through SDS-PAGE, and transferred to nitrocellulose membranes as described previously [11]. Subsequently, the membrane was incubated with primary antibodies overnight followed by horseradish peroxidase-linked secondary antibody for 1 h. The target protein was visualized by chemiluminescence. The band intensity was quantified using VisionWork LS image acquisition and analysis software (UVP, Upland, CA). The following antibodies were used: MAP4K1 (1:200 dilution), Xpress (1:5000 dilution), c-Myc (1:1000 dilution), phospho-JNK (1:1000 dilution), phospho-c-Jun (Ser-73) (1:1000 dilution), phospho-ERK (1:1000 dilution),JNK (1:1000 dilution), ERK (1:1000 dilution), and c-Jun (1:1000 dilution). MAP4K1 antibody was purchased from Santa Cruz Biotechnology (Santa Cruz, CA) and Xpress antibody was from Life Technologies (Grand Island, NY), while the rest of the antibodies were purchased from Cell Signaling (Danvers, MA).
2.3. Site-specific mutagenesis
The −792 bp to −51 bp 5′-flanking region of human map4k1 promoter was generated by PCR and ligated into pGL3-basic vector (Promega, Madison, WI) as described previously [18]. The consensus sequence of the c-Myc binding site at −536 bp on the map4k1 promoter was mutated from CACGTG to TATATA (mutated nucleotides are underlined) using wild-type pMAP4K1(792)-LUC as the template and the produced mutant was named as 536m. The site-specific mutagenesis was performed using QuickChange II XL Site-Directed Mutagenesis kit (Agilent Technologies, Santa Clara, CA). Dominant negative Tcf4 construct (pcDNA4-dnTcf4) was generated using wild-type Tcf4 cDNA as the template. The first 92 bp were deleted using QuickChange II XL Site-Directed Mutagenesis kit (Agilent Technologies) and sub-cloned into pcDNA4/HisMax vector (Life Technologies). The wild-type Tcf4 cDNA was purchased from Origene (Rockville, MD). All constructs were verified by DNA sequencing.
2.4. Cell transfection and luciferase activity assays
For map4k1 promoter activity assays, 3×104 cells were transiently transfected with 0.2 µg of pMAP4Kl(792)-LUC (or 536m) along with 10 ng of pRL-SV40 using jetPRIME transfection reagent (Polyplus-Transfection Inc., New York, NY). For the specificity assay, 0.1 µg of pcDNA, pCMV-Myc, or pCMV-β-gal and 0.2 µg of pMAP4K1(792)-LUC were transfected along with 10 ng of pRL-SV40 using jetPRIME transfection reagent as above. For AP-1 dependent transcription assays, 3×104 cells were transfected with various amounts of dnTcf4 expression plasmid (pcDNA-dnTcf4) and 0.2 µg of 4× AP-1-Luc plasmid [11] and 10 ng of pRL-SV40 plasmid. After 48 h, the cells were lysed in 1 × lysis buffer (Promega) and the luciferase activity was determined as previously described [20].
For over-expression of c-Myc, 2 µg or otherwise indicated of c-Myc expression plasmid (Origene) was transiently transfected into HEK293 cells (2×105 cells/60 mm dish) using Fugene HD reagent (Promega). After 72 h, cells were collected for RNA extraction or cell lysates.
2.5. Real-time PCR (qPCR)
The total RNA isolation and real-time PCR were performed as described previously [19]. Briefly, after synthesis of the first strand cDNA using the Superscript First-Strand Kit (Invitrogen), mRNA levels of map4k1, c-myc, or GAPDH were quantified by real-time PCR in a LightCycler 480 (Roche Applied Science, Indianapolis, IN). The PCR cycling was performed at 95 °C for 6 min followed by 40 cycles of denaturation (95 °C for 15 s), annealing (61 °C for 30 s), and extension (72 °C for 20 s). To determine the specificity of the PCR, the amplified products were subjected to melt-curve analysis using the standard machine method. The target mRNA level was normalized to the internal control, GAPDH, using the formula ΔCT = CT (target) - CT (GAPDH) (CT: threshold cycle). The level of target gene expression in control cells was designated as 100%. The relative expression levels were calculated using the equation 100×2−[average ΔCT (test)−average ΔCT (control)] [28]. The primers used for amplifying map4k1, c-myc, and GAPDH were purchased from SA Biosciences (Frederick, MD).
2.6. Chromatin immunoprecipitation (ChIP) assay
GEO-shLacZ and GEO-shPdcd4 cells as well as HT29-shLacZ and HT29-shPdcd4 cells were grown to 70–80% confluence and fixed with formaldehyde. ChIP assay was performed using ChlP-IT™ Express kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. The c-Myc antibody was used and the preimmune mouse serum was used as the negative control. The immunoprecipitated DNA was quantitated by qPCR. The following primers were used to amplify human map4ld promoter: forward, 5′-CATCTGCCTGGATACCTGTG; reverse, 5′-TCTCCATTAGGTTCCGGTCT. The input DNA not subjected to immunoprecipitation was used as control in PCR reaction. PCR products were analyzed onto 2% agarose/Tris-borate EDTA gels.
2.7. Knock-down of c-Myc and expression of dnTcf4
GE0-shPdcd4 cells (5 × 105 cells on a 60 mm plate) were transfected with 4.4 µl of 10 µM siRNA (Santa Cruz Biotechnology) (or 0.8 µg of pcDNA-dnTcf4) along with 0.4 µg of pMACS Kk.II plasmid (Miltenyi Biotec, Auburn, CA) using jetPRlME transfection reagent (Polyplus-Transfection Inc.) according to the manufacturer's protocol. Twenty-four hours post-transfection, the transfected cells were enriched by H-2Kk antibody conjugated magnetic beads according to the manufacturer's protocol (Miltenyi Biotec). The eluted cells were then cultured for additional 48 h for qPCR, Western blot, or transfection assays.
2.8. Statistical analysis
Statistical analyses were performed using one-way ANOVA (http://faculty.vassar.edu/lowry/anova1u.html). Data are shown as the mean±standard deviation (SD) with at least three replicates (n≥3). Differences were considered statistically significant at the P≤0.05 level. Each experiment was repeated at least twice to confirm the results.
3. Results
3.1. Pdcd4 knockdown stimulates MAP4K1 expression and activates the JNK signaling pathway
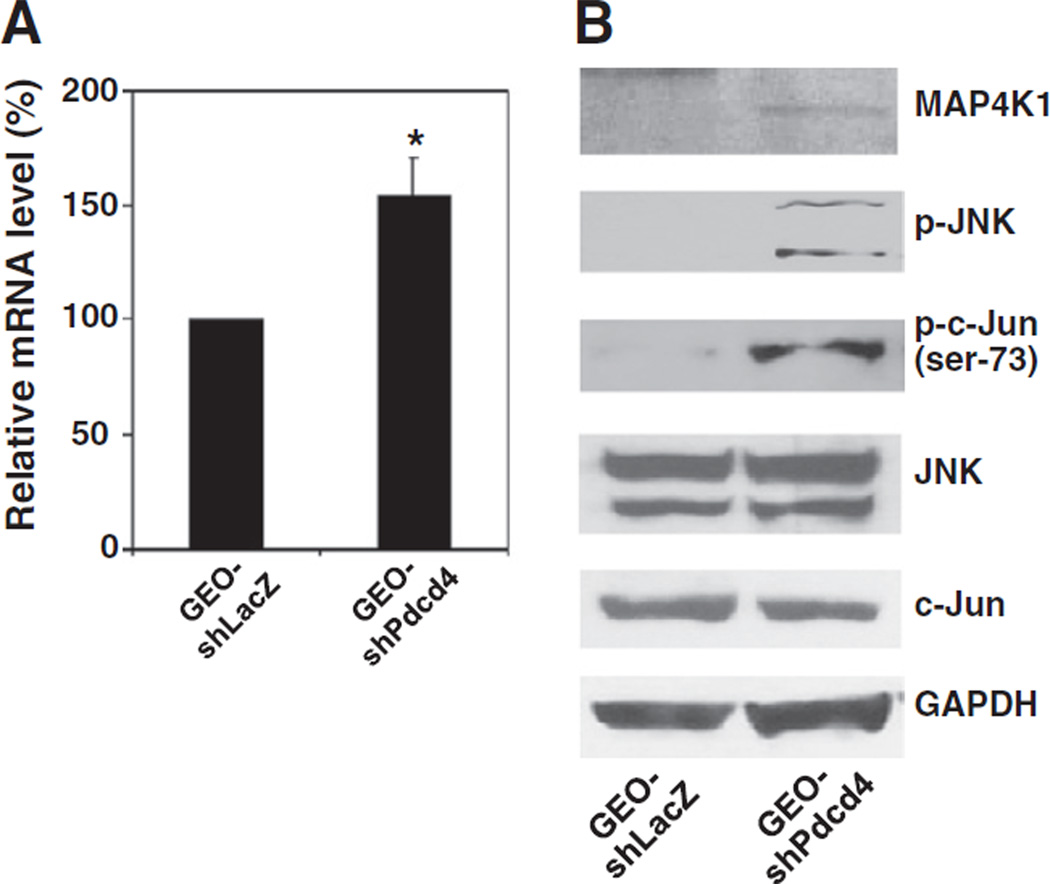
Previously, we have demonstrated that over-expression of Pdcd4 resulted in inhibition of MAP4K1 expression [18]. To test whether knockdown of Pdcd4 up-regulates MAP4K1 expression and activates JNK and c-Jun, the stable Pdcd4 knockdown (GEO-shPdcd4) and vector control (GEO-shLacZ) GEO cells were used [20]. The total mRNAs from these cells were isolated and the map4k1 mRNA levels were compared between GEO-shLacZ and GEO-shPdcd4 cells by qPCR using MAP4K1 specific primers. As shown in Fig. 1A, the map4k1 mRNA expression was increased by approximately 50% by Pdcd4 knockdown. In addition, the protein level of MAP4K1 also increased in GEO-shPdcd4 cells comparing to the GEO-shLacZ cells (Fig. 1B). MAP4K1 is a kinase upstream of JNK that regulates the activation of JNK [22]. In order to determine whether Pdcd4 knockdown activates the JNK signaling pathway, the Western blot analysis was performed. The phospho-JNK and phospho-c-Jun (ser-73) were expressed at a much higher level in GEO-shPdcd4 cells than in GEO-shLacZ cells in which they were barely detectable, whereas the levels of total JNK and c-Jun proteins were similar in GEO-shLacZ and GEO-shPdcd4 cells (Fig. 1B). These results suggest that Pdcd4 knockdown elevates MAP4K1 expression and activates the JNK signaling pathway. It is noteworthy that the level of phospho-c-Jun at Ser-63 is similar in both control and Pdcd4 knockdown cells [19]. Thus, Ser-73 is probably the primary phosphorylation site in the activated c-Jun in Pdcd4 knockdown cells. The differential phosphorylation at Ser-63 and Ser-73 of c-Jun has been reported previously [29].
Fig. 1.
Pdcd4 knockdown stimulates MAP4K1 expression and activates the JNK signaling pathway. (A) The mRNA level of map4k1 is up-regulated in Pdcd4 knockdown cells. The mRNA levels of map4k1 and GAPDH were determined by qPCR using total RNA isolated from GEO-shLacZ and GEO-shPdcd4 cells. The ratio of map4k1/GAPDH in GEO-shLacZ cells is designated as 100%. Three independent experiments were performed with triplicates for each sample. The data are shown and expressed as mean ± standard deviation (SD). The asterisk indicates a significant difference as determined by one-way ANOVA (P<0.01). (B) The protein level of MAP4K1 is increased and its downstream targets are activated in Pdcd4 knockdown cells. Western blot analysis was performed using antibodies against MAP4K1, phospho-JNK, phospho-c-Jun (ser73), JNK, c-Jun, and GAPDH.
3.2. Over-expression of c-Myc stimulates MAP4K1 expression and activates c-Jun
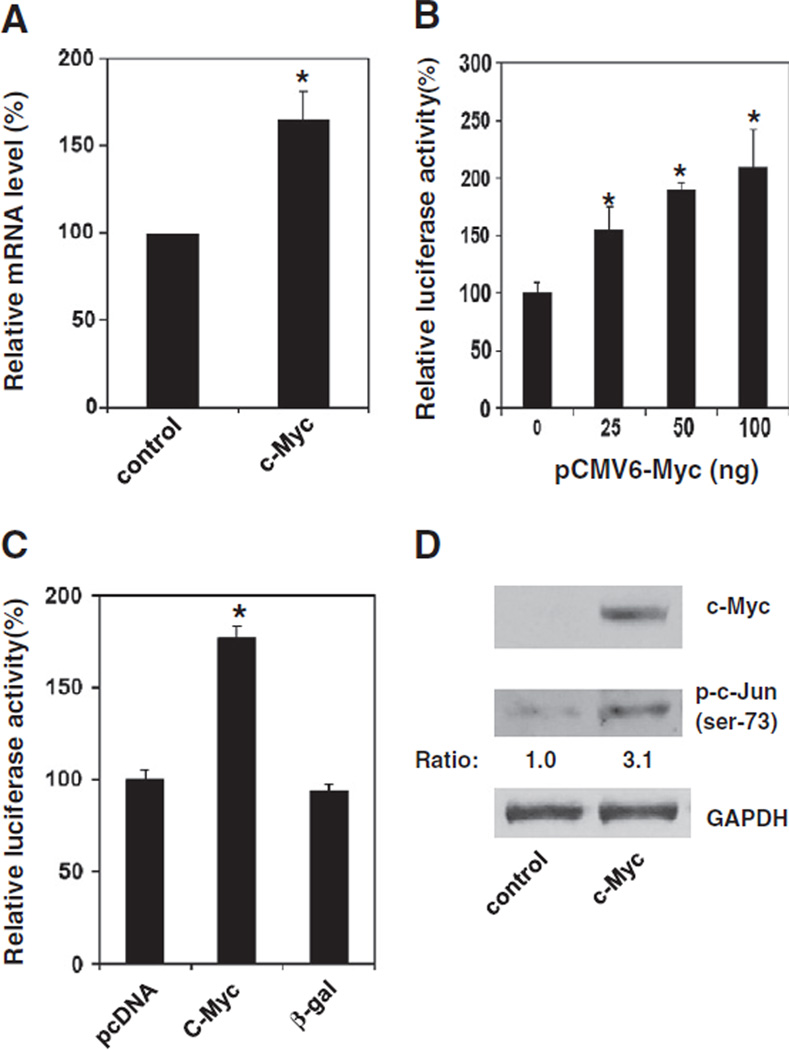
Recently, we reported that knockdown of Pdcd4 up-regulated c-Myc expression [20]. Since c-Myc regulates the expression of numerous genes, we thus hypothesize that up-regulation of c-Myc contributes to the stimulation of MAP4K1 expression. To test this, the c-Myc expression plasmid (pCMV6-Myc) was transiently transfected into HEK293 cells and the MAP4K1 expression and c-Jun activation were examined. After 72 h, the cells were harvested, and total RNA and cell lysate were prepared. The level of map4k1 mRNA was assayed by qPCR. As shown in Fig. 2A, the mRNA level of map4k1 is approximately 50% higher in the cells transfected with pCMV6-Myc (c-Myc) than in the cells transfected with empty vector (control). To test whether c-Myc also activates map4k1 promoter activity, the map4k1 promoter luciferase construct [pMAP4Kl(792)-LUC] was transfected along with pCMV6-Myc into HEK293 cells. After 48 h, cells were lysed and the luciferase activity was assayed. The −792 nt to −51 nt (relative to the initiation ATG) in the 5′ flanking region of human map4k1 promoter was amplified by PCR and ligated into pGL3-basic vector and named as pMAP4Kl(792)-LUC. Transient co-transfection of pCMV6-Myc and pMAP4Kl(792)-LUC plas-mids stimulated map4k1 promoter activity in a concentration dependent manner (Fig. 2B). The map4k1 promoter was activated by approximately 2-folds when 100 ng of pCMV6-Myc plasmid was transfected. Activation of map4k1 promoter by c-Myc is specific as transient expression of β-galactosidase did not stimulate map4k1 promoter activity (Fig. 2C). To test whether c-Myc regulates c-Jun activation, the level of phospho-c-Jun was analyzed by Western blot analysis using phospho-c-Jun (ser-73) antibody. The level of phospho-c-Jun was approximately 3-folds higher in c-Myc expressing cells than that in control cells (Fig. 2D), revealing that over-expression of c-Myc not only stimulates MAP4K1 expression but also activates c-Jun.
Fig. 2.
Over-expression of c-Myc stimulates MAP4K1 expression and activates c-Jun. (A) Over-expression of c-Myc increases map4k1 mRNA The mRNA levels of map4k1 and GAPDH were determined by qPCR using total RNA isolated from HEK293 cells transfected with either control or c-Myc expression plasmid for 72 h. The ratio of map4kl/GAPDH in cells transfected with the control vector is designated as 100%. Two independent experiments were performed with 3 replicates for each sample. The data are shown and expressed as mean±SD. The asterisk indicates a significant difference as determined by one-way ANOVA (P<0.01). (B) c-Myc enhances the promoter activity of MAP4K1. Increasing amounts (0–100 ng) of pCMV6-Myc plasmid and pMAP4Kl-LUC (0.2 µg) along with 10 ng of pRL-SV40 were transfected into HEK293 cells. The total DNA was maintained at 0.3 µg by adding the empty vector pcDNA 3.1 DNA The activity of cells transfected with 0 ng of pCMV6-Myc is designated as 100%. Three independent experiments were performed with 5 replicates for each sample. The represented data are shown and expressed as mean±SD (n=5). The asterisk denotes a significant difference compared to transfection with 0 µg of pCMV6-Myc as determined by one-way ANOVA (P<0.005). (C) Stimulation of map4k1 promoter by c-Myc is specific. The pcDNA pCMV-Myc, or pCMV-β-gal (0.1 µg) and pMAP4Kl-LUC (0.2 µg) along with 10 ng of pRL-SV40 were transfected into HEK293 cells as described in (B). The activity of cells transfected with pcDNA and pMAP4Kl-LUC is designated as 100%. The asterisk denotes a significant difference as determined by one-way ANOVA (P<0.005). (D) Over-expression of c-Myc elevates c-Jun phosphorylation. Cell lysate from HEK293 cells transfected with either control or c-Myc expression plasmid for 72 h was used. Western blot analysis was performed using antibodies against c-Myc, phospho-c-Jun (ser-73), and GAPDH. The ratio of phospho-c-Jun (ser-73)/GAPDH in control cells is designated as 1.0.
3.3. The c-Myc binding site at −536 of the map4k1 promoter responds to c-Myc regulation
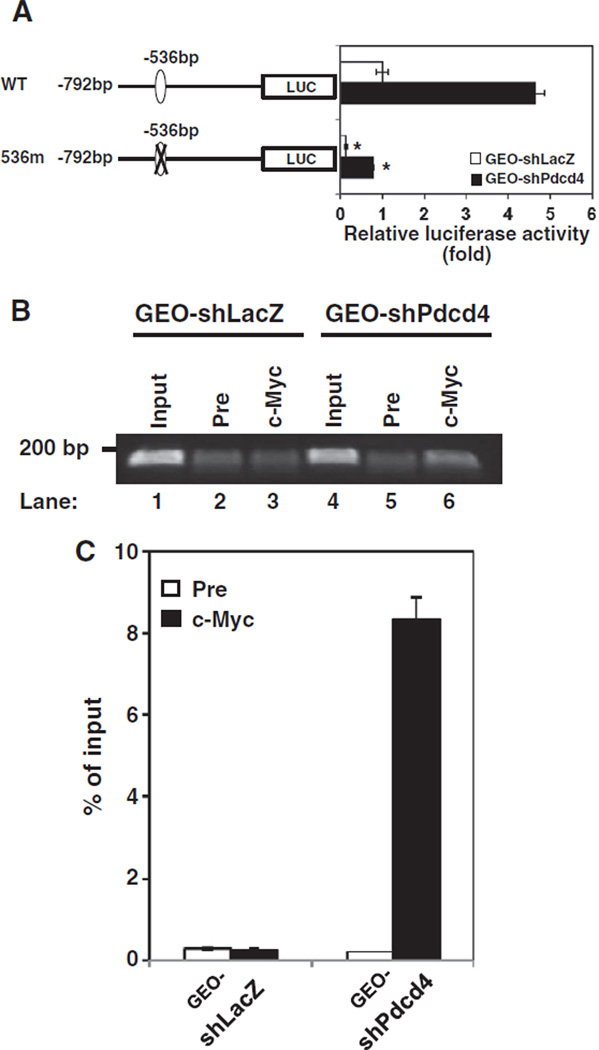
To investigate how MAP4K1 expression is regulated in the Pdcd4 knockdown cells, the pMAP4Kl(792)-LUC was transfected into GEO-shLacZ and GEO-shPdcd4 cells. The luciferase activity in GEO-shPdcd4 cells was approximately 4.5-folds of that seen in GEO-shLacZ cells (Fig. 3A, WT), indicating that map4k1 promoter activity ( −792 bp to −51 bp) is enhanced by Pdcd4 knockdown. This region of map4k1 promoter contains a potential c-Myc binding site located at −536 bp (Fig. 3A). To test whether this potential c-Myc binding site mediates the stimulation of map4k1 promoter activity in Pdcd4 knockdown cells, we mutated the c-Myc binding site and transfected the mutated construct (536m) into GEO-shLacZ and GEO-shPdcd4 cells. The 536m exhibited an approximately 6-fold reduction of map4k1 promoter activity in the GEO-shPdcd4 cells (Fig. 3A, filled bars). A similar reduction was also observed when the 536m was transfected into GEO-shLacZ cells (Fig. 3A, open bars), suggesting that this c-Myc binding site is essential for MAP4K1 expression. Transfection of 536m into HT29-shPdcd4 cells (HT29 cells with Pdcd4 knockdown) also showed a dramatic reduction of map4k1 promoter activity (Supplementary Fig. 1). Since the c-Myc protein level in GEO-shPdcd4 cells is approximately 2.5-folds higher than that in GEO-shLacZ cells [20], it is expected that more c-Myc molecules bind to the map4k1 promoter in GEO-shPdcd4 than in GEO-shLacZ cells. To test this, ChIP assays were performed to examine the binding of c-Myc to the map4k1 promoter using map4k1 primers which amplified the c-Myc binding site at position −536 bp. The input chromatin without immunoprecipitation was used as the control in PCR reaction. A high level of PCR products (185 bp) was observed using c-Myc antibody precipitated chromatin from lysates of GEO-shPdcd4 cells (Fig. 3B, lane 6), wherein the band intensity of the PCR product was approximately 20-folds higher than that of the pre-immune serum precipitated chromatin (Fig. 3C). However, using c-Myc antibody precipitated chromatin from GEO-shLacZ cell lysates, the same primers only generated a low level of PCR products (Fig. 3B, lane 3), whose band intensity is similar to that of the pre-immune serum precipitated chromatin (Fig. 3C). Similar results were also observed when c-Myc antibody was used to precipitate chromatin from HT29-shLacZ and HT29-shPdcd4 cell lysates (Supplementary Fig. 2). These results directly show that c-Myc binds to the map4k1 promoter to stimulate MAP4K1 expression in Pdcd4 knockdown cells.
Fig. 3.
The c-Myc binding site at−536 of the map4k1 promoter responds to c-Myc regulation. (A) Pdcd4 knockdown stimulates map4k1 promoter activity. The pMAP4Kl(792)-LUC (WT) or 536m promoter construct (0.2 µg) was transfected into GEO-shLacZ and GEO-shPdcd4 cells along with 10 ng of pRL-SV40. The activity of GEO-shLacZ cells transfected with WT is designated as 1. Three independent experiments were performed with 5 replicates for each sample. The represented data are shown and expressed as mean±SD (n=5). The asterisk denotes a significant difference compared with cells transfected with WT as determined by one-way ANOVA (P<0.0001). (B and C) c-Myc directly binds to the map4k1 promoter in Pdcd4 knock-down cells. ChIP assays were performed with cell lysates from either GEO-shLacZ or GEO-shPdcd4 cells using control preimmune IgG (Pre) or anti-c-Myc antibody (c-Myc). The DNAs from cell lysate (input) and ChIP enriched were quantified by qPCR using primers for amplifying the c-Myc binding site at −536 bp on the promoter of MAP4K1. The representative PCR products were resolved onto 2% agarose gels (B). The level of the target gene in immunoprecipitated DNA of each sample is compared to that in input chromatin, which is equivalent to 100% (C).
3.4. Down-regulation of c-Myc reverses MAP4K1 expression in Pdcd4 knockdown cells
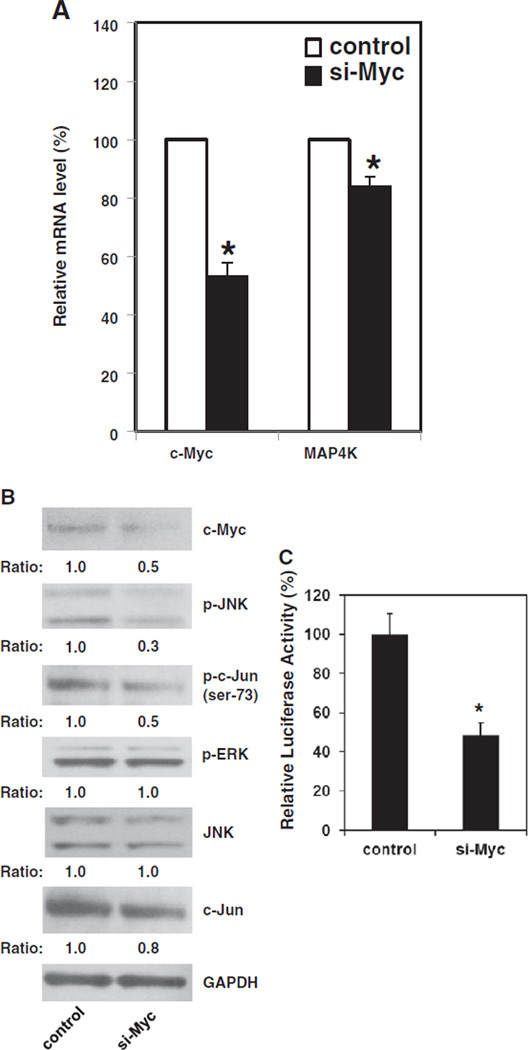
If c-Myc contributes to MAP4K1 expression in Pdcd4 knockdown cells, knockdown of c-Myc should inhibit MAP4K1 expression. In order to enhance the population of c-Myc knockdown cells, pMACS Kk.II plasmid was co-transfected with c-myc siRNA into GEO-shPdcd4 cells. The pMACS Kk.II plasmid produces a mouse MHC class I H-2Kk protein on the cell membrane with a truncated cytoplasmic domain. Twenty-four hours post-transfection, the cells with successful transfection were enriched by H-2Kk antibody conjugated magnetic beads. As shown in Fig. 4A, transient transfection of c-myc siRNA resulted in reduction of approximately 50% of c-myc mRNA The level of map4k1 mRNA is about 20% lower in the cells transfected with c-myc siRNA (si-Myc) than in the cells transfected with scramble siRNA (control), suggesting that down-regulation of c-Myc reverses MAP4K1 expression in Pdcd4 knockdown cells. Although knockdown of c-Myc inhibits about 20% of map4k1 mRNA expression, this inhibition is significantly enough to affect the activation of downstream targets. The level of phospho-JNK and phospho-c-Jun in the si-Myc cells was approximately 30% and 50% of that observed in the control cells, respectively (Fig. 4B). In contrast, the total JNK protein level was similar between control and si-Myc cells (Fig. 4B). In addition, the levels of phospho-ERK were similar between si-Myc and control cells, suggesting that knockdown of c-Myc did not affect the ERK signaling pathway. Interestingly, the total c-Jun protein level was slightly decreased in the si-Myc cells, which might be due to the feedback inhibition of c-Jun expression by inactivating c-Jun [30]. Moreover, knockdown of c-Myc also inhibited approximately 50% of AP-1 dependent transcription (Fig. 4C). These results suggest that c-Myc enhances MAP4K1 expression which contributes to the activation of JNK, c-Jun, and AP-1 dependent transcription in Pdcd4 knockdown cells.
Fig. 4.
Down-regulation of c-Myc reverses MAP4K1 expression in Pdcd4 knock-down cells. The cells transfected with c-myc siRNA (si-Myc) or scramble siRNA (control) along with pMACS K+.II plasmid were collected for extracting RNA or making cell ly-sates. The c-Myc knockdown cells were enriched by H-2Kk antibody conjugated beads as described in Materials and methods. (A) The mRNA level of map4k1 decreased in c-Myc knock-down cells. The total RNAs from control and si-Myc cells were reverse transcribed and subjected to qPCR. The ratio of c-myc/GAPDH and map4k1/GAPDH in control cells is designated as 100%. Two independent experiments were performed with 3 replicates for each sample. The data are shown and expressed as mean ± SD. The asterisk indicates a significant difference compared with control cells as determined by one-way ANOVA (P<0.05). (B) c-Myc knockdown decreases the phosphorylation of JNK and c-Jun (ser-73). The cell lysates from control and si-Myc cells were subjected to Western blot analysis using various antibodies as indicated. The ratio of target protein/GAPDH in control cells is designated as 1.0. (C) Knockdown of c-Myc inhibits AP-1 dependent transcription. The relative luciferase activity in control cells is designated as 100%. Three independent experiments were performed with 5 replicates for each sample. The represented data are shown and expressed as mean ± SD (n = 5). The asterisk indicates a significant difference as determined by one-way ANOVA (p<0.005).
3.5. β-catenin/Tcf dependent transcription regulates MAP4K1 expression, JNK activation, and AP-1 dependent transcription
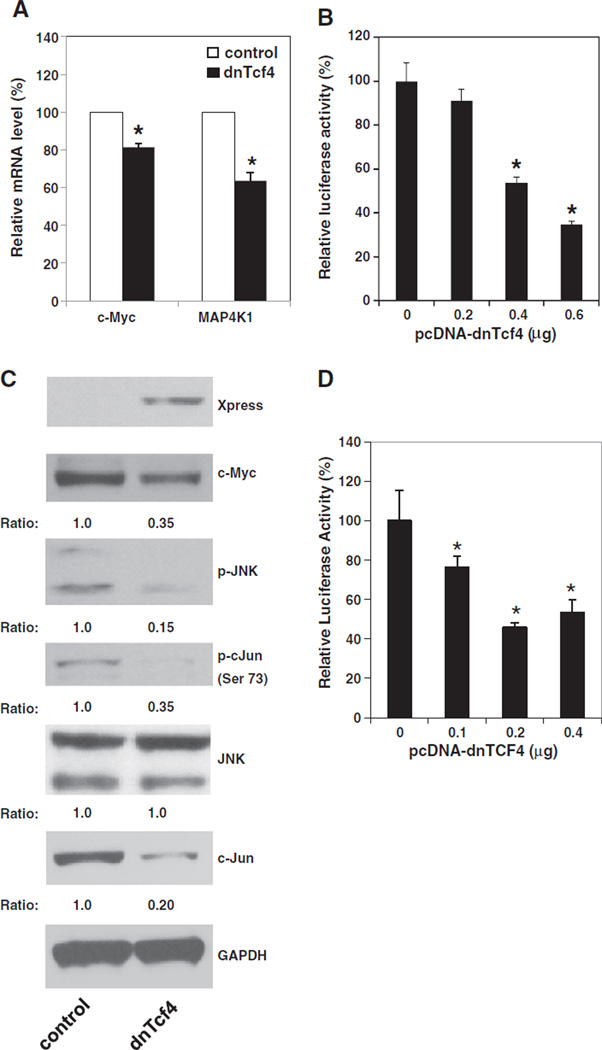
To study the functional significance of β-catenin/Tcf dependent transcription in regulating MAP4K1 expression, we tested whether dominant negative Tcf4 (dnTcf4) inhibits the expression of MAP4K1 and activation of the JNK signaling pathway. The dnTcf4 expression plasmid (pcDNA4-dnTcf4) and pMACS Kk.ll plasmids were transfected into GEO-shPdcd4 cells and the transfected cells were enriched by H-2Kk antibody conjugated beads. The mRNA levels of c-myc and map4k1 in control and dnTcf4 expressing (dnTcf4) cells were determined by qPCR The dnTcf4 is lacking the β-catenin interaction domain and expression of dnTcf4 has been known to inhibit the β -catenin dependent transcription [31]. As shown in Fig. 5A, over-expression of dnTcf4 cDNA decreased by approximately 20% and 40% in c-myc and map4k1 mRNA levels, respectively, indicating that dnTcf4 suppressed c-Myc and MAP4K1 expression. To further confirm that dnTcf4 regulates MAP4K1 expression, the pcDNA4-dnTcf4 was transfected along pMAP4Kl(792)-LUC into GEO-shPdcd4 cells. Expression of dnTcf4 inhibited map4k1 promoter activity in a dose dependent manner (Fig. 5B). The map4k1 promoter activity was reduced to approximately 35% when 0.6 µg of pcDNA4-dnTcf4 plasmid was transfected. To further investigate the effects of over-expressing dnTcf4 on the JNK signaling pathway, a series of Western blotting analyses was performed. The c-Myc protein expression was suppressed by about 3-folds by dnTcf4 (Fig. 5C). The levels of phospho-JNK and phospho-c-Jun in dnTcf4 cells were approximately 15% and 35% of that seen in control cells, respectively (Fig. 5C). The total JNK protein levels were similar between control and dnTcf4 cells while the total c-Jun levels were suppressed in the dnTcf4 cells. Moreover, over-expression of dnTcf4 cDNA resulted in the inhibition of AP-1 dependent transcription (Fig. 5D). Transfection of 0.2 or 0.4 µg of pcDNA4-dnTcf4 plasmid displayed approximately 50% inhibition. It has been known that c-Myc is a target of the β-catenin-dependent transcription in the Pdcd4 knockdown cells [20]. Thus, these results suggest that the β-catenin/Tcf4 complex regulates the c-Myc expression to mediate the MAP4K1 expression in Pdcd4 knockdown cells resulting in the change of JNK activity and AP-1 dependent transcription.
Fig. 5.
β-catenin/Tcf dependent transcription regulates MAP4K1 expression, JNK activation, and AP-1 dependent transcription. (A) Over-expression of dnTcf4 inhibits the expression of MAP4K1. GEO-shPdcd4 cells transfected with pcDNA4/Max (control) or pcDNA4-dnTcf4 (dnTcf4) plasmid along with pMACS K+.II plasmid were collected for extracting RNA or making cell lysates. The dnTcf expressing cells were enriched by H-2Kk antibody conjugated beads as described in Materials and methods. Total RNA was isolated and used in qPCR to examine the mRNA level of c-myc and map4k1. The ratio of c-myc/GAPDH and map4k1/GAPDH in cells transfected with pcDNA4/Max is designated as 100%. Two independent experiments were performed with 3 replicates for each sample. The data are shown and expressed as mean ± SD. The asterisk indicates a significant difference compared with control cells as determined by one-way ANOVA (P<0.05). (B) Over-expression of dnTcf4 inhibits map4k1 promoter activity. Indicated amount of pcDNA4-dnTcf4 plasmid was transfected into GEO-shPdcd4 cells while the total DNA for each transfection was maintained at 0.6 µg by adding pcDNA4/Max vector DNA. The activity of GEO-shPdcd4 cells transfected with 0 µg of dnTcf4 expression plasmid is designated as 100%. Three independent experiments were performed with 5 replicates for each sample. The represented data are shown and expressed as mean ± SD (n = 5). The asterisk denotes a significant difference compared with cells transfected with 0 µg of dnTcf4 expression plasmid as determined by one-way ANOVA (P<0.001). (C) Over-expression of dnTcf4 inhibits the activation of the JNK pathway. Western blot analysis was performed using the cell lysates from (A) with the indicated antibodies. The ratio of target protein/GAPDH in control cells is designated as 1.0. (D) Over-expression of dnTcf4 suppresses AP-1 dependent transcription. Indicated amount of pcDNA4-dnTcf4 plasmid was transfected into GEO-shPdcd4 cells while the total DNA for each transfection was maintained at 0.4 µg by adding pcDNA4/Max vector DNA The activity of GEO-shPdcd4 cells transfected with 0 µg of dnTcf4 expression plasmid is designated as 100%. Three independent experiments were performed with 5 replicates for each sample. The represented data are shown and expressed as mean ± SD (n = 5). The asterisk denotes a significant difference compared with cells transfected with 0 µg of dnTcf4 expression plasmid as determined by one-way ANOVA (P<0.01).
4. Discussion
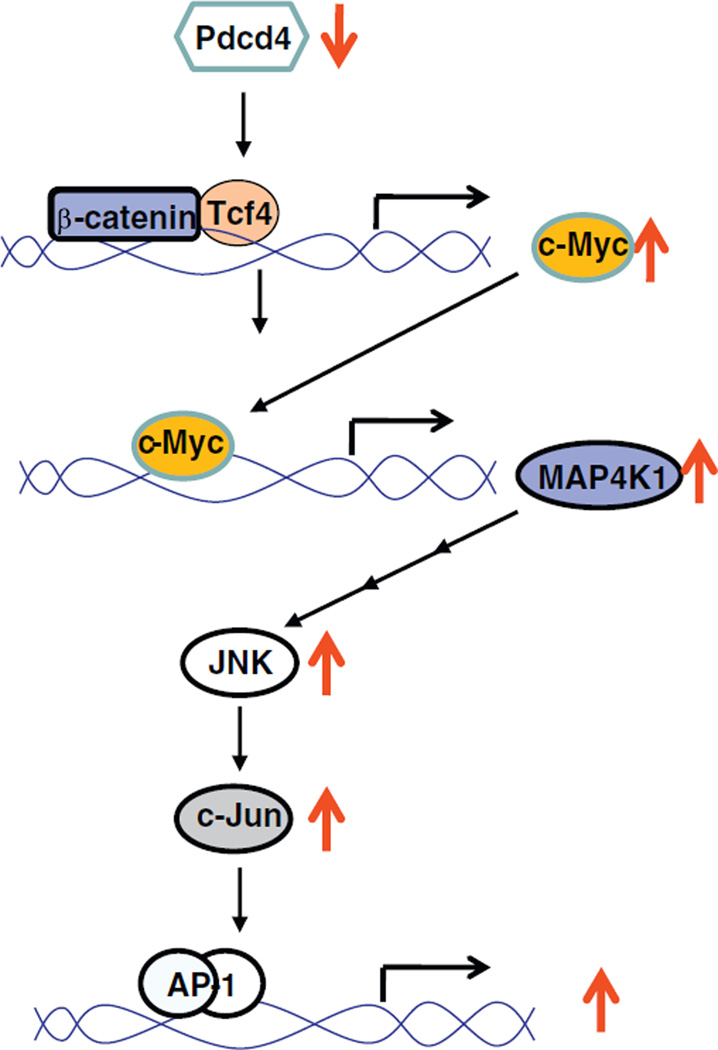
In this study, we demonstrated that MAP4K1 expression is upregulated by elevating c-Myc expression. Knockdown of c-Myc expression results in inhibition of MAP4K1 expression, c-Jun activation, and AP-1 dependent transcription. In addition, over-expression of dnTcf4 suppresses c-Myc and MAP4K1 expression as well as the activation of c-Jun and AP-1 dependent transcription. Our results suggest that activation of β-catenin/Tcf dependent transcription by Pdcd4 knockdown up-regulates MAP4K1 expression and activates the JNK signaling pathway through c-Myc (Fig. 6).
Fig. 6.
A model depicting the correlation between β-catenin and AP-1 dependent transcription in Pdcd4 knock-down cells.
Elevated or deregulated expression of c-Myc has been detected in a wide range of human cancers, and is often associated with aggressive, poorly differentiated tumors [24]. In human colon cancer, c-Myc expression is frequently elevated at both early and late stages of colon carcinogenesis [32]. Although it has been well understood that c-Myc regulates cell cycle progression and cell proliferation, several studies also implicated that c-Myc might be involved in tumor cell invasion and metastasis. For example, c-myc mRNA levels were higher in metastatic lesions than in primary lesions [33,34]. Over-expression of c-Myc can reverse the inhibitory effect of F box only protein 8 on tumor invasion [35]. In addition, we recently demonstrated that c-Myc contributes to colon tumor cell invasion induced by Pdcd4 knockdown because down-regulation of c-Myc results in inhibition of invasion in the Pdcd4 knockdown cells [20]. These findings suggest that c-Myc is an important regulator of tumor cell invasion.
How does c-Myc regulate colon tumor cell invasion? The present study extends this mechanistic understanding to now implicate that MAP4K1 expression is regulated by c-Myc. Knockdown of Pdcd4 up-regulates MAP4K1 expression (Fig. 1) and stimulates MAP4K1 promoter activity (Fig. 3). Over-expression of c-Myc increases MAP4K1 mRNA level (Fig. 2A), while c-Myc knockdown decreases MAP4K1 mRNA level (Fig. 4A). Mutation of the c-Myc binding site on the MAP4K1 promoter decreases the promoter activity dramatically (Fig. 3A and Supplementary Fig. 1). In addition, the regulation of MAP4K1 expression by c-Myc is further supported by the direct binding of c-Myc to the promoter region of MAP4K1 (Fig. 3B,C, and Supplementary Fig. 2). These findings collectively indicate that MAP4K1 expression is regulated by c-Myc. MAP4K1, a kinase three steps upstream of JNK, regulates JNK activation. In turn, JNK regulates the activation of c-Jun by phosphorylating it at Ser-63 and Ser-73 and subsequently activates AP-1 dependent transcription. Our data also suggest that regulation of the JNK signaling pathway by c-Myc is specific since knockdown of c-Myc did not affect ERK phosphorylation (Fig. 4B). Ectopic expression of dominant negative MAP4K1 in which methionine substituted the place of lysine 46 inhibits c-Jun phosphorylation, AP-1 dependent transcription, and invasion [18]. Conversely, overexpression of MAP4K1 cDNA increases phosphorylation of c-Jun [18]. AP-1 is a transcription factor complex composed of Jun-Jun homodimers or Jun-Fos heterodimers. The Jun protein family includes c-Jun, JunB, and JunD. The Fos protein family contains c-Fos, Fra-1, Fra-2, and FosB. Immunohistochemical studies of human colon cancer tissues revealed that c-Jun and Fra-1 expression is frequently elevated in adenoma, adenocarcinoma, and neuroendocrine carcinoma [36]. Activation of AP-1 activity by over-expression of Jun or Fos proteins enhances invasion and metastasis [30]. In addition, inhibition of AP-1 activity by the dominant negative c-Jun, TAM67, suppresses the invasive ability of a keratinocyte [37], fibroblast [38], and squamous carcinoma [39]. Thus, activation of AP-1 dependent transcription by Pdcd4 knockdown is likely to contribute, at least in part, to the promotion of colon tumor cell invasion through elevation of MAP4K1 expression, which is regulated by c-Myc.
The Tcf transcription factor family consists of four members, Tcf1, Tcf3, Tcf4, and LEF1. In the presence of Wnt signaling, β-catenin translocates into the nucleus and binds with a member of the Tcf family to form a β-catenin/Tcf complex resulting in the activation of transcription of the β-catenin/Tcf target genes, including c-Myc [40]. The finding that expression of dnTcf4 attenuates JNK phosphorylation, c-Jun phosphorylation, and AP-1 transactivation (Fig. 5) suggests that β-catenin/Tcf dependent transcription affects the JNK signaling pathway. On the other hand, it has been suggested that JNK/c-Jun regulates β-catenin/Tcf dependent transcription since the expression of Tcf4 is elevated in JNK1 transgenic mice while depletion of c-Jun expression significantly reduces Tcf4 expression [41]. These findings reveal a feedback mechanism of regulation between the Wnt signaling pathway and the JNK/c-Jun signaling pathway. It is noteworthy that the map4k1 mRNA level decreases more in dnTcf4 expressing cells (Fig. 5A) than in c-Myc knockdown cells (Fig. 4A). This finding suggests that other target(s) of β-catenin/Tcf dependent transcription besides c-Myc may regulate MAP4K1 expression, which needs to be further investigated.
In conclusion, our results show that elevation of c-Myc expression by Pdcd4 knockdown stimulates MAP4K1 expression resulting in the activation of JNK, and c-Jun, and transactivation of AP-1. These findings provide a molecular explanation of how Pdcd4 knockdown activates AP-1 dependent transcription and connects β-catenin/Tcf dependent transcription and AP-1 dependent transcription.
Supplementary Material
Acknowledgements
This study was supported by a National Institute of Health grant (RO1CA 129015).
Abbreviations
- Pdcd4
programmed cell death 4
- MAP4K1
mitogen-activated protein kinase kinase kinase kinase 1
- AP-1
activator protein-1
- TPA
12-O-tetradecanoylphorbol-13-acetate
- DMBA
7,12-dimethylbenz(a)anthracene
- TAK1
transforming growth factor β-activated kinase 1
- MKK4
mitogen-activated kinase kinase 4
- JNK
Jun N-terminal kinase
- ERK
extracellular signal-regulated kinases
- ChIP
chromatin immunoprecipitation
- siRNA
small interfering RNA
Footnotes
Appendix A. Supplementary data
Supplementary data to this article can be found online at http://dx.doi.org/10.1016/j.bbamcr.2012.07.004.
References
- 1.Shibahara K, Asano M, Ishida Y, Aoki T, Koike T, Honjo T. Isolation of a novel mouse gene MA-3 that is induced upon programmed cell death. Gene. 1995;166:297–301. doi: 10.1016/0378-1119(95)00607-9. [DOI] [PubMed] [Google Scholar]
- 2.Afonja O, Juste D, Das S, Matsuhashi S, Samuels HH. Induction of PDCD4 tumor suppressor gene expression by RAR agonists, antiestrogen and HER-2/neu antagonist in breast cancer cells. Evidence for a role in apoptosis. Oncogene. 2004;23:8135–8145. doi: 10.1038/sj.onc.1207983. [DOI] [PubMed] [Google Scholar]
- 3.Zhang H, Ozaki I, Mizuta T, Hamajima H, Yasutake T, Eguchi Y, Ideguchi H, Yamamoto K, Matsuhashi S. Involvement of programmed cell death 4 in transforming growth factor-beta1-induced apoptosis in human hepatocellular carcinoma. Oncogene. 2006;25:6101–6112. doi: 10.1038/sj.onc.1209634. [DOI] [PubMed] [Google Scholar]
- 4.Guo X, Li W, Wang Q, Yang HS. AKT activation by Pdcd4 knockdown up-regulates cyclin D1 expression and promotes cell proliferation. Genes Cancer. 2011;2:818–828. doi: 10.1177/1947601911431082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wei N, Liu SS, Chan KK, Ngan HY. Tumour suppressive function and modulation of programmed cell death 4 (PDCD4) in ovarian cancer. PLoS One. 2012;7:e30311. doi: 10.1371/journal.pone.0030311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wei ZT, Zhang X, Wang XY, Gao F, Zhou CJ, Zhu FL, Wang Q, Gao Q, Ma CH, Sun WS, Fu QZ, Chen YH, Zhang LN. PDCD4 inhibits the malignant phenotype of ovarian cancer cells. Cancer Sci. 2009;100:1408–1413. doi: 10.1111/j.1349-7006.2009.01210.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Nakashima M, Hamajima H, Xia J, Iwane S, Kwaguchi Y, Eguchi Y, Mizuta T, Fujimoto K, Ozaki I, Matsuhashi S. Regulation of tumor suppressor PDCD4 by novel protein kinase C isoforms. Biochim. Biophys. Acta. 2010;1803:1020–1027. doi: 10.1016/j.bbamcr.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 8.Bitomsky N, Bohm M, Klempnauer KH. Transformation suppressor protein Pdcd4 interferes with JNK-mediated phosphorylation of c-Jun and recruitment of the coactivator p300 by c-Jun. Oncogene. 2004;23:7484–7493. doi: 10.1038/sj.onc.1208064. [DOI] [PubMed] [Google Scholar]
- 9.Singh P, Marikkannu R, Bitomsky N, Klempnauer KH. Disruption of the Pdcd4 tumor suppressor gene in chicken DT40 cells reveals its role in the DNA-damage response. Oncogene. 2009;28:3758–3764. doi: 10.1038/onc.2009.239. [DOI] [PubMed] [Google Scholar]
- 10.Eto K, Goto S, Nakashima W, Ura Y. S.I. Abe, Loss of programmed cell death 4 induces apoptosis by promoting the translation of procaspase-3 rnRNA. Cell Death Differ. 2012;19:573–581. doi: 10.1038/cdd.2011.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yang HS, Jansen AP, Nair R, Shibahara K, Verma AK, Cmarik JL, Colburn NH. A novel transformation suppressor, Pdcd4, inhibits AP-1 transactivation but not NF-kappaB or ODC transactivation. Oncogene. 2001;20:669–676. doi: 10.1038/sj.onc.1204137. [DOI] [PubMed] [Google Scholar]
- 12.Yang HS, Knies JL, Stark C, Colburn NH. Pdcd4 suppresses tumor phenotype in JB6 cells by inhibiting AP-1 transactivation. Oncogene. 2003;22:3712–3720. doi: 10.1038/sj.onc.1206433. [DOI] [PubMed] [Google Scholar]
- 13.Jansen AP, Camalier CE, Colburn NH. Epidermal expression of the translation inhibitor programmed cell death 4 suppresses tumorigenesis. Cancer Res. 2005;65:6034–6041. doi: 10.1158/0008-5472.CAN-04-2119. [DOI] [PubMed] [Google Scholar]
- 14.Schmid T, Jansen AP, Baker AR, Hegamyer G, Hagan JP, Colburn NH. Translation inhibitor Pdcd4 is targeted for degradation during tumor promotion. Cancer Res. 2008;68:1254–1260. doi: 10.1158/0008-5472.CAN-07-1719. [DOI] [PubMed] [Google Scholar]
- 15.Leupold JH, Yang HS, Colburn NH, Asangani I, Post S, Allgayer H. Tumor suppressor Pdcd4 inhibits invasion/intravasation and regulates urokinase receptor (u-PAR) gene expression via Sp-transcription factors. Oncogene. 2007;26:4550–4562. doi: 10.1038/sj.onc.1210234. [DOI] [PubMed] [Google Scholar]
- 16.Nieves-Alicea R, Colburn NH, Simeone AM, Tari AM. Programmed cell death 4 inhibits breast cancer cell invasion by increasing tissue inhibitor of metalloproteinases-2 expression. Breast Cancer Res. Treat. 2009;114:203–209. doi: 10.1007/s10549-008-9993-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Santhanam AN, Baker AR, Hegamyer G, Kirschmann DA, Colburn NH. Pdcd4 repression of lysyl oxidase inhibits hypoxia-induced breast cancer cell invasion. Oncogene. 2010;29:3921–3932. doi: 10.1038/onc.2010.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang HS, Matthews CP, Clair T, Wang Q, Baker AR, Li CC, Tan TH, Colburn NH. Tumorigenesis suppressor Pdcd4 down-regulates mitogen-activated protein kinase kinase kinase kinase 1 expression to suppress colon carcinoma cell invasion. Mol. Cell. Biol. 2006;26:1297–1306. doi: 10.1128/MCB.26.4.1297-1306.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wang Q, Sun Z, Yang HS. Downregulation of tumor suppressor Pdcd4 promotes invasion and activates both beta-catenin/Tcf and AP-1 -dependent transcription in colon carcinoma cells. Oncogene. 2008;27:1527–1535. doi: 10.1038/sj.onc.1210793. [DOI] [PubMed] [Google Scholar]
- 20.Wang Q, Sun ZX, Allgayer H, Yang HS. Downregulation of E-cadherin is an essential event in activating beta-catenin/Tcf-dependent transcription and expression of its target genes in Pdcd4 knockdown cells. Oncogene. 2010;29:128–138. doi: 10.1038/onc.2009.302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma W, Xia C, Ling P, Qiu M, Luo Y, Tan TH, Liu M. Leukocyte-specific adaptor protein Grap2 interacts with hematopoietic progenitor kinase 1 (HPK1) to activate JNK signaling pathway in T lymphocytes. Oncogene. 2001;20:1703–1714. doi: 10.1038/sj.onc.1204224. [DOI] [PubMed] [Google Scholar]
- 22.Zhou G, Lee SC, Yao Z, Tan TH. Hematopoietic progenitor kinase 1 is a component of transforming growth factor beta-induced c-Jun N-terminal kinase signaling cascade. J. Biol. Chem. 1999;274:13133–13138. doi: 10.1074/jbc.274.19.13133. [DOI] [PubMed] [Google Scholar]
- 23.Kiefer F, Tibbies LA, Anafi M, Janssen A, Zanke BW, Lassam N, Pawson T, R Woodgett J, Iscove NN. HPK1, a hematopoietic protein kinase activating the SAPK/JNK pathway. EMBO J. 1996;15:7013–7025. [PMC free article] [PubMed] [Google Scholar]
- 24.Pelengaris S, Khan M, Evan G. c-MYC: more than just a matter of life and death. Nat. Rev. Cancer. 2002;2:764–776. doi: 10.1038/nrc904. [DOI] [PubMed] [Google Scholar]
- 25.Wolfer A, Ramaswamy S. MYC and metastasis. Cancer Res. 2011;71:2034–2037. doi: 10.1158/0008-5472.CAN-10-3776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wasylishen AR, Penn LZ. Myc: the beauty and the beast. Genes Cancer. 2010;1:532–541. doi: 10.1177/1947601910378024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Knoepfler PS. Myc goes global: new tricks for an old oncogene. Cancer Res. 2007;67:5061–5063. doi: 10.1158/0008-5472.CAN-07-0426. [DOI] [PubMed] [Google Scholar]
- 28.Portereiko MF, Lloyd A, Steffen JG, Punwani JA, Otsuga D, Drews GN. AGL80 is required for central cell and endosperm development in Arabidopsis. Plant Cell. 2006;18:1862–1872. doi: 10.1105/tpc.106.040824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Morton S, Davis RJ, McLaren A, Cohen P. A reinvestigation of the multisite phosphorylation of the transcription factor c-Jun. EMBO J. 2003;22:3876–3886. doi: 10.1093/emboj/cdg388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Eferl R, Wagner EF. AP-1: a double-edged sword in tumorigenesis. Nat. Rev. Cancer. 2003;3:859–868. doi: 10.1038/nrc1209. [DOI] [PubMed] [Google Scholar]
- 31.Graham NA, Asthagiri AR. Epidermal growth factor-mediated T-cell factor/lymphoid enhancer factor transcriptional activity is essential but not sufficient for cell cycle progression in nontransformed mammary epithelial cells. J. Biol. Chem. 2004;279:23517–23524. doi: 10.1074/jbc.M314055200. [DOI] [PubMed] [Google Scholar]
- 32.Smith DR, Myint T, Goh HS. Over-expression of the c-myc proto-oncogene in colorectal carcinoma. Br. J. Cancer. 1993;68:407–413. doi: 10.1038/bjc.1993.350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Corzo C, Corominas JM, Tusquets I, Salido M, Bellet M, Fabregat X, Serrano S, Sole F. The MYC oncogene in breast cancer progression: from benign epithelium to invasive carcinoma. Cancer Genet. Cytogenet. 2006;165:151–156. doi: 10.1016/j.cancergencyto.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 34.Onoda N, Maeda K, Chung YS, Yano Y, Matsui-Yuasa I, Otani S, Sowa M. Overexpression of c-myc messenger RNA in primary and metastatic lesions of carcinoma of the stomach. J. Am. Coll. Surg. 1996;182:55–59. [PubMed] [Google Scholar]
- 35.Cho HJ, Oh YJ, Kwon J, Kwon JY, Kim KS, Kim H. c-Myc stimulates cell invasion by inhibiting FBX8 function. Mol. Cells. 2010;30:355–362. doi: 10.1007/s10059-010-0134-8. [DOI] [PubMed] [Google Scholar]
- 36.Zhang W, Hart J, McLeod HL, Wang HL. Differential expression of the AP-1 transcription factor family members in human colorectal epithelial and neuroendocrine neoplasms. Am. J. Clin. Pathol. 2005;124:11–19. doi: 10.1309/T1H2Y2CHWY7PD2BN. [DOI] [PubMed] [Google Scholar]
- 37.Dong Z, Crawford HC, Lavrovsky V, Taub D, Watts R, Matrisian LM, Colburn NH. A dominant negative mutant of jun blocking 12-0-tetradecanoylphorbol-13-acetate-induced invasion in mouse keratinocytes. Mol. Carcinog. 1997;19:204–212. doi: 10.1002/(sici)1098-2744(199707)19:3<204::aid-mc8>3.0.co;2-d. [DOI] [PubMed] [Google Scholar]
- 38.Lamb RF, Hennigan RF, Turnbull K, Katsanakis KD, MacKenzie ED, Birnie GD, Ozanne BW. AP-1-mediated invasion requires increased expression of the hyaluronan receptor CD44. Mol. Cell. Biol. 1997;17:963–976. doi: 10.1128/mcb.17.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yuspa SH. The pathogenesis of squamous cell cancer: lessons learned from studies of skin carcinogenesis. J. Dermatol. Sci. 1998;17:1–7. doi: 10.1016/s0923-1811(97)00071-6. [DOI] [PubMed] [Google Scholar]
- 40.He TC, Sparks AB, Rago C, Hermeking H, Zawel L, da Costa LT, Morin PJ, Vogelstein B, Kinzler KW. Identification of c-MYC as a target of the APC pathway. Science. 1998;281:1509–1512. doi: 10.1126/science.281.5382.1509. [DOI] [PubMed] [Google Scholar]
- 41.Sancho R, Nateri AS, de Vinuesa AG, Aguilera C, Nye E, Spencer-Dene B, Behrens A. JNK signalling modulates intestinal homeostasis and tumourigenesis in mice. EMBO J. 2009;28:1843–1854. doi: 10.1038/emboj.2009.153. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.