Abstract
The subcellular localizations and toxicity of conjugated polymer nanoparticles (CPNs) are dependent on the chemical structure of the side chain and backbone structures. Primary amine-containing CPNs exhibit high Golgi localization with no toxicity. Incorporation of short ethylene oxide and tertiary amine side chains contributes to decreased Golgi localization and increased toxicity, respectively.
Semiconducting conjugated polymer nanoparticles (CPNs) and conjugated polyelectrolytes (CPEs) are emerging fluorescent biomaterials for cellular labelling,1 sensing,2 therapeutic,3 and delivery4 of biological substances. Conjugated Polymers’ (CPs) excellent photophysical properties including high molar absorptivity, quantum yield, and energy transfer efficiency make them suitable for the various biological applications.5 Well-established synthetic methods also allow facile modifications of both π-electron conjugated backbones and side chains with various sensing or targeting units. By treating non-aqueous soluble CPs under various particle formation conditions, non-toxic soft nanoparticles have been fabricated and used for cellular labelling and nucleic acid delivery.6
Understanding cellular interactions and entry pathways of CPNs is paramount to improving overall labelling and delivery efficiency. Depending on the entry pathways, the materials and its payloads (i.e., drugs or genes) will be trafficked into different organelles, which will significantly influence the overall efficiency.7 For example, carriers entrapped in endosomes or lysosomes trafficked via a certain type of endocytosis will experience recycling of the contents back to the cell surface and degradation processes in acidic lysosomes, lowering overall labelling and delivery efficiency.8 Meanwhile, exogenous materials trafficked by non-destructive organelles such as caveosomes to Golgi apparatus (i.e., caveolae-mediated endocytosis) have high intracellular retention.9 Delivery via macropinocytosis also can avoid lysosomal degradation routes because macropinosomes do not fuse with the lysosomes, and the membranes of macropinosomes are highly leaky.10 Therefore, systematic investigation to understand and modulate the cellular interaction and pathways will have significant impact on designing efficient labelling and delivery vehicles.
Previously, we demonstrated that CPNs fabricated by treating a CP containing both short ethylene oxide (EO) and primary amine (e.g., P1, Fig. 1) with organic acids followed by dialysis exhibit efficient cellular labelling and delivery of small interfering RNA without toxic effects.4a,b Mechanistic studies further indicate that CPNs use both energy dependent and independent entry pathways. Among the energy dependent pathways, CPNs enter cancer cells via caveolae-mediated endocytosis as one of entry pathways.11 It is not clear why the CPNs use the specific entry pathway; however, the positive charges and hydrophobicity of CPNs play important roles for interaction with various serum proteins and the cell membranes, which will significantly influence the subsequent cellular uptake.12 It is also known that materials having high surface-to-volume ratios exhibit size, shape, and functional group-dependent cellular interactions and subsequent entry.13
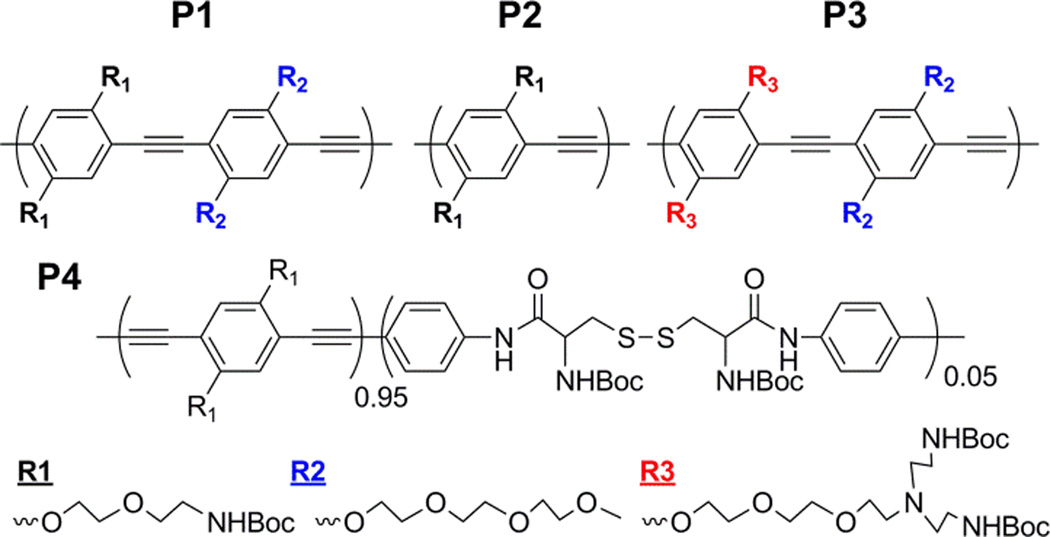
Fig. 1.
Chemical structures of CPs. Poly(p-phenyleneethynylenes) (PPEs) with different side chains (P1-P3) and poly(p-phenylenebutadiynylene) (PPB) containing a small amount of flexible unit in the backbone (P4) were synthesized and compared for cellular behaviours.
Based on the results and observations, we hypothesized that chemical modifications in the side chains of CPs will change the subcellular localizations of CPNs, because the modulated surface properties will influence the cellular interactions of CPNs and their subsequent entry into cells.
To test the hypothesis, we synthesized four CPNs with different side chain (P1-3) and backbone structures (P4) (Fig. 1). Since the cellular interactions and entry processes of nanomaterials are collectively influenced by the physicochemical properties, it is important to keep other physicochemical properties constant when a specific parameter is tested. Because of the particle formation mechanism (i.e., molecular weight independent phase inverse precipitation driven by aqueous insolubility of CPs),14 the shapes and hydrodynamic radii of CPNs are relatively constant.15 CPN-2 was designed and synthesized to check the EO side chain effects on the toxicity and localization by removing the EO unit from the repeating unit of CPN-1. CPN-3 was synthesized to increase amine density using branched amine side chains containing tertiary amines. CPN-4 was synthesized to compare the backbone flexibility effects while amine density was maintained close to that of CPN-2. Synthesis of P4 was reported in our recent publication.16 All polymers were treated with a series of organic acids followed by dialysis, affording CPNs that are physically stable in water. Physicochemical properties of CPNs are listed in Table 1. Since aggregation behaviours are concentration dependent, the concentrations of all CPN solutions were adjusted to be 0.5 mM. Non-EO containing CPN-2 and CPN-4 exhibited slightly larger hydrodynamic radii than those of CPN-1 and CPN-3 fabricated with EO containing polymers. The difference in hydrodynamic radius among the CPNs is expected to have minimal effects on the cellular interaction and subcellular localization due to the polydisperse nature of CPNs. Zeta potentials of CPNs were determined to be ~+42–46 mV, except for CPN-1 exhibiting ~+20 mV (Table 1).
Table 1.
Physicochemical properties of CPNs.
| CPN | Type | Mna (kDa) |
PDIb | λmax,absc (nm) |
λmax, emd (nm) |
Hydrodynamic radius (nm)e |
PDIe | Zeta Potential (mV)f |
|---|---|---|---|---|---|---|---|---|
| 1 | P1(PPE) | 16.4 | 1.49 | 433 | 496 | 61 ±6.7 | 0.27 ±0.02 | +20 ±0.4 |
| 2 | P2(PPE) | 11.8 | 1.43 | 427 | 492 | 71 ±7.9 | 0.29 ±0.02 | +42 ±5.1 |
| 3 | P3(PPE) | 10.7 | 1.64 | 420 | 496 | 58 ±3.4 | 0.33 ±0.06 | +44 ±1.1 |
| 4 | P4(PPB) | 22.3 | 2.28 | 444 | 500 | 87 ±6.1 | 0.51 ±0.03 | +46 ±2.3 |
Determined by gel permeation chromatography in THF relative to polystyrene standard.
Polydispersity index (PDI) = Mw/Mn
Measured in water.
Measured in water, excitation wavelength 400 nm.
Measured by DLS at 500 µM in water. Mean±standard deviation.
Electrophoretic measurement at pH 7.0. Mean±standard deviation.
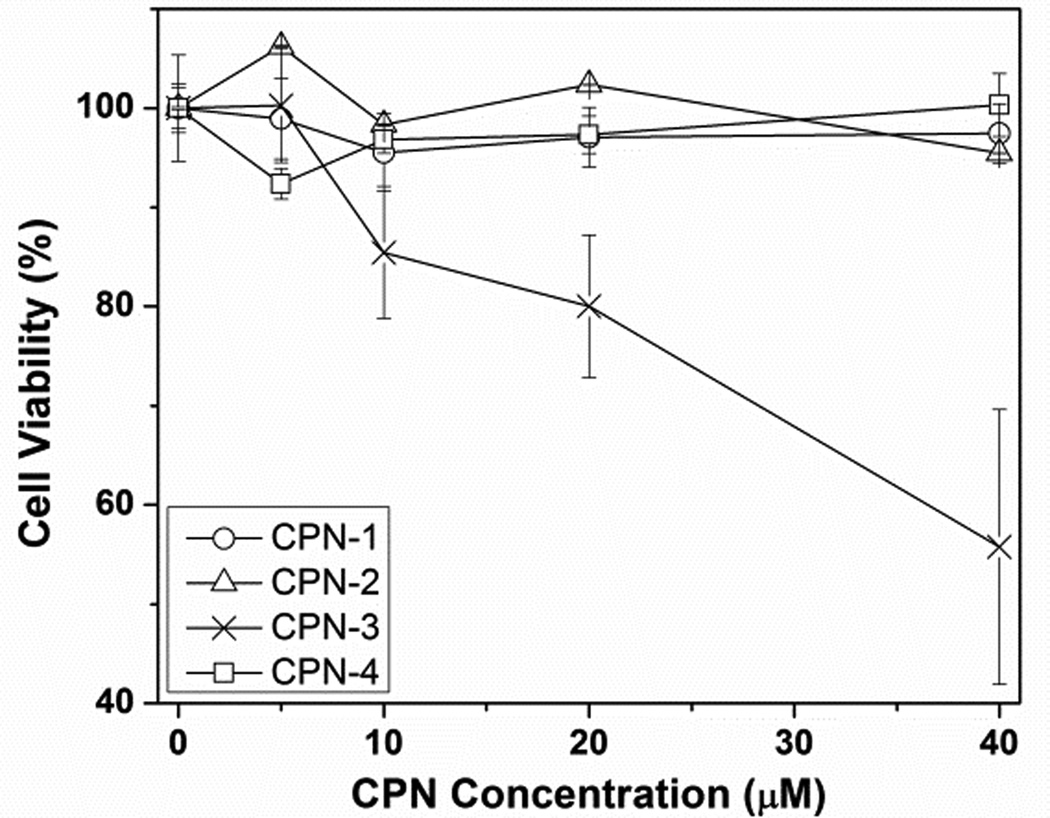
To test how the side chain structure influences cellular toxicity, CPNs were incubated with human cervical carcinoma cells (HeLa) overnight at various concentrations. Zeta potentials of CPNs had no direct correlation to the toxicity, but the chemical structure (i.e., type and density of amine) of the side chains was related to the cellular toxicity. As shown in Fig. 2, CPN-3 containing the highest amine density, including tertiary amines, exhibited substantial toxicity starting from 10 µM, while no cell viability inhibition was observed up to 40 µM from the primary amine containing CPNs, whether they contain EO side chains or flexible backbones (i.e., CPN-1, -2, and -4). Compared to CPN-2, which contains the same amount of primary amines per repeating unit as CPN-3, toxicity of CPN-3 can be attributed to both increased amine density and the high buffering capacity of tertiary amines. Membrane disruption properties of synthetic carriers containing tertiary amines have been used to increase payload escape from the endosomes or lysosomes; however, these classes of materials often cause toxicity issues.17
Fig. 2.
Cellular toxicity of CPNs measured by cell viability inhibition at various concentrations
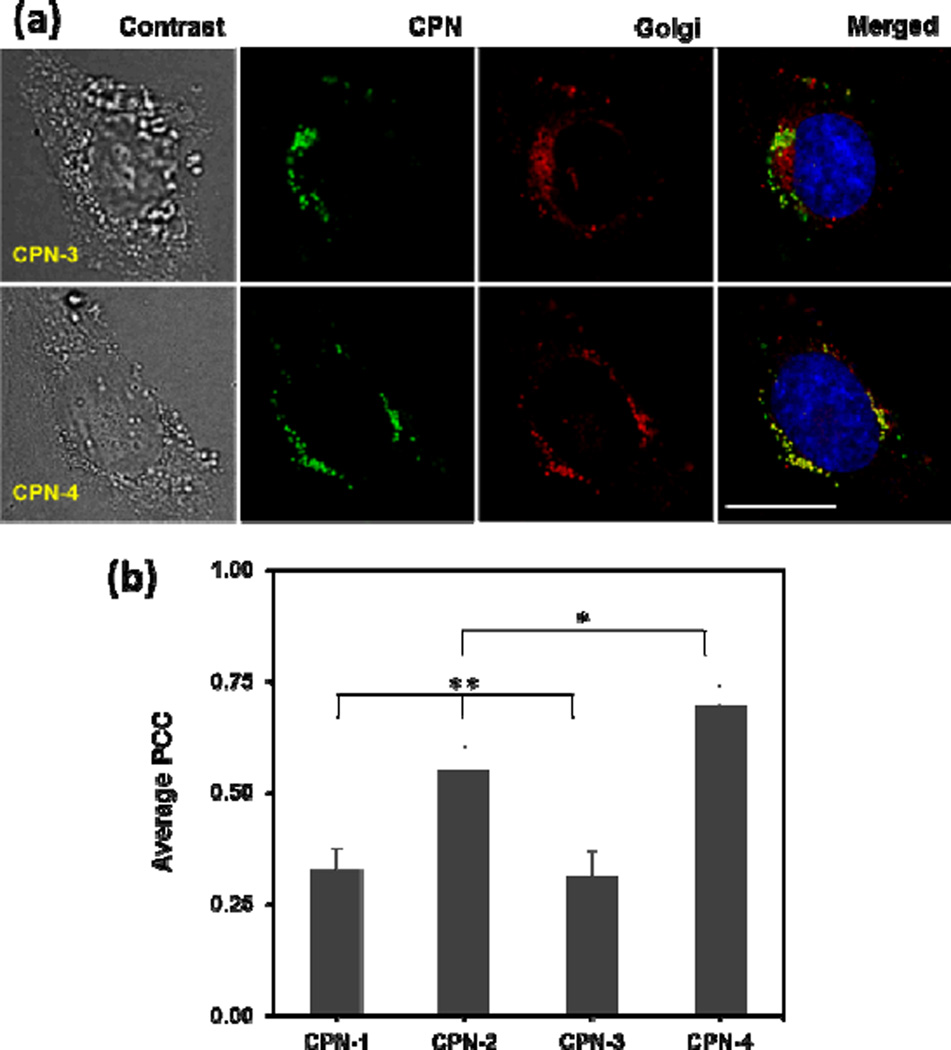
Subcellular localization of CPNs was monitored by fluorescent microscopic imaging. Overnight incubation of HeLa cells with CPNs (green) were co-stained with pHRhodo Dextran (10kDa) (red) and BODIPY-TR C5-ceramide-BSA complex (red) for labelling acidic organelles (i.e., endosomes and lysosomes) and Golgi apparatus, respectively (Fig. 3a and ESI). CPNs were mainly found at the perinuclear regions (punctuated green dots) and exhibited overlaps with both pHRhodo and BODIPY. Co-localization patterns with the Golgi were clearly distinguishable among CPNs having different side chain or backbone structures (Fig. 3a), while overlapping patterns with pHRhodo were relatively uniform (ESI). CPN-2 and CPN-4, which only contain primary amine side chains, exhibit high Golgi localizations (Fig. 3a), while CPN-1 and CPN-3, which contain both EO and amine side chains, exhibit a relatively low Golgi overlaps.
Fig. 3.
(a) Microscopic images of HeLa cells incubated with CPN-3 and CPN-4, followed by Golgi (red) and nucleus (blue) staining. The scale bar is 20 µm. CPN-4 exhibits higher overlap with Golgi than CPN-3 does. (b) Quantitative analysis of co-localization using the PCC algorithm. Co-localization with Golgi is dependent on the side chain and backbone structures. The error bar represents ±standard deviation (n=3). * <0.05 when CPN-4 compared with CPN-2. ** <0.0005 when CPN-1 and CPN-3 compared with CPN-2 and CPN-4 (n=3).
To obtain quantitative co-localization information, all images were further analysed using the Pearson's Correlation Coefficient (PCC) method. The PCC method gauges the level of overlap by measuring the pixel-by-pixel covariance in the signals of two images. Because the PCC method uses normalized signals by subtracting the mean intensity from each pixel's intensity value, PCC is independent of signal levels (probe brightness) and signal offset (background).18 PCC values of 0 and 1 correspond to uncorrelated and perfectly linear correlated images, respectively. Instead of picking small, subjective regions of interest within an image, three independent images of an entire cell were selected and analysed to increase the analysis objectivity. As shown in Fig. 3b, average PCC values were dependent on the side chain and backbone structures of the CPNs. The CPNs with only amine side chains exhibited higher Golgi localization than the CPNs containing both EO and amine side chains. In addition, CPNs fabricated with a semi-flexible CP exhibited the highest Golgi localization. One-way ANOVA followed by the Tukey means separation method confirmed that Golgi co-localization of CPN-2 and CPN-4 was statistically significant (p<0.003) from CPN-1 and CPN-3. The Golgi co-localization between CPN-2 and CPN-4 was also statistically significant (p<0.05).
In conclusion, we demonstrated that CPNs are promising biomaterials with tunable physicochemical properties. The side chain and backbone structures of CPNs are closely related to toxicity and subcellular localization. Therefore, cellular interaction and cellular entry pathways of CPNs can be fine-tuned to improve labelling and delivery efficiency. Combining the excellent intrinsic fluorescent natures of CPNs, which are useful for labelling and monitoring biological substances, the tunable physicochemical properties and the related biophysical properties make CPNs excellent biomaterials. The concept we demonstrated here will lead to the development of novel multifunctional materials for labelling, sensing, and delivery. Using the highly non-destructive delivery pathway and biodegradability of CPN-4, we are currently investigating the delivery of small RNA molecules to target cells.
Supplementary Material
Acknowledgments
This work was supported by National Institute of Health/National Institute of General Medical Sciences Grant No. SC1GM092778 and R25GM61347.
Footnotes
† Electronic Supplementary Information (ESI) available: detailed synthetic procedures, characterizations, and co-localization analyses. See DOI: 10.1039/b000000x/
Notes and references
- 1.Kim IB, Shin H, Garcia AJ, Bunz UHF. Bioconjugate Chem. 2007;18:815. doi: 10.1021/bc0603440. [DOI] [PubMed] [Google Scholar]; Feng XL, Tang YL, Duan XR, Liu LB, Wang SJ. Mater. Chem. 2010;20:1312. [Google Scholar]; Lee K, Lee J, Jeong EJ, Kronk A, Elenitoba-Johnson KSJ, Lim MS, Kim J. Adv. Mater. 2012;24:2479. doi: 10.1002/adma.201103895. [DOI] [PubMed] [Google Scholar]; Wu CF, Chiu DT. Angew. Chem. Int. Ed. 2013;52:3086. doi: 10.1002/anie.201205133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Disney MD, Zheng J, Swager TM, Seeberger PH. J. Am. Chem. Soc. 2004;126:13343. doi: 10.1021/ja047936i. [DOI] [PubMed] [Google Scholar]; Fan CH, Wang S, Hong JW, Bazan GC, Plaxco KW, Heeger AJ. Proc. Natl. Acad. Sci. USA. 2003;100:6297. doi: 10.1073/pnas.1132025100. [DOI] [PMC free article] [PubMed] [Google Scholar]; Ho HA, Dore K, Boissinot M, Bergeron MG, Tanguay RM, Boudreau D, Leclerc MJ. Am. Chem. Soc. 2005;127:12673. doi: 10.1021/ja053417j. [DOI] [PubMed] [Google Scholar]; Achyuthan KE, Bergstedt TS, Chen L, Jones RM, Kumaraswamy S, Kushon SA, Ley KD, Lu L, McBranch D, Mukundan H, Rininsland F, Shi X, Xia W, Whitten DGJ. Mater. Chem. 2005;15:2648. [Google Scholar]; Sun CJ, Gaylord BS, Hong JW, Liu B, Bazan GC. Nat. Protoc. 2007;2:2148. doi: 10.1038/nprot.2007.307. [DOI] [PubMed] [Google Scholar]; Liu B, Bazan GC. Chem. Mater. 2004;16:4467. [Google Scholar]
- 3.Wang Y, Jett SD, Crum J, Schanze KS, Chi EY, Whitten DG. Langmuir. 2013;29:781. doi: 10.1021/la3044889. [DOI] [PMC free article] [PubMed] [Google Scholar]; Zhu CL, Yang Q, Lv FT, Liu LB, Wang S. Adv. Mater. 2013;25:1203. doi: 10.1002/adma.201204550. [DOI] [PubMed] [Google Scholar]; Xing CF, Liu LB, Tang HW, Feng XL, Yang Q, Wang S, Bazan GC. Adv. Funct. Mater. 2011;21:4058. [Google Scholar]
- 4.Moon JH, Mendez E, Kim Y, Kaur A. Chem. Commun. 2011;47:8370. doi: 10.1039/c1cc10991j. [DOI] [PubMed] [Google Scholar]; Silva AT, Alien N, Ye CM, Verchot J, Moon JH. BMC Plant Biol. 2010;10:291. doi: 10.1186/1471-2229-10-291. [DOI] [PMC free article] [PubMed] [Google Scholar]; Feng XL, Lv FT, Liu LB, Yang Q, Wang S, Bazan GC. Adv. Mater. 2012;24:5428. doi: 10.1002/adma.201202145. [DOI] [PubMed] [Google Scholar]
- 5.Thomas SW, Joly GD, Swager TM. Chem. Rev. 2007;107:1339. doi: 10.1021/cr0501339. [DOI] [PubMed] [Google Scholar]; Zhu CL, Liu LB, Yang Q, Lv FT, Wang S. Chem. Rev. 2012;112:4687. doi: 10.1021/cr200263w. [DOI] [PubMed] [Google Scholar]
- 6.Moon JH, McDaniel W, MacLean P, Hancock LE. Angew. Chem. Int. Ed. 2007;46:8223. doi: 10.1002/anie.200701991. [DOI] [PubMed] [Google Scholar]; Wu CF, Szymanski C, McNeill J. Langmuir. 2006;22:2956. doi: 10.1021/la060188l. [DOI] [PubMed] [Google Scholar]; Baier MC, Huber J, Mecking SJ. Am. Chem. Soc. 2009;131:14267. doi: 10.1021/ja905077c. [DOI] [PubMed] [Google Scholar]
- 7.Duncan R, Richardson SCW. Mol. Pharm. 2012;9:2380. doi: 10.1021/mp300293n. [DOI] [PubMed] [Google Scholar]; Sahay G, Alakhova DY, Kabanov AVJ. Controlled Release. 2010;145:182. doi: 10.1016/j.jconrel.2010.01.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Rejman J, Bragonzi A, Conese M. Mol. Ther. 2005;12:468. doi: 10.1016/j.ymthe.2005.03.038. [DOI] [PubMed] [Google Scholar]
- 9.Pelkmans L, Helenius A. Traffic. 2002;3:311. doi: 10.1034/j.1600-0854.2002.30501.x. [DOI] [PubMed] [Google Scholar]
- 10.Khalil IA, Kogure K, Futaki S, Harashima H. J. Biol. Chem. 2006;281:3544. doi: 10.1074/jbc.M503202200. [DOI] [PubMed] [Google Scholar]
- 11.Lee J, Twomey M, Machado C, Gomez G, Doshi M, Gesquiere AJ, Moon JH. Macromol. Biosci. doi: 10.1002/mabi.201300030. In print. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Nel AE, Madler L, Velegol D, Xia T, Hoek VEMV, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nature Mater. 2009;8:543. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]; Fernando LP, Kandel PK, Yu J, McNeill J, Ackroyd PC, Christensen KA. Biomacromolecules. 2010;11:2675. doi: 10.1021/bm1007103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tan SJ, Jana NR, Gao SJ, Patra PK, Ying JY. Chem. Mater. 2010;22:2239. [Google Scholar]
- 14.Ko YJ, Mendez E, Moon JH. Macromolecules. 2011;44:5527. doi: 10.1021/ma200661h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Pancake-like shapes of CPNs on a mica surface were observed from atomic force microscopy imaging. See Macromol. Biosci. 2013 [Google Scholar]
- 16.Vokatá T, Moon JH. Macromolecules. 2013;46:1253. doi: 10.1021/ma3019975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee C-C, Liu Y, Reineke TM. Bioconjugate Chem. 2008;19:428. doi: 10.1021/bc7001659. [DOI] [PubMed] [Google Scholar]
- 18.Dunn KW, Kamocka MM, McDonald JH. Am. J. Physiol.-Cell Ph. 2011;300:C723. doi: 10.1152/ajpcell.00462.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.