To the Editor:
The ability to add or delete genes to the genome of genetic model organisms is essential. Previously, we developed methods based on the Mos1 transposon1 to make targeted transgene insertions (Mos1-mediated Single Copy transgene Insertions, MosSCI2) and targeted deletions (Mos1-mediated deletions, MosDEL3) in Caenorhabditis elegans, the latter reported in your pages. Here, we present new reagents that improve the efficiency, facilitate the selection for transgenic strains and expand the set of MosSCI insertion sites.
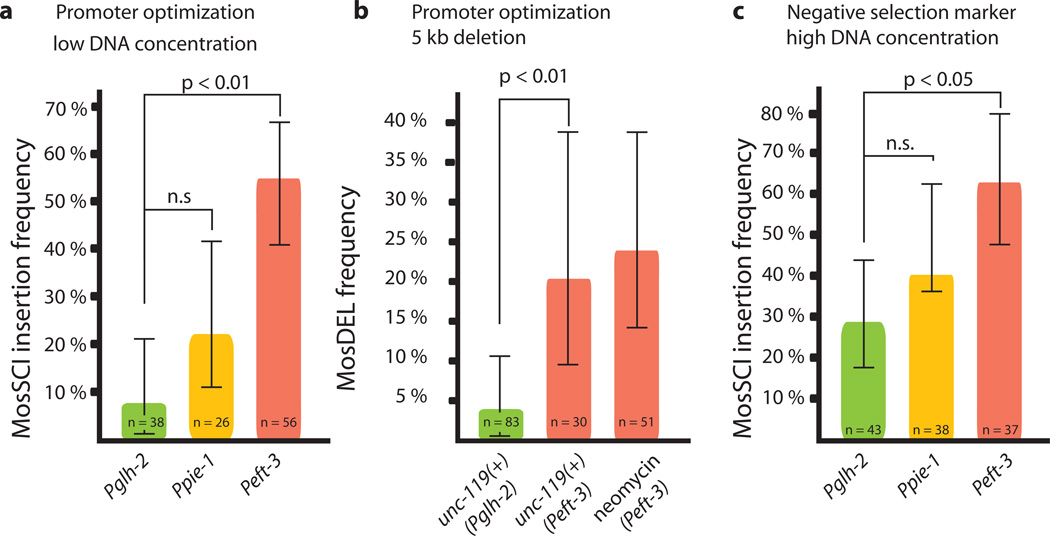
The Mos1 transposase is expressed from a helper plasmid co-injected with template DNA. Increased transposase expression would be expected to improve both single copy insertions and targeted gene deletions. We tested several promoters driving transposase expression for their effect on MosSCI and MosDEL efficiency (Fig. 1a and Supplementary Fig. 1). Relative to the glh-2 promoter, the most effective promoter (eft-3) resulted in a more than 6-fold improvement in transgene insertion efficiency (from 8% to 54% of injected animals) and gene deletion efficiency (from 3%, n=66 injected animals2 to 20%, n=30 injected animals, Fig. 1b).
Figure 1.
Improvements to Mos1-based genome manipulation. (a) A plasmid expressing transposase under the indicated promoters was coinjected with low DNA concentration (32.5 ng/ul) of a 4.4 kb transgene. The plot shows insertion frequency into the ttTi5605 locus. (b) The plot shows the frequency of a 5 kb targeted deletion of dpy-13. Pglh-2 data from Frøkjær-Jensen et al (2010)2) using the indicated selection markers (see Supplementary Methods for discussion). (c) Insertion frequency with higher total DNA concentrations (~100 ng/ul) and in the presence of the negative selection marker peel-1. Error bars, 95 % confidence intervals, significance was determined with Fischer’s exact test
An effective, inducible negative selection marker would facilitate identification of transgenic strains. We developed a negative selection marker (Phsp16-2∷peel-1) based on the toxin peel-1 4. Array animals carrying the peel-1 plasmid are killed by a two-hour heat-shock at 34°C with approximately 10% false positives (2/19 transgenic animals) (Fig. 1c and Supplementary Fig. 2). A positive selection marker is critical for identifying transgenic animals with insertions or deletions and we have used unc-119(+) extensively. Recently, antibiotic selection markers have been developed for nematode transgenesis5,6. Targeted dpy-13 deletions were generated with Neomycin/G418 selection at frequencies comparable to unc-119 selection (24%, 12/51 injected animals, Fig. 1b), see Supplementary Methods for a discussion of the recommended use of selection markers.
Multiple insertion sites are important for generating complex genotypes. We have expanded the number of MosSCI insertion sites from two to six (Supplementary Fig. 3) with a full set of outcrossed strains containing the Mos1 insertion and targeting vectors (three-way Gateway or multiple cloning site compatible) based on unc-119 selection and for one site, unc-18 selection (Table 1). All sites readily generate MosSCI inserts and express in somatic tissue. Three of the insertion sites (ttTi4348 I, ttTi5605 II and cxTi10816 IV) express robustly in the germline from a ubiquitous promoter (Supplementary Fig. 4). Because MosSCI reagents are important for expression in the germline, we generated an expression vector that coexpresses GFP-histone for confirmation of expression (Supplementary Fig. 5). All strains are available from the Caenorhabditis elegans Genetics Center (CGC) and targeting plasmids (targeting, transposase and negative selection vectors) from Addgene.
Table 1.
MosSCI insertion sites.
| Locus | Genetic position |
Insertion strain1 |
Gateway vector2 |
MCS vector |
Germline expression3 |
Insertion frequency4 |
Balancer strain |
|
|---|---|---|---|---|---|---|---|---|
| unc-119 | ttTi4348 | I: −5.32 | EG6701 | pCFJ210 | pCFJ352 | yes | 25% (3/12) | EG6173 |
| ttTi4391 | I: 7.93 | EG6702 | pCFJ604 | pCFJ353 | no | 29% (4/21) | EG6171 | |
| ttTi5605 | II: 0.77 | EG6699 | pCFJ150 | pCFJ350 | yes | 43% (6/14) | EG6070 | |
| cxTi10816 | IV: 1.41 | EG6703 | pCFJ212 | pCFJ356 | yes | 20% (2/10) | EG6401 | |
| cxTi10882 | IV: −0.05 | EG6700 | pCFJ201 | pCFJ351 | variable | 29% (4/14) | EG5568 | |
| ttTi14024 | X:22.84 | EG6705 | pCFJ606 | pCFJ355 | no | 21% (3/14) | EG6109 | |
| unc-18 | ttTi4348 | I: −5.32 | EG6032 | pCFJ448 | pCFJ676 | yes | N.D. | EG6173 |
4× outcrossed strain, distributed with extrachromosomal unc-119 rescue to facilitate handling and maintenance
pDESTR4-R3, three-way Gateway Compatible vector
Based on germline expression of Pdpy-30∷GFP transgene
Insertion frequency of Pdpy-30∷GFP∷H2B transgene
Supplementary Material
Acknowledgements
CFJ was funded by postdoctoral stipends from the Lundbeck foundation and the Carlsberg foundation. This research was funded by the National Institutes of Health (1R01GM095817) and by the Howard Hughes Medical Institute.
Footnotes
Competing financial interests:
Yes, the authors declare competing financial interests.
References
- 1.Robert V, Bessereau JL. EMBO J. 2007;26:170–183. doi: 10.1038/sj.emboj.7601463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Frøkjær-Jensen C, et al. Nat. Genetics. 2008;40:1375–1383. [Google Scholar]
- 3.Frøkjær-Jensen C, et al. Nature Methods. 2010;7:451–453. [Google Scholar]
- 4.Seidel HS, Ailion M, Li J, van Oudenaarden A, Rockman MV, Kruglyak L. PLoS Biology. 2011;9:e1001115. doi: 10.1371/journal.pbio.1001115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Giordano-Santini, et al. Nat. Methods. 2010;7:721–723. doi: 10.1038/nmeth.1494. [DOI] [PubMed] [Google Scholar]
- 6.Semple JI, Garcia-Verdugo R, Lehner B. Nat. Methods. 2010;7:725–727. doi: 10.1038/nmeth.1495. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.