Abstract
Tuberculosis is one of the world's most prevalent infectious diseases. The causative agent, M. tuberculosis, asymptomatically infects more than 30% of the world population and causes 8 million cases of active disease and 2 million deaths annually. Its pathogenic success stems from its ability to block phagolysosome biogenesis and subsequent destruction in the host macrophages. Recently, our laboratory has uncovered autophagy as a new means of overcoming this block and promoting the killing of mycobacteria. Here we describe the methods to study autophagy during M. tuberculosis infection of macrophages. The described assays can be used to investigate and identify factors important for autophagic elimination of mycobacteria that could potentially provide new therapeutic targets to defeat this disease.
1. Introduction
Mycobacterium tuberculosis is the causative agent of tuberculosis, the leading global cause of infectious disease-associated morbidity and mortality in humans. Approximately 2 billion people or one-third of the world population are latently infected, and nearly 2 million deaths from 8 million cases of active tuberculosis yearly are associated with the disease (WHO, 2008). In most cases, the mycobacteria can be contained by the host immune system and infected individuals are in the majority of cases asymptomatic for long periods of time (Russell, 2001; Kaufmann et al., 2006). However, nearly 10% of infected individuals will eventually develop active disease in their lifetime due to weakening of the immune system caused by many factors such as immunosuppression, malnutrition, or old age (Chan and Flynn, 2004). The typical 10%-lifetime risk increases substantially to an annual risk of 10% if an individual is coinfected with human immunodeficiency virus (HIV) (Corbett et al., 2003). Antibiotics can be used to treat the disease. However, as mycobacteria are slow growing, the treatment requires several months to completely eliminate the microbe (Budha et al., 2008). In addition, new strains that are resistant to all available drugs (dubbed XDR) have emerged (Ducati et al., 2006). Currently, there are no new drugs available or vaccines effective in adults.
The success of M. tuberculosis as a human pathogen is contributed by its ability to survive and replicate in macrophages (Vergne et al., 2004). After the mycobacteria are inhaled into the lung, the bacteria are phagocytosed into phagosomes by alveolar macrophages (Russell, 2007). To avoid destruction by the host, the mycobacteria interfere with macrophage membrane trafficking and block the fusion of the phagosome with lysosomes (Deretic et al., 2004; Vergne et al., 2004). This blocking event is characterized by the lack of complete acidification and acquisition of lysosomal components by the mycobacterial phagosome (Russell et al., 2002) and is mediated by several mycobacterial secreted factors including specific lipids (Beatty et al., 2000; Vergne et al., 2003). This results in mycobacterial survival, disease latency, and the risk of reactivation later on in life (Russell et al., 2002). Recently, however, induction of host autophagy was shown to overcome this block and eliminate mycobacteria in macrophages (Gutierrez et al., 2004; Singh et al., 2006; Delgado et al., 2008).
Autophagy is an evolutionarily conserved intracellular process of cytoplasmic degradation. It begins with the sequestration of cytoplasmic contents into a double-membrane bound organelle called an autophagosome (Levine and Klionsky, 2004; Mizushima, 2004). Autophagosomes then fuse with lysosomes to acquire lysosomal hydrolases resulting in the degradation of its engulfed constituents (Klionsky et al., 2007; Mizushima and Klionsky, 2007). Autophagy has been linked to many cellular functions. These include the maintenance of intracellular metabolism, the removal of defective organelles and protein aggregates, the elimination of pathogens such as bacteria, parasites, and viruses, and the processing of self and foreign proteins for antigen presentation (Talloczy et al., 2002; Gutierrez et al., 2004; Nakagawa et al., 2004; Ogawa et al., 2005; Schmid and Münz, 2005; Ling et al., 2006; Rubinsztein, 2006; Sakai et al., 2006; Singh et al., 2006; Williams et al., 2006; Bernales et al., 2007; Devenish, 2007; Schmid et al., 2007; Liang et al., 2008). In the context of M. tuberculosis infection, induction of autophagy in macrophages by starvation or rapamycin treatment has been shown to overcome the phagolysosome biogenesis block resulting in increases in mycobacterial phagosome acidification and lysosomal component acquisition (Gutierrez et al., 2004). These effects are inhibitable by known pharmacological inhibitors of autophagy such as 3-methyladenine (3-MA) and wortmannin (Gutierrez et al., 2004) and Beclin 1and Atg5 siRNA knockdown (Harris et al., 2007; Delgado et al., 2008). The outcome is the reduction in M. tuberculosis survival in macrophages. A model by which autophagic induction leads to the fusion of mycobacterial phagosomes with autophagic organelles and hence the acquisition of lysosomal contents to overcome the phagolysosome biogenesis block and promote the killing of mycobacteria has been proposed (Harris, 2006). Moreover, a specific delivery of mycobactericidal agents, such as fragments of ubiquitin, to the M. tuberculosis phagosome has been shown following induction of autophagy (Alonso et al., 2007). However, the factors and mechanisms involved remain largely uncharacterized. In this chapter, we describe assays to monitor autophagy during M. tuberculosis infection. The assays include the monitoring of mycobacterial phagosome acidification, mycobacterial phagosome acquisition of a lysosomal protease, and mycobacterial elimination on autophagic induction. In addition, methods for siRNA-mediated knockdown of proteins in macrophages are outlined. The combination of these assays are well suited and can be used to characterize important factors involved in autophagic elimination of mycobacteria which could potentially pave the way for pharmacological development to combat this noxious disease.
2. Methods
In the following text, we describe methods to monitor autophagy during M. tuberculosis infection in murine RAW264.7 macrophages. These methods can be adapted to other murine and human macrophage cell lines and primary cells (Harris, 2006).
2.1. Autophagic killing assay and the use of inhibitors
2.1.1. Materials
2.1.1.1. Growth and use of RAW264.7 cell line
Murine RAW264.7 macrophage cell line (ATCC, No. TIB-71).
Dulbecco's modified Eagle's medium (DMEM) (Gibco), 10% fetal bovine serum (FBS; Hyclone), and 4 mM L-glutamine (Gibco) (Full medium): Add 100 mL of FBS and 10 mL of 200 mM L-glutamine to 890 mL of DMEM. Filter through 0.2-mm filter. Store the medium at 4 °C.
175-cm2 tissue culture flasks (Greiner Bio-One).
Disposable cell scraper (Sarstedt).
Earle's balanced salt solution (EBSS) (Starvation medium) (Sigma).
12-well tissue culture plates.
2.1.1.2. Growth and preparation of mycobacteria
Middlebrook albumin dextrose catalase (ADC) enrichment (BD Biosciences).
20% Tween 80 (Sigma): Prepare 20% v/v stock at 4 °C and filter through 0.2-mm filter. Store at 4 °C.
Middlebrook 7H9 broth (Difco): Dissolve 4.7 g of powder in 900 mL of deionized water with 2 mL of glycerol (EMD Chemicals) and autoclave. Temper to room temperature. Add 100 mL of ADC and 2.5 mL of 20% Tween 80. Store at 4 °C.
Middlebrook 7H11 agar (Difco): Dissolve 21 g of powder in 900 mL deionized water with 5 mL of glycerol (EMD Chemicals) and autoclave. Temper to 50 °C. Add 100 mL of ADC and 2.5 mL of 20% Tween 80 and dispense into Petri dishes. Store at 4 °C.
Sterile, 150-mL tissue-culture roller bottles.
Autoclaved 7-mL Dounce glass homogenizer (Kontes Scientific Glassware).
Phosphate-buffered saline (PBS, 10X) (Gibco): Prepare 1X PBS by adding 100 mL into 900 mL of deionized water and filter through 0.2-mm filter. Store at room temperature.
PBS-Tween: Add 100 mL of 10X PBS and 2.5 mL of 20% Tween 80 into 897.5 mL of deionized water. Filter through 0.2-mm filter and store at 4 °C.
96-well tissue culture plates.
Polyethylene bags (Fisher).
2.1.1.3. Inhibitor preparation
Rapamycin (FW ¼ 914.19 g/mol) (LC Laboratories, R-5000): Prepare 50 mg/mL stock solution by dissolving the powder with 100% DMSO. Aliquot and store at −20 °C. For the experiment, prepare working solution by diluting the stock solution 1:1000 in complete DMEM to yield a final concentration of 50 mg/mL.
3-methyladenine (FW ¼ 149.16 g/mol) (Sigma, M9281]): Prepare 100 mM stock solution by dissolving 14.916 mg of powder in 1 mL of deionized water. Heat to 60–80 °C. Prepare a working solution by adding 100 mL of 100 mM stock into 900 mL of starvation media, EBSS, to obtain a final concentration of 10 mM. Always prepare fresh solution for each experiment.
Wortmannin (FW 428.47 g/mol) (Sigma, W1628): Prepare 1 mM stock solution by dissolving 1 mg of powder in 2.064 mL 100% DMSO. From this solution, prepare 100 mM stock solution by performing a 1:10 dilution in 1X PBS. Prepare a working solution by diluting the 100 mM stock solution 1:1000 in EBSS to achieve a final concentration of 100 nM. Always prepare fresh solution for each experiment.
2.1.2. Methods
2.1.2.1. Macrophage cell preparation
Murine RAW264.7 macrophages are maintained in complete DMEM at 37 °C, 5% CO2.
Subculture RAW264.7 cells to reach 70%–80% confluency by scraping off the cells into 12 mL complete DMEM. Pipet up and down to obtain a single cell suspension. Transfer 1.5 mL of cells into a 175-cm2 tissue culture flask containing 25 mL of complete DMEM. Incubate for approximately 2 d at 37 °C, 5% CO2.
One day before the experiment, scrape off the cells into 12 mL of complete DMEM and determine cell density using trypan blue staining and a hemocytometer.
Dilute cells to 3 × 105 cells/mL in complete DMEM and dispense 1 mL into each well of a 12-well plate. Plate 3 wells (triplicate) for each experimental condition. Incubate the plate at 37 °C, 5% CO2 overnight.
2.1.2.2. Infection of macrophages
Mycobacterium tuberculosis var. bovis BCG (BCG) culture is maintained in Middlebrook 7H9 broth supplemented with 10% ADC, 0.05% Tween 80, and 0.2% glycerol at 37 °C on a roller.
Transfer 10 mL of a log-phase BCG culture into a 15-mL conical tube. Centrifuge to pellet mycobacteria at 2500 rpm for 5 min at room temperature.
Remove and discard the supernatant fraction and wash the cell pellet once with 10 mL of 1X PBS.
Spin at 2500 rpm for 5 min and remove and discard the supernatant fraction.
Resuspend the mycobacteria pellet in 6 mL of complete DMEM and transfer the suspension to a 7-mL Dounce homogenizer. Homogenize 35 times to generate a single cell suspension.
Measure OD600 of the 1:10 dilution of the homogenized culture.
Prepare mycobacterial inoculum in complete DMEM at the concentration of 3 × 106 BCG/mL (MOI ¼ 10) using the following formula:(3 × 106); (total mL needed for infection)/(OD600 × 109) ¼ mL homogenate needed.
Remove medium from cells and add 1 mL of 3 × 106 BCG/mL inoculum into each well of RAW264.7 macrophages.
Spin at 1200 rpm for 5 min at room temperature to settle mycobacteria onto macrophages. Incubate the plate for 1 h at 37 °C, 5% CO2.
2.1.2.3. Autophagic induction and inhibitor treatment
For complete medium control (Full medium) and rapamycin treatment, remove the medium containing mycobacteria from the wells. Quickly wash each well three times with 2 mL of complete DMEM to remove noninternalized mycobacteria and add 1 mL of the respective media/treatment into each well (see Notes 1 and 2).
For starvation, 3-methyladenine, and wortmannin treatments, remove the medium containing mycobacteria from the wells. Quickly wash each well 3 times with 2 mL of EBSS and add 1 mL of the respective media/treatment into the well.
Incubate the plate for 2 h at 37 °C, 5% CO2 (see Note 3).
For wortmannin treatment, at 1 h after incubation, remove the old media/treatment and add freshly prepared media/treatment to each well.
2.1.2.4. Recovery of mycobacteria and data analysis
After 2 h of incubation, wash each well twice with 1 mL of 1X PBS.
Add 0.5 mL of cold deionized water to each well and incubate the plate at 4 °C for 10–15 min to lyse the cells and release intracellular mycobacteria.
Prepare a dilution plate by adding 180 mL of PBS-Tween into each well of a 96-well plate.
Pipet the cell lysate up and down to mix, and transfer 20 mL into a well of the dilution plate containing 180 mL of PBS-Tween. This is the 10−1 dilution. Perform 10-fold serial dilutions to obtain 10−2, 10−3, and 10−4 dilutions. Repeat the process for each well. Since there are three wells (triplicate) for each treatment, there will be 12 dilutions total per each condition.
Inoculate 5 mL of each dilution onto Middlebrook 7H11 agar plates. To save plates, 12 dilutions can be inoculated onto different slots on the same plate.
Let the plate stand for 5 min to absorb the inoculum. Put plates in a plastic bag. Close the bag tightly (see Note 4). Incubate the plates at 37 °C for 14 d. Colonies should be visible at this time.
Count colonies from dilutions that yield good resolution (1–100 visible colonies).
Repeat the entire assay two more times to obtain data from three different experiments for analysis and plotting (Fig. 21.1A).
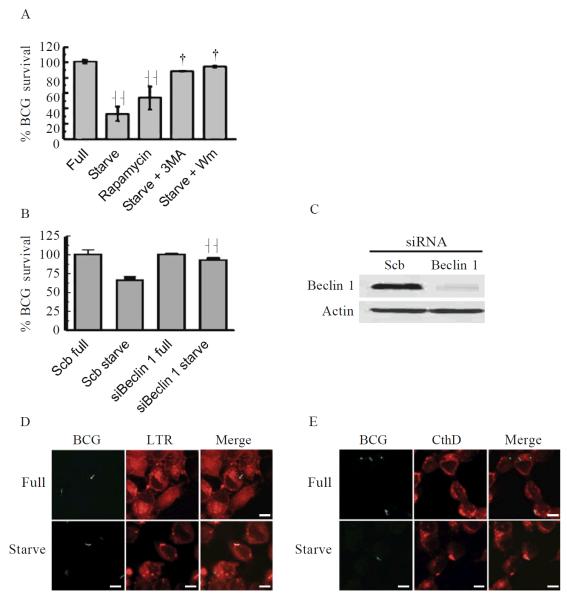
Figure 21.1.
Autophagy increases M. tuberculosis phagosome maturation and eliminates mycobacteria in macrophages. A. Autophagy reduces mycobacterial survival. RAW264.7 macrophages were infected with BCG for 1 h and incubated with or without starvation medium in the presence or absence of rapamycin, 3-methyladenine (3MA), or wortmannin (Wm) for 2 h. Cells were lysed to determine mycobacterial viability. B. and C. Beclin 1 is important for autophagic killing of mycobacteria. RAW264.7 cells were transfected with siRNAs against Beclin 1or scramble control. Protein knockdown was allowed to proceed for 48 h. Mycobacterial viability was determined as in A. Immunoblot analysis was performed to validate Beclin 1knockdown level using Actin as a loading control. D. and E. Autophagic induction enhances acidification and acquisition of a lysosomal protease by the mycobacterial phagosome. RAW264.7 cells were transfected with siRNAs against proteins of interest. Cells were infected with BCG and stained with LysoTracker Red or antibodies against cathepsin D. Quantitative analysis of percent colocalization was performed. Confocal images of cells transfected with siRNAs scramble control subjected to starvation treatment are shown as examples. Data, means ± SEM from three different experiments, **p ∷: 0.01, {p 2' 0.05. Fig. 21.1A is modified from Gutierrez et al. (2004). Figs. 21.1B and 21.C are modified from Delgado et al. (2008).
2.2. siRNA-mediated protein knockdown supplying the autophagic killing
2.2.1. Materials
2.2.1.1. Growth and use of RAW264.7 cell line
Murine RAW264.7 macrophage cell line (ATCC, No. TIB-71).
Dulbecco's modified Eagle's medium (DMEM) (Gibco), 10% fetal bovine serum (FBS; Hyclone), and 4 mM L-glutamine (Gibco) (Full medium): Add 100 mL of FBS and 10 mL of 200 mM L-glutamine to 890 mL of DMEM. Filter through 0.2-mm filter. Store the medium at 4 °C.
175-cm2 and 75-cm2 tissue culture flasks (Greiner Bio-One).
Disposable cell scraper (Sarstedt).
12-well tissue culture plates.
2.2.1.2. Transfection of macrophages with siRNAs
Amaxa Nucleofector Device and Nucleofector solution V (Amaxa).
siRNA duplexes (Dharmacon) designed to specifically target a gene of interest.
2.2.1.3. Immunoblot analysis of siRNA-mediated protein knockdown
BCA protein assay kit (Pierce).
Immunoblotting lysis buffer: 50 mM Tris-HCl, pH 7.4, 150 mM NaCl, 1% Nonidet-P40, 0.25% Na-deoxycholate, 1 mM EDTA, 1mM PMSF, 1 mg/mL aprotinin, 10 mg/mL leupeptin, 1 mg/mL pepstatin, 1 mM Na3VO4, and 1 mM NaF.
12% precast BioRad minigel (BioRad).
MiniProtean 3 Western blot system (BioRad).
SuperSignal West Dura Extended Duration Substrate for western blot detection (Pierce).
2.2.2. Methods
2.2.2.1. Transfection of macrophages with siRNAs
Murine RAW264.7 macrophages are maintained in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS) and 4 mM L-glutamine at 37 °C, 5% CO2.
Subculture the cells at 1:3 dilution 24 h before transfection by scraping off the cells into 12 mL of complete DMEM. Pipette up and down to obtain single cell suspension. Transfer 4 mL of cells into a 175-cm2 tissue culture flask containing 25 mL of complete DMEM. Incubate cells overnight at 37 °C, 5% CO2.
Transfection of cells with siRNAs is performed using an Amaxa Nucleofector Device according to the manufacturer's protocol (see Note 5). Scrape cells into 12 mL of complete DMEM and aliquot 3 mL of cells (approximately 2 × 107 cells) into 15-mL conical tubes.
Pellet the cells at 700 rpm for 5 min at room temperature.
Completely remove DMEM from cells using a vacuum suction system.
Resuspend cells with 100 mL of Nucleofector Solution V containing 1.5 mg of siRNAs and transfer the cell suspension into a cuvette.
Nucleoporate the cells using program D-032.
Transfer cells into a 75-cm2 tissue culture flask containing 12 mL of complete DMEM. Incubate cells for 5–6 h at 37 °C, 5% CO2.
Remove the old medium and add fresh complete DMEM to cells.
At 24 h after transfection, scrape cells into 15-mL conical tubes and determine the cell density using trypan blue staining and a hemocytometer.
For the killing assay outlined in section 2, dilute cells to obtain a density of 3 × 105 cells/mL in complete DMEM and dispense 1 mL into each well of a 12-well plate. Plate 3 wells (triplicate) for each experimental condition. Incubate the plate at 37 °C, 5% CO2 overnight. Plate the rest of the cells back into a 75-cm2 tissue culture flask. Incubate the flasks overnight at 37 °C, 5% CO2.
Perform killing assay (see section 2) at 48 h after transfection (Fig. 21.1B) (see Note 6).
2.2.2.2. Immunoblot analysis of siRNA-mediated protein knockdown
Collect cells from each knockdown condition at 48 h after transfection from the respective 75-cm2 tissue culture flask by scraping off the cells into 15-mL conical tubes.
Pellet the cells at 700 rpm for 5 min at room temperature.
Wash the cells once with 1X PBS, pellet by centrifugation, and remove and discard the supernatant fraction.
Lyse the cells in 0.5 mL of cold lysis buffer on ice for 30 min. Transfer the lysate into 1.5-mL microcentrifuge tubes. Spin at 13,000 rpm for 15 min at 4 °C. Transfer the supernatant fraction containing the protein lysate into new 1.5-mL microcentrifuge tubes.
Determine the protein concentration using BCA protein assay according to the manufacturer's protocol.
Prepare and load 50 mg of total protein per each condition using the standard method of Laemmli and the MiniProtean 3 Western blot system according to the manufacturer's protocol.
Block blots with 5% nonfat dry milk in 1X PBS containing 0.1% Tween 20 for 1 h at room temperature.
Incubate blots with primary antibodies according to the manufacturer's protocol followed by the appropriate horseradish peroxidase-conjugated secondary antibodies.
Develop blots with SuperSignal West Dura Extended Duration Substrate according to the manufacturer's protocol and standard autoradiography (Fig. 21.1C).
2.3. LysoTracker colocalization analysis to monitor the acidification of the M. tuberculosis phagosome
2.3.1. Materials
2.3.1.1. Growth and use of RAW264.7 cell line
Murine RAW264.7 macrophage cell line (ATCC, No. TIB-71).
Dulbecco's modified Eagle's medium (DMEM) (Gibco), 10% fetal bovine serum (FBS; Hyclone), and 4 mM L-glutamine (Gibco) (Full medium): Add 100 mL of FBS and 10 mL of 200 mM L-glutamine to 890 mL of DMEM. Filter through 0.2-mm filter. Store the medium at 4 °C.
175-cm2 tissue culture flasks (Greiner Bio-One).
Disposable cell scraper (Sarstedt).
Earle's balanced salt solution (EBSS) (Starvation medium) (Sigma).
12-well tissue culture plates.
Circular microscope cover glasses (18 mm in diameter) (Fisher).
LysoTracker Red DND-99 1 mM stock solution in DMSO (Molecular Probes).
Fixing solution: Dissolve 2 g of paraformaldehyde in 100 mL of 1X PBS. Heat to 50–60 °C inside a chemical hood until the powder is solubilized. Cool the solution to room temperature, aliquot, and store at −20 °C.
PermaFluor mounting medium (Thermo Scientific, 434990).
2.3.1.2. Growth and preparation of mycobacteria
Middlebrook albumin dextrose catalase (ADC) enrichment (BD Biosciences).
20% Tween 80 (Sigma): Prepare 20% v/v stock at 4 °C and filter through 0.2-mm filter. Store at 4 °C.
Middlebrook 7H9 broth (Difco): Dissolve 4.7 g of powder in 900 mL deionized water with 2 mL of glycerol (EMD Chemicals, Inc.) and autoclave. Temper to room temperature. Add 100 mL of ADC and 2.5 mL of 20% Tween 80. Store at 4 °C.
Sterile, 150-mL tissue-culture roller bottles.
Autoclaved 7-mL Dounce glass homogenizer (Kontes Scientific Glassware).
Phosphate-buffered saline (PBS, 10X) (Gibco): Prepare 1X PBS by adding 100 mL in 900 mL of deionized water and filter through 0.2-mm filter and store at room temperature.
Alexa 488 carboxylic acid succinimidyl ester (Molecular Probes): Prepare 1 mg/mL stock solution by adding 1 mL of 100% DMSO to 1 mg of powder. Store at −20 °C.
2.3.2. Methods
2.3.2.1. Macrophage cell preparation
Dispense 1 mL of cells (3 × 105 cells/mL) into each well of a 12-well plate containing a sterile microscope cover glass. Plate 2 wells for each experimental condition. Incubate the plate at 37 °C, 5% CO2 overnight.
For siRNA-mediated protein knockdown and macrophage cell preparation, see section 2. Plate 3 × 105 cells/well in duplicate for each siRNA knockdown/experimental condition into a 12-well plate containing sterile microscope cover glasses. Incubate the plate at 37 °C, 5% CO2 overnight.
2.3.2.2. M. tuberculosis var. bovis BCG (BCG) labeling with Alexa 488
Transfer 10 mL of a log-phase BCG culture into a 15-mL conical tube. Centrifuge to pellet mycobacteria at 2500 rpm for 5 min at room temperature.
Remove and discard the supernatant fraction and wash once with 10 mL of 1X PBS.
Spin at 2500 rpm for 5 min and remove and discard the supernatant fraction.
Resuspend the pellet fraction in 1 mL of 1X PBS and transfer the cell suspension into a 1.5-mL microcentrifuge tube. Add 10 mL of Alexa 488 caboxylic acid succinimidyl ester stock solution to the tube to achieve a final concentration of 10 mg/mL. Wrap the tube with foil and incubate at room temperature for 45–60 min on a shaker.
Pellet mycobacteria at 8000 rpm for 3 min at room temperature. Remove the supernatant and wash twice with 1 mL of 1X PBS.
Resuspend the mycobacteria pellet in 6 mL of complete DMEM. Transfer to a 7-mL Dounce homogenizer and homogenize 35 times to generate single cell suspension.
Measure OD600 of the 1:10 dilution of the homogenized culture.
Prepare mycobacterial inoculum in complete DMEM at the concentration of 3 × 106 BCG/mL (MOI ¼ 10) using the following formula:(3 × 106); (total mL needed for infection)/(OD600 × 109) ¼ mL homogenate needed.
Add LysoTracker Red 1 mM stock solution to the inoculum to yield a final concentration of 0.25 mM.
2.3.2.3. Infection of macrophages and autophagic induction
At 2 h before infection, remove the old medium from the macrophages and stain the lysosomes with LysoTracker Red by adding 1 mL of complete DMEM containing 0.25 mM LysoTracker Red into each well. Incubate the plate at 37 °C, 5% CO2.
After 2 h of incubation, remove the medium from the macrophages and add 1 mL of Alexa 488-labeled 3 × 106 BCG/mL inoculum into each well.
Spin at 1,200 rpm for 5 min to settle mycobacteria onto macrophages. Incubate the plate at 37 °C, 5% CO2 for 15 min (pulse period).
Remove the inoculum and quickly wash each well 3 times with complete DMEM. Add 1 mL of complete DMEM containing 0.25 mM LysoTracker Red into each well and incubate the plate at 37 °C, 5% CO2 for 60 min (chase period).
To induce autophagy, remove complete DMEM from the respective wells and quickly wash 3 times with 2 mL of starvation medium, EBSS. Add 1 mL of EBSS containing 0.25 mM LysoTracker Red into these wells and put the plate back into the incubator. Incubate for 2 h.
2.3.2.4. Immunofluorescence confocal microscopy
After 2 h of incubation, fix the cells with 1 mL of 2% paraformaldehyde/PBS for 10 min at room temperature.
Wash the cells 3 times for 5 min each with 1X PBS.
Mount coverslips onto microscope slides using PermaFluor mounting medium.
Collect 0.7-mm-thick optical sections using a 63x oil objective on a LSM META 510 (Carl Zeiss) and prepare images using Zeiss LSM Image Browser.
Count at least 50 phagosomes per each condition and calculate the percentage of LysoTracker Red-BCG colocalization.
Repeat the entire assay two more times to obtain data from three different experiments for analysis and plotting (Fig. 21.1D).
2.4. Monitoring lysosomal protease delivery to the M. tuberculosis phagosome
2.4.1. Materials
2.4.1.1. Growth and use of RAW264.7 cell line
Murine RAW264.7 macrophage cell line (ATCC, No. TIB-71).
Dulbecco's modified Eagle's medium (DMEM) (Gibco), 10% fetal bovine serum (FBS; Hyclone), and 4 mM L-glutamine (Gibco) (Full medium): Add 100 mL of FBS and 10 mL of 200 mM L-glutamine to 890 mL of DMEM. Filter through 0.2-mm filter. Store the medium at 4 °C.
175-cm2 tissue culture flasks (Greiner Bio-One).
Disposable cell scraper (Sarstedt).
Earle's balanced salt solution (EBSS) (Starvation medium) (Sigma).
12-well tissue culture plates.
Circular microscope cover glasses (18 mm in diameter) (Fisher).
Fixing solution: Dissolve 2 g of paraformaldehyde in 100 mL of 1X PBS. Heat to 50–60 °C inside a chemical hood until the powder is solubilized. Cool the solution to room temperature, aliquot, and store at −20 °C.
Permeabilization solution: 0.1% (v/v) Triton-X 100 in 1X PBS.
Blocking solution: Dissolve 3 g of bovine serum albumin Fraction V (BSA) (Sigma) in 1X PBS containing 2% (v/v) of the appropriate serum matching the host of the secondary antibody.
Primary antibody: goat anticathepsin D antibody (R&D Systems).
Secondary antibody: Alexa 568 donkey antigoat IgG (Molecular Probes).
PermaFluor mounting medium (Thermo Scientific, [Cat. No. 434990]).
2.4.1.2. Growth and preparation of mycobacteria
Middlebrook albumin dextrose catalase (ADC) enrichment (BD Biosciences).
20% Tween 80 (Sigma): Prepare 20% v/v stock at 4 °C and filter through 0.2-mm filter. Store at 4 °C.
Middlebrook 7H9 broth (Difco): Dissolve 4.7 g of powder in 900 mL deionized water with 2 mL of glycerol (EMD Chemicals) and autoclave. Temper to room temperature. Add 100 mL of ADC and 2.5 mL of 20% Tween 80. Store at 4 °C.
Sterile, 150-mL tissue-culture roller bottles.
Autoclaved 7-mL Dounce glass homogenizer (Kontes Scientific Glassware).
Phosphate-buffered saline (PBS, 10X) (Gibco): Prepare 1X PBS by adding 100 mL in 900 mL of deionized water. Filter through 0.2-mm filter and store at room temperature.
Alexa 488 carboxylic acid succinimidyl ester (Molecular Probes): Prepare 1 mg/mL stock solution by adding 1 mL of 100% DMSO to 1 mg of powder. Store at −20 °C.
2.4.2. Methods
2.4.2.1. Macrophage cell preparation
2.4.2.2. M. tuberculosis var. bovis BCG (BCG) labeling with Alexa 488 (see section 2.3.2.2., omit LysoTracker red in step 9)
2.4.2.3. Infection of macrophages and autophagic induction
Remove the old medium from macrophages and add 1 mL of Alexa 488-labeled 3 × 106 BCG/mL inoculum into each well.
Spin at 1200 rpm for 5 min to settle mycobacteria onto macrophages. Incubate the plate at 37 °C, 5% CO2 for 15 min (pulse period).
Remove the inoculum and quickly wash each well 3 times with 1 mL of complete DMEM. Add 1 mL of complete DMEM into each well and incubate the plate at 37 °C, 5% CO2 for 60 min (chase period).
To induce autophagy, remove complete DMEM from the respective wells and quickly wash 3 times with 2 mL of starvation medium, EBSS. Add 1 mL of EBSS into the wells. Incubate the plate for 4 h at 37 °C, 5% CO2 (see Note 7).
2.4.2.4. Immunofluorescence confocal microscopy
Remove the media from each well and fix the cells with 1 mL of 2% paraformaldehyde/PBS for 10 min at room temperature.
Wash the cells 3 times for 5 min each with 1X PBS.
Permeabilize the cells with 1 mL of 0.1% Triton X-100/PBS for 3 min at room temperature.
Wash 3 times for 5 min each with 1X PBS.
Block the cells for 1 h at room temperature using the blocking solution.
Prepare primary antibody in blocking solution according to the manufacturer's protocol.
Invert cover slips onto 50-uL droplets of primary antibody/blocking solution dispensed onto Parafilm placed over a hard surface.
Cover samples, seal with Parafilm, and incubate overnight at 4 °C.
Transfer the cover slips back into the plate and wash the cover slips 3 times for 5 min each with 1X PBS.
Prepare appropriate secondary antibody in blocking solution at 1:500 dilution.
Add 1 mL into each well and incubate for 2 h at room temperature.
Wash 3 times for 5 min each with 1X PBS.
Mount coverslips onto microscope slides using PermaFluor mounting medium.
Collect 0.7-mm-thick optical sections using a 63x oil objective on a LSM META 510 (Carl Zeiss) and prepare images using Zeiss LSM Image Browser.
Count at least 50 phagosomes per each condition and calculate the percentage of cathepsin D-BCG colocalization.
Repeat the entire assay two more times to obtain data from three different experiments for analysis and plotting (Fig. 21.1E).
ACKNOWLEDGMENTS
We thank Sharon S. Master and Alexander S. Davis for sharing techniques and materials and Esteban A. Roberts for critical reading of the manuscript. This work is supported by NIH grants A1069345, A145148, and A142999.
Footnotes
To prevent premature autophagic induction, it is necessary to wash the cells using complete DMEM instead of using 1X PBS.
To prevent cell detachment during the assay, it is important to wash the cells gently by adding the media along the side of the plastic well and aspirate off the media from the well using a vacuum suction system with a pipette tip attached to the end of a glass pipette to reduce the suction force.
The incubation time depends on the experimental conditions being tested. For autophagic killing of mycobacteria, 4 h incubation is routinely used for the assay.
Putting the plates in a plastic bag helps to prevent the plates from drying out and from contamination with other microorganisms.
Optimized transfection protocols for many cell types are available from the manufacturer's website (http://www.amaxa.com).
The incubation time to obtain the desirable knockdown level of a protein of interest needs to be predetermined for each individual protein.
The incubation time depends on the experimental conditions being tested. For analysis of autophagic-mediated acidification of M. tuberculosis phagosomes using LysoTracker Red staining, 2 h is found to be optimum. For analysis of autophagic-mediated delivery of a lysosomal protease into the M. tuberculosis phagosome using cathepsin D staining, 4 h is found to be best.
REFERENCES
- Alonso S, Pethe K, Russell DG, Purdy GE. Lysosomal killing of Mycobacterium mediated by ubiquitin-derived peptides is enhanced by autophagy. Proc. Natl. Acad. Sci. USA. 2007;104:6031–6036. doi: 10.1073/pnas.0700036104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beatty WL, Rhoades ER, Ullrich HJ, Chatterjee D, Heuser JE, Russell DG. Trafficking and release of mycobacterial lipids from infected macrophages. Traffic. 2000;1:235–247. doi: 10.1034/j.1600-0854.2000.010306.x. [DOI] [PubMed] [Google Scholar]
- Bernales S, Schuck S, Walter P. ER-phagy: selective autophagy of the endoplasmic reticulum. Autophagy. 2007;3:285–287. doi: 10.4161/auto.3930. [DOI] [PubMed] [Google Scholar]
- Budha NR, Lee RE, Meibohm B. Biopharmaceutics, pharmacokinetics and pharmacodynamics of antituberculosis drugs. Curr. Med. Chem. 2008;15:809–825. doi: 10.2174/092986708783955509. [DOI] [PubMed] [Google Scholar]
- Chan J, Flynn J. The immunological aspects of latency in tuberculosis. Clin. Immunol. 2004;110:2–12. doi: 10.1016/s1521-6616(03)00210-9. [DOI] [PubMed] [Google Scholar]
- Corbett EL, Watt CJ, Walker N, Maher D, Williams BG, Raviglione MC, Dye C. The growing burden of tuberculosis: global trends and interactions with the HIV epidemic. Arch. Intern. Med. 2003;163:1009–1021. doi: 10.1001/archinte.163.9.1009. [DOI] [PubMed] [Google Scholar]
- Delgado MA, Elmaoued RA, Davis AS, Kyei G, Deretic V. Toll-like receptors control autophagy. EMBO J. 2008;27:1110–1121. doi: 10.1038/emboj.2008.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deretic V, Vergne I, Chua J, Master S, Singh SB, Fazio JA, Kyei G. Endosomal membrane traffic: Convergence point targeted by Mycobacterium tuberculosis and HIV. Cell. Microbiol. 2004;6:999–1009. doi: 10.1111/j.1462-5822.2004.00449.x. [DOI] [PubMed] [Google Scholar]
- Devenish RJ. Mitophagy: Growing in intricacy. Autophagy. 2007;3:293–294. doi: 10.4161/auto.4273. [DOI] [PubMed] [Google Scholar]
- Ducati RG, Ruffino-Netto A, Basso LA, Santos DS. The resumption of consumption–a review on tuberculosis. Mem. Inst. Oswaldo Cruz. 2006;101:697–714. doi: 10.1590/s0074-02762006000700001. [DOI] [PubMed] [Google Scholar]
- Gutierrez MG, Master SS, Singh SB, Taylor GA, Colombo MI, Deretic V. Autophagy is a defense mechanism inhibiting BCG and Mycobacterium tuberculosis survival in infected macrophages. Cell. 2004;119:753–766. doi: 10.1016/j.cell.2004.11.038. [DOI] [PubMed] [Google Scholar]
- Harris J, De Haro SA, Master SS, Keane J, Roberts EA, Delgado M, Deretic V. T helper 2 cytokines inhibit autophagic control of intracellular Mycobacterium tuberculosis. Immunity. 2007;27:505–517. doi: 10.1016/j.immuni.2007.07.022. [DOI] [PubMed] [Google Scholar]
- Harris J, De Haro S, Deretic V. Autophagy and Mycobacterium tuberculosis. In: Deretic V, editor. Autophagyin Immunity and Infection. WILEY-VCH, Weinheim; 2006. pp. 129–138. [Google Scholar]
- Kaufmann SH, Baumann S, Nasser Eddine A. Exploiting immunology and molecular genetics for rational vaccine design against tuberculosis. Int. J. Tuberc. Lung. Dis. 2006;10:1068–1079. [PubMed] [Google Scholar]
- Klionsky DJ, Cuervo AM, Seglen PO. Methods for monitoring autophagyfrom yeast to human. Autophagy. 2007;3:181–206. doi: 10.4161/auto.3678. [DOI] [PubMed] [Google Scholar]
- Levine B, Klionsky DJ. Development by self-digestion: molecular mechanisms and biological functions of autophagy. Dev. Cell. 2004;6:463–477. doi: 10.1016/s1534-5807(04)00099-1. [DOI] [PubMed] [Google Scholar]
- Liang C, E X, Jung JU. Downregulation of autophagy by herpesvirus Bcl-2 homologs. Autophagy. 2008;4:268–272. doi: 10.4161/auto.5210. [DOI] [PubMed] [Google Scholar]
- Ling YM, Shaw MH, Ayala C, Coppens I, Taylor GA, Ferguson DJ, Yap GS. Vacuolar and plasma membrane stripping and autophagic elimination of Toxoplasma gondii in primed effector macrophages. J. Exp. Med. 2006;203:2063–2071. doi: 10.1084/jem.20061318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizushima N. Methods for monitoring autophagy. Int. J. Biochem. Cell. Biol. 2004;36:2491–2502. doi: 10.1016/j.biocel.2004.02.005. [DOI] [PubMed] [Google Scholar]
- Mizushima N, Klionsky DJ. Protein turnover via autophagy: implications for metabolism. Annu. Rev. Nutr. 2007;27:19–40. doi: 10.1146/annurev.nutr.27.061406.093749. [DOI] [PubMed] [Google Scholar]
- Nakagawa I, Amano A, Mizushima N, Yamamoto A, Yamaguchi H, Kamimoto T, Nara A, Funao J, Nakata M, Tsuda K, Hamada S, Yoshimori T. Autophagydefends cells against invading group A Streptococcus. Science. 2004;306:1037–1040. doi: 10.1126/science.1103966. [DOI] [PubMed] [Google Scholar]
- Ogawa M, Yoshimori T, Suzuki T, Sagara H, Mizushima N, Sasakawa C. Escape of intracellular Shigella from autophagy. Science. 2005;307:727–731. doi: 10.1126/science.1106036. [DOI] [PubMed] [Google Scholar]
- Rubinsztein DC. The roles of intracellular protein-degradation pathways in neurodegeneration. Nature. 2006;443:780–786. doi: 10.1038/nature05291. [DOI] [PubMed] [Google Scholar]
- Russell DG. Mycobacterium tuberculosis: here today, and here tomorrow. Nat. Rev. Mol. Cell Biol. 2001;2:569–577. doi: 10.1038/35085034. [DOI] [PubMed] [Google Scholar]
- Russell DG. Who puts the tubercle in tuberculosis? Nat Rev Microbiol. 2007;5:39–47. doi: 10.1038/nrmicro1538. [DOI] [PubMed] [Google Scholar]
- Russell DG, Mwandumba HC, Rhoades EE. Mycobacterium and the coat of many lipids. J. Cell Biol. 2002;158:421–426. doi: 10.1083/jcb.200205034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakai Y, Oku M, van der Klei IJ, Kiel JAWK. Pexophagy: autophagic degradation of peroxisomes. Biochim. Biophys. Acta. 2006;1763:1767–1775. doi: 10.1016/j.bbamcr.2006.08.023. [DOI] [PubMed] [Google Scholar]
- Schmid D, Münz C. Immune surveillance of intracellular pathogens via autophagy. Cell. Death. Differ. 2005;12(Suppl. 2):1519–1527. doi: 10.1038/sj.cdd.4401727. [DOI] [PubMed] [Google Scholar]
- Schmid D, Pypaert M, Münz C. Antigen-loading compartments for major histocompatibility complex class II molecules continuously receive input from autophagosomes. Immunity. 2007;26:79–92. doi: 10.1016/j.immuni.2006.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SB, Davis AS, Taylor GA, Deretic V. Human IRGM induces autophagyto eliminate intracellular mycobacteria. Science. 2006;313:1438–1441. doi: 10.1126/science.1129577. [DOI] [PubMed] [Google Scholar]
- Tallóczy Z, Jiang W, Virgin HW, IV, Leib DA, Scheuner D, Kaufman RJ, Eskelinen E-L, Levine B. Regulation of starvation- and virus-induced autophagy by the eIF2a kinase signaling pathway. Proc. Natl. Acad. Sci. USA. 2002;99:190–195. doi: 10.1073/pnas.012485299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I, Chua J, Deretic V. Tuberculosis toxin blocking phagosome maturation inhibits a novel Ca2þ/calmodulin-PI3K hVPS34 cascade. J. Exp. Med. 2003;198:653–659. doi: 10.1084/jem.20030527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vergne I, Chua J, Singh SB, Deretic V. Cell biology of mycobacterium tuberculosis phagosome. Annu. Rev. Cell Dev. Biol. 2004;20:367–394. doi: 10.1146/annurev.cellbio.20.010403.114015. [DOI] [PubMed] [Google Scholar]
- WHO Global tuberculosis control-surveillance, planning, and financing. 2008 http://www.who.int/tb/publications/global_report/2008/summary/en/index.html.
- Williams A, Jahreiss L, Sarkar S, Saiki S, Menzies FM, Ravikumar B, Rubinsztein DC. Aggregate-prone proteins are cleared from the cytosol by autophagy:Therapeutic implications. Curr. Top. Dev. Biol. 2006;76:89–101. doi: 10.1016/S0070-2153(06)76003-3. [DOI] [PubMed] [Google Scholar]