Abstract
Hexavalent chromium [Cr(VI)] generated during industrial processes is carcinogenic. Although much is known about the deleterious effects caused by reactive oxygen species generated during the reduction of Cr(VI) after its absorption by biological systems, the precise mechanisms underlying Cr(VI) cytotoxicity remain poorly defined. Here, we analyzed, at the global proteome scale, the perturbation of protein expression in GM00637 human skin fibroblast cells upon exposure to potassium dichromate (K2Cr2O7). We were able to quantify ~ 4600 unique proteins, among which ~ 400 exhibited significant alterations in expression levels upon a 24-hr treatment with 0.5 μM K2Cr2O7. Pathway analysis revealed the Cr(VI)-induced perturbation of cholesterol biosynthesis, G-protein signaling, inflammatory response, and selenoprotein pathways. In particular, we discovered that the K2Cr2O7 treatment led to pronouncedly elevated expression of a large number of enzymes involved in de novo cholesterol biosynthesis. Real-time PCR analysis revealed the increased mRNA expression of selected genes involved in cholesterol biosynthesis. Consistently, K2Cr2O7 treatment resulted in marked increases in cellular cholesterol level in multiple cell lines. Moreover, the Cr(VI)-induced growth inhibition of cultured human cells could be rescued by a cholesterol-lowering drug, lovastatin. Together, we demonstrated, for the first time, that Cr(VI) may exert its cytotoxic effect, at least partly, through the up-regulation of enzymes involved in de novo cholesterol biosynthesis and the resultant increase of cholesterol level in cells.
Keywords: Hexavalent chromium, human skin fibroblast, quantitative proteomics, mass spectrometry, SILAC, cholesterol biosynthesis
Introduction
Heavy metal toxication has become a leading aspect of environmental pollution over the past century. Generally speaking, heavy metals exert their cytotoxic effects mainly through imitating the action of essential elements and forming complexes or ligands with cellular components which cause malfunction and eventually death of the affected individual.1 As a representative heavy metal, chromium is an indispensable source for metallurgical, chemical and refractory industries; however, it is also a well documented occupational and environmental health hazard.2 Unlike other toxic heavy metals, such as lead, cadmium and arsenic, chromium toxicity depends on its oxidation state.3 Among the different oxidation states, hexavalent chromium [Cr(VI)] is carcinogenic, although trivalent chromium [Cr(III)] in trace amounts is an essential dietary nutrient.4 Exposure to Cr(VI) occurs primarily through inhalation, dermal contact and ingestion. Previous studies have shown that Cr(VI) species are more readily absorbed by biological systems than Cr(III), because Cr(VI) can be easily uptaken into cells through non-specific phosphate and sulfate ion transporters.5, 6
Cr(VI) is an oxidizing agent, which undergoes facile reduction to yield Cr(III) in plasma and cells. This process appears to be somewhat paradoxical. On one hand, the reduction of Cr(VI) to Cr(III) is the first defense against Cr(VI) after exposure. For instance, inhaled Cr(VI) can be reduced to Cr(III) in the lower respiratory tract by the epithelial lining fluid and pulmonary alveolar macrophages, which minimizes the cellular uptake of chromium species.7 On the other hand, Cr(VI) toxicity also arises mainly from its reduction, but this occurs intracellularly after it enters the cells. Under physiological conditions, Cr(VI) can be reduced by cellular reductants, including ascorbic acid, glutathione, and enzymes (e.g. glutathione reductase) to produce Cr(V), Cr(IV) and ultimately, Cr(III).4 These reduction processes also generate other reactive intermediates including thiyl radicals and hydroxyl radicals.4 The reactive oxygen species (ROS) generated during Cr(VI) reduction can modify DNA to form DNA-chromium complexes, DNA strand breaks, DNA-DNA cross-links and DNA-protein cross-links.8 They can also attack proteins and membrane lipids resulting in the disruption of cellular integrity and functions.9
Although intracellular reduction of Cr(VI) is known to be important for its toxicity, more studies are needed to systematically explore the exact modes of action. DNA microarray has been used for exploring Cr(VI)-induced changes in gene expression by providing a quantitative measurement of gene transcription.10 However, microarray analysis can only assess the elevated or depressed mRNA expression levels and it provides little information about changes in protein expression, considering that protein expression is also affected by translational regulation and, more importantly, protein turnover.11
Recent advances in mass spectrometry instrumentation with improved mass accuracy and resolution render it possible to conduct rapid and high-throughput identification and quantification of proteins at the entire proteome scale.12 Here we utilized a mass spectrometry-based quantitative proteomic technique using stable isotope labeling by amino acids in cell culture (SILAC)13 to elucidate the cellular mechanisms targeted by Cr(VI) via interrogating the toxicant-induced perturbation of the whole proteome. With this method, we were able to quantify a total of 4607 unique proteins, among which 270 and 127 were significantly up- and down-regulated, respectively. Importantly, our study revealed that, apart from several other cellular pathways, Cr(VI) induced up-regulation of many enzymes involved in cholesterol biosynthesis. This unprecedented finding, confirmed by follow-up studies, allowed us to unveil that Cr(VI) may exert its cytotoxic effect, at least in part, by stimulating cholesterol biosynthesis.
Materials and Methods
Cell culture
All reagents unless otherwise noted were purchased from Sigma-Aldrich (St. Louis, MO) and all culture medium unless otherwise stated were from ATCC (Manassas, VA). GM00637 and HCT-116 cells were generously provided by Profs. Gerd P. Pfeifer (the City of Hope) and Frances Sladek (University of California Riverside), respectively. HEK293T cells were obtained from ATCC. Control GM00637 and HEK293T cells were cultured in Dulbecco's Modified Eagle's Medium (DMEM, ATCC) supplemented with 10% fetal bovine serum (FBS, Invitrogen, Carlsbad, CA) and 100 IU/ml penicillin (ATCC). HCT-116 cells were cultured under the same conditions except that McCoy's 5a medium (ATCC) was used. Cells were maintained in a humidified atmosphere with 5% CO2 at 37°C, with medium renewal at every 1–2 days depending on cell densities. MTT assay was performed using a kit from Roche (Basel, Switzerland) and absorbance reading was taken using a Victor 2 plate reader (Perkin Elmer, Waltham, MA, Figure S1).
DMEM medium without lysine or arginine (DMEM minus L-lysine and L-arginine) was purchased from Cambridge Isotope Laboratories (Andover, MA). The complete light and heavy DMEM medium were prepared by adding light or heavy ([13C6,15N2]-L-lysine and [13C6]-L-arginine, Sigma) lysine and arginine to the above medium at amounts according to ATCC formulation. Cells were cultured in light or heavy DMEM medium containing dialyzed FBS (Invitrogen, Grand Island, NY) for at least 10 days (more than 5 cell doublings) to ensure complete stable isotope incorporation.
Potassium dichromate treatment and sample preparation
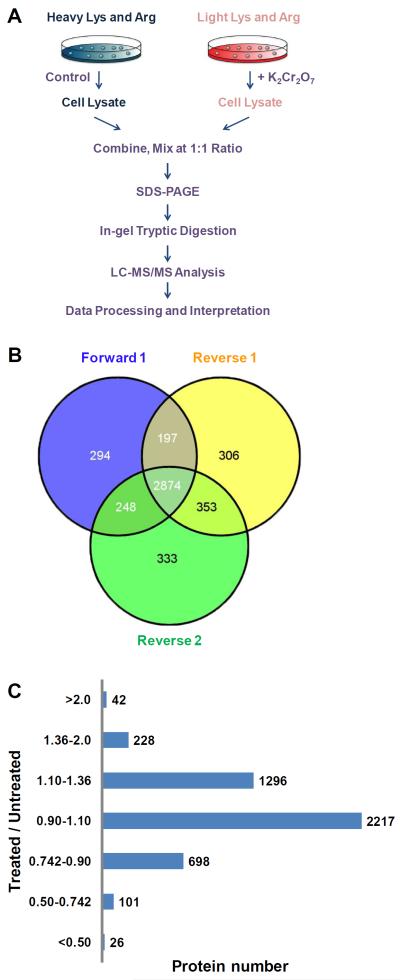
GM00637 cells, cultured in 75 cm2 flasks at a density of ~ 5×106 cells per flask in light or heavy DMEM medium, were treated with 0.5 μM K2Cr2O7 for 24 hrs. The cells were subsequently harvested by centrifugation at 300 g at 4°C for 5 min, washed for three times with ice-cold phosphate-buffered saline (PBS), and lysed with 2% SDS solution on ice with vortexing every other 5 min for a total of 30 min. The mixture was subsequently boiled for 5 min. The resulting cell lysate was centrifuged at 16,000 g at 4°C for 5 min and supernatant collected. The protein concentration in the cell lysate was measured using Bicinchoninic Acid Kit for Protein Determination (Sigma). In forward SILAC, the lysate of light labeled, K2Cr2O7-treated cells and that of the heavy labeled, control cells were combined at 1:1 ratio (w/w), whereas the heavy labeled, K2Cr2O7-treated cell lysate was mixed equally with the light labeled, control lysate in the reverse SILAC experiment (Figure 1A).
Figure 1.
An SILAC-based quantitative proteomics approach to reveal the potassium dichromate-induced perturbation of global protein expression. (A) A flowchart illustrating forward SILAC combined with LC-MS/MS for the comparative analysis of protein expression in GM00637 cells upon potassium dichromate treatment. (B) A summary of the number of proteins quantified from three independent SILAC experiments. (C) The distribution of expression ratios (treated/untreated) for proteins that were quantified in at least one set of SILAC experiment.
SDS-PAGE separation and in-gel digestion
The above equi-mass mixture of light and heavy lysates were denatured by heating to 95°C in Laemmli loading buffer and then separated on a 12% SDS-PAGE gel with a 4% stacking gel. The resulting gel was stained with Coomassie blue and equally cut into 20 slices. The proteins in each individual gel slices were reduced in-gel with dithiothreitol (DTT) and alkylated with iodoacetamide (IAA). The processed proteins were subsequently digested with trypsin (Promega, Madison, WI) at 37°C overnight. Subsequently, peptides were extracted from gels with a solution containing 5% acetic acid in H2O and then CH3CN/H2O (1:1, v/v).
Desalting and purification
The protein digests were desalted and purified using OMIX C18 pipette tips (Agilent, Santa Clara, CA). Formic acid was added to the peptide samples until the solution pH reached ~4. The OMIX tips were hydrated in CH3CN/H2O (1:1, v/v) and solvent discarded, followed by equilibration with 0.1% formic acid. Thereafter, pre-treated samples were aspirated and dispensed into OMIX tip. After rinsing the tip with 0.1% formic acid, bound peptides were eluted with CH3CN/H2O (1:1, v/v). The resulting peptide solution was dried in SpeedVac concentrator and stored at −80°C until further analysis.
LC-MS/MS for protein identification and quantification
On-line LC-MS/MS analyses were conducted on an LTQ-Orbitrap Velos mass spectrometer equipped with a nanoelectrospray ionization source and coupled with an EASY n-LCII HPLC system (Thermo, San Jose, CA). HPLC separation was carried out automatically using a homemade trapping column (150 μm×50 mm) and a separation column (75 μm×200 mm, packed with ReproSil-Pur C18-AQ resin, 3 μm in particle size and 100 Å in pore size, Dr. Maisch HPLC GmbH, Germany). Initially, the peptide mixture was loaded onto the trapping column with a mobile phase of 0.1% formic acid in H2O at a flow rate of 3.0 μL/min. The peptides were then separated using a 120-min linear gradient of 2–40% CH3CN in 0.1% formic acid at a flow rate of 230 nL/min. The LTQ-Orbitrap Velos mass spectrometer was operated in the positive-ion mode, and the spray voltage was set at 1.8 kV. All MS/MS spectra were acquired in a data-dependent scan mode, where one full-scan MS was followed with twenty MS/MS scans. The full-scan MS spectra (from m/z 350 to 2000) were acquired with a resolution of 60,000 at m/z 400 after accumulation to a target value of 500,000. The twenty most abundant ions detected in MS at a threshold above 500 counts were selected for further fragmentation by collision-induced dissociation at a normalized collision energy of 35%.
Data processing
The identification and quantification of global proteome were achieved by searching the LC-MS/MS data using Maxquant14, Version 1.2.2.5 against the International Protein Index database, version 3.68 with 87,083 entries to which contaminants and reverse sequences were added. The maximum number of miss-cleavages for trypsin was set at two per peptide. Cysteine carbamidomethylation and methionine oxidation were included as fixed and variable modifications, respectively. The search was performed with the tolerances in mass accuracy of 10 ppm and 0.6 Da for MS and MS/MS, respectively. In addition, only proteins with at least two distinct peptides being discovered from LC-MS/MS analysis were considered reliably identified.
The normalized protein expression ratio reported in the present study was determined by Maxquant, with the assumption that the expression levels of the majority of proteins remained unchanged upon K2Cr2O7 treatment. The required false positive rate was set to be 1% at both peptide and protein levels. The minimal required peptide length was set at six amino acids. The quantification was based on three independent SILAC and LC-MS/MS experiments, which included one forward and two reverse SILAC labelings. The threshold of significant protein expression changes was determined by the corresponding significant A value for every protein ratio using Perseus 4.0 (Figure S2).14 Only those proteins with alteration in expression levels being greater than 1.36 or less than 0.742 fold, as revealed by SILAC labeling experiments, were considered significantly changed. Subsequent pathway and GO analysis of significantly altered proteins was conducted using Gene Map Annotator and Pathway Profiler (GenMAPP-CS).15
RNA extraction and quantitative real-time PCR analysis
Total RNA was extracted using the RNeasy Mini Kit (QIAGEN, Valencia, CA) and reverse transcribed by employing M-MLV reverse transcriptase (Promega, Madison, WI) and a poly(dT) primer. Quantitative real-time PCR was performed with iQ SYBR Green Supermix kit (Bio-Rad, Hercules, CA) on a Bio-Rad MyiQ thermal cycler, and gene-specific primers are listed in Supporting Table S1. The comparative cycle threshold (Ct) method (ΔΔCt) was utilized for the relative quantification of gene expression level,16 and GAPDH gene was used as internal control. The mRNA level of each gene was normalized to that of the internal control.
Extraction and determination of the cellular cholesterol level
Cells were collected and washed for 3 times with ice-cold PBS and extracted with chloroform:methanol:water (2:1.1:0.9, v/v/v), following previously published procedures.17 The bottom chloroform layer was then washed three times with a methanol-water mixture (5:4, v/v), collected, and solvent removed using a SpeedVac. The cholesterol level was measured using HPLC as recently described.18 Cholesterol amount was normalized against the total protein content determined by the Bradford Assay (Bio-Rad).
Results and Discussion
Potassium dichromate treatment, protein identification, and quantification
It is generally believed that intracellular reduction of Cr(VI) to Cr(III) can give rise to ROS, which are capable of inducing DNA damage and triggering inflammatory response. However, it remains possible that Cr(VI) also exerts its cytotoxic effects by altering other cellular pathways. To explore this, we set out to employ an unbiased quantitative proteomic approach to assess the Cr(VI)-induced perturbation of the entire proteome of GM00637 human skin fibroblast cells. We first determined the optimal dose of K2Cr2O7 by examining the dose-dependent survival rate of GM00637 cells. On the basis of MTT assay, we observed a less than 7% cell death after a 24-hr treatment with 0.5 μM K2Cr2O7, whereas a significant reduction in cell viability (by ~30%) was induced by a 24-hr treatment with 2 μM K2Cr2O7 (Figure S1). Thus, 0.5 μM K2Cr2O7 was selected for the subsequent experiments to minimize the apoptosis-induced alteration in protein expression.
GM00637 cells were cultured in light or heavy medium for more than 5 cell doublings. After treatment with 0.5 μM K2Cr2O7 for 24 hrs, the cells were lysed, and the lysates combined and subsequently fractionated with SDS-PAGE. After in-gel digestion, the proteins were identified and quantified using LC-MS/MS. To obtain reliable quantification results, we conducted SILAC experiments in triplicate, with one set of forward and two sets of reverse labelings (Figure 1A).
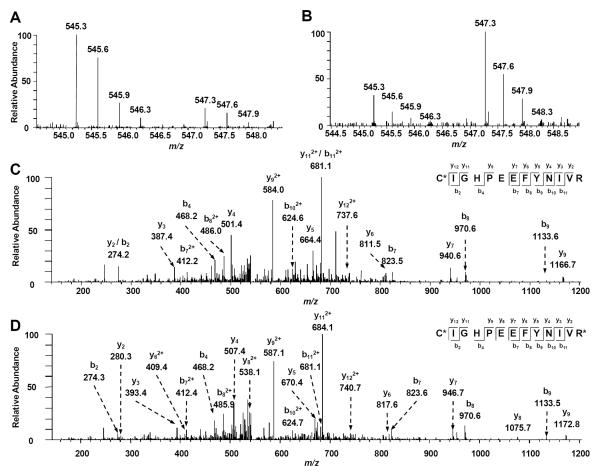
Protein identification and quantification were achieved by searching the mass spectrometric data using MaxQuant. Figure 2 depicts the representative LC-MS/MS results of the peptide CIGHPEEFYNIVR from squalene synthase. The MS results, for both the forward and reverse SILAC labeling samples, revealed the significantly higher level of this peptide in K2Cr2O7-treated cells than in control cells (Figure 2A&B), supporting the increased expression of the protein from which the peptide was derived. In addition, the MS/MS results supported unambiguously the identification of this peptide (Figure 2C&D).
Figure 2.
Representative ESI-MS and MS/MS data revealing the Cr (VI) induced up regulation of squalene synthase. Shown are the MS for the [M+2H]2+ ions of squalene synthase peptide C*IGHPEEFYNIVR and C*IGHPEEFYNIVR* (“C*” represents carbamidomethylated cysteine and “R*” designates the heavy labeled arginine) from the forward (A) and reverse (B) SILAC experiments. Depicted in (C) and (D) are the MS/MS for the [M+2H]2+ ions of C*IGHPEEFYNIVR and C*IGHPEEFYNIVR*, respectively.
The LC-MS and MS/MS results from the three SILAC labeling experiments enabled us to identify 5469 proteins, among which 4607 were quantified (Table S2). Of these proteins, 2784, 798, and 933 could be quantified in all three SILAC labeling experiments, only in two labeling experiments, and only in one labeling experiment, respectively (Figure 1B).
Pathway and GO analysis for significantly changed proteins upon Cr(VI) treatment
The distribution of the changes in protein expression levels induced by K2Cr2O7 treatment is shown in Figure 1C. The majority of the 4607 quantified proteins did not display significant alterations in expression levels. To screen for significantly changed proteins, we first used Perseus to calculate the significant A value for each protein ratio. As shown in Figure S1, the chart is plotted by significant A versus log10(Ratio). Proteins with significant A values of < 0.05 were considered significantly changed. By using this criterion, proteins exhibiting changes that are greater than 1.360- or less than 0.742-fold are considered significantly altered upon K2Cr2O7 treatment, and a total of 270 and 127 proteins were significantly up- and down-regulated, respectively.
All significantly changed proteins were included for pathway analysis using GenMAPP. Among the perturbed pathways identified, only one was down-regulated and the rest up-regulated. We also used GenMAPP to analyze our data from the perspective of GO annotation, such as biological process, cellular component and molecular function (Tables S4&S5). From the GenMAPP analysis results, we chose the top five significant pathways based on their importance and Z scores (Tables S4&S5). Next we discuss further several pathways that are altered by Cr(VI) exposure.
Potassium dichromate-induced perturbation of cholesterol biosynthesis
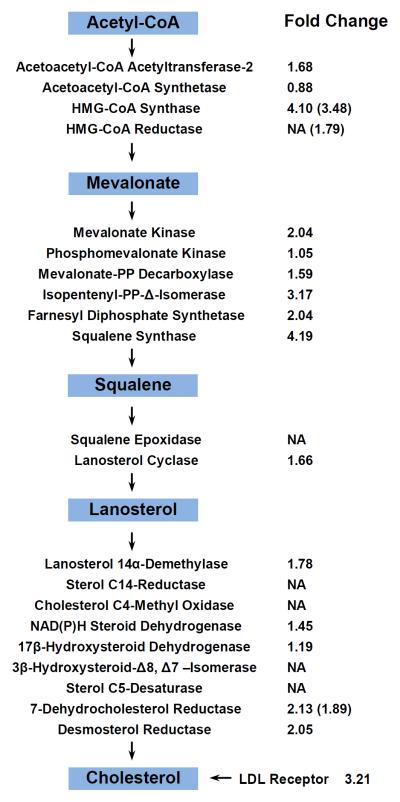
GenMAPP pathway and GO analysis demonstrated that the cholesterol biosynthesis pathway was significantly perturbed following Cr(VI) exposure (Figure 3). In this vein, we were able to quantify 16 enzymes involved in the cholesterol biosynthesis pathway, 13 of which were significantly up-regulated (by 1.4–4.2 fold, Figure 3). Additionally, the level of low-density lipoprotein (LDL) receptor, which mediates the cellular uptake of LDL cholesterol,19 is also up-regulated by 3.2 fold.
Figure 3.
The key intermediary steps in de novo cholesterol biosynthesis. The fold changes of quantified enzymes are shown. NA, not available (i.e., not quantified in SILAC experiments). The fold changes of mRNA expression level measured by real-time PCR are shown in the parenthesis, and the values represent mean of results obtained from three independent experiments.
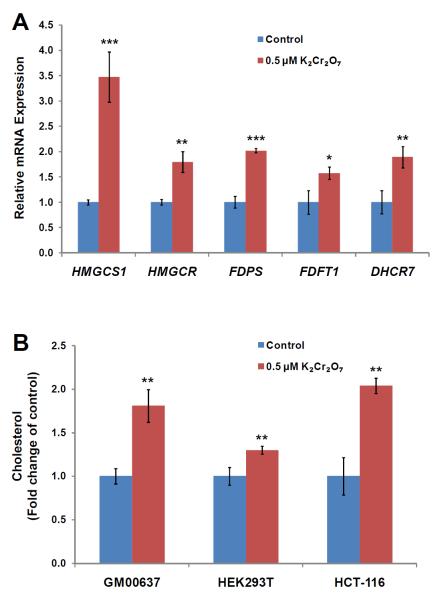
Sterol regulatory element binding proteins (SREBPs) are the major known transcription factors responsible for regulating the above-mentioned low-density lipoprotein receptor and enzymes associated with cholesterol biosynthesis.20 Upon proteolytic maturation, the N-terminal domain of SREBPs is released and translocated to the nucleus, where it binds the promoter/enhancer regions of genes, like HMGCS1 and HMGCR, involved in cholesterol biosynthesis and LDL uptake, thereby initiating their transcription.21 To assess whether the up-regulation of the cholesterol biosynthesis enzymes occurred at the transcriptional level through SREBP activation, we performed qRT-PCR analysis of several key SREBP target genes, some of which are involved in cholesterol biosynthesis. Our results revealed that the relative mRNA expression levels of all five genes tested (HMGCS1, HMGCR, FDPS, FDFT1 and DHCR7) were significantly increased (by 1.5–3.5 fold, Figure 4A), supporting that the elevated expression of these enzymes at protein levels occurs mainly through a transcriptional mechanism via SREBP activation.
Figure 4.
Potassium dichromate perturbed de novo cholesterol biosynthesis. (A) Shown are the histograms of gene expression levels of proteins involved in cholesterol biosynthesis in GM00637 cells that were untreated or treated with 0.5 μM potassium dichromate for 24 hrs. (B) Effect of a 24-hr treatment with 0.5 μM potassium dichromate on cholesterol content in GM00637, HEK293T and HCT-116 cells. Cholesterol content is expressed as fold change of the value from control. The values represent mean ± S.D. of results obtained from three independent experiments. `*', p < 0.05; `**', p < 0.01; `***', p < 0.001. The p values were calculated by using unpaired two-tailed t-test.
Previous studies suggested that oxidative stress could result in increased expression or enhanced activity of SREBPs. In this vein, Waris et al.22 observed that viral infection-induced oxidative stress could stimulate the activation of SREBPs thereby augmenting cholesterol biosynthesis. These authors further demonstrated that oxidative stress induced by hepatitis C virus infection results in the activation of phosphatidylinositol 3-kinase-Akt (PI3K-Akt) pathway, which subsequently transactivates SREBPs.22 Our quantitative proteomic data unveiled that phosphatidylinositol 3-kinase 85 kDa regulatory subunit beta and protein kinase B (Akt) were up-regulated by 1.52 and 1.49 fold, respectively (Table S3). Therefore, the Cr(VI)-induced up-regulation of enzymes involved in cholesterol biosynthesis likely arises from the Cr(VI)-induced oxidative stress, the subsequent activation of the PI3K-Akt pathway and the ensuing activation of SREBPs.
Cr(VI) led to increased intracellular cholesterol level and Cr(VI)-induced inhibition of cell proliferation could be rescued by lovastatin
Considering that a large number of enzymes involved in cholesterol biosynthesis were up-regulated, we reason that Cr(VI) may induce elevated cellular cholesterol production. To test this, we measured the cholesterol levels in GM00637 human skin fibroblast, HEK293T human embryonic kidney epithelial cells, and HCT-116 human colon carcinoma cells with and without K2Cr2O7 treatment. Our results showed that a 24-hr treatment with 0.5 NM K2Cr2O7 led to statistically significant increase of the cellular cholesterol content (by 1.3–2.0 fold) in these three cell lines (Figure 4B). These results are in line with the Cr(VI)-induced up-regulation of enzymes involved in cholesterol biosynthesis, as revealed by quantitative proteomic experiment. Together, the above findings demonstrated that Cr(VI) stimulates endogenous cholesterol biosynthesis.
Cholesterol is an essential component for mammalian cell membrane and is required for proper membrane permeability, fluidity, protein function and organelle identity.23 During cholesterol biosynthesis, a series of metabolites are produced and utilized in membrane assembly, cell signaling, protein synthesis and cell-cycle progression.24 However, unregulated, especially high cholesterol level is known to be associated with cardiovascular disease and cancer promotion.25
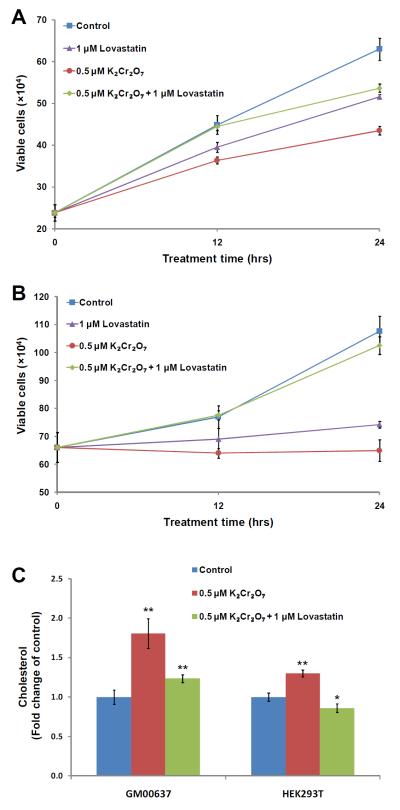
We reasoned that the elevated production of cellular cholesterol may be one of the major factors contributing to the toxic effects of Cr(VI). To explore this possibility, we assessed the Cr(VI)-induced perturbation of cell growth and examined whether such perturbation can be rescued by a cholesterol-lowering drug. Thus, we used lovastatin to inhibit HMG-CoA reductase (HMGCR), which is a rate-limiting enzyme in cholesterol biosynthesis.26 It turned out that exposure to Cr(VI) resulted in pronounced growth inhibition of GM00637 and HEK293T cells and this inhibition could indeed be rescued by the addition of lovastastin (Figure 5A&B). Along this line, the cellular cholesterol returned to a level that is similar to that of control cells after a 24-hr co-treatment with 0.5 μM Cr(VI) and 1 μM lovastatin (Figure 5C). These results demonstrated that the Cr(VI)-induced growth inhibition of GM00637 and HEK293T cells could be mainly attributed to the elevated expression of enzymes involved in cholesterol biosynthesis and the resultant over-production of endogenous cholesterol.
Figure 5.
Potassium dichromate-induced cell proliferation can be rescued by external addition of lovastatin. The cell viability of GM00637 (A) and HEK293T (B) determined by trypan blue exclusion assay after 12 and 24 hrs of treatment with 0 μM, 0.5 μM K2Cr2O7 in the presence or absence of 1 μM lovastatin. (C) Shown are the cholesterol levels in GM00637 and HEK293T cells that were untreated or treated for 24 hrs with 0.5 μM potassium dichromate, alone or in combination with 1 μM lovastatin.
Elevated G protein signaling pathway upon potassium dichromate treatment
Pathway analysis also revealed that the guanine nucleotide binding protein (G proteins) signaling pathway is significantly up-regulated upon Cr(VI) treatment. G protein signaling is a pivotal transmembrane signaling pathway which is responsible for regulating a myriad of physiological responses.27 Briefly, G protein signaling pathway starts with the activation of G protein-coupled receptors (GPCRs) through binding of extracellular signaling molecules. GPCR activation may trigger a multitude of downstream signal transduction pathways mediated by their membrane-bound partners, the G proteins.28 One is through the activation of Gαs subunit, which triggers the activation of adenylate cyclase (AC) to produce cyclic-adenosine monophosphate (cAMP) and eventually the activation of the transcription factor of cAMP-responsive element-binding protein (CREBP). Another G protein, Gαq, regulates the activity of its downstream effector phospholipase C-β, which is responsible for catalyzing the cleavage of phosphatidylinositol 4,5-bisphosphate into the second messengers inositol 1,4,5-trisphosphate (IP3) and diacylglycerol (DAG), thereby giving rise to protein kinase C (PKC) activation.29 Along this line, our quantitative proteomic data showed that several proteins involved in the G protein signaling pathway, including CREBP1, PKC, and mitogen-activated protein kinase 3, were significantly up-regulated by 1.43, 1.96 and 1.40 fold, respectively (Table S3).
Elevated GPCRs and relevant G protein signaling pathways after Cr(VI) treatment may result in malfunction of cells and promote aberrant cell proliferation through the crosstalk between GPCRs and growth factor receptors.28 Thus, the stimulation of diverse downstream signal transduction pathways may lead to changes in gene expression, cell migration and proliferation, and ultimately, cancer progression.29
Potassium dichromate-induced alteration of inflammatory response pathways and inhibition of selenoprotein expression
Pathway analysis also showed that inflammatory response and interleukin 5 (IL-5) signaling pathways were up-regulated after Cr(VI) exposure (Table S4). Our quantitative proteomic results revealed the increased expression of several collagen proteins as well as kinases involved in IL-5 signaling. In the latter respect, the aforementioned phosphatidylinositol 3-kinase 85 kDa regulatory subunit beta and protein kinase B (Akt), as well as Janus kinase 1, mitogen-activated protein kinase 3, and protein kinase C beta were all up-regulated following K2Cr2O7 treatment (Table S3). This finding is in accordance with a previous study demonstrating that Cr(VI) could activate the Akt, NF-κB and MAPK pathways in kerotinocytes, which is accompanied with the increased production of cytokines, including tumor necrosis factor-α (TNF-α) and IL-1α.30 ROS are known to activate transcription factors, such as NF-κB and AP-1, which are responsible for regulating the expression levels of numerous inflammatory pathway genes.31, 32 Thus, the Cr(VI)-induced activation of inflammatory response may reflect cellular ROS generation arising from Cr(VI) reduction.
GenMAPP analysis also revealed the down-regulation of two selenoproteins, i.e., 15kDa selenoprotein (Sep15, by 0.56 fold) and selenoprotein S (Sep S, by 0.59 fold, Table S3). Along this line, previous studies showed that exposure to silver nanoparticles or monomethylarsonous acid could lead to inhibition of selenoprotein synthesis.33, 34 Selenoproteins function to protect cells from oxidative damage and apoptosis by preventing the deleterious consequences of accumulation of misfolded proteins that has been linked to immune and inflammatory processes.35 Thus, diminished expression of these two selenoproteins may exaggerate the cellular inflammatory response with increased release of cytokines like tumor necrosis factor-α (TNF-α) after Cr(VI) treatment.36
Conclusions
Cr(VI) is a major heavy metal pollutant that is responsible for inducing different types of cancers among exposed population.6 By using SILAC combined with LC-MS/MS analysis, we were able to achieve a quantitative assessment of the K2Cr2O7-induced perturbation of the proteome of GM00637 cells. The quantitative information facilitated us to elucidate the mechanisms through which Cr(VI) exerts its deleterious effect. In the present study, we were able to identify 5469 proteins, with 4607 quantified from three sets of SILAC experiments. Among them, 270 and 127 proteins were significantly up- and down-regulated, respectively, upon a 24-hr treatment with 0.5 μM K2Cr2O7. Particularly, we demonstrated, for the first time, that Cr(VI) may exert its cytotoxic effect, at least in part, through elevating endogenous cholesterol biosynthesis. Additionally, the stimulation of inflammatory response and GPCR signaling pathways, as well as the suppression of selenoproteins may also contribute to the cytotoxic effects of Cr(VI). The novel insight into the cytotoxicity of Cr(VI) provides the basis for the future development of effective approaches for the therapeutic intervention following Cr(VI) exposure.
Supplementary Material
Synopsis.
By using SILAC together with LC-MS/MS analysis, we assessed the K2Cr2O7-induced perturbation of the global proteome of GM00637 cells. This facilitated us to elucidate the mechanisms through which Cr(VI) exerts its deleterious effect. Particularly we provided evidence to support that Cr(VI) may confer its cytotoxic effect through elevating endogenous cholesterol biosynthesis. Additionally, Cr(VI) treatment stimulated inflammatory response and GPCR signaling, but suppressed the expression of selenoproteins.
Acknowledgment
This work was supported by the National Institutes of Health (R01 ES019873).
Footnotes
Supporting Information Available. A detailed list of all quantified proteins, all significantly changed proteins, GO ontology and pathway analysis results, and primers used for real-time PCR. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- (1).Shanker AK. Trace Elements as Contaminants and Nutrients. John Wiley & Sons, Inc.; 2008. pp. 523–553. [Google Scholar]
- (2).WHO Chromium (Environmental Health Criteria 61) International Programme on Chemical Safety Geneva, Switzerland. 1990 [Google Scholar]
- (3).ATSDR . Toxicological Profile for Chromium. 2012. [PubMed] [Google Scholar]
- (4).Ding M, Shi X. Molecular mechanisms of Cr(VI)-induced carcinogenesis. Mol. Cell. Biochem. 2002:234–235. 293–300. [PubMed] [Google Scholar]
- (5).Kerger BD, Paustenbach DJ, Corbett GE, Finley BL. Absorption and elimination of trivalent and hexavalent chromium in humans following ingestion of a bolus dose in drinking water. Toxicol. Appl. Pharmacol. 1996;141:145–58. doi: 10.1006/taap.1996.0271. [DOI] [PubMed] [Google Scholar]
- (6).ATSDR . Toxicological Profile for Chromium. 2000. [PubMed] [Google Scholar]
- (7).Dayan AD, Paine AJ. Mechanisms of chromium toxicity, carcinogenicity and allergenicity: review of the literature from 1985 to 2000. Hum. Exp. Toxicol. 2001;20:439–51. doi: 10.1191/096032701682693062. [DOI] [PubMed] [Google Scholar]
- (8).Wise SS, Holmes AL, Wise JP., Sr. Hexavalent chromium-induced DNA damage and repair mechanisms. Rev. Environ. Health. 2008;23:39–57. doi: 10.1515/reveh.2008.23.1.39. [DOI] [PubMed] [Google Scholar]
- (9).De Mattia G, Bravi MC, Laurenti O, De Luca O, Palmeri A, Sabatucci A, Mendico G, Ghiselli A. Impairment of cell and plasma redox state in subjects professionally exposed to chromium. Am. J. Ind. Med. 2004;46:120–5. doi: 10.1002/ajim.20044. [DOI] [PubMed] [Google Scholar]
- (10).Ye J, Shi X. Gene expression profile in response to chromium-induced cell stress in A549 cells. Mol. Cell. Biochem. 2001;222:189–97. [PubMed] [Google Scholar]
- (11).Izzotti A, Bagnasco M, Cartiglia C, Longobardi M, De Flora S. Proteomic analysis as related to transcriptome data in the lung of chromium(VI)-treated rats. Int. J. Oncol. 2004;24:1513–22. [PubMed] [Google Scholar]
- (12).Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422:198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- (13).Ong S-E, Blagoev B, Kratchmarova I, Kristensen DB, Steen H, Pandey A, Mann M. Stable Isotope Labeling by Amino Acids in Cell Culture, SILAC, as a Simple and Accurate Approach to Expression Proteomics. Mol. Cell. Proteomics. 2002;1:376–386. doi: 10.1074/mcp.m200025-mcp200. [DOI] [PubMed] [Google Scholar]
- (14).Cox J, Mann M. MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 2008;26:1367–1372. doi: 10.1038/nbt.1511. [DOI] [PubMed] [Google Scholar]
- (15).Salomonis N, Hanspers K, Zambon A, Vranizan K, Lawlor S, Dahlquist K, Doniger S, Stuart J, Conklin B, Pico A. GenMAPP 2: new features and resources for pathway analysis. BMC Bioinformatics. 2007;8:217. doi: 10.1186/1471-2105-8-217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (16).Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(−ΔΔCt) Method. Methods. 2001;25:402–8. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- (17).Osada K, Ravandi A, Kuksis A. Rapid analysis of oxidized cholesterol derivatives by high-performance liquid chromatography combined with diode-array ultraviolet and evaporative laser light-scattering detection. J. Am. Oil Chem. Soc. 1999;76:863–871. [Google Scholar]
- (18).Dong X, Xiong L, Jiang X, Wang Y. Quantitative proteomic analysis reveals the perturbation of multiple cellular pathways in jurkat-T cells induced by doxorubicin. J. Proteome. Res. 2010;9:5943–51. doi: 10.1021/pr1007043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (19).Defesche JC. Low-density lipoprotein receptor--its structure, function, and mutations. Semin. Vasc. Med. 2004;4:5–11. doi: 10.1055/s-2004-822993. [DOI] [PubMed] [Google Scholar]
- (20).Brown MS, Goldstein JL. A proteolytic pathway that controls the cholesterol content of membranes, cells, and blood. Proc. Natl. Acad. Sci. USA. 1999;96:11041–8. doi: 10.1073/pnas.96.20.11041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (21).Horton JD, Goldstein JL, Brown MS. SREBPs: activators of the complete program of cholesterol and fatty acid synthesis in the liver. J. Clin. Invest. 2002;109:1125–31. doi: 10.1172/JCI15593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (22).Waris G, Felmlee DJ, Negro F, Siddiqui A. Hepatitis C virus induces proteolytic cleavage of sterol regulatory element binding proteins and stimulates their phosphorylation via oxidative stress. J. Virol. 2007;81:8122–30. doi: 10.1128/JVI.00125-07. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- (23).Alberts B JA, Lewis J. Molecular Biology of the Cell. 4th edition Garland Science; New York: 2002. [Google Scholar]
- (24).Herold G, Jungwirth R, Rogler G, Geerling I, Stange EF. Influence of cholesterol supply on cell growth and differentiation in cultured enterocytes (CaCo-2) Digestion. 1995;56:57–66. doi: 10.1159/000201223. [DOI] [PubMed] [Google Scholar]
- (25).Tete S, Nicoletti M, Saggini A, Maccauro G, Rosati M, Conti F, Cianchetti E, Tripodi D, Toniato E, Fulcheri M, Salini V, Caraffa A, Antinolfi P, Frydas S, Pandolfi F, Conti P, Potalivo G, Theoharides TC. Nutrition and cancer prevention. Int. J. Immunopathol. Pharmacol. 2012;25:573–81. doi: 10.1177/039463201202500303. [DOI] [PubMed] [Google Scholar]
- (26).Wong WW, Dimitroulakos J, Minden MD, Penn LZ. HMG-CoA reductase inhibitors and the malignant cell: the statin family of drugs as triggers of tumor-specific apoptosis. Leukemia. 2002;16:508–19. doi: 10.1038/sj.leu.2402476. [DOI] [PubMed] [Google Scholar]
- (27).Venkatakrishnan AJ, Deupi X, Lebon G, Tate CG, Schertler GF, Babu MM. Molecular signatures of G-protein-coupled receptors. Nature. 2013;494:185–94. doi: 10.1038/nature11896. [DOI] [PubMed] [Google Scholar]
- (28).Lappano R, Maggiolini M. G protein-coupled receptors: novel targets for drug discovery in cancer. Nat. Rev. Drug Discov. 2011;10:47–60. doi: 10.1038/nrd3320. [DOI] [PubMed] [Google Scholar]
- (29).Dorsam RT, Gutkind JS. G-protein-coupled receptors and cancer. Nat. Rev. Cancer. 2007;7:79–94. doi: 10.1038/nrc2069. [DOI] [PubMed] [Google Scholar]
- (30).Wang BJ, Sheu HM, Guo YL, Lee YH, Lai CS, Pan MH, Wang YJ. Hexavalent chromium induced ROS formation, Akt, NF-kappaB, and MAPK activation, and TNF-alpha and IL-1alpha production in keratinocytes. Toxicol. Lett. 2010;198:216–24. doi: 10.1016/j.toxlet.2010.06.024. [DOI] [PubMed] [Google Scholar]
- (31).Bubici C, Papa S, Dean K, Franzoso G. Mutual cross-talk between reactive oxygen species and nuclear factor-kappa B: molecular basis and biological significance. Oncogene. 2006;25:6731–48. doi: 10.1038/sj.onc.1209936. [DOI] [PubMed] [Google Scholar]
- (32).Reuter S, Gupta SC, Chaturvedi MM, Aggarwal BB. Oxidative stress, inflammation, and cancer: how are they linked? Free Radic. Biol. Med. 2010;49:1603–16. doi: 10.1016/j.freeradbiomed.2010.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (33).Meno SR, Nelson R, Hintze KJ, Self WT. Exposure to monomethylarsonous acid (MMAIII) leads to altered selenoprotein synthesis in a primary human lung cell model. Toxicol. Appl. Pharmacol. 2009;239:130–136. doi: 10.1016/j.taap.2008.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (34).Srivastava M, Singh S, Self WT. Exposure to silver nanoparticles inhibits selenoprotein synthesis and the activity of thioredoxin reductase. Environ. Health Persp. 2012;120:56–61. doi: 10.1289/ehp.1103928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- (35).Lu J, Holmgren A. Selenoproteins. J. Biol. Chem. 2009;284:723–7. doi: 10.1074/jbc.R800045200. [DOI] [PubMed] [Google Scholar]
- (36).Huang Z, Rose AH, Hoffmann PR. The role of selenium in inflammation and immunity: from molecular mechanisms to therapeutic opportunities. Antioxid. Redox. Signal. 2012;16:705–43. doi: 10.1089/ars.2011.4145. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.