Abstract
AIM: To further analyse cancer involvement of basic transcription factor 3 (BTF3) after detection of its upregulation in gastric tumor samples.
METHODS: BTF3 transcription rates in human gastric tumor tissue samples (n = 20) and adjacent normal tissue (n = 18) specimens as well as in the gastric cancer cell lines AGS, SGC-7901, MKN-28, MKN-45 and MGC803 were analyzed via quantitative real-time polymerase chain reaction. The effect of stable BTF3 silencing via infection with a small interfering RNA (siRNA)-BTF3 expressing lentivirus on SGC-7901 cells was measured via Western blotting analysis, proliferation assays, cell cycle and apoptosis profiling by flow cytometry as well as colony forming assays with a Cellomic Assay System.
RESULTS: A significant higher expression of BTF3 mRNA was detected in tumors compared to normal gastric tissues (P < 0.01), especially in section tissues from female patients compared to male patients, and all tested gastric cancer cell lines expressed high levels of BTF3. From days 1 to 5, the relative proliferation rates of stable BTF3-siRNA transfected SGC7901 cells were 82%, 70%, 57%, 49% and 44% compared to the control, while the percentage of cells arrested in the G1 phase was significantly decreased (P = 0.000) and the percentages of cells in the S (P = 0.031) and G2/M (P = 0.027) phases were significantly increased. In addition, the colony forming tendency was significantly decreased (P = 0.014) and the apoptosis rate increased from 5.73% to 8.59% (P = 0.014) after BTF3 was silenced in SGC7901 cells.
CONCLUSION: BTF3 expression is associated with enhanced cell proliferation, reduced cell cycle regulation and apoptosis and its silencing decreased colony forming and proliferation of gastric cancer cells.
Keywords: Basic transcription factor 3, Gastric cancer, Small interfering RNA, Proliferation, Apoptosis, Cell cycle
Core tip: After we found that basic transcription factor 3 (BTF3) transcription rates in human gastric tumor tissue samples were significantly higher than in adjacent normal tissues, we extended our study on gastric cancer cell lines. We silenced BTF3 in SGC7901 cells, which led to 82%, 70%, 57%, 49% and 44% proliferation rates of the control within the first 5 d after infection with a small interfering RNA-BTF3 containing lentivirus. After BTF3 silencing, the percentage the G1 phase arrested SGC7901 cells was decreased (P = 0.000), the percentages of cells in the S (P = 0.031) and G2/M (P = 0.027) phases were increased and the apoptosis rate increased from 5.73% to 8.59% (P = 0.014).
INTRODUCTION
Basic transcription factor (BTF3) is a 27 kD protein that in humans is encoded by the BTF3 gene[1,2] and evolutionarily conserved in a variety of cells[3]. BTF3 was initially discovered as a member of the general transcription machinery and functions as a transcriptional initiation factor from proximal promoter elements by forming a stable complex with RNA polymerases[4,5]. There are two ubiquitously expressed isoforms of the BTF3 gene that encode BTF3a and BTF3b proteins. The BTF3a is the transcriptional active form of BTF3, while the isoform BTF3b, which lacks the first 44 amino acids of the BTF3a N-terminus is transcriptionally inactive, although it is able to bind to the RNA polymerase II[6]. However, previous studies indicated that BTF3 was not in fact essential for specific, in vitro initiation of transcription, but its biological importance was shown by the fact that mouse embryos, homozygous for a loss of function mutation in the BTF3 gene, died at the early stage of development indicating its important role during development[7]. In addition, BTF3 was up-regulated strongly in mouse pregnancy, indicating an involvement in alveolar growth[8]. In cancer, it has been reported that the BTF3 gene has been overexpressed in colorectal cancer, glioblastomas and hepatocellular carcinomas[9-12]. In the pancreatic ductal adenocarcinoma, the median level of the BFT3 and BFT3a mRNA was increased by 1.3 and 4.6 folds compared to the normal tissues, respectively. Down-regulation of the BTF3 expression using small interfering RNA (siRNA) resulted in reduced expression of several cancer-associated genes, including ephrin receptor B2, which is mainly expressed during development and involved in tumor cell survival[13,14] and heparanase 2 an extracellular matrix degrading enzyme involved in cell adhesion. In addition, ataxia-telangectasia mutated gene, which is implicated in cell cycle arrest[15], DNA repair[16] or apoptosis and the oncogene V-abl Abelson murine leukemia viral oncogene homolog 2, a nuclear protein tyrosine kinase for cell differentiation, cell division and cell adhesion were also had reduced expression[17]. By BFT3 silencing, up-regulated genes were k-ras oncogene-associated gene, related ras viral oncogene homolog 2, nuclear factor kappa-B, murine retrovirus integration site 1 and mucosal vascular addressing cell adhesion molecule 1, all known to be involved in tumor development[18]. In addition, BTF3 interacts with either 17β-estradiol or epidermal growth factor activated estrogen receptor α (ERα) via its AF1 domain in the breast cancer cell line MCF-7 and up-regulates transcriptional responses of ERα reporter genes[19,20]. In this study, we analyzed the expression of BTF3 mRNA and protein in gastric tumors and normal samples and compared BTF3 expressions in different gastric tumor cell lines. Finally we used siRNA-BTF3 to down-regulate BTF3 expressions in gastric tumor cells and measured the changes of proliferation and apoptosis to investigate the relationship between BTF3 and gastric cancer.
MATERIALS AND METHODS
Patients
Human tissue samples of gastric tumor (n = 20) and adjacent normal tissue (n = 18) specimens were obtained from patients with a median age of 63 years (range, 35-83 years) who received gastric resections from November 2011 to March 2012 in the Second Xiangya Hospital of Central South University, Changsha, China. All samples were confirmed histologically. Freshly removed tissues (within 5 min after surgical excision) were: (1) fixed in paraformaldehyde solution for 12-24 h and then paraffin embedded for histological analysis; (2) kept in RNAlater (Ambion Ltd., Huntingdon, Cambridgeshire, United Kingdom) for RNA analysis; or (3) snap-frozen in liquid nitrogen and maintained at -80 °C for protein analysis. All studies were approved by The Human Ethics Committee of the First Affiliated Hospital of Hunan Normal University in China, and written informed consent was obtained from all patients.
Cell culture
Gastric cancer cell lines (AGS, SGC-7901, MKN-28, MKN-45 and MGC803) were grown in complete DMEM medium (Gibco, New York, United States), supplemented with 10% fetal calf serum (Gibco, New York, United States) and 5 U/mL penicillin, and incubated at 37 °C in 5% CO2 atmosphere.
Plasmid and vector construction
The selected and optimized siRNA (sense: GCCGAAGAAGCCTGGGAATCA, anti-sense: TGATTCCCAGGCTTCTT CGGC) against human BTF3 and negative control (sense: TTCTCCGAACG TGTCACGT, anti-sense: ACGTGACACGTTCGGAGAA) were used in generating the lentiviral vector. Basically, the BTF3 siRNA transcript template was synthesized, digested with Age I/EcoRI enzymes, and then ligated with pGCSIL-GFP vector. After checking by agarose electrophoresis, successful BTF3-siRNA-pGCIL-GFP recombinants were amplified, purified and sequenced. In order to evaluate the inhibitive effect of BTF3 sequences, the confirmed pGCSIL-GFP recombinants were transfected into 293T cells (data not shown). The expressed BTF3 protein was detected by Western blotting.
Quantitative real-time polymerase chain reaction
Cells were split during the log-phase growth before plating into the 6-well plate with complete medium, and incubated at 37 °C with 5% CO2. An appropriate amount of lentiviral vectors was added to cells when 30% confluence was reached. Total RNA was extracted 5 d after treatment from cells with GFP expression by Trizol (Invitrogen, San Diego, United States). RNA (2 μg) was reversely transcribed using M-MLV-RTase (Promega, Madison, United States) following manufacturer’s instructions. Real-time polymerase chain reaction (PCR) was performed using the tested primer set (Table 1) and SYBR Master Mixture (TAKARA, DRR041B) following the manufacturer’s instructions with a TAKARA’s TP800 Real Time PCR Instrument. Following the normalization with glyceraldehyde-3-phosphate dehydrogenase (GAPDH) gene, relative quantification of BTF3 expression was done by the comparative CT method (2-ΔΔC method).
Table 1.
Primer sequences
| Primer | Sequence |
| GAPDH-F | TGACTTCAACAGCGACACCCA |
| GAPDH-R | CACCCTGTTGCTGTAGCCAAA |
| BTF3-F | GCGAACACTTTCACCATTACAG |
| BTF3-R | AACTTCATCATCATCATCCTCTCC |
BTF3: Basic transcription factor 3; GAPDH: Glyceraldehyde-3-phosphate dehydrogenase.
Total protein extraction, sodium dodecyl sulfate-polyacrylamide gel electrophoresis and immunoblotting
Cells were harvested 36-48 h following the transfection. Pellets were washed twice with cold phosphate-buffered saline (PBS) (Gibco, New York, United States) and then cells were lysed on ice for 10-15 min using proper lysis buffer [1 mol/L Tris-HCl 100 mmol/L, 2% 2-mercaptoethanol, 20% glycerol, 4% sodium dodecyl sulfate (SDS) (Life Technologies, New York, United States)]. After sonication, total protein extraction was performed by centrifugation at 1200 g for 15 min at 4 °C. Protein was separated using SDS-polyacrylamide gel electrophoresis (SDS-PAGE), with 4% separating and 3%-15% gradient stacking gel. After SDS-PAGE, protein samples were wet transferred to polyvinylidenfluorid (PVDF) (Life Technologies, New York, United States) membranes for immunoblotting. To detect the target protein, the PVDF membrane was incubated with appropriate primary antibodies, either 2 h at the room temperature or overnight in the cold room (Mouse Anti-GFP Santa-Cruz SC-9996, 1:2000, Mouse anti-GAPDH Santa-Cruz SC-32233 1:5000), following 3 times of 10 min TBS-Tween 20 buffer (Life Technologies, New York, United States) washes, membranes were incubated with secondary antibodies for 2 h at room temperature (Goat Anti-Mouse IgG Santa-Cruz SC-2005 1:5000). After further 3 times washing, target proteins were detected using ECL™ Western blotting system (Amersham Cat. No. RPN2135).
Flow cytometric assays
Cells were harvested by Trypsin/EDTA (Life Technologies, New York, United States) solution from the 6-cm dishes at 80% confluence followed by setting up the designed assays. Pellets were then washed with PBS and collected into 5 mL centrifuge tubes in triplicate. After fixation using pre-cooled 4 °C 70% ethanol for 1 h, cells were washed again and then stained with propidium iodide (FACSCalibur, BD, New York, United States).
Colony forming assay
Five hundred cells were plated per well in 96-well plates in triplicate. After the colony number reached more than 5 in most of the wells, the colonies were scanned using Cellomics system following the manufacturer’s protocol. The colony size and cell numbers within each colony were analyzed by Cellomics system (Thermo, Philadelphia, United States). The data were then statistically analyzed.
Statistical analysis
Results of all calculations were expressed as mean ± SD. Analysis of variance (ANOVA) with multiple comparisons using Bonferroni’s test or one-way ANOVA was performed where appropriate. The difference between two groups was analyzed by unpaired t test GraphPad Prism (version 3.02-2000). The results of statistical tests were considered significant if the P value was < 0.05 and < 0.01.
RESULTS
Level of BTF3 expression in gastric cancer and tumor cell lines
A quantitative real-time PCR was performed to identify the transcription levels of the BTF3 gene in 20 human gastric tumors and the relative tumor free margins (n = 18) as healthy tissue control. A significantly higher transcription of BTF3 mRNA was detected in tumors compared to normal gastric tissues (Figure 1A, P < 0.01), especially in female patients’ section tissues compared to male patients (Figure 1B) and between the gastric body tumor and adjacent normal tissue, but no significant difference was detectable between gastric antrum or cardia tumor and adjacent normal tissues (Figure 1C). In order to further confirm that the BTF3 gene was widely expressed in our gastric tumor cell lines, quantitative real-time PCR was performed in five different cell lines AGS, SGC-7901, MKN-28, MKN-45 and MGC803. SGC-7901, MKN-45 and MGC803 which are all poorly differentiated adenocarcinomas, while the MKN-28 and AGS are high and moderately differentiated cell lines. The expected products of BTF3 mRNA were detected in all the above five cell lines (Figure 1D).
Figure 1.
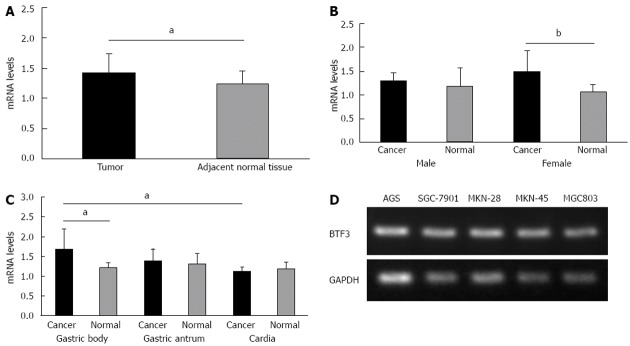
Level of basic transcription factor 3 expression in gastric cancer and tumor cell lines. A: Bar charts represent the mRNA levels of basic transcription factor 3 (BTF3) in gastric tumor and adjacent normal tissue measured by quantitative real-time polymerase chain reaction (PCR); B: BTF3 mRNA expression data from (A) further classified by gender; C: BTF3 expression pattern among gastric body, gastric antrum and cardia tumors; D: The expression of BTF3 mRNA was detected by quantitative real-time PCR among different cell lines. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the reference gene. Black bars represent tumors and gray bars normal tissues. The difference in BTF3 expression, aP < 0.05, bP < 0.01 between groups was assessed by unpaired t test and considered significantly different.
Silencing of BTF3 by siRNA
Based on the significant BTF3 up-regulation from the clinical data, we sought that the regulation of BTF3 in gastric tissue might be related to cancer initiation promotion and progression. To investigate this, the gene was silenced using lentiviral vectors expressing the specifically designed synthetic siRNA-BTF3. In pilot experiments, the siRNA-BTF3 was successfully transfected into human SGC7901 and 293T cell lines for mRNA expression profiles and protein analyses via GFP expression to estimate the transfection efficiency (Figure 2A). In SGC7901 cells, a significant down-regulation of the BTF3 gene was measured after siRNA-BTF3 application (Figure 2B). The BTF3 transcription in siRNA transfected cells was 12.4% of the negative control transfected cells (P = 0.0066). The BTF3 protein levels in SGC7901 cells, detected by Western blotting 48 h after siRNA-BTF3 transfection, were reduced compared to the control (Figure 2C).
Figure 2.
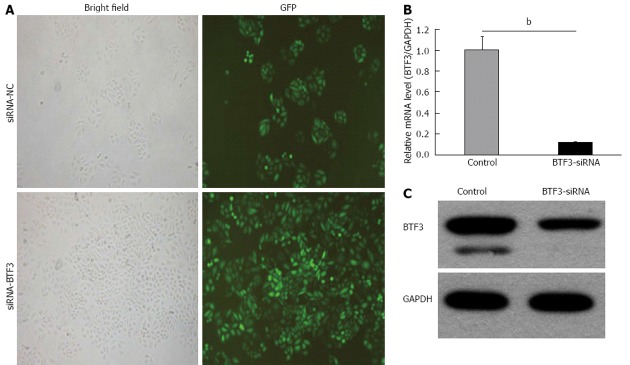
Silencing of basic transcription factor 3 using small interfering RNA. A: Transfection rates of small interfering RNA (siRNA)-normal control (NC)/siRNA-basic transcription factor 3 (BTF3)-green fluorescent protein (GFP) vectors in human SGC7901 cells, estimated by bright field and fluorescence microscope; B: Bar charts represent quantitative real-time polymerase chain reaction data of BTF3 mRNA levels in control and BTF3-siRNA transfected SGC7901 cells (bP < 0.01 between groups, unpaired t test); C: Immuno-blotting analysis using BTF3-specific antibodies to detect BTF3 protein expressions in control and BTF3-siRNA transfected SGC7901 cells. Glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as internal control.
BTF3 silencing effect in SGC7901 cells
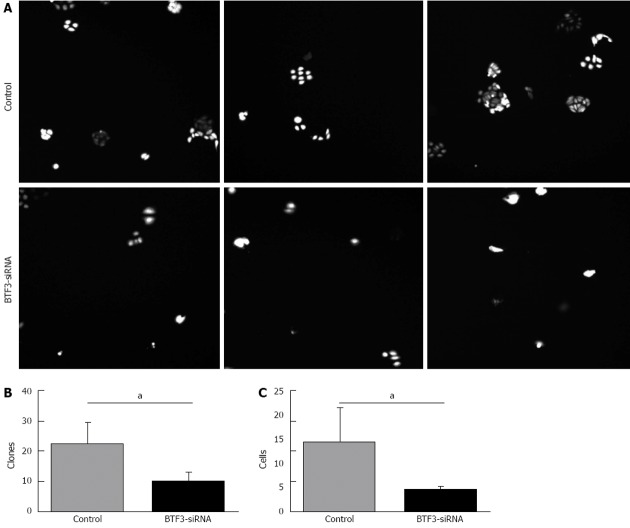
In order to identify the effect on gastric tumor cells, proliferation, cell cycle, apoptosis and colony forming assays were performed with siRNA-BTF3 transfected SGC7901 cells. The silencing of BTF3 significantly slowed down the SGC7901 cell growth in the proliferation assay (Figure 3). On day 1, the siRNA transfected cells already started to show an 18% slower proliferation rate compared with the negative controls. The difference became increasingly significant and peaked at the end of the proliferation assay on day 5, on which the proliferation rate in the siRNA transfected cells was only 44% of the negative control. From days 1 to 5, the relative proliferation rates of siRNA transfected SGC7901 were 82%, 70%, 57%, 49% and 44%, respectively. In summary, an obvious effect on proliferation of SGC7901 was induced by silencing BTF3 (Figure 3, BTF3-siRNA). A cell cycle profile of the siRNA-BTF3 transfected cells using PI-FACS and Annexin V showed that the percentage of cells arrested in the G1 phase was significantly decreased, while the percentages of cells at the S and G2/M phases were significantly increased (Figure 4). The knockdown of BTF3 in SGC7901 cells also induced an increased level of apoptosis. Cells with good transfection efficiency were harvested on day 5 for flow cytometry (Figure 5). The negative control group which was transfected with empty vectors showed 5.73% and 0.76% apoptosis, while the apoptosis rate of siRNA-BTF3 transfected group was 8.59% and 1.1% (Figure 5B and D). The down-regulation of BTF3 increased the apoptosis rate by nearly 2 folds. To detect the long-term effect on SGC7901 cells following BTF3 silencing, a colony forming assay was performed. Colonies were scanned and analyzed using the Cellomic Assay System. Cells after BTF3 silencing showed a significantly lower colony forming ability compared with the negative controls (Figure 6A). In the silenced group, 14 d following the initiation of the assay in average 10 colonies were detected, whereas a significantly higher number of 22 colonies (mean) was detected in the negative group (P = 0.014) (Figure 6B). In addition, the number of cells in each siRNA-BTF3 transfected SGC7901 cell colony was also reduced compared with the control.
Figure 3.
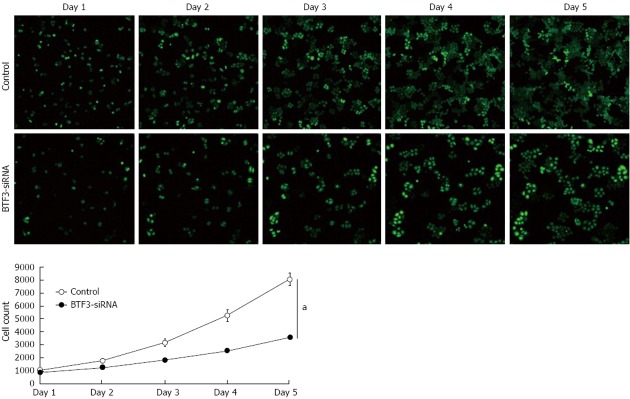
Cell proliferation assay. Fluorescent images of control and small interfering RNA (siRNA)-basic transcription factor 3 (BTF3) transfected cell proliferation assays. Cells were counted based on the green fluorescent protein signals during 5 d. BTF3-siRNA chart of the cell proliferation assay using two-way analysis of variance. aP < 0.05 between groups.
Figure 4.
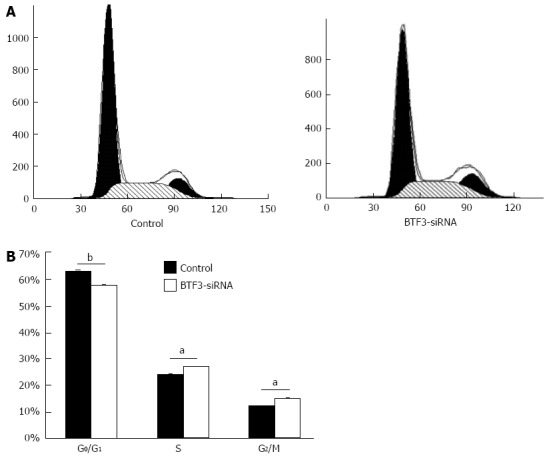
Flow cytometric assays. A: Cell-cycle stages of control and basic transcription factor 3 (BTF3)-small interfering RNA (siRNA) transfected cells were analyzed by flow cytometry. Data are presented as a histogram, with cell number (y-axis) plotted against DNA content (x-axis); B: Cells arrested at different cell-cycle stages were plotted as bar charts. aP < 0.05, bP < 0.01 between groups was assessed by unpaired t test and comparisons were considered significantly different.
Figure 5.
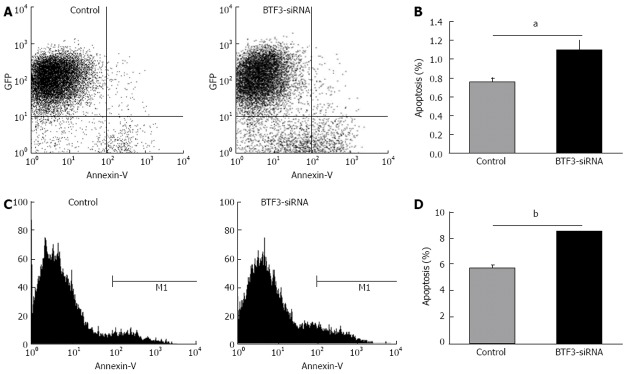
Apoptosis analyses by flow cytometry. A and C: Flow cytometric analysis of control and basic transcription factor 3 (BTF3)-small interfering RNA (siRNA) transfected cells; B and D: Apoptosis rates plotted as bar charts. aP < 0.05, bP < 0.01 between groups. GFP: Green fluorescent protein.
Figure 6.
Colony forming abilities identified by Cellomic Assay System assays. A: Colonies formed from control and basic transcription factor 3 (BTF3)-small interfering RNA (siRNA) transfected cells were imaged via the Cellomic Assay System; B: Number of clones plotted in a bar chart; C: Colony cell numbers of control and BTF3-siRNA transfected cells plotted in a bar chart. aP < 0.05 between groups were analysed with unpaired t test.
DISCUSSION
In cells, multiple protein complexes are required to be orderly assembled on proximal promoter elements such as the TATA box and CAAT box sequences in order to initiate the gene transcription. At the very early stage of the transcription initiation, transcription factor class II D (TFIID), which belongs to the transcription factor class II (TFII) family is required to be stably bound to the transcription factor assembling TATA box. The BTF3 protein, which works as an additional TFII related protein, does not directly associate with the proximal promoter, but forms a stable complex with RNA polymerase II and is part of the gene transcription initiation complex[4,5]. Several studies have reported that expression pattern of transcription factors is frequently changed in gastric tumors. For example, the transcription factor Sp1 has been reported to be higher in malignant gastric tissues, and might serve as an independent prognostic factor, by influencing the tumor infiltration and progression[21]. Another transcription factor, the E2F transcription factor 1, which plays a critical role in cell cycle regulation and other biological processes of the cells, is also up-regulated in human gastric cancer tissues, but further analysis revealed that its overexpression slowed down the cancer cell growth rate[22]. Other transcription related genes were reported to be differentially expressed in gastric tumors compared to normal tissues. Some genes like the antitumor genes GATA-4 and GATA-5 were down-regulated[23,24], whereas others like the oncogenes Forkhead box protein M1[25], and phosphorylated Forkhead box protein O1, which are related to angiogenesis[26] were up-regulated. In addition, the activating transcription factor 4[27], which is cell protective and thereby leading to multidrug resistance, also had enhanced expression. In the present study, we found that the expression of BTF3 is up-regulated in 20 gastric tumor samples compared with normal tissues. We further investigated the expression levels of BTF3 in different malignant gastric tumor cell lines and found uniform expression levels among them (Figure 2). This is in agreement with previous information, because high BTF3 protein levels in gastric cancers have been reported in the Human Protein Atlas[28]. Hence BTF3 is a transcription factor and related to apoptosis[29-32], we thought that the expression level of BTF3 might be important for cell cycle check points, proliferation and further potentially linked with human tumor development and progression. In order to elucidate this, we silenced BTF3 in SGC7901 cells (Figure 3). As a result, the cell cycle arrest shifted from the G1 to the G2/M and S phases with a significantly increased apoptosis rate, which was also reflected in our colony forming assay with significantly less cell growth after BTF3 silencing (Figure 6). This data indicated that BTF3 silencing might induce G2/M check point failure, which led to the inhibition of cell cycle completion and enhanced apoptotic activity.
In summary, our data indicated that BTF3 has a potential link with gastric cancer development and progression. Low expression or silencing of BTF3 might inhibit tumor growth and be beneficial for cancer treatment. However, the clinical data revealed that the tumors had different BTF3 levels possibly due to the different stages of the gastric tumors examined.
COMMENTS
Background
It has been reported that the basic transcription factor 3 (BTF3) gene has been overexpressed in colorectal cancer, glioblastomas as well as pancreatic ductal adenocarcinoma. Down-regulation of the BTF3 expression using small interfering RNA (siRNA) resulted in changed expression of several cancer associated genes involved in tumor cell survival, cell adhesion and cell cycle.
Research frontiers
BTF3 was upregulated in all gastric cancer samples measured in this study. The authors demonstrated that silencing of BTF3 in SGC7901 cells led to a significant decline of proliferation due to enhanced cell cycle arrest and a concomitant higher apoptosis rate. The long-term effect of BTF3 silencing led to a lower amount of colony forming with less cell survival.
Innovations and breakthroughs
In gastric cancer tissues, enhanced expression of BTF3 was reported in the Human Protein Atlas, but the exact role of BTF3 in gastric cancer was not further analyzed. In this study, BTF3 was found to play an essential role in gastric cancer cell proliferation.
Applications
BTF3 was upregulated in all gastric cancer tissues derived from surgical interventions. Silencing of BTF3 led to less proliferation with an enhanced apoptosis rate in SGC7901 cells. The findings may be useful for the diagnosis and treatment of gastric cancer in the future.
Terminology
BTF3 was initially discovered as a member of the general transcription machinery and functions as a transcriptional initiation factor from proximal promoter elements by forming a stable complex with RNA polymerases, and mouse embryos, homozygous for a loss of function mutation in the BTF3 gene, died at the early stage of development. In later developmental stages, the role of BTF3 has been described as transcriptional regulator and modulator of apoptosis.
Peer review
The authors analyzed the BTF3 expression in human gastric cancer tissues and compared the rates with expression in adjacent non-tumor tissues. As a result, all cancer tissues showed enhanced BTF3 expression. Further analysis via stable silencing of BTF3 in gastric cancer SGC7901 cells revealed that inhibiting BTF3 activity led to lower proliferation and enhanced apoptosis rates. The results are interesting and BTF3 might be a target gene for gastric cancer treatment.
Footnotes
Supported by Science and Technology Project of Hunan Province, China, No. 2013FJ3151
P- Reviewer Aurello P S- Editor Gou SX L- Editor Ma JY E- Editor Li JY
References
- 1.Kanno M, Chalut C, Egly JM. Genomic structure of the putative BTF3 transcription factor. Gene. 1992;117:219–228. doi: 10.1016/0378-1119(92)90732-5. [DOI] [PubMed] [Google Scholar]
- 2.Zheng XM, Black D, Chambon P, Egly JM. Sequencing and expression of complementary DNA for the general transcription factor BTF3. Nature. 1990;344:556–559. doi: 10.1038/344556a0. [DOI] [PubMed] [Google Scholar]
- 3.Potashkin J, Wentz-Hunter K, Callaci J. BTF3 is evolutionarily conserved in fission yeast. Biochim Biophys Acta. 1996;1308:182–184. doi: 10.1016/0167-4781(96)00114-5. [DOI] [PubMed] [Google Scholar]
- 4.Cavallini B, Huet J, Plassat JL, Sentenac A, Egly JM, Chambon P. A yeast activity can substitute for the HeLa cell TATA box factor. Nature. 1988;334:77–80. doi: 10.1038/334077a0. [DOI] [PubMed] [Google Scholar]
- 5.Zheng XM, Moncollin V, Egly JM, Chambon P. A general transcription factor forms a stable complex with RNA polymerase B (II) Cell. 1987;50:361–368. doi: 10.1016/0092-8674(87)90490-9. [DOI] [PubMed] [Google Scholar]
- 6.Parvin JD, Shykind BM, Meyers RE, Kim J, Sharp PA. Multiple sets of basal factors initiate transcription by RNA polymerase II. J Biol Chem. 1994;269:18414–18421. [PubMed] [Google Scholar]
- 7.Deng JM, Behringer RR. An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res. 1995;4:264–269. doi: 10.1007/BF01969120. [DOI] [PubMed] [Google Scholar]
- 8.Davies CR, Morris JS, Griffiths MR, Page MJ, Pitt A, Stein T, Gusterson BA. Proteomic analysis of the mouse mammary gland is a powerful tool to identify novel proteins that are differentially expressed during mammary development. Proteomics. 2006;6:5694–5704. doi: 10.1002/pmic.200600202. [DOI] [PubMed] [Google Scholar]
- 9.Roy L, Laboissière S, Abdou E, Thibault G, Hamel N, Taheri M, Boismenu D, Lanoix J, Kearney RE, Paiement J. Proteomic analysis of the transitional endoplasmic reticulum in hepatocellular carcinoma: an organelle perspective on cancer. Biochim Biophys Acta. 2010;1804:1869–1881. doi: 10.1016/j.bbapap.2010.05.008. [DOI] [PubMed] [Google Scholar]
- 10.Wang CJ, Frånbergh-Karlson H, Wang DW, Arbman G, Zhang H, Sun XF. Clinicopathological significance of BTF3 expression in colorectal cancer. Tumour Biol. 2013;34:2141–2146. doi: 10.1007/s13277-013-0745-8. [DOI] [PubMed] [Google Scholar]
- 11.Odreman F, Vindigni M, Gonzales ML, Niccolini B, Candiano G, Zanotti B, Skrap M, Pizzolitto S, Stanta G, Vindigni A. Proteomic studies on low- and high-grade human brain astrocytomas. J Proteome Res. 2005;4:698–708. doi: 10.1021/pr0498180. [DOI] [PubMed] [Google Scholar]
- 12.Dunican DS, McWilliam P, Tighe O, Parle-McDermott A, Croke DT. Gene expression differences between the microsatellite instability (MIN) and chromosomal instability (CIN) phenotypes in colorectal cancer revealed by high-density cDNA array hybridization. Oncogene. 2002;21:3253–3257. doi: 10.1038/sj.onc.1205431. [DOI] [PubMed] [Google Scholar]
- 13.Pasquale EB. Eph receptors and ephrins in cancer: bidirectional signalling and beyond. Nat Rev Cancer. 2010;10:165–180. doi: 10.1038/nrc2806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surawska H, Ma PC, Salgia R. The role of ephrins and Eph receptors in cancer. Cytokine Growth Factor Rev. 2004;15:419–433. doi: 10.1016/j.cytogfr.2004.09.002. [DOI] [PubMed] [Google Scholar]
- 15.Westphal CH, Schmaltz C, Rowan S, Elson A, Fisher DE, Leder P. Genetic interactions between atm and p53 influence cellular proliferation and irradiation-induced cell cycle checkpoints. Cancer Res. 1997;57:1664–1667. [PubMed] [Google Scholar]
- 16.Lee JH, Paull TT. Activation and regulation of ATM kinase activity in response to DNA double-strand breaks. Oncogene. 2007;26:7741–7748. doi: 10.1038/sj.onc.1210872. [DOI] [PubMed] [Google Scholar]
- 17.Liu LX, Liu ZH, Jiang HC, Qu X, Zhang WH, Wu LF, Zhu AL, Wang XQ, Wu M. Profiling of differentially expressed genes in human gastric carcinoma by cDNA expression array. World J Gastroenterol. 2002;8:580–585. doi: 10.3748/wjg.v8.i4.580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kusumawidjaja G, Kayed H, Giese N, Bauer A, Erkan M, Giese T, Hoheise JD, Friess H, Kleeff J. Basic transcription factor 3 (BTF3) regulates transcription of tumor-associated genes in pancreatic cancer cells. Cancer Biol Ther. 2007;6:367–376. doi: 10.4161/cbt.6.3.3704. [DOI] [PubMed] [Google Scholar]
- 19.el-Tanani MK, Green CD. Transcription factor, BTF3, and the AF-1 function of the estrogen receptor. Biochem Soc Trans. 1998;26:S252. doi: 10.1042/bst026s252. [DOI] [PubMed] [Google Scholar]
- 20.Green CD, Thompson PD, Johnston PG, El-Tanani MK. Interaction between transcription factor, basal transcription factor 3, and the NH2-terminal domain of human estrogen receptor alpha. Mol Cancer Res. 2007;5:1191–1200. doi: 10.1158/1541-7786.MCR-07-0123. [DOI] [PubMed] [Google Scholar]
- 21.Zhang J, Zhu ZG, Ji J, Yuan F, Yu YY, Liu BY, Lin YZ. Transcription factor Sp1 expression in gastric cancer and its relationship to long-term prognosis. World J Gastroenterol. 2005;11:2213–2217. doi: 10.3748/wjg.v11.i15.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Xiao Q, Li L, Xie Y, Tan N, Wang C, Xu J, Xia K, Gardner K, Li QQ. Transcription factor E2F-1 is upregulated in human gastric cancer tissues and its overexpression suppresses gastric tumor cell proliferation. Cell Oncol. 2007;29:335–349. doi: 10.1155/2007/147615. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Haveri H, Westerholm-Ormio M, Lindfors K, Mäki M, Savilahti E, Andersson LC, Heikinheimo M. Transcription factors GATA-4 and GATA-6 in normal and neoplastic human gastrointestinal mucosa. BMC Gastroenterol. 2008;8:9. doi: 10.1186/1471-230X-8-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Akiyama Y, Watkins N, Suzuki H, Jair KW, van Engeland M, Esteller M, Sakai H, Ren CY, Yuasa Y, Herman JG, et al. GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol Cell Biol. 2003;23:8429–8439. doi: 10.1128/MCB.23.23.8429-8439.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li Q, Zhang N, Jia Z, Le X, Dai B, Wei D, Huang S, Tan D, Xie K. Critical role and regulation of transcription factor FoxM1 in human gastric cancer angiogenesis and progression. Cancer Res. 2009;69:3501–3509. doi: 10.1158/0008-5472.CAN-08-3045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim SY, Yoon J, Ko YS, Chang MS, Park JW, Lee HE, Kim MA, Kim JH, Kim WH, Lee BL. Constitutive phosphorylation of the FOXO1 transcription factor in gastric cancer cells correlates with microvessel area and the expressions of angiogenesis-related molecules. BMC Cancer. 2011;11:264. doi: 10.1186/1471-2407-11-264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhu H, Xia L, Zhang Y, Wang H, Xu W, Hu H, Wang J, Xin J, Gang Y, Sha S, et al. Activating transcription factor 4 confers a multidrug resistance phenotype to gastric cancer cells through transactivation of SIRT1 expression. PLoS One. 2012;7:e31431. doi: 10.1371/journal.pone.0031431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Uhlen M, Oksvold P, Fagerberg L, Lundberg E, Jonasson K, Forsberg M, Zwahlen M, Kampf C, Wester K, Hober S, et al. Towards a knowledge-based Human Protein Atlas. Nat Biotechnol. 2010;28:1248–1250. doi: 10.1038/nbt1210-1248. [DOI] [PubMed] [Google Scholar]
- 29.Thakur D, Saxena R, Singh V, Haq W, Katti SB, Singh BN, Tripathi RK. Human beta casein fragment (54-59) modulates M. bovis BCG survival and basic transcription factor 3 (BTF3) expression in THP-1 cell line. PLoS One. 2012;7:e45905. doi: 10.1371/journal.pone.0045905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Brockstedt E, Otto A, Rickers A, Bommert K, Wittmann-Liebold B. Preparative high-resolution two-dimensional electrophoresis enables the identification of RNA polymerase B transcription factor 3 as an apoptosis-associated protein in the human BL60-2 Burkitt lymphoma cell line. J Protein Chem. 1999;18:225–231. doi: 10.1023/a:1020636308270. [DOI] [PubMed] [Google Scholar]
- 31.Li R, Liu XL, Du QF, Zhang S, Luo RC, Zhou SY. Proteome analysis of apoptotic K562 cells induced by harringtonine. Zhonghua Xueyexue Zazhi. 2004;25:323–327. [PubMed] [Google Scholar]
- 32.Pham VC, Pitti R, Anania VG, Bakalarski CE, Bustos D, Jhunjhunwala S, Phung QT, Yu K, Forrest WF, Kirkpatrick DS, et al. Complementary proteomic tools for the dissection of apoptotic proteolysis events. J Proteome Res. 2012;11:2947–2954. doi: 10.1021/pr300035k. [DOI] [PubMed] [Google Scholar]