Abstract
The bacterial RNase P holoenzyme catalyzes the formation of the mature 5′-end of tRNAs and is composed of an RNA and a protein subunit. Among the two folding domains of the RNase P RNA, the catalytic domain (C-domain) contains the active site of this ribozyme. We investigated specific binding of the Bacillus subtilis C-domain with the B.subtilis RNase P protein and examined the catalytic activity of this C-domain–P protein complex. The C-domain forms a specific complex with the P protein with a binding constant of ∼0.1 µM. The C-domain–P protein complex and the holoenzyme are equally efficient in cleaving single-stranded RNA (∼0.9 min–1 at pH 7.8) and substrates with a hairpin–loop 3′ to the cleavage site (∼40 min–1). The holoenzyme reaction is much more efficient with a pre-tRNA substrate, binding at least 100-fold better and cleaving 10–500 times more efficiently. These results demonstrate that the RNase P holoenzyme is functionally constructed in three parts. The catalytic domain alone contains the active site, but has little specificity and affinity for most substrates. The specificity and affinity for the substrate is generated by either the specificity domain of RNase P RNA binding to a T stem–loop-like hairpin or RNase P protein binding to a single-stranded RNA. This modular construction may be exploited to obtain RNase P-based ribonucleoprotein complexes with altered substrate specificity.
INTRODUCTION
Bacterial RNase P is responsible for the generation of the mature 5′-ends of all tRNAs in the cell (1,2). The RNase P holoenzyme is composed of an RNA of ∼330–420 nt and a protein of ∼120 amino acids. The Bacillus subtilis RNase P RNA (P RNA) contains two folding domains that also have distinct functions (3–6): one binds the T stem–loop region of a pre-tRNA substrate (the specificity domain or S-domain) and the other contains the active site (the catalytic domain or C-domain). Previous work has identified the primary function of the S-domain as providing specificity and affinity to this ribozyme reaction.
The P protein in the holoenzyme has been shown to bind a single-stranded region in the 5′-leader of a pre-tRNA substrate (7). This ability of P protein binding to a single-stranded RNA turns the holoenzyme into an excellent enzyme that cleaves a wide range of substrates as simple as single-stranded RNA (8). In previous studies on the holoenzyme it was thought that both domains of P RNA are required for P protein binding to P RNA (9–11). Therefore, it is not known how the holoenzyme is partitioned functionally.
This work demonstrates that the catalytic domain of B.subtilis P RNA alone forms a specific complex with B.subtilis P protein and that the C-domain–P protein complex is a very efficient enzyme (Fig. 1). The C-domain–P protein complex cleaves single-stranded and hairpin–loop RNA substrates with the same efficiency as the holoenzyme. For cleavage of a pre-tRNA substrate the holoenzyme is much more efficient and much less sensitive to variations in the 5′-leader length/sequence. These results indicate that a ribonucleoprotein enzyme as exemplified by the RNase P holoenzyme is functionally constructed in multiple parts.
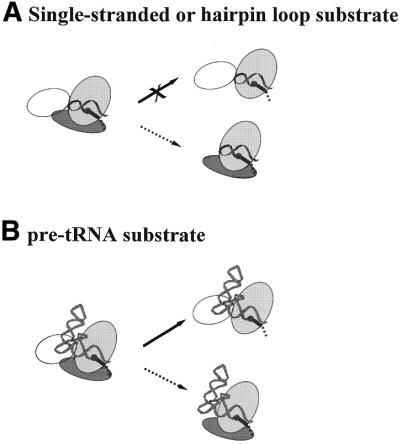
Figure 1.
Effects of P protein depletion [previous results summarized in Loria and Pan (19)] (solid arrows) or S-domain deletion (this work, dashed arrows) on catalytic activity. (A) P RNA cleaves a single-stranded or a hairpin–loop substrate at least 10 000-fold less efficiently under high salt conditions (data not shown). In contrast, the C-domain–P protein complex has the same catalytic efficiency for a hairpin–loop or a single-stranded RNA substrate as the holoenzyme. (B) P RNA effectively binds and cleaves a pre-tRNA substrate. The C-domain–P protein complex also binds and cleaves a pre-tRNA substrate, but less efficiently than a hairpin–loop substrate.
MATERIALS AND METHODS
Preparation of the RNA and P protein
The C-domain of the B.subtilis P RNA, containing nt 240–409 + 1–85, was constructed as described (5). The RNA was prepared by the standard in vitro transcription method using T7 RNA polymerase (12). The single-stranded and hairpin–loop substrates were chemically synthesized using 2′-ortho-ester-protected phosphoramidites (Dharmacon Research, Boulder, CO). The pre-tRNA substrates were prepared by enzymatic ligation (13) of a synthetic oligonucleotide to an RNA transcript composed of nt 10–76 of yeast tRNAPhe (14). The B.subtilis P protein was prepared by overexpression as described by Fierke and co-workers (15).
Hydroxyl radical protection of the C-domain–P protein complex
Hydroxyl radical protection was carried out using the standard Fe(II)–EDTA method in 20 mM Tris–HCl pH 7.5, 10 mM MgCl2 at 37°C (16,17). Briefly, the C-domain alone in buffer was heated at 85°C for 2 min, followed by incubation for 3 min at room temperature. Mg2+ was added to designated concentrations. The mixture was incubated for 5 min at 50°C. An equimolar ratio of P protein was added. The mixture was further incubated for 5 min at 37°C. Ascorbic acid and dithiothreitol were added to final concentrations of 1 and 5 mM, respectively. The reaction was immediately initiated by addition of 1 mM Fe(II), 1.2 mM EDTA. The reaction proceeded for 30 min at 37°C and was quenched upon addition of 10 mM thiourea. The reaction mixture was separated on polyacrylamide gels containing 7 M urea and the hydroxyl radical cleavage products quantitated by phosphorimaging.
In this work the C-domain and P protein were always kept at a 1:1 molar ratio to prevent non-specific binding by P protein. Under these conditions the fraction of the C-domain protected (y) as a function of total RNA concentration (x) can be described by applying equilibria equations:
y = 1 + Kd/2x – √ (Kd2/4x2 + Kd/x) 1a
where Kd is the binding constant of the complex. Another way to present this type of data is to plot the apparent equilibrium constant Keq [= y/(1 – y)2] as a function of total RNA concentration:
y/(1 – y)2 = Keq = x/Kd 1b
Kinetics of the cleavage reaction
All reactions were performed under single turnover conditions at 0.02–25 µM enzyme and <2 nM 32P-labeled substrate. The C-domain–P protein complex was reconstituted as described above. The 5′-32P-labeled RNA substrates were renatured as described previously (14). The cleavage reaction was initiated upon mixing an equal volume of the ribozyme and the substrate. Aliquots were taken at designated time points and mixed with an excess of 9 M urea, 100 mM EDTA to stop the cleavage reaction. The reaction products were separated from the unreacted substrates on denaturing gels containing 7 M urea. The amounts of products and substrates were determined by phosphorimaging using a Fuji Phosphorimager. Reaction rates were obtained by fitting the amount of cleavage products over time to a single exponential.
RESULTS AND DISCUSSION
The C-domain forms a specific complex with P protein
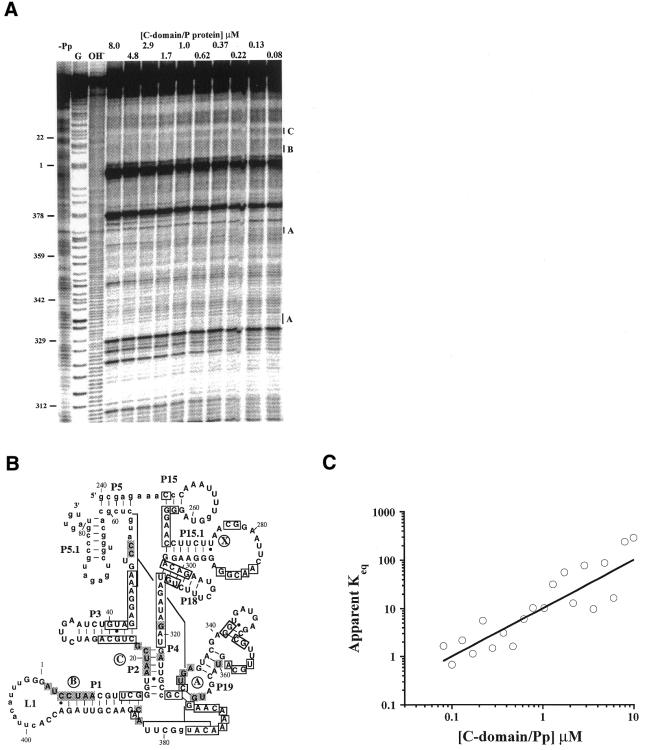
Hydroxyl radical protection was performed to determine whether the C-domain forms a specific complex with P protein (Fig. 2). Hydroxyl radicals cleave the solvent-accessible ribose–phosphate positions in RNA. Extensive protection was observed upon folding of the C-domain alone (Fig. 2A, lane –Pp). Since the P protein has a strong tendency to bind RNA non-specifically (10,11,15,18), a 1:1 molar ratio of C-domain and P protein was maintained to minimize protections arising from non-specific binding by P protein (data not shown).
Figure 2.
The B.subtilis C-domain forms a specific complex with the B.subtilis P protein in the absence of substrate. (A) Hydroxyl radical protection at varying concentrations of C-domain and P protein, always at a 1:1 molar ratio. All reactions contained 20 mM Tris–HCl pH 7.5, 10 mM MgCl2 and the reaction time was 30 min at 37°C. G, partial nuclease T1 digestion; OH–, partial alkaline hydrolysis; –Pp, 8 µM C-domain alone. (B) The protected residues upon P protein binding superimposed on the phylogenetically derived secondary structure (33). Only residues that have protection factors of >1.5 are considered to be protected. Residues protected upon P protein binding are shaded. Residues protected upon Mg2+-induced folding are boxed (5). Region X (residues 270–280) was only protected in the holoenzyme (11). Nucleotides shown in lower case could not be analyzed either due to gel resolution or due to autolytic cleavage (around nt 405). (C) Determination of the binding constant of the C-domain–P protein complex [apparent Keq = y/(1 – y)2, curve fitted to equation 1b]. The protection factors (PF) for regions A–C are averaged. The fraction of C-domain bound with P protein corresponds to 1/PF and is plotted against the C-domain and P protein concentration, always at a 1:1 molar ratio.
Additional protections at specific regions in the C-domain are observed upon addition of the P protein (Fig. 2A, compare lane –Pp or 0.08 with 8; Fig. 2B, shaded nucleotides). Because it is not possible to distinguish direct protein contacts and conformational changes in the C-domain on the basis of footprinting alone, only regions that had more than five protected residues are further discussed. Protected regions A and C are present in the C-domain–P protein complex and in the holoenzyme (11), suggesting that they are contact sites for P protein. Region B in the C-domain–P protein complex could not be analyzed in the holoenzyme due to its proximity to the 5′-end of P RNA. Region X (residues 270–280) was protected in the holoenzyme, but no protection was present in the C-domain–P protein complex. Therefore, protection of region X in the holoenzyme is likely derived from either interdomain RNA–RNA interactions or from additional protein binding that can only occur in the presence of the S-domain.
The binding constant (Kd) of the C-domain–P protein complex can be obtained from the extent of protection as a function of C-domain and P protein concentration (Fig. 2C, where the slope = 1/Kd). Because the RNA and protein are always equimolar, the fraction of C-domain bound with P protein is fitted by 1b, assuming that the complex contains one C-domain and one P protein subunit. The Kd of the C-domain–P protein complex is 0.099 ± 0.015 µM, ∼200-fold weaker than the Kd of the holoenzyme (table 3 in Talbot and Altman; 18).
In summary, the C-domain forms a specific complex with the P protein. Compared to the holoenzyme, this specific C-domain–P protein complex has decreased regions of protection and lower binding affinity.
Catalytic activity of the C-domain–P protein complex
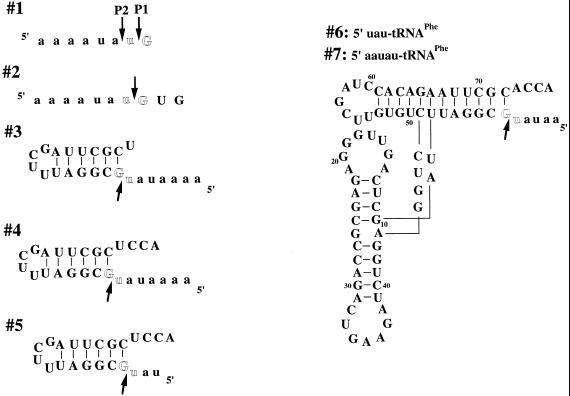
The catalytic activity of the C-domain–P protein complex was demonstrated briefly in a previous study but no detailed analyses were carried out (19). In order to compare substrate recognition and catalytic efficiency of the C-domain–P RNA complex and the holoenzyme, cleavage of seven substrates was determined (Fig. 3 and Table 1). All seven substrates had similar 5′-leader sequences but different RNA structure on the 3′-side of the cleavage site: single-stranded (1 and 2), hairpin–loop (3–5) or tRNA (6 and 7).
Figure 3.
Substrates used in this study with the intended cleavage site shown between the highlighted residues. The observed cleavage sites are indicated by arrows. The 5′-leader regions are shown in lower case.
Table 1. Catalysis by the C-domain–P protein complex.
| Substrate |
kcl (min–1)a |
K1/2 (µM)b |
| 1 | 0.057 ± 0.002 (P2) | 7.1 ± 0.6 |
| 0.012 ± 0.002 (P1) | ||
| 2 | 0.92 ± 0.04 | 5.6 ± 0.6 |
| 3 | 31 ± 5 | 4.6 ± 1.8 |
| 4 | 35 ± 3 | 1.8 ± 0.5 |
| 5 | 17 ± 3 | 13 ± 4 |
| 6 | 0.90 ± 0.06 | 1.9 ± 0.4 |
| 7 | 8.1 ± 0.4 | 0.70 ± 0.15 |
aCleavage rate at saturating enzyme concentration under single turnover conditions. Conditions for all substrates: 50 mM Tris–HCl pH 7.78, 10 mM MgCl2, 2% glycerol, 37°C.
bThe enzyme concentration at kcl/2.
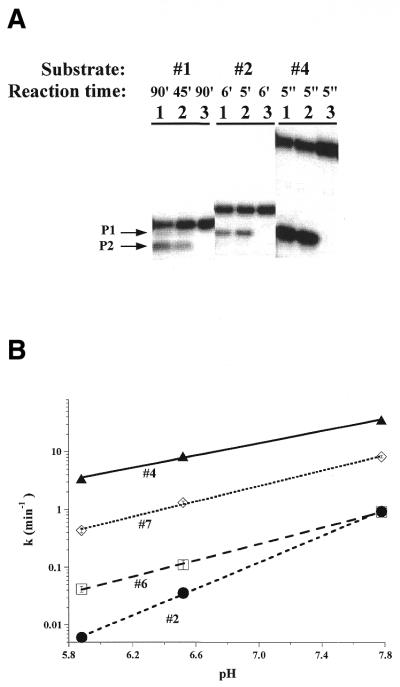
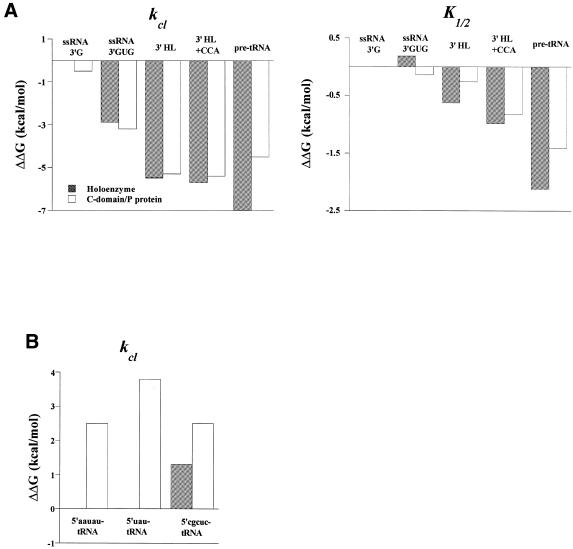
The C-domain–P protein complex accurately and efficiently cleaved all seven substrates (Fig. 4 and Table 1). Catalysis by the C-domain–P protein complex and by the holoenzyme had remarkable similarities. Both complexes cleaved all substrates at the same sites (see for example Fig. 4A), suggesting similar specificities in substrate binding, and showed the same pH dependencies (see for example Fig. 4B), suggesting a similar rate limiting step in the catalysis. Both the cleavage rate (kcl) and the ribozyme concentration at kcl/2 (K1/2) are within 2-fold (ΔΔG < 0.4 kcal/mol) for the single-stranded and the hairpin–loop substrates (Fig. 5A).
Figure 4.
(A) Cleavage of 5′-32P-labeled substrates 1, 2 and 4 by 1 µM holoenzyme (lane 1) and 1 µM C-domain–P protein complex (lane 2) under single turnover conditions. The control reaction was performed with 1 µM P protein alone (lane 3). All reactions contained 20 mM Tris–HCl pH 7.5, 10 mM MgCl2, 2% glycerol. (B) pH dependence of the cleavage rate at saturating concentrations of the C-domain–P protein complex (>10 µM) for substrates 2, 4, 6 and 7. The pH dependencies for cleavage of substrates 2 and 4 have slopes of 1.1 and 0.6 for the C-domain–P protein complex, similar to those for the holoenzyme (1.0 for substrate 2 and 0.7 for substrate 4; 8).
Figure 5.
(A) Changes in the cleavage rate (kcl) and Michaelis constant (K1/2) upon successive addition of RNA structure/nucleotides 3′ to the cleavage site. The holoenzyme data are taken from Loria and Pan (8). The ΔΔG values [left, ΔΔG = –RT ln(k2–7/k1(P1)); right, ΔΔG = –RT ln(K2–7/K1)] are normalized to the cleavage site P1 of substrate 1 in the holoenzyme reaction. The pre-tRNA substrate data is for substrate 7, 5′-aauau-tRNAPhe. (B) Changes in the cleavage rate upon altering the 5′-leader of pre-tRNAPhe substrates. The ΔΔG values are normalized to 5′-aauau-tRNAPhe in the holoenzyme reaction. The data for the 5′-cgcuc-tRNAPhe are taken from Loria and Pan (19).
The C-domain–P protein complex cleaved pre-tRNA substrates significantly less efficiently compared to the holoenzyme (Fig. 5B). The quantitative difference in the cleavage rates, however, strongly depended on the length and sequence of the 5′-leader. When the 5′-leader was 5′-aauau (substrate 7) the holoenzyme had an ∼50-fold faster cleavage rate (ΔΔG ≈ 2.4 kcal/mol). Truncation of 5′-aauau to 5′-uau (substrate 6) made no difference for the holoenzyme reaction, but decreased the cleavage rate by the C-domain–P protein ∼10-fold (ΔΔG ≈ 3.8 kcal/mol). Changing 5′-aauau to 5′-cgcuc decreased the cleavage rate by the holoenzyme ∼8-fold, but had no effect on the C-domain–P protein reaction (ΔΔG ≈ 1.2 kcal/mol). These results show that holoenzyme interactions with the T stem–loop of the tRNA generate faster rates, less sensitivity to the 5′-leader length and a stronger preference for an A/U-rich 5′-leader sequence. This result is consistent with a preference for the 5′-leader sequence and length of the natural B.subtilis tRNA precursors (20).
As for binding of pre-tRNA substrates, the K1/2 values measured in Mg2+ from single turnover reactions did not approximate the binding constant as determined by pulse–chase experiments (8; data not shown). However, the K1/2 values measured from single turnover reactions carried out in Ca2+ can be used to approximate the binding constant of a pre-tRNA substrate. Compared to the holoenzyme, the C-domain–P protein complex bound the 5′-aauau-tRNA substrate at least 100-fold weaker in Ca2+ (data not shown).
In summary, the S-domain–T stem–loop interaction increased the binding affinity of a pre-tRNA by >100-fold and increased the cleavage rate by up to 500-fold. The catalytic activities of the C-domain–P protein complex and the holoenzyme were essentially identical for any other substrate that only interacts with the C-domain and the P protein.
Modular construction for function of a ribonucleoprotein enzyme
This work shows that bacterial RNase P is a modular enzyme, functionally partitioned among the two P RNA domains and the P protein. The catalytic domain contains the active site and is a very efficient ribozyme for substrates that can interact with the L1 loop (21), although it cleaves pre-tRNA and other substrates very poorly. The specificity domain contains the binding site for a specific hairpin–loop, e.g. the T stem–loop in tRNA or hairpin–loops in several selected substrates (22), and it can specifically bind a pre-tRNA substrate at micromolar affinity (unpublished results). The P protein contains a substrate-binding site for single-stranded RNA plus two other potential binding sites, presumably for binding to P RNA (23).
Starting from the C-domain, the addition of either the S-domain or the P protein or both produces different enzymes. The C-domain linked with the S-domain is P RNA, a ribozyme capable of specific recognition and cleavage of a pre-tRNA substrate, but works very poorly with single-stranded and hairpin–loop substrates (Fig. 1A and B, top). For P RNA the efficiency and specificity is conferred by the S-domain interaction with a portion of the substrate 3′ to the cleavage site. Association of the C-domain and P protein produces the C-domain–P protein complex, a ribonucleoprotein enzyme capable of cleaving single-stranded and hairpin–loop substrates but which cannot distinguish pre-tRNA from hairpin–loop substrates (Fig. 1A and B, bottom). For the C-domain–P protein complex the efficiency and a large portion of the specificity is conferred by the P protein interaction with a portion of the substrate 5′ to the cleavage site. The desired specificity and catalytic efficiency for a pre-tRNA substrate is only achieved by combination of all three modules to produce the holoenzyme.
Our results establish a parallel for ribonucleoprotein and protein enzymes. Examples of functional division in protein enzymes abound. The restriction enzyme FokI contains two domains: one responsible for specific binding of its recognition site, the other responsible for endonucleolytic cleavage 5–9 bp from the recognition site (24,25). Many aminoacyl-tRNA synthetases contain more than one domain: one responsible for recognition of anticodon nucleotides, the other responsible for aminoacylation (26–31). The Tn5 transposase contains three domains: one binds to DNA, another cleaves the DNA strand and the third is responsible for protein–protein contacts in the synaptic complex (32).
The modular construction of RNase P function may be exploited to obtain RNase P-based ribonucleoprotein enzymes with altered specificity. We have previously demonstrated that the S-domain can be substituted by a randomly chosen RNA structure and that this altered ribozyme efficiently cleaves only RNA substrates that contain matching structures to the S-domain replacement (6). Likewise, the sequence of the 5′-leader binding-site in the P protein may be changed and a C-domain–altered P protein complex may efficiently cleave only substrates that contain matching structures to the altered binding site in the P protein mutant.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr X.-W.Fang and other members of the Pan laboratory for helpful discussions. We also thank Dr C.Correll and the reviewers for insightful comments on the manuscript. This work was supported by a grant from the NIH (GM52993).
References
- 1.Frank D.N. and Pace,N.R. (1998) Ribonuclease P: unity and diversity in a tRNA processing ribozyme. Annu. Rev. Biochem., 67, 153–180. [DOI] [PubMed] [Google Scholar]
- 2.Altman S. and Kirsebom,L. (1999) Ribonuclease P. In Gesteland,R.F., Cech,T.R. and Atkins,J.F. (eds), The RNA World. Cold Spring Harbor Laboratory Press, Cold Spring Harbor, NY, pp. 351–380.
- 3.Loria A. and Pan,T. (1996) Domain structure of the ribozyme from eubacterial ribonuclease P. RNA, 2, 551–563. [PMC free article] [PubMed] [Google Scholar]
- 4.Massire C., Jaeger,L. and Westhof,E. (1998) Derivation of the three-dimensional architecture of bacterial ribonuclease P RNAs from comparative sequence analysis. J. Mol. Biol., 279, 773–793. [DOI] [PubMed] [Google Scholar]
- 5.Fang X., Pan,T. and Sosnick,T.R. (1999) A thermodynamic framework and cooperativity in the tertiary folding of a Mg2+-dependent ribozyme. Biochemistry, 38, 16840–16846. [DOI] [PubMed] [Google Scholar]
- 6.Mobley E.M. and Pan,T. (1999) Design and isolation of ribozyme–substrate pairs using RNase P-based ribozymes containing altered substrate binding sites. Nucleic Acids Res., 27, 4298–4304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Niranjanakumari S., Stams,T., Crary,S.M., Christianson,D.W. and Fierke,C.A. (1998) Protein component of the ribozyme ribonuclease P alters substrate recognition by directly contacting precursor tRNA. Proc. Natl Acad. Sci. USA, 95, 15212–15217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Loria A. and Pan,T. (2000) The 3′ substrate determinants for the catalytic efficiency of the Bacillus subtilis RNase P holoenzyme suggest autolytic processing of the RNase P RNA in vivo. RNA, 6, 1413–1422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Vioque A., Arnez,J. and Altman,S. (1988) Protein–RNA interactions in the RNase P holoenzyme from Escherichia coli. J. Mol. Biol., 202, 835–848. [DOI] [PubMed] [Google Scholar]
- 10.Talbot S.J. and Altman,S. (1994) Kinetic and thermodynamic analysis of RNA–protein interactions in the RNase P holoenzyme from Escherichia coli. Biochemistry, 33, 1406–1411. [DOI] [PubMed] [Google Scholar]
- 11.Loria A., Niranjanakumari,S., Fierke,C.A. and Pan,T. (1998) Recognition of a pre-tRNA substrate by the Bacillus subtilis RNase P holoenzyme. Biochemistry, 37, 15466–15473. [DOI] [PubMed] [Google Scholar]
- 12.Milligan J.F., Groebe,D.R., Witherell,G.W. and Uhlenbeck,O.C. (1987) Oligoribonucleotide synthesis using T7 RNA polymerase and synthetic DNA templates. Nucleic Acids Res., 15, 8783–8798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moore M.J. and Sharp,P.A. (1992) Site-specific modification of pre-mRNA: the 2′ hydroxyl groups at the splice site. Science, 256, 992–997. [DOI] [PubMed] [Google Scholar]
- 14.Loria A. and Pan,T. (1997) Recognition of the T stem–loop of a pre-tRNA substrate by the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry, 36, 6317–6235. [DOI] [PubMed] [Google Scholar]
- 15.Niranjanakumari S., Kurz,J.C. and Fierke,C.A. (1998) Expression, purification and characterization of the recombinant RNase P protein component from Bacillus subtilis. Nucleic Acids Res., 26, 3090–3096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Celander D.W. and Cech,T.R. (1991) Visualizing the higher order folding of a catalytic RNA molecule. Science, 251, 401–407. [DOI] [PubMed] [Google Scholar]
- 17.Pan T. (1995) Higher order folding and domain analysis of the ribozyme from Bacillus subtilis ribonuclease P. Biochemistry, 34, 902–909. [DOI] [PubMed] [Google Scholar]
- 18.Talbot S.J. and Altman,S. (1994) Gel retardation analysis of the interaction between C5 protein and M1 RNA in the formation of the ribonuclease P holoenzyme from Escherichia coli. Biochemistry, 33, 1399–1405. [DOI] [PubMed] [Google Scholar]
- 19.Loria A. and Pan,T. (1999) The cleavage step of ribonuclease P catalysis is determined by ribozyme-substrate interactions both distal and proximal to the cleavage site. Biochemistry, 38, 8612–8620. [DOI] [PubMed] [Google Scholar]
- 20.Kunst F., Ogaswara,N., Moszer,I., Albertini,A.M., Alloui,G., Azevedo,V., Bertero,M.G., Bessieres,P., Bolotin,A., Borchert,S. et al (1997) The complete genome sequence of the gram-positive bacterium Bacillus subtilis. Nature, 390, 249–256. [DOI] [PubMed] [Google Scholar]
- 21.Pan T. and Jakacka,M. (1996) Multiple substrate binding sites in the ribozyme from Bacillus subtilis RNase P. EMBO J., 15, 2249–2255. [PMC free article] [PubMed] [Google Scholar]
- 22.Odell L., Huang,V., Jakacka,M. and Pan,T. (1998) Interaction of structural modules in substrate binding by the ribozyme from Bacillus subtilis RNase P. Nucleic Acids Res., 26, 3717–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stams T., Niranjanakumari,S., Fierke,C.A. and Christianson,D.W. (1998) Ribonuclease P protein structure: evolutionary origins in the translational apparatus. Science, 280, 752–755. [DOI] [PubMed] [Google Scholar]
- 24.Wah D.A., Bitinaite,J., Schildkraut,I. and Aggarwal,A.K. (1998) Structure of FokI has implications for DNA cleavage. Proc. Natl Acad. Sci. USA, 95, 10564–10569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aggarwal A.K. and Wah,D.A. (1998) Novel site-specific DNA endonucleases. Curr. Opin. Struct. Biol., 8, 19–25. [DOI] [PubMed] [Google Scholar]
- 26.Ibba M., Curnow,A.W. and Soll,D. (1997) Aminoacyl-tRNA synthesis: divergent routes to a common goal. Trends Biochem. Sci., 22, 39–42. [DOI] [PubMed] [Google Scholar]
- 27.Whelihan E.F. and Schimmel,P. (1997) Rescuing an essential enzyme–RNA complex with a non-essential appended domain. EMBO J., 16, 2968–2974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chihade J.W. and Schimmel,P. (1999) Assembly of a catalytic unit for RNA microhelix aminoacylation using nonspecific RNA binding domains. Proc. Natl Acad. Sci. USA, 96, 12316–12321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sugiura I., Nureki,O., Ugaji-Yoshikawa,Y., Kuwabara,S., Shimada,A., Tateno,M., Lorber,B., Giege,R., Moras,D., Yokoyama,S. and Konno,M. (2000) The 2.0 Å crystal structure of Thermus thermophilus methionyl-tRNA synthetase reveals two RNA-binding modules. Strucure Fold. Des., 8, 197–208. [DOI] [PubMed] [Google Scholar]
- 30.Cura V., Moras,D. and Kern,D. (2000) Sequence analysis and modular organization of threonyl-tRNA synthetase from Thermus thermophilus and its interrelation with threonyl-tRNA synthetases of other origins. Eur. J. Biochem., 267, 379–393. [DOI] [PubMed] [Google Scholar]
- 31.Woese C.R., Olsen,G.J., Ibba,M. and Soll,D. (2000) Aminoacyl-tRNA synthetases, the genetic code and the evolutionary process. Microbiol. Mol. Biol. Rev., 64, 202–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Davies D.R., Goryshin,I.Y., Reznikoff,W.S. and Rayment,I. (2000) Three-dimensional structure of the Tn5 synaptic complex transposition intermediate. Science, 289, 77–85. [DOI] [PubMed] [Google Scholar]
- 33.Haas E.S., Brown,J.W., Pitulle,C. and Pace,N.R. (1994) Further perspective on the catalytic core and secondary structure of ribonuclease P RNA. Proc. Natl Acad. Sci. USA, 91, 2527–2531. [DOI] [PMC free article] [PubMed] [Google Scholar]