Abstract
The autoimmune lymphoproliferative syndrome (ALPS) is characterized by early-onset lymphadenopathy, splenomegaly, immune cytopenias, and an increased risk for B cell lymphomas. Most ALPS patients harbor mutations in the FAS gene, which regulates lymphocyte apoptosis. These are commonly missense mutations affecting the intracellular region of the protein and have a dominant-negative effect on the signaling pathway. However, analysis of a large cohort of ALPS patients revealed that ~30% have mutations affecting the extracellular region of FAS, and among these, 70% are nonsense, splice site, or insertions/ deletions with frameshift for which no dominant-negative effect would be expected. We evaluated the latter patients to understand the mechanism(s) by which these mutations disrupted the FAS pathway and resulted in clinical disease. We demonstrated that most extracellular-region FAS mutations induce low FAS expression due to nonsense-mediated RNA decay or protein instability, resulting in defective death-inducing signaling complex formation and impaired apoptosis, although to a lesser extent as compared with intracellular mutations. The apoptosis defect could be corrected by FAS overexpression in vitro. Our findings define haploinsufficiency as a common disease mechanism in ALPS patients with extracellular FAS mutations.
The autoimmune lymphoproliferative syndrome (ALPS) is characterized by early-onset development of benign lymphadenopathy and splenomegaly, multilineage cytopenias due to autoimmune peripheral destruction and splenic sequestration of blood cells, and increased risk for B cell lymphomas (1–3). Patients typically accumulate a hallmark population of mature TCRαβ+ T cells that are CD4 and CD8 negative (4, 5). ALPS is caused by defects in proteins involved in the FAS pathway of lymphocyte apoptosis. Most (~65%) patients have mutations in the FAS (TNFRSF6/APO1/CD95) gene, whereas a minority has mutations in the gene encoding FAS ligand (FASL) or caspase-10 (6–12). Germline mutations in CASP8 or somatic mutations in NRAS or KRAS cause ALPS-related syndromes currently classified separately (12–15).
The FAS gene contains nine exons spanning 26 kb on chromosome 10q24.1 (16). The first five exons encode the extracellular portion of the protein containing three cysteine-rich domains that are involved in receptor trimerization and FASL binding required for triggering of the apoptotic signal. Exon 6 codes for the transmembrane domain, and the intracellular portion is encoded by exons 7–9. The FAS death domain (DD), an 85-aa-long structure encoded by exon 9, is required for FAS-induced apoptosis of lymphocytes under physiological conditions (17).
The majority of FAS defects associated with ALPS are heterozygous missense mutations that affect the intracellular DD, allowing for the expression of a defective protein with a dominant-negative effect on the signaling pathway (7, 18). These mutations demonstrate high penetrance for clinical symptoms including refractory cytopenias and an increased risk for lymphoma (2, 18). However, evaluation of a large cohort of ALPS patients at our center has revealed a significant subpopulation harboring mutations affecting the extracellular region of the protein, and these are commonly nonsense, or insertions, deletions, and splice site mutations with frameshift (Fig. 1). These mutations are predicted to abolish protein expression and hence are not expected to result in dominant-negative interference, and the mechanism by which this affects FAS apoptosis signaling is not clearly defined. FAS haploinsufficiency is one possible disease mechanism in ALPS associated with extracellular mutations, but currently there are only very limited data supporting this mechanism (19, 20). In fact, FAS haploinsufficiency was clearly demonstrated in only one ALPS patient to date, such that the prevalence, long-term clinical impact, and biochemical consequences are unknown (19).
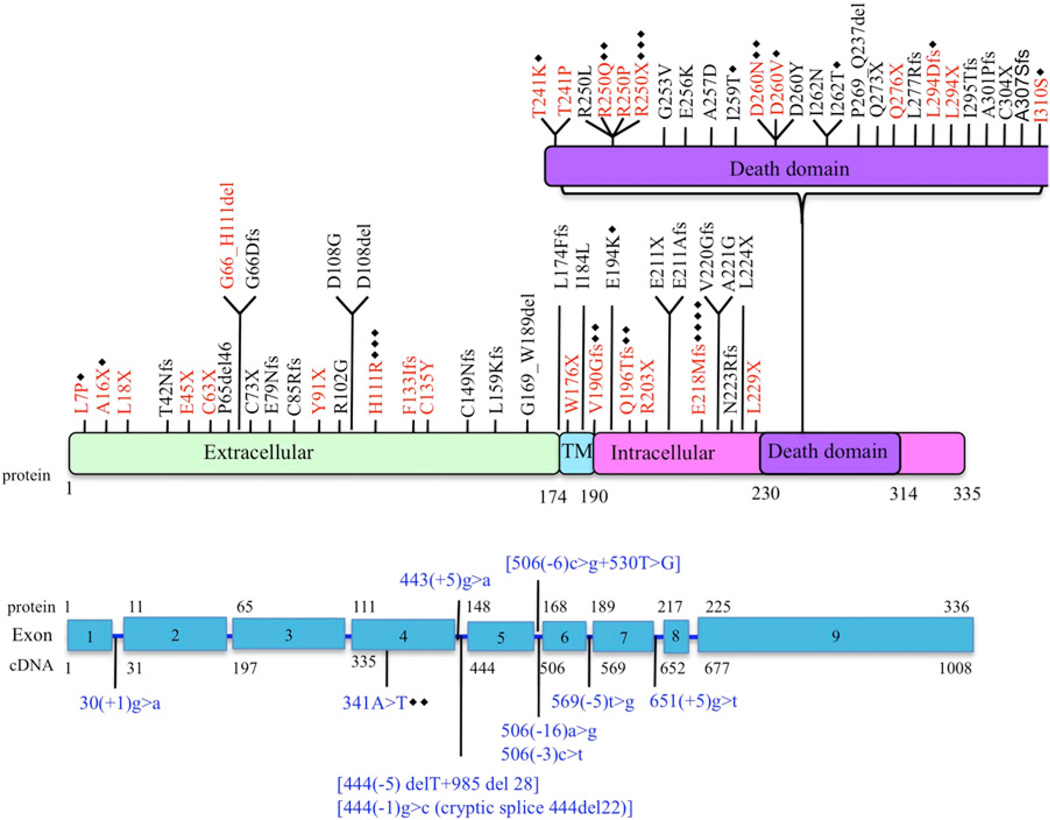
FIGURE 1.
Schematic representation of FAS mutations in ALPS patients. Red text indicates mutations evaluated in this study. Blue text indicates complex mutations. Black diamonds represent the number of additional families with same mutation. TM, transmembrane.
To understand the mechanism by which extracellular region FAS mutations disrupt the apoptotic machinery, we analyzed the functional impact of 29 representative mutations across all regions of FAS using patient-derived B cell lines. We demonstrate that most (8 of the 11) mutations affecting the extracellular regions of the FAS receptor we studied are associated with haploinsufficiency. These mutations induced less severe disruption of the FAS-mediated apoptosis-signaling platform when compared with missense intracellular mutations, and the apoptotic defect could be rescued in vitro by FAS overexpression. These findings define haploinsufficiency as an ALPS-causing disease mechanism, in addition to the well-documented dominant-negative interference seen with intracellular mutations.
Materials and Methods
Patient samples, cell culture, and gene sequencing
All patients were studied at the National Institutes of Health under Institutional Review Board-approved protocols (93-I-0063 and 95-I-0066). Genomic DNA samples were PCR-amplified and sequenced as previously described (21). For mRNA sequencing, cDNA was prepared from EBV-transformed B cell lines using the RNeasy plus mini kit (Qiagen). To rule out the presence of contaminating DNA, RNA samples were subjected to PCR amplification using intron-specific primers; the absence of amplified product (DNA) was confirmed by denaturing agarose gel electrophoresis (data not shown). cDNA was PCR amplified (forward: 5′-GTG-AGGGAAGCGGTTTACGAGTGA-3′; and reverse: 5′-AGTGGGGTTAG-CCTGTGGATAGAC-3′), and the products were subjected to sequencing (primer sequences available upon request). To determine the ratio of mutant to wild-type FAS transcripts, cDNA was amplified and cloned into the pcDNA 3.0-HA vector (modified from pcDNA3.0 [Invitrogen]) with EcoRI and XhoI enzyme sites. After transformation, we picked 10–20 colonies and performed sequencing. Three samples with DD mutations (D260N, Q276X, L294Dfs) were used as controls. For functional studies, EBV-transformed B cell lines from the patients were cultured in RPMI 1640 media with 10% FBS, 100 units/ml penicillin, 100 µg/ml streptomycin, and 2 mM l-glutamine (Life Technologies, Invitrogen).
Apoptosis measurement
EBV-transformed B cells (200,000/well) were aliquoted in triplicate into 96-well plates and cultured with or without APO-1–3 (1 µg/ml) (ENZO Life Sciences) in the presence of protein A (1 µg/ml). After 24 h, cell loss was determined by measuring the loss of the mitochondrial transmembrane potential using 3,3′-dihexyloxacarbocyanine iodide (DiOC6) (Calbiochem, EMC Biosciences) staining. Briefly, cells were incubated with 40 nM DiOC6 for 15 min at 37°C, and live cells (DiOC6 high) were counted by flow cytometry (BD FACSCanto II; BD Biosciences) by constant time acquisition. The percentage of cell loss was calculated according to the following formula: (number of live cells without APO-1–3 treatment – number of live cells with APO-1–3 treatment/number of live cells without APO-1–3 treatment) × 100.
FAS cell surface expression
To determine FAS (CD95) expression, EBV-transformed B cells (0.5 × 106 cells/sample) were washed with PBS and incubated with 10 µg/ml PE-CD95 (BD Biosciences) or IgG control for 30 min at 4°C (dark) in 100 µl 5% FCS in PBS. After washing with PBS two times, 10,000 live cells were analyzed by flow cytometry. Absolute number of FAS molecules on the cell surface was established by developing a mean equivalent soluble fluorochrome standard curve using QuantiBRITE PE (BD Biosciences) beads run in parallel for each experiment, according to the manufacturer’s protocol.
Immunoprecipitation
EBV-transformed B cells (3 × 106 cells/sample) were cultured with or without APO-1–3 (0.5 µg/ml) in the presence of protein A (1 µg/ml) for 20 min at 37°C. Immunoprecipitation was performed according to the manufacturer’s instructions (Pierce Classic IP Kit; Thermo Scientific). Briefly, after exposure, cells were washed with cold PBS, lysed, and spun down (13,000 rpm, 4°C, 20 min). Supernatants were incubated with anti-FAS Ab (A–20; Santa Cruz Biotechnology) and protein A/G agarose for 2 h, after which the immune complexes were washed, and proteins were separated by electrophoresis on 4–12% NuPAGE Bis–Tris gels (Invitrogen), transferred to nitrocellulose membranes, and probed using the anti–Fas-associated DD protein (FADD) (BD Biosciences) or anti–caspase-8 (Cell Signaling Technology) Abs. To quantitate changes in protein level, the ECL films were scanned using a Quantity One scanner (Bio-Rad).
FAS transfection
EBV-transformed B cells (3 × 106 cells /sample) were transfected with 5 µg GFP (pmaxGFP) or 5 µg YFP-FAS (pEYFP-N1-FAS) using the Amaxa Human B-cell Nucleofector kit (Program U-015; Amaxa). Twenty-four hours after transfection, medium was replaced with fresh complete medium. After 48 h of transfection, expression of FAS on the cell surface was determined by flow cytometry as described above, gating on live GFP-or YFP-transfected cells. To evaluate apoptosis, the transfected cell lines were treated with vehicle or APO-1–3 in the presence of protein A (as above) and live GFP+ or YFP+ cells counted as described above.
Biomarkers
The quantification of IL-10, soluble Fas ligand, vitamin B12, IL-18, TNF-α, and TCRαβ+CD4−CD8− T cells on patient samples was performed as previously described (22).
Statistical analysis
Data are represented as the mean ± SEM, except where noted. The statistical analyses were performed by unpaired Student t test or Mann– Whitney rank-sum test. Differences were considered significant when p < 0.05.
Results
FAS cell-surface expression in ALPS-associated FAS mutations
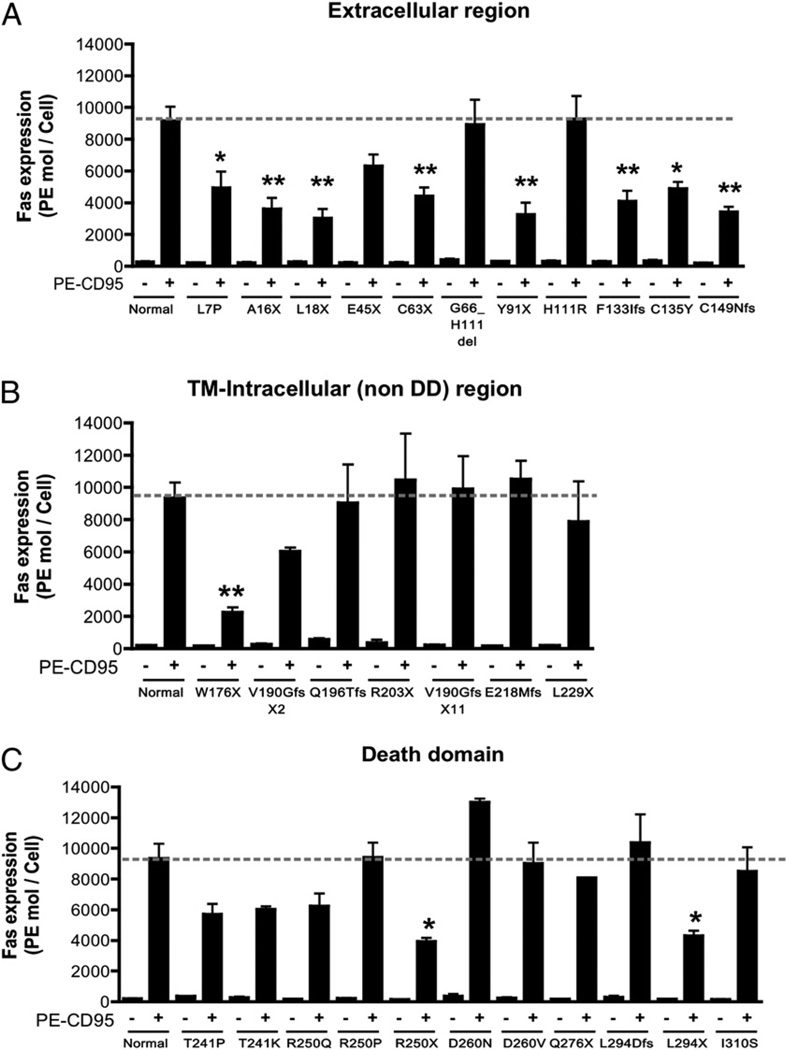
We reviewed genetic data from 108 ALPS probands and identified 84 distinct mutations (27 extracellular, 5 transmembrane, 24 intracellular non-DD, and 28 intracellular DD mutations) (Fig. 1). The majority of the non-DD mutations were either nonsense (11 of 57) or splice site or insertions/deletions with predicted frameshifts (36 of 57). More importantly, most (19 of 27) of the mutations affecting the extracellular regions of the protein were also nonsense, insertions/deletions, or splice site with frameshifts. We selected 29 mutations across all regions to determine their functional effect (Fig. 1, Table I). To assess the impact of the different FAS mutations on the expression of FAS, we determined the number of FAS molecules on the cell surface of patient-derived EBV-transformed B cell lines using a flow cytometry-based assay. The majority of the mutations affecting the extracellular region (8 out of 11) demonstrated significantly reduced FAS expression (Fig. 2A, Supplemental Fig. 1). The mutation W176X, which abolishes the entire transmembrane region, also resulted in low FAS expression (Fig. 2B). The mutation H111R and the inframe deletion G66_H111 affect the FASL-binding domain and were not expected to result in lower protein expression. As predicted, most mutations affecting intracellular regions of the FAS protein did not significantly change cell-surface FAS expression (Fig. 2B, 2C, Supplemental Fig. 1). Exceptions included the intracellular nonsense mutations R250X and L294X, which significantly reduced FAS surface expression (Fig. 2B, 2C, Supplemental Fig. 1). Taken together, these data suggested haploinsufficiency as a possible disease mechanism in ALPS caused by FAS extracellular region mutations.
Table I.
Summary of FAS mutations and their effects
| Identification No. |
cDNA | Protein | Location | Surface Fas (% Normal) |
Cell Loss (% Normal) |
FADD Recruitment (% Normal) |
Caspase-8 Recruitment (% Normal) |
RNA Stability (% Mutant Clones)a |
|---|---|---|---|---|---|---|---|---|
| 127 | c.20T > C | p.L7P | Exon 1 | 54 | 74 | 117 | 98 | Stable (44.5) |
| 220 | c.46_47delGC | p.A16X | Exon 2 | 39 | 65 | 45 | 46 | Stable (15) |
| 74 | c.53T > G | p.L18X | Exon 2 | 33 | 1 | 21 | 30 | Stable (31) |
| 153 | c.133G > T | p.E45X | Exon 2 | 69 | 75 | 99 | 82 | Unstable (0) |
| 50 | c.189T > A | p.C63X | Exon 2 | 48 | 73 | 85 | 71 | Unstable (0) |
| 111 | c.197(−1)g > a | p.G66_H111del | Intron 2 | 98 | 70 | 89 | 70 | Stable |
| 89 | c.273C > A | p.Y91X | Exon 3 | 36 | 19 | 36 | 30 | Unstable (8) |
| 180 | c.332A > G | p.H111R | Exon 3 | 101 | 47 | 83 | 66 | Stable |
| 77 | c.397_398delTTinsA | p.F133IfsX54 | Exon 4 | 45 | 42 | 63 | 42 | Stable (15) |
| 205 | c.404G > A | p.C135Y | Exon 4 | 53 | 62 | 49 | 50 | Stable (22) |
| 34 | c.444(−1)g > c | p.C149NfsX32 | Intron 4 | 37 | 21 | 64 | 42 | Unstable (0) |
| 175 | c.528G > A | p.W176X | Exon 6 | 24 | 77 | 51 | 33 | Stable (6) |
| 4 | c.569(−2)a > c | p.V190GfsX2 | Intron 6 | 64 | 31 | 80 | 70 | Stable |
| 45 | c.585_595del11 | p.Q196TfsX12 | Exon 7 | 97 | 24 | 14 | 24 | Stable |
| 72 | c.607A > T | p.R203X | Exon 7 | 112 | 26 | 15 | 24 | Stable |
| 149 | c.651(+2)t > a | p.V190GfsX11 | Intron 7 | 106 | 17 | 14 | 15 | Stable |
| 62 | c.676(+2)t > c | p.E218MfsX4 | Intron 8 | 109 | 9 | 13 | 17 | Stable |
| 98 | c.686delT+690_694del5 | p.L229X | Exon 9 | 84 | 10 | 13 | 16 | Stable |
| 3 | c.721A > C | p.T241P | Exon 9 | 63 | 32 | 28 | 27 | Stable |
| 110 | c.722C->A | p.T241K | Exon 9 | 67 | 28 | 6 | 14 | Stable |
| 29 | c.749G > A | p.R250Q | Exon 9 | 69 | 6 | 24 | 22 | Stable |
| 31 | c.749G > C | p.R250P | Exon 9 | 104 | 63 | 70 | 57 | Stable |
| 55 | c.748C > T | p.R250X | Exon 9 | 44 | 8 | 21 | 15 | Stable (37.5) |
| 197 | c.778G > A | p.D260N | Exon 9 | 145 | 29 | 11 | 11 | Stable (28.6) |
| 137 | c.779A > G | p.D260V | Exon 9 | 100 | 59 | 8 | 13 | Stable |
| 33 | c.826C > T | p.Q276X | Exon 9 | 90 | 36 | 18 | 15 | Stable (60) |
| 121 | c.879_880delAT | p.L294DfsX2 | Exon 9 | 115 | 44 | 74 | 48 | Stable (60) |
| 30 | c.880delT | p.L294X | Exon 9 | 48 | 54 | 38 | 25 | Stable (28) |
| 17 | c.929T > G | p.I310S | Exon 9 | 94 | 71 | 98 | 92 | Stable |
Stable: both wild-type and mutant forms were detected; Unstable: >90% of the clones were wild-type.
FIGURE 2.
Cell-surface expression of FAS on EBV-transformed B cells. A–C, Cells (0.5 × 106) were stained for FAS expression using 10 µg/ml PE-conjugated anti-CD95 or PE-conjugated isotype-matched IgG for 30 min at 4°C. After washing two times with PBS, 10,000 live cells were analyzed by flow cytometry. FAS expression was quantified using QuantiBRITE PE (BD Biosciences). Data were represented as means ± SE of three to four separate experiments. *p < 0.05, **p < 0.01 by Student t test for comparison with normal cell lines.
Low FAS expression is associated with apoptosis dysfunction due to impaired death-inducing signaling complex formation
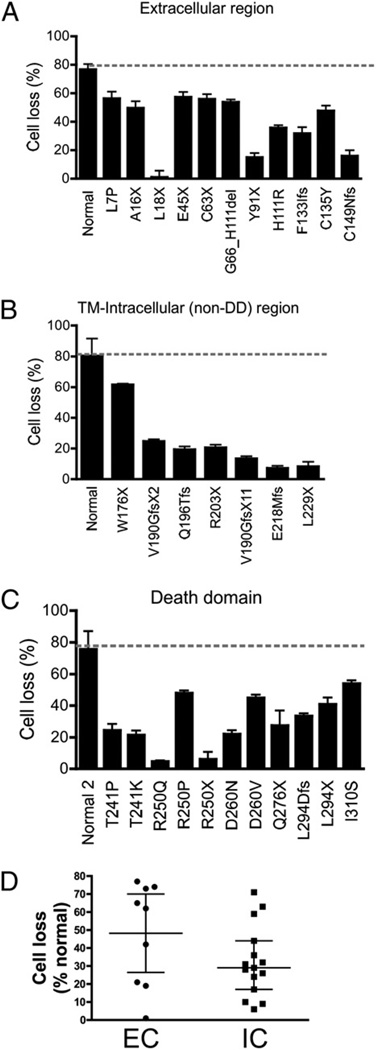
We next evaluated the functional impact of the 29 selected mutations in the FAS pathway by the treatment of EBV-transformed B cell lines with the agonistic anti-FAS Ab APO-1–3 followed by cross-linking. All cell lines with extracellular, transmembrane, or intracellular mutations showed significant resistance to FAS-mediated cell death when compared with control cell lines (Fig. 3A–C). The apoptotic defect could also be observed on primary cultured T cells (Supplemental Table I). Mutations in the extracellular domains resulted in milder apoptotic defects when compared with intracellular mutations, with median cell losses of 62 and 29%, respectively (Fig. 3D). However, this difference did not reach statistical significance (p = 0.17), likely due to the large data spread in the extracellular group and the limited sample number in each group.
FIGURE 3.
Sensitivity of EBV-transformed B cells to CD95-induced cell death. A–C, EBV-transformed B cells from ALPS patients and three different normal controls were stimulated with APO-1–3 (1 µg/ml) and protein A (1 µg/ml) for 24 h. Cell death was determined by measuring the loss of the mitochondrial transmembrane potential using DiOC6 by flow cytometry. D, Comparison of the degree of apoptotic defect between cells with extracellular (EC) mutations (including W176X) affecting FAS expression (n = 9) and intracellular (IC) mutations with normal FAS expression (n = 16), with numbers normalized to the value in normal cells. The data in A–C were represented as means ± SE of three separate experiments. The data in D were presented as medians and interquartile ranges, and medians were compared using Mann–Whitney U test. All results from ALPS patients showed significant reduction in FAS-mediated cell death compared with normal cell lines.
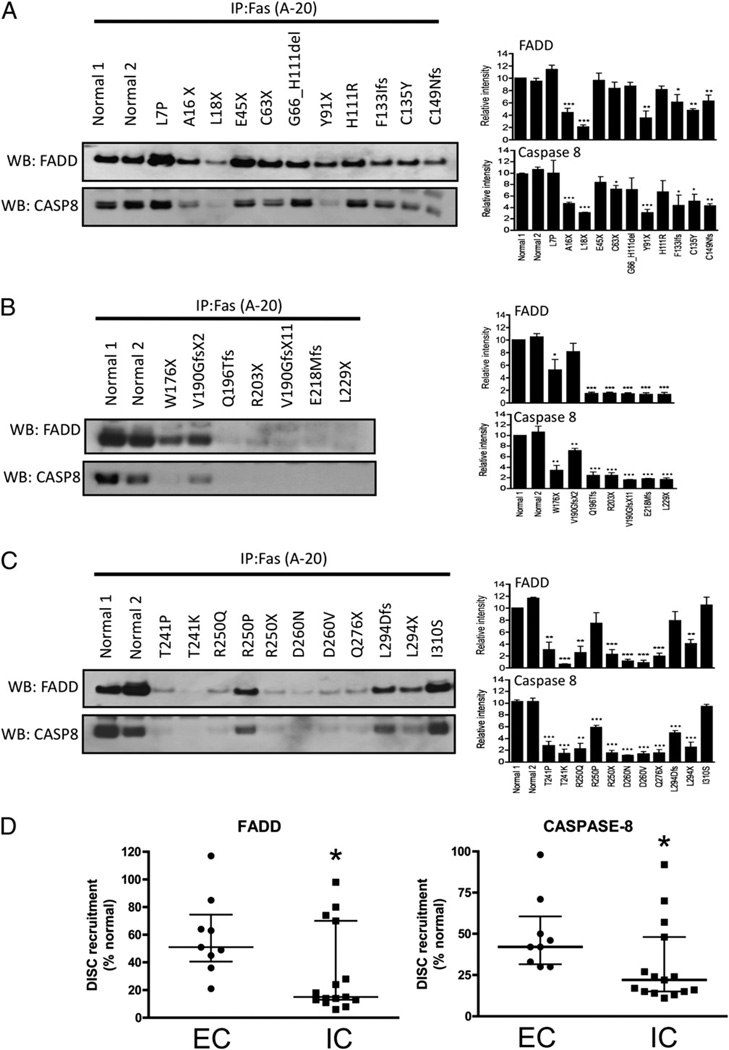
To further dissect the impact of FAS mutations on the downstream signaling pathways, we evaluated death-inducing signaling complex (DISC) formation in response to FAS activation. This complex includes FADD together with caspase-8/10 and is formed within seconds following FAS stimulation, resulting in the processing and activation of caspase-8/10 with propagation of the apoptotic signal. Following FAS activation with APO-1–3 and protein A, immunoprecipitated protein complexes were examined by Western blotting to measure the levels of coimmunoprecipitated FADD and caspase-8 bound to the intracellular portion of the FAS receptor. Compared to mutations affecting the intracellular region (Fig. 4B, 4C), the extracellular mutations (Fig. 4A) showed consistent but more limited impairment in FADD (p = 0.032) and caspase-8 (p = 0.02) recruitment to the signaling complex, based on multiple experiments (Fig. 4D). These data demonstrate that extracellular mutations with low FAS expression attenuate downstream signaling and impair apoptosis, although to a lesser extent when compared with the intracellular mutations, and confirm interference of apoptosis by haploinsufficiency at the molecular level.
FIGURE 4.
DISC formation in EBV-transformed B cells of ALPS patients. A–C, Cells were stimulated with APO-1–3 (0.5 µg/ml) or isotype control IgG in the presence of protein A (1 µg/ml) for 20 min. After lysis, cell lysates were incubated with anti-FAS Ab and protein A/G agarose for 2 h. The immunocomplexes were then subjected to Western blotting using anti-FADD and anti–caspase-8 (CASP8). Caspase-8 band represents cleaved caspase-8 (~43 kDa). Because FAS size overlaps with IgG H chain, we could not show immunoprecipitated FAS. D, Comparison of the degree of recruitment of FADD or caspase-8 to the FAS receptor upon stimulation between cells with extracellular mutations affecting FAS expression (EC; n = 9) and intracellular mutations with normal FAS expression (IC; n = 16). All numbers were normalized to the value in normal cells. Densitometry data (A–C) are presented as means ± SE of (n = 3) separate experiments and compared by Student t test. The data in D were presented as medians and interquartile ranges, and medians were compared using Mann–Whitney U test. *p < 0.05, **p < 0.01, ***p , 0.001.
Different mechanisms mediate low FAS expression in ALPS patients
To gain insight into the mechanism(s) responsible for low cell-surface FAS protein expression associated with extracellular FAS mutations, we first evaluated mRNA stability of mutant and wild-type alleles by performing FAS cDNA cloning followed by sequencing. Although rare mRNAs harboring premature stop codons escape the degradation pathway, most transcripts containing a premature stop codon followed by a spliceable intron undergo nonsense-mediated mRNA decay (NMD) (23). Accordingly, we detected almost exclusively wild-type FAS cDNA in cells with the E45X, C63X, Y91X, and C149Nfs mutations (Table I), suggesting these mutated transcripts underwent NMD. However, the nonsense mutations A16X, L18X, W176X, R250X, L294X, and F133Ifs, and, as expected, the missense L7P and C135Y, allowed both wild-type and mutant allele expression at varying ratios, suggesting that alternative mechanisms other than NMD also lead to low FAS expression seen with specific extracellular FAS mutations.
Ectopic expression of L18X and F133Ifs in 293 T cells resulted in small amounts of truncated proteins that did reach the cell surface (Supplemental Fig. 2A, 2B). L16X is believed to behave in a similar manner. W176X also resulted in a short protein missing the transmembrane region that did not anchor on the cell surface (Supplemental Fig. 2A, 2B). Interestingly, ectopic expression of R250X and L294X resulted in subnormal surface expression of the mutant FAS (Supplemental Fig. 2A), similar to that observed in the EBV B cell lines (Fig. 2). These findings suggest that these two latter mutations may result in a combination of haploinsufficiency and dominant-negative inhibition.
L7P is located in the signal peptide, and it is plausible that impaired intracellular trafficking to the endoplasmic reticulum and Golgi apparatus prevents normal surface FAS expression, as it has been demonstrated for other proteins (24, 25). However, we could not detect any clear accumulation of FAS in the endoplasmic reticulum or Golgi by confocal microscopy (data not shown). These experiments were complicated by the fact that the mutation is heterozygous, such that 50% of the proteins are expected to traffic normally. Lastly, C135Y affects a cysteine residue within the third cysteine-rich domain, possibly resulting in an abnormally folded protein. Ectopic expression of this mutant did result in surface FAS expression, but at lower levels as compared with controls (Supplemental Fig. 2A).
Taken together, these data suggest that haploinsufficiency associated with extracellular mutations can diminish FAS expression by inducing NMD, protein instability, and, potentially, abnormal intracellular trafficking.
Clinical and laboratory findings in patients with haploinsufficient alleles
We next analyzed the impact of the different FAS mutations on clinical and laboratory parameters in ALPS patients. Extracellular mutations are known to result in lower penetrance and variable expressivity, and these were not readdressed in this study (18, 26). We evaluated recently described biomarkers associated with FAS mutations, including percentage of TCRαβ+CD4−CD8− T cells as well as levels of soluble FASL, IL-10, IL-18, TNF-α, and vitamin B12 (27, 28). No statistically significant differences were noticed between patients with null extracellular or missense intracellular mutations (Supplemental Fig. 3). However, one additional clinical finding that appeared to distinguish the two groups was the absence of lymphomas in patients with extracellular FAS mutations. Among >189 patients from 108 families with FAS mutations in the National Institutes of Health cohort, all 21 lymphomas documented to date occurred in patients with intracellular mutations (29). Additionally, lymphoma occurred in one patient with a somatic mutation in NRAS (14). This finding suggests that the residual apoptotic function observed in haploinsufficiency-associated extracellular mutations may translate clinically not only into lower penetrance and expressivity but also into a lower risk for lymphomagenesis.
Overexpression of FAS in haploinsufficient samples corrects the apoptotic defect
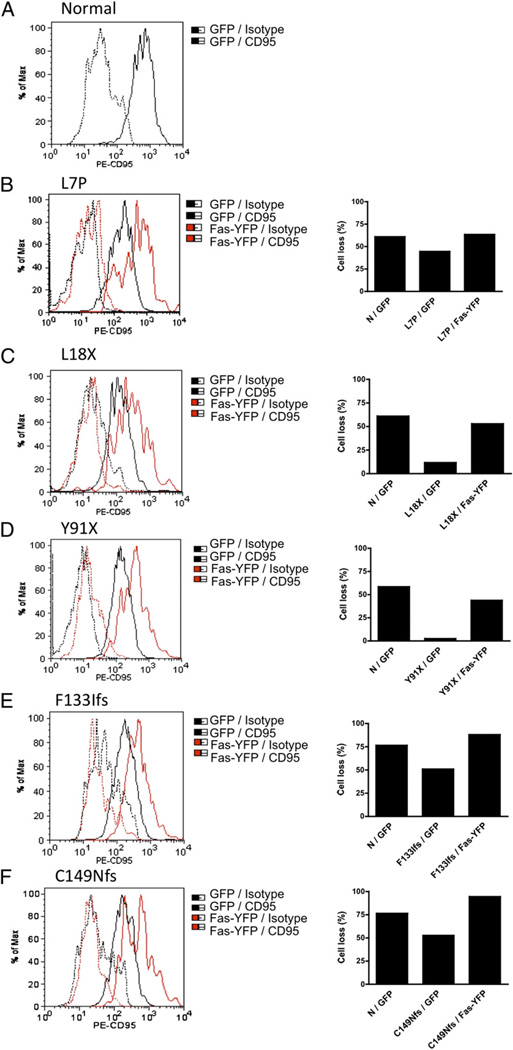
To conclusively demonstrate haploinsufficiency as a disease mechanism, we transfected YFP-tagged wild-type FAS or an empty GFP-expressing plasmid into selected EBV-transformed cell lines that showed reduced FAS expression and reduced sensitivity to FAS-induced cell death. Transfection efficiency ranged from 10– 25% (data not shown). Forty-eight hours after transfection, we measured cell-surface FAS expression by flow cytometry on live-gated GFP+- or YFP+-transfected cells (Fig. 5). Cell-surface FAS expression was increased after transfection with YFP-FAS, and this corrected the apoptotic defect, as compared with control GFP-transfected cells and normal controls. These results suggest that the level of surface FAS expression is a crucial determinant of the sensitivity to FAS-induced cell death.
FIGURE 5.
Overexpression of wild-type FAS into FAS-haploinsufficient samples. A–F, Normal and patient EBV-transformed B cells (3 × 106 cells/sample) were transfected with 5 µg GFP (pEGFP-C1) or 5 µg YFP-FAS (pEYFP-N1-wt FAS) using Amaxa Human B-cell Nucleofector. After 48 h of transfection, over-expressed FAS on the cell surface was determined by flow cytometry using PE-CD95, and cells were stimulated with APO-1–3 (1 µg/ml) and protein A (1 µg/ ml) for 24 h. Cell death was determined by flow cytometry. Analysis was performed by gating on live GFP- or YFP-FAS–transfected cells. Data were presented as means of duplicate experiments. N, normal control.
Discussion
Missense mutations affecting the DD of the FAS gene are the most common genetic abnormality detected in patients with ALPS and disrupt the apoptosis pathway by dominant-negative interference (7, 18, 30). This occurs because FAS is present on the cell surface as a preassociated homotrimeric receptor, such that in the presence of a heterozygous mutation, seven out of eight trimers will contain at least one mutant copy of the FAS protein, and this prevents adequate signaling (31). However, analysis of our extensive cohort of patients revealed that a significant proportion of ALPS patients harbor null mutations in the extracellular portion of the FAS protein. These mutations were not expected to have a dominant-negative effect, as no mutant protein expression was anticipated, and alternative mechanisms were explored. We analyzed the impact of disease-associated, naturally occurring FAS mutations on the apoptotic signaling pathway and associated clinical and laboratory findings in ALPS patients. This was, to our knowledge, the first comprehensive effort to understand the functional consequences of a large group of non-DD ALPS-associated FAS mutations.
Our findings demonstrate that haploinsufficiency is associated with nonsense or frameshift FAS mutations, primarily located in gene regions encoding for the extracellular portion of the FAS protein. From 11 extracellular mutations associated with a defect in apoptosis, only 3 did not result in decreased FAS expression. Among these is E45X, a truncation that did not reduce surface FAS levels to statistical significance and correspondingly did not affect FAS signaling in any measurable way. It is possible that even a mild decrease in FAS expression may result in dysfunction in vivo, although the underlying mechanism for disease in this patient remains to be fully understood. The recurrent mutation H111R and the in-frame G66_H111 deletion also did not reduce FAS expression but are predicted to affect the FASL binding site, providing a possible explanation for the clinical and cellular phenotype. Curiously, despite having documented apoptosis defects for all mutations tested, no impairment in DISC formation could be detected for the extracellular mutations L7P, E45X, H111R, G66_H111del, and the intracellular I310S. This may reflect a low sensitivity of the immunoprecipitation assay used in the study to very mild but still clinically significant functional defects.
Previous case reports lend support to the concept that haploinsufficiency is a pathogenetic mechanism in ALPS. Vaishnaw et al. (20) suggested that haploinsufficiency was at play in two patients with a nonsense mutation in the extracellular domain, and Roesler et al. (19) demonstrated haploinsufficiency in another ALPS patient with a transmembrane mutation that prevented FAS expression. Studies in lpr mice, which harbor spontaneous mutations in the Fas gene, also indirectly support our findings. Although commonly considered an exclusively recessive phenotype, a heterozygous lpr mutation does induce the development of autoimmunity in certain backgrounds, with autoantibody production, glomerulonephritis, sialoadenitis, and lymphoid accumulation (32, 33). At odds with human disease, lpr heterozygous mice do not appear to develop TCRαβ+CD4−CD8− T cell accumulation, but the mutation was crossed to only a limited number of genetic backgrounds as compared with the outbred human population (32, 33).
Haploinsufficiency has been commonly reported for genes involved in nonlinear signaling processes, such as DNA transcription and assembly of macromolecular complexes (34). Accordingly, productive FAS signaling requires the assembly of large signaling platforms on the cell surface (35). This is thought to be required to concentrate procaspase-8 in close proximity allowing for self-cleavage, activation, and propagation of the apoptotic signal (36, 37). Thus, lower amounts of surface FAS may prevent the formation of a critical threshold of these complexes resulting in a disrupted apoptotic signal. Indeed, we showed that DISC formation is adversely affected in most of these patients, although to a lesser extent than in patients with intracellular mutations.
The milder nature of the apoptotic defect seen in haploinsufficiency-associated FAS mutations can be molecularly explained by the fact that the intact allele will allow expression of normal FAS proteins on the cell surface, which will presumably preassociate through their preligand assembly domain and form functional trimers, albeit at levels ~50% of that seen in healthy controls. In contrast, missense intracellular mutations with dominant-negative effect will result in the incorporation of mutant proteins in seven out of eight FAS trimers on the cells surface, rendering them nonfunctional.
The residual FAS function seen in ALPS patients with haploinsufficiency-associated extracellular mutations can potentially explain the incomplete clinical penetrance and variable expressivity, typical also of other diseases associated with haploinsufficient alleles (18, 20, 30, 34). More strikingly, the absence of lymphoma cases to date in patients with FAS extracellular (EC) mutations suggests that this level of residual FAS function may be sufficient for tumor suppression, placing these patients at a lower cancer risk (3, 29). However, further long-term follow-up of ALPS patients at our and other centers will be necessary to fully substantiate this initial observation.
It is plausible that modifying factors, including genetic, epigenetic, or environmental, may make a larger contribution to disease phenotype in patients with EC mutations as compared with more severe dominant-negative mutations. Along these lines, Magerus-Chatinet et al. (38) demonstrated in recent work that a group of ALPS patients with low-penetrance EC mutations present with a somatic event in the second FAS allele in double-negative T cells, and this was associated with the presence of clinical symptoms.
Lastly, one can also speculate that the definition of haploinsufficiency as a common disease mechanism in ALPS makes gene therapy a future possibility for this group of patients based on the finding that increasing FAS cell-surface expression can re-establish normal apoptosis. In contrast, the current approach of viral vector gene insertion would likely not be effective in correcting the apoptotic defect associated with dominant-negative FAS mutations.
Supplementary Material
Acknowledgments
We thank the patients and their families for contributions to the study. We also thank Richard Siegel for the FAS-YFP plasmid construct.
This work was supported by the National Institutes of Health Intramural Program.
Abbreviations used in this article
- ALPS
autoimmune lymphoproliferative syndrome
- DD
death domain
- DiOC6
3,3′-dihexyloxacarbocyanine iodide
- DISC
death-inducing signaling complex
- EC
extracellular
- FADD
Fas-associated death domain protein
- FASL
FAS ligand
- NMD
nonsense-mediated mRNA decay.
Footnotes
H.S.K. performed research, analyzed data, made the figures, and wrote the paper; I.C. performed analysis of the biomarkers; J.E.N. performed genomic DNA sequencing analysis; V.K.R. and J.D. were responsible for patient care; T.A.F. supervised research; and J.B.O. designed and supervised the project and wrote the paper.
The online version of this article contains supplemental material.
Disclosures
The authors have no financial conflicts of interest.
References
- 1.Sneller MC, Wang J, Dale JK, Strober W, Middelton LA, Choi Y, Fleisher TA, Lim MS, Jaffe ES, Puck JM, et al. Clincal, immunologic, and genetic features of an autoimmune lymphoproliferative syndrome associated with abnormal lymphocyte apoptosis. Blood. 1997;89:1341–1348. [PubMed] [Google Scholar]
- 2.Le Deist F, Emile JF, Rieux-Laucat F, Benkerrou M, Roberts I, Brousse N, Fischer A. Clinical, immunological, and pathological consequences of Fas-deficient conditions. Lancet. 1996;348:719–723. doi: 10.1016/S0140-6736(96)02293-3. [DOI] [PubMed] [Google Scholar]
- 3.Straus SE, Jaffe ES, Puck JM, Dale JK, Elkon KB, Rösen-Wolff A, Peters AM, Sneller MC, Hallahan CW, Wang J, et al. The development of lymphomas in families with autoimmune lymphoproliferative syndrome with germline Fas mutations and defective lymphocyte apoptosis. Blood. 2001;98:194–200. doi: 10.1182/blood.v98.1.194. [DOI] [PubMed] [Google Scholar]
- 4.Sneller MC, Straus SE, Jaffe ES, Jaffe JS, Fleisher TA, Stetler-Stevenson M, Strober W. A novel lymphoproliferative/autoimmune syndrome resembling murine lpr/gld disease. J. Clin. Invest. 1992;90:334–341. doi: 10.1172/JCI115867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bleesing JJ, Brown MR, Straus SE, Dale JK, Siegel RM, Johnson M, Lenardo MJ, Puck JM, Fleisher TA. Immunophenotypic profiles in families with autoimmune lymphoproliferative syndrome. Blood. 2001;98:2466–2473. doi: 10.1182/blood.v98.8.2466. [DOI] [PubMed] [Google Scholar]
- 6.Rieux-Laucat F, Le Deist F, Hivroz C, Roberts IA, Debatin KM, Fischer A, de Villartay JP. Mutations in Fas associated with human lymphoproliferative syndrome and autoimmunity. Science. 1995;268:1347–1349. doi: 10.1126/science.7539157. [DOI] [PubMed] [Google Scholar]
- 7.Fisher GH, Rosenberg FJ, Straus SE, Dale JK, Middleton LA, Lin AY, Strober W, Lenardo MJ, Puck JM. Dominant interfering Fas gene mutations impair apoptosis in a human autoimmune lymphoproliferative syndrome. Cell. 1995;81:935–946. doi: 10.1016/0092-8674(95)90013-6. [DOI] [PubMed] [Google Scholar]
- 8.Wang J, Zheng L, Lobito A, Chan FK, Dale J, Sneller M, Yao X, Puck JM, Straus SE, Lenardo MJ. Inherited human Caspase 10 mutations underlie defective lymphocyte and dendritic cell apoptosis in autoimmune lymphoproliferative syndrome type II. Cell. 1999;98:47–58. doi: 10.1016/S0092-8674(00)80605-4. [DOI] [PubMed] [Google Scholar]
- 9.Wu J, Wilson J, He J, Xiang L, Schur PH, Mountz JD. Fas ligand mutation in a patient with systemic lupus erythematosus and lymphoproliferative disease. J. Clin. Invest. 1996;98:1107–1113. doi: 10.1172/JCI118892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Del-Rey M, Ruiz-Contreras J, Bosque A, Calleja S, Gomez-Rial J, Roldan E, Morales P, Serrano A, Anel A, Paz-Artal E, Allende LM. A homozygous Fas ligand gene mutation in a patient causes a new type of autoimmune lymphoproliferative syndrome. Blood. 2006;108:1306–1312. doi: 10.1182/blood-2006-04-015776. [DOI] [PubMed] [Google Scholar]
- 11.Bi LL, Pan G, Atkinson TP, Zheng L, Dale JK, Makris C, Reddy V, McDonald JM, Siegel RM, Puck JM, et al. Dominant inhibition of Fas ligand-mediated apoptosis due to a heterozygous mutation associated with autoimmune lymphoproliferative syndrome (ALPS) Type Ib. BMC Med. Genet. 2007;8:41. doi: 10.1186/1471-2350-8-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Oliveira JB, Bleesing JJ, Dianzani U, Fleisher TA, Jaffe ES, Lenardo MJ, Rieux-Laucat F, Siegel RM, Su HC, Teachey DT, Rao VK. Revised diagnostic criteria and classification for the autoimmune lymphoproliferative syndrome (ALPS): report from the 2009 NIH International Workshop. Blood. 2010;116:e35–e40. doi: 10.1182/blood-2010-04-280347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chun HJ, Zheng L, Ahmad M, Wang J, Speirs CK, Siegel RM, Dale JK, Puck J, Davis J, Hall CG, et al. Pleiotropic defects in lymphocyte activation caused by caspase-8 mutations lead to human immunodeficiency. Nature. 2002;419:395–399. doi: 10.1038/nature01063. [DOI] [PubMed] [Google Scholar]
- 14.Oliveira JB, Bidère N, Niemela JE, Zheng L, Sakai K, Nix CP, Danner RL, Barb J, Munson PJ, Puck JM, et al. NRAS mutation causes a human autoimmune lymphoproliferative syndrome. Proc. Natl. Acad. Sci. USA. 2007;104:8953–8958. doi: 10.1073/pnas.0702975104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Niemela JE, Lu L, Fleisher TA, Davis J, Caminha I, Natter M, Beer LA, Dowdell KC, Pittaluga S, Raffeld M, Rao VK, Oliveira JB. Somatic KRAS mutations associated with a human non-malignant syndrome of autoimmunity and abnormal leukocyte homeostasis. Blood. 2011;117:2883–2886. doi: 10.1182/blood-2010-07-295501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Behrmann I, Walczak H, Krammer PH. Structure of the human APO-1 gene. Eur. J. Immunol. 1994;24:3057–3062. doi: 10.1002/eji.1830241221. [DOI] [PubMed] [Google Scholar]
- 17.Chinnaiyan AM, O’Rourke K, Tewari M, Dixit VM. FADD, a novel death domain-containing protein, interacts with the death domain of Fas and initiates apoptosis. Cell. 1995;81:505–512. doi: 10.1016/0092-8674(95)90071-3. [DOI] [PubMed] [Google Scholar]
- 18.Jackson CE, Fischer RE, Hsu AP, Anderson SM, Choi Y, Wang J, Dale JK, Fleisher TA, Middelton LA, Sneller MC, et al. Autoimmune lymphoproliferative syndrome with defective Fas: genotype influences penetrance. Am. J. Hum. Genet. 1999;64:1002–1014. doi: 10.1086/302333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Roesler J, Izquierdo JM, Ryser M, Rösen-Wolff A, Gahr M, Valcarcel J, Lenardo MJ, Zheng L. Haploinsufficiency, rather than the effect of an excessive production of soluble CD95 (CD95DeltaTM), is the basis for ALPS Ia in a family with duplicated 3′ splice site AG in CD95 intron 5 on one allele. Blood. 2005;106:1652–1659. doi: 10.1182/blood-2004-08-3104. [DOI] [PubMed] [Google Scholar]
- 20.Vaishnaw AK, Orlinick JR, Chu JL, Krammer PH, Chao MV, Elkon KB. The molecular basis for apoptotic defects in patients with CD95 (Fas/Apo-1) mutations. J. Clin. Invest. 1999;103:355–363. doi: 10.1172/JCI5121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Niemela JE, Hsu AP, Fleisher TA, Puck JM. Single nucleotide polymorphisms in the apoptosis receptor gene TNFRSF6. Mol. Cell. Probes. 2006;20:21–26. doi: 10.1016/j.mcp.2005.05.004. [DOI] [PubMed] [Google Scholar]
- 22.Caminha I, Fleisher TA, Hornung RL, Dale JK, Niemela JE, Price S, Davis J, Perkins K, Dowdell KC, Brown MR, Rao VK, Oliveira JB. Using biomarkers to predict the presence of FAS mutations in patients with features of the autoimmune lymphoproliferative syndrome. J. Allergy Clin. Immunol. 125:946–949. doi: 10.1016/j.jaci.2009.12.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chang YF, Imam JS, Wilkinson MF. The nonsense-mediated decay RNA surveillance pathway. Annu. Rev. Biochem. 2007;76:51–74. doi: 10.1146/annurev.biochem.76.050106.093909. [DOI] [PubMed] [Google Scholar]
- 24.Siggaard C, Rittig S, Corydon TJ, Andreasen PH, Jensen TG, Andresen BS, Robertson GL, Gregersen N, Bolund L, Pedersen EB. Clinical and molecular evidence of abnormal processing and trafficking of the vasopressin preprohormone in a large kindred with familial neurohypophy-seal diabetes insipidus due to a signal peptide mutation. J. Clin. Endocrinol. Metab. 1999;84:2933–2941. doi: 10.1210/jcem.84.8.5869. [DOI] [PubMed] [Google Scholar]
- 25.Birney E, Stamatoyannopoulos JA, Dutta A, Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. ENCODE Project Consortium; NISC Comparative Sequencing Program; Baylor College of Medicine Human Genome Sequencing Center; Washington University Genome Sequencing Center; Broad Institute; Children’s Hospital Oakland Research Institute. 2007. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rieux-Laucat F, Blachère S, Danielan S, De Villartay JP, Oleastro M, Solary E, Bader-Meunier B, Arkwright P, Pondaré C, Bernaudin F, et al. Lymphoproliferative syndrome with autoimmunity: A possible genetic basis for dominant expression of the clinical manifestations. Blood. 1999;94:2575–2582. [PubMed] [Google Scholar]
- 27.Caminha I, Fleisher TA, Hornung RL, Dale JK, Niemela JE, Price S, Davis J, Perkins K, Dowdell KC, Brown MR, Rao VK, Oliveira JB. Using biomarkers to predict the presence of FAS mutations in patients with features of the autoimmune lymphoproliferative syndrome. J. Allergy Clin. Immunol. 2010;125:946–949. doi: 10.1016/j.jaci.2009.12.983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Magerus-Chatinet A, Stolzenberg MC, Loffredo MS, Neven B, Schaffner C, Ducrot N, Arkwright PD, Bader-Meunier B, Barbot J, Blanche S, et al. FAS-L, IL-10, and double-negative CD4-CD8-TCR alpha/beta+ T cells are reliable markers of ALPS associated with FAS loss of function. Blood. 2009;113:3027–3030. doi: 10.1182/blood-2008-09-179630. [DOI] [PubMed] [Google Scholar]
- 29.Rao VK, Price S, Davis J, Perkins K, Gill F, Pittaluga S, Fleisher T, Jaffe E. Development of lymphomas in families with autoimmune lymphoproliferative syndrome (ALPS) Pediatr. Blood Cancer. 2010;54:813. (Abstr. 171) [Google Scholar]
- 30.de Villartay JP, Rieux-Laucat F, Fischer A, Le Deist F. Clinical effects of mutations to CD95 (Fas): relevance to autoimmunity? Springer Semin. Immunopathol. 1998;19:301–310. doi: 10.1007/BF00787227. [DOI] [PubMed] [Google Scholar]
- 31.Siegel RM, Frederiksen JK, Zacharias DA, Chan FK, Johnson M, Lynch D, Tsien RY, Lenardo MJ. Fas preassociation required for apoptosis signaling and dominant inhibition by pathogenic mutations. Science. 2000;288:2354–2357. doi: 10.1126/science.288.5475.2354. [DOI] [PubMed] [Google Scholar]
- 32.Carlsten H, Tarkowski A, Jonsson R, Nilsson LA. Expression of heterozygous lpr gene in MRL mice. II. Acceleration of glomerulonephritis, sialadenitis, and autoantibody production. Scand. J. Immunol. 1990;32:21–28. doi: 10.1111/j.1365-3083.1990.tb02887.x. [DOI] [PubMed] [Google Scholar]
- 33.Ogata Y, Kimura M, Shimada K, Wakabayashi T, Onoda H, Katagiri T, Matsuzawa A. Distinctive expression of lprcg in the heterozygous state on different genetic backgrounds. Cell. Immunol. 1993;148:91–102. doi: 10.1006/cimm.1993.1093. [DOI] [PubMed] [Google Scholar]
- 34.Veitia RA, Birchler JA. Dominance and gene dosage balance in health and disease: why levels matter! J. Pathol. 2010;220:174–185. doi: 10.1002/path.2623. [DOI] [PubMed] [Google Scholar]
- 35.Siegel RM, Muppidi JR, Sarker M, Lobito A, Jen M, Martin D, Straus SE, Lenardo MJ. SPOTS: signaling protein oligomeric transduction structures are early mediators of death receptor-induced apoptosis at the plasma membrane. J. Cell Biol. 2004;167:735–744. doi: 10.1083/jcb.200406101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Muzio M, Chinnaiyan AM, Kischkel FC, O’Rourke K, Shevchenko A, Ni J, Scaffidi C, Bretz JD, Zhang M, Gentz R, et al. FLICE, a novel FADD-homologous ICE/CED-3-like protease, is recruited to the CD95 (Fas/ APO-1) death—inducing signaling complex. Cell. 1996;85:817–827. doi: 10.1016/s0092-8674(00)81266-0. [DOI] [PubMed] [Google Scholar]
- 37.Muzio M, Stockwell BR, Stennicke HR, Salvesen GS, Dixit VM. An induced proximity model for caspase-8 activation. J. Biol. Chem. 1998;273:2926–2930. doi: 10.1074/jbc.273.5.2926. [DOI] [PubMed] [Google Scholar]
- 38.Magerus-Chatinet A, Neven B, Stolzenberg MC, Daussy C, Arkwright PD, Lanzarotti N, Schaffner C, Cluet-Dennetiere S, Haerynck F, Michel G, et al. Onset of autoimmune lymphoproliferative syndrome (ALPS) in humans as a consequence of genetic defect accumulation. J. Clin. Invest. 2011;121:106–112. doi: 10.1172/JCI43752. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.