Abstract
Polo-like kinase 3 (Plk3) is an important mediator of the cellular responses to genotoxic stresses. In this study, we examined the physiologic function of Plk3 by generating Plk3-deficient mice. Plk3−/− mice displayed an increase in weight and developed tumors in various organs at advanced age. Many tumors in Plk3−/− mice were large in size, exhibiting enhanced angiogenesis. Plk3−/− mouse embryonic fibroblasts were hypersensitive to the induction of hypoxiainducible factor-1α (HIF-1α) under hypoxic conditions or by nickel and cobalt ion treatments. Ectopic expression of the Plk3-kinase domain (Plk3-KD), but not its Polo-box domain or a Plk3-KD mutant, suppressed the nuclear accumulation of HIF-1α induced by nickel or cobalt ions. Moreover, hypoxia-induced HIF-1α expression was tightly associated with a significant down-regulation of Plk3 expression in HeLa cells. Given the importance of HIF-1α in mediating the activation of the “survival machinery” in cancer cells, these studies strongly suggest that enhanced tumorigenesis in Plk3-null mice is at least partially mediated by a deregulated HIF-1 pathway.
Introduction
Polo-like kinases (Plk) are a family of proteins highly conserved in terms of their structures and functions (1–4). In mammals, the Plk family consists of four members, namely, Plk1, Plk2, Plk3 (also termed Prk and Fnk), and Plk4 (5–9). Besides the kinase domain (KD) at the NH2 terminus, Plks share a highly conserved amino acid sequence at the COOH terminus termed the Polo-box domain (PBD; ref. 10). Mammalian Plk3 was originally identified as an immediate-early response gene product (5, 8). Plk3 is involved in regulating a variety of molecular and cellular events, including DNA damage responses and cell cycle control (4). Plk3 is rapidly activated by reactive oxygen species and several other genotoxic signals (11, 12). Plk3 seems to target p53 when cells are under oxidative stresses (11, 12). Overexpression of wild-type Plk3 or its PBD induces rapid apoptosis (13). Interestingly, despite the structural similarities between Plk1 and Plk3, the latter behaves differently from that of Plk1 in terms of its regulation during the cell cycle and response to stimulation by growth factors and stresses (4, 14, 15). Whereas Plk1 levels gradually accumulate peaking at mitosis, Plk3 levels remain rather constant during the cell cycle.
To understand the role of Plk3 in controlling cell proliferation and oncogenesis, we generated and characterized Plk3 knockout mice. We found that ageing Plk3−/− mice displayed an increase in weight and developed tumors in multiple organs. Plk3−/− mouse embryonic fibroblasts (MEF) were hypersensitive to the induction of hypoxia-inducible factor-1α (HIF-1α) under hypoxic conditions, which was correlated with an elevated level of the angiogenic factor vascular endothelial growth factor-A (VEGF-A). Ectopic expression of the Plk3-KD suppressed the nuclear accumulation of HIF-1α induced by nickel or cobalt ions. Moreover, hypoxia-induced HIF-1α expression was tightly associated with a significant reduction in Plk3 levels in HeLa cells. Combined, our studies strongly suggest that suppression of tumor development by Plk3 is mediated partly through regulating the HIF-1 signaling pathway.
Materials and Methods
Cell culture
The HeLa, NIH3T3, and HEK293 cell lines were obtained from the American Type Culture Collection. Cells were cultured under 5% CO2 in dishes or on Lab-Tek II chamber slides (Fisher Scientific) in DMEM supplemented with 10% fetal bovine serum (FBS) and antibiotics (100 µg of penicillin and 50 µg of streptomycin sulfate per mL). Primary wild-type and Plk3−/− MEFs were derived from embryonic day 14.5 embryos produced from Plk3+/− intercrosses. MEF cells were cultured under 5% CO2 in DMEM supplemented with 15% FBS and antibiotics (as above).
Targeted disruption of the murine Plk3 gene
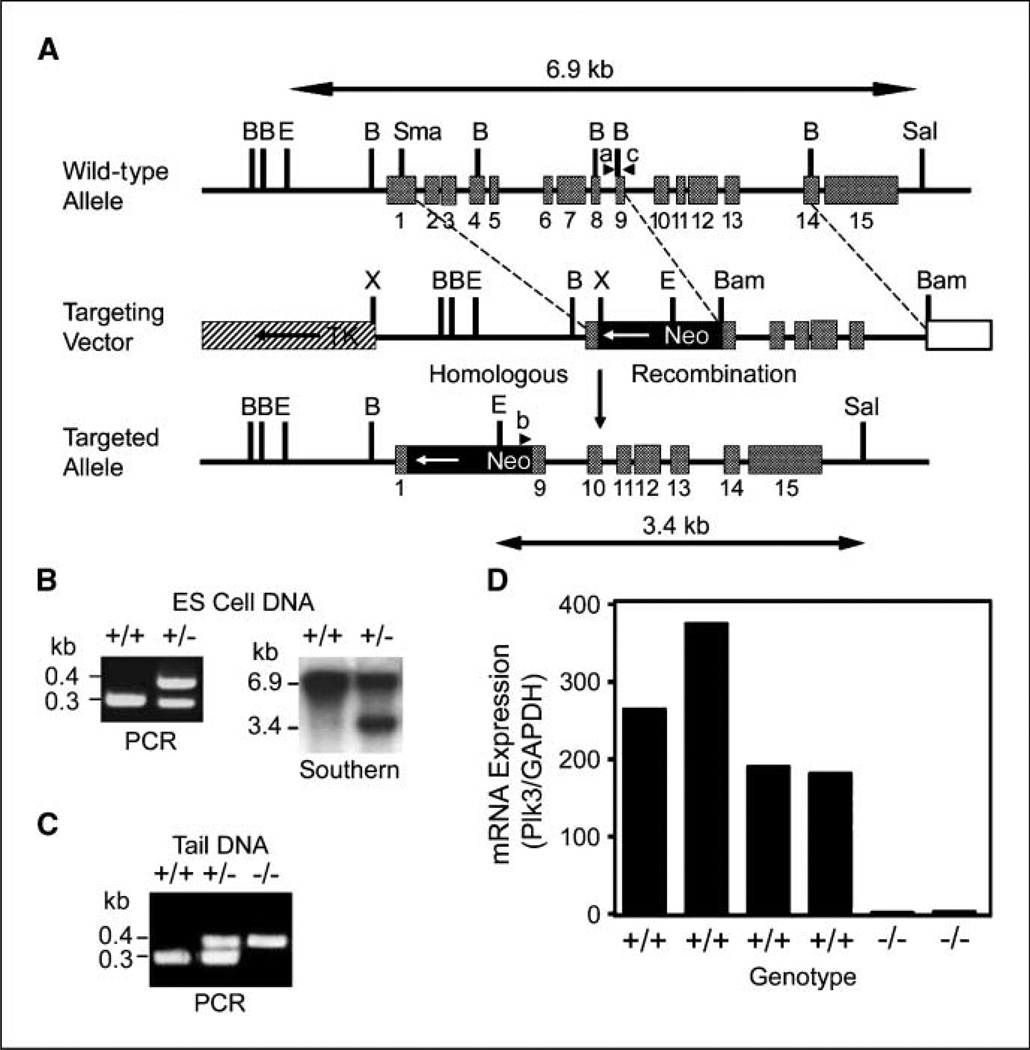
First, an ~ 6.9-kbp EcoRI-SalI genomic DNA fragment containing the entire Plk3 gene was isolated from a mouse 129/SvJ bacterial artificial chromosome library (Genome Systems, Inc.) and sequenced. A SmaI/BglII fragment was isolated and cloned into the Bam HI and Xho I sites that flank the neomycin resistance gene in the MJK+ knockout targeting vector (gift from Dr. Steven Potter, Children’s Hospital Medical Center, Cincinnati, OH). The targeting construct was sent to the University of Cincinnati Gene-Targeted Mouse Service where it was transfected into the J1 129 embryonic stem (ES) cell line. ES cell clones were selected with G418 and then screened for homologous recombination using PCR and Southern blot analysis. For these analyses, genomic DNA was isolated using the DNeasy gDNA Extraction kit (Qiagen). The primer pairs for detecting the wild-type allele were as follows: 5′-GTCAGGTGCCAGTGGGCT-GAGGGTATAG-3′ (forward) and 5′-GACCGATGGATGTGCGCATGAGGC-CAC-3′ (reverse). The primer pairs for detecting the mutant allele were as follows: 5′-GTCACGTCCTGCACGACGCGAGCTGCG-3′ (forward) and 5′-GACCGATGGATGTGCGCATGAGGCCAC-3′ (reverse). PCR was performed for 35 cycles using the following conditions: denaturing at 94°C for 30 s, annealing at 66°C for 30 s, and extension at 72°C for 30 s. For Southern blotting, genomic DNA was isolated as described above and then digested with EcoRI and SalI. Hybridization analysis was conducted as described (16). Targeted ES clones were injected into C57BL/6 blastocysts to generate chimeras. Mice heterozygous for targeted Plk3 alleles were obtained through further breeding and identified using either Southern or PCR. Homozygous Plk3-null mice were obtained through standard breeding schemes. An independent mouse line lacking Plk3 was also obtained through a gene-trapping method as described (17) in collaboration with Lexicon Genetics, Inc.
RNA isolation and real-time quantitative reverse transcription-PCR analysis
Heart tissue was obtained from mice of differing genotypes and RNA was isolated as described (18). Quantitative real-time PCR was performed using murine Plk3-specific and glyceraldehyde-3-phosphate dehydrogenase–specific primers and fluorescence-labeled probes (Applied Biosystems Assay-on-Demand products) as previously described (18).
Plasmid constructs and transfection
The wild-type human Plk3, mutant human Plk3, and human Plk1 proteins were expressed as enhanced green fluorescent protein (EGFP) fusion proteins. Constructs of pGFP-Plk3-full-length (amino acids 1–646), pGFP-Plk3-PBD (amino acids 312–646), pGFP-Plk3-KD (amino acids 1–334), and pGFP-Plk1-KD (amino acids 1–307) were generated by inserting appropriate PCR fragments into the multiple cloning site of pEGFP-N3 (BD Biosciences Clontech) as described (19). pGFP-Plk3-KDT219D where Thr219 was replaced with aspartic acid was generated using GFP-Plk3-KD as a template via site-directed mutagenesis. The wild-type and mutant murine Plk3 proteins were expressed as hemagglutinin (HA) epitope-tagged proteins. The pcDNA1neo-Plk3-HA, wild-type construct was previously described (20). Mutagenesis was performed using this plasmid as template and the PCR overlap extension method. The mutagenic oligonucleotides used to generate the pcDNA1neo-Plk3-4LA-HA construct were as follows: 5′-GTCCCTGAGAGCACGAGCAGCGTAGCGCGCTCGCTGCCGCGCGTCTGGGGAGCAGCCCAGCTG-3′ (forward) and 5′-TCCCCAGACGCGCGGCAGCGAGCGCGCTACGCTGCTC-GTGCTCTCAGGGACCAAAGCCCTGCT-3′ (reverse). The underlined nucleotides are the mutated Plk3 sequences. The remaining constructs encoding Plk3 single leucine to alanine substitutions were generated in the same way using the appropriate shorter oligonucleotides. All mutations were confirmed by DNA sequence analysis.
HeLa cells cultured in dishes or on chamber slides were transfected with plasmid constructs for 24 h using Lipofectamine 2000 (Invitrogen). Transfected cells were then treated with or without NiCl2 (0.5 mmol/L) or CoCl2 (0.3 mmol/L) for 4 h before fixing and staining with antibodies to HIF-1α and GFP for fluorescence microscopic analysis. NIH3T3 and HEK293 cells were transfected using Lipofectamine 2000 as described above. Each transfection experiment was repeated at least thrice.
Immunoblotting
HeLa cells or MEFs were lysed as described (21). Cytoplasmic and nuclear proteins were isolated from HeLa cells as described (22). NIH3T3 and HEK293 cells were lysed as previously described (20). Equal amounts (50 µg) of protein lysates were analyzed by SDS-PAGE followed by immunoblotting with antibodies to Plk3 (1:1,000;BD Biosciences), HIF-1α (1:1,000;Bethyl Laboratories), HIF-1β (1:1,000;BD Biosciences), GFP (1:10,000;DAKO), Plk1 (1:1,000;Zymed), lamin B (1:1,000; Santa Cruz Biotechnology), β-actin (1:1,500;Sigma), and the HA epitope (1:1,000;Cell Signaling). Specific signals were detected with horseradish peroxidase–conjugated goat secondary antibodies (Cell Signaling) and enhanced chemiluminescence reagents (Pierce Biotechnology).
Fluorescence microscopy
Cells were quickly washed with PBS and fixed in 4% paraformaldehyde for 10 min at room temperature. Fixed cells were treated with 0.1% Triton X-100 in PBS for 10 min and then washed thrice with ice-cold PBS. After blocking with 2% bovine serum albumin (BSA) in PBS for 15 min on ice, cells were incubated for 1 h at room temperature with the mouse monoclonal GFP antibody (1:50;Santa Cruz Biotechnology), HA antibody (1:50;Cell Signaling), and mouse HIF-1α IgG (1:50;Santa Cruz Biotechnology) in a 2% BSA solution. Cells were then washed with PBS and incubated with Rhodamine Red-X-conjugated antirabbit IgGs and/or FITC-conjugated anti-mouse IgGs (Jackson Immuno-Research) at room temperature for 1 h in the dark. Cells were finally stained with 4′,6-diamidino-2-phenylindole (DAPI;1 µg/mL; Fluka) for 5 min. Fluorescence microscopy was performed and images were captured using a digital camera (Optronics) using Optronics MagFire and Image-Pro Plus software.
Immunohistochemistry
Lung tumor sections from Plk3+/+ and Plk3−/− mice were evaluated for angiogenesis using a blood vessel staining kit (Millipore) in which von Willebrand factor (vWF) was used as an endothelial cell marker. The tumor slides were stained with an antibody against vWF according to instructions provided by the manufacturer. Slides were viewed under microscope and the tumor blood vessel density was determined by counting the number of blood vessels in six random microscopic fields.
ELISA
Conditioned media from Plk3+/+ and Plk3−/− MEFs treated with nickel ions for various times were collected. VEGF-A levels in the conditioned medium were quantitated using a mouse VEGF-A ELISA kit (Bender MedSystems) according to instructions provided by the manufacturer. The significance of statistical differences between two groups was analyzed using Student’s t test.
Results
Given that deregulated expression of mammalian Plks is closely associated with cell proliferation and oncogenesis (3, 8), we generated Plk3 knockout mice and examined their phenotypic characteristics. We first designed a targeting construct in which a neomycin resistance cassette was inserted between mouse Plk3 exons 1 and 9. The neomycin resistance gene was flanked by Plk3 gene sequences at both the 5′ and 3′ ends (Fig. 1A). The mutant allele was expected to be devoid of exons 2 to 8 after homologous recombination (Fig. 1A). Mouse ES cells were transfected with the linearized targeting vector and stable, G418-resistant transfectants were selected. An analysis of DNA isolated from stable ES cell transfectants by PCR revealed that one Plk3 allele was mutated (Fig. 1B). Mutation of the Plk3 allele was confirmed by Southern blot analysis (Fig. 1B). Whereas a 6.9-kbp fragment was detected in wild-type ES cell DNA, an additional fragment of 3.4 kbp was detected in DNA isolated from mutant ES cells, consistent with the predicted sizes for these alleles (Fig. 1A and B). Two independent 129/SvJ-derived ES cell clones were injected into C57BL/6 blastocysts, which were subsequently implanted into pseudopregnant female mice. The resulting chimeric mice were backcrossed to wild-type C57BL/6 animals. Disruption of the Plk3 gene was confirmed by a PCR strategy using mouse tail DNA (Fig. 1C). Heart RNA was isolated from four wild-type and two Plk3-null mice and subjected to quantitative real-time PCR analysis. Plk3 mRNA expression was not detected in Plk3-null mouse tissue (Fig. 1D). Combined, these results indicate that our mice were Plk3 deficient.
Figure 1.
Disruption of mouse Plk3 locus. A, schematic presentation of mouse Plk3 gene structure showing the relative sizes and positions of 15 exons. Structures of the targeting vector and the targeted mutant allele are also shown. TK, thymidine kinase gene; Neo, neomycin resistance gene. Major restriction sites are shown. B, Bg/II; E, EcoRI; X, XbaI. ab, and c, positions of primers used for genotyping by PCR. B, genotyping of ES cell clones via PCR (left) and Southern blot hybridization analysis (right). C, genotyping of Plk3+/+Plk3+/−, and Plk−/− mice using PCR. D, quantitative analysis of Plk3 mRNA expression in heart tissue obtained from four wild-type and two Plk3−/− mice. GAPDH, glyceraldehyde-3-phosphate dehydrogenase.
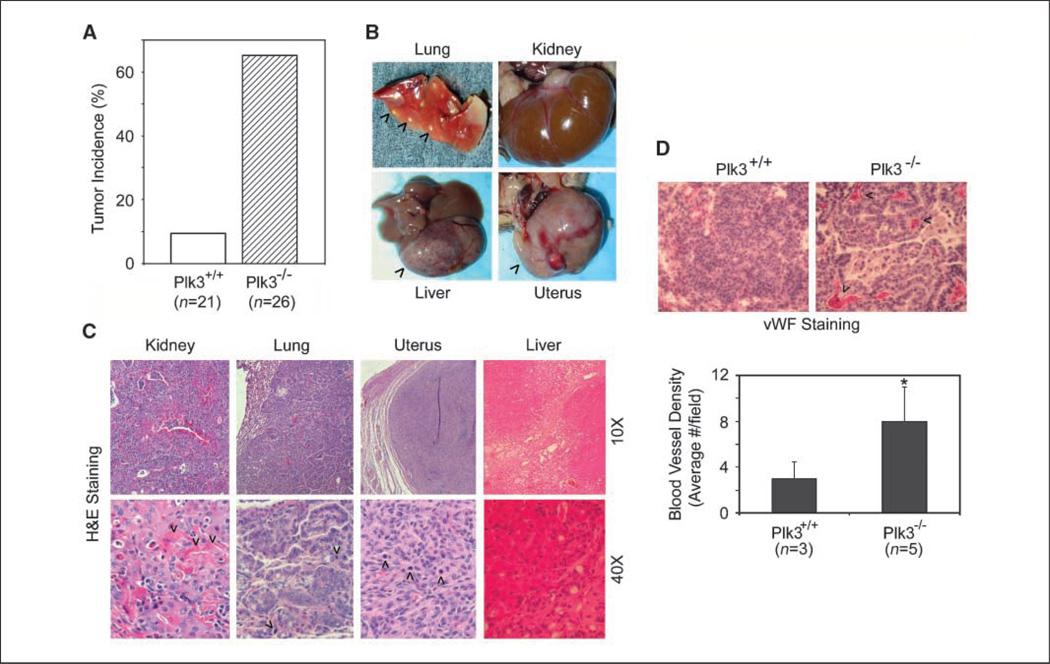
During the first year of breeding, we obtained >300 live animals from crosses of Plk3+/− mice. Genotype analysis of live births from Plk3+/− intercrosses revealed that wild-type animals, heterozygotes, and homozygotes were born at a ratio of 1:1.8:0.85. The slightly lower numbers than expected for heterozygotes and homozygotes suggest that there were some disadvantages for embryonic development of Plk3-deficient mice. Because Plk2−/− newborn mice are smaller than wild-type or Plk2+/− littermates (23), we measured the weight of newborn mice of Plk3+/− intercrosses and followed their postnatal weight up to 2 months. We did not observe a significant weight difference between Plk3+/+, Plk3+/−, and Plk3−/− mice (data not shown). However, Plk3−/− mice at 20 months of age were somewhat larger than their age- and sex-matched littermates (Supplementary Fig. S1A). Because a previous report indicated that Plk4 haploinsufficiency results in enhanced carcinogenesis (24), we studied tumor development in Plk3-deficient mice. No significantly elevated levels of tumor formation were observed in young (<12 months) Plk3+/− or Plk3−/− mice. In contrast, ageing Plk3−/− mice (>18 months) developed tumors in various organs at a much enhanced rate compared with that of wild-type littermates (Fig. 2A and B). Whereas the incidence of lung cancer was the highest, other cancers including those of kidney, liver, and uterus were also observed (Fig. 2B, arrowheads; Supplementary Table S1).
Figure 2.
Plk3 deficiency results in enhanced tumorigenesis. A, tumor incidences of age-matched wild-type and Plk3−/− mice. B, representative liver, lung, uterus, and kidney tumors that developed in aged Plk3−/− mice. Arrowheads, tumors. C, H&E staining of various tumors from Plk3−/− mice. D, sections of representative lung tumors that developed in either wild-type (Plk3+/+) or Plk3−/− mice were stained with the vWF antibody. Arrows, top, some vWF-positive areas/foci; bottom, tumor blood vessel density manifested as vWF-positive areas/foci of six random microscopic fields from representative tumor sections was quantitated. *, significant statistical difference (P < 0.05).
Histologic analysis revealed that kidney typically developed adenocarcinoma composed of tubuloglandular architecture with heavy inflammatory infiltration. Numerous eosinophilic crystalloids filled tumor cells near the luminal surface (Fig. 2C, arrowheads). The adenocarcinoma cells exhibited large nuclei with prominent nucleoli and moderate to abundant eosinophilic cytoplasm (Fig. 2C). Lung tumors were mostly adenocarcinomas and the tumor cells had crowded large hyperchromatic nuclei with prominent nucleoli and scant basophilic cytoplasm (Fig. 2C, arrowheads). Uterine tumors were leiomyosarcomas, which were well circumscribed but not encapsulated; also, the tumors were highly cellular with cytologic atypia and active mitosis (Fig. 2C, arrowheads). Liver cancer exhibited bridging necrosis with dilated sinus and congested vessels. These tumors had regenerative hepatocytes with prominent nucleoli, binuclei or multiple nuclei, and active bile duct hyperplasia with mild cholestasis (Fig. 2C).
Many tumors in Plk3−/− mice were large in size and vascularized (Fig. 2B, arrowheads), suggesting active tumor angiogenesis. To confirm this possibility, we stained paired lung tumor sections with an antibody to vWF, an endothelial cell marker. We observed that tumor blood vessel density, manifested as vWF-positive areas/foci, was much higher in Plk3−/− tumors than that of Plk3+/+tumors (Fig. 2D).
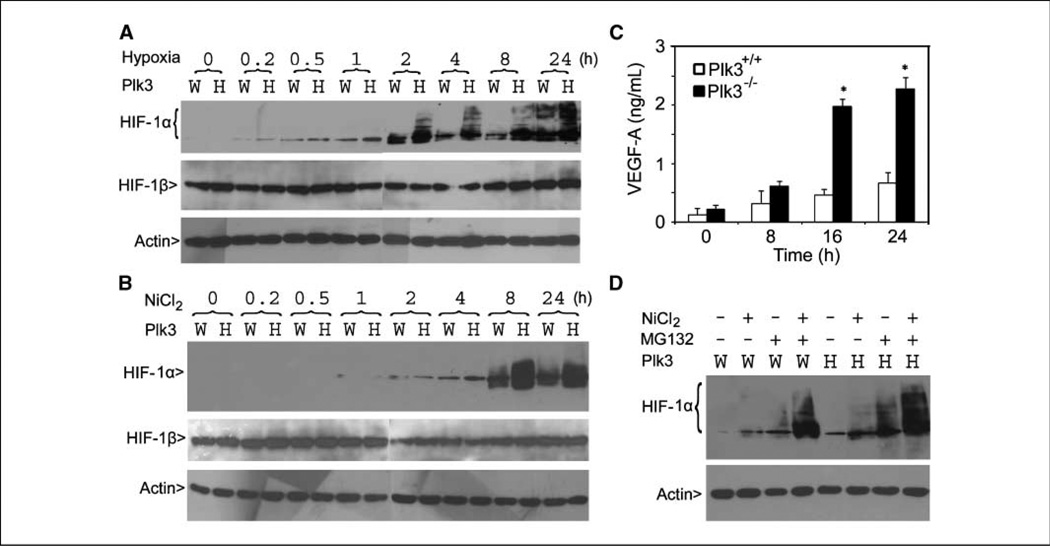
Plk3 activation is highly responsive to oxidative stress (11);we therefore examined HIF-1α expression in cultured MEFs because this transcription factor plays a critical role in supporting cancer angiogenesis. Paired MEFs were maintained under hypoxic conditions (1% O2) for various times. Western blot analysis revealed low HIF-1α expression in untreated Plk3+/+ and Plk3−/− MEFs but HIF-1α expression was rapidly induced after exposure to hypoxia (Fig. 3A). However, HIF-1α levels were induced to a greater extent in Plk3−/− MEFs compared with Plk3+/+ MEFs. HIF-1 β expression, on the other hand, was not induced in wild-type nor Plk3−/− MEFs (Fig. 3A). The increase in HIF-1α levels was correlated with the appearance of slow-mobility HIF-1α forms in Plk3−/− MEFs, which were especially prominent in cells exposed to hypoxia for 24 h (Fig. 3A). Although HIF-1α was also induced in wild-type MEFs after an extended exposure to hypoxia, the magnitude of induction in these cells was much smaller than that observed in Plk3-null MEFs (Fig. 3A).
Figure 3.
Superinduction of HIF-1 α in Plk3−/− MEFs under hypoxic conditions. A, paired wild-type (W) and Plk3−/− (H) MEFs were cultured under hypoxic conditions for various times as indicated. Equal amounts of cell lysates were blotted for HIF-1α, HIF-1β, and β-actin. B, paired wild-type and Plk3−/− MEFs were treated with NiCl2 for various times as indicated. Equal amounts of cell lysates were blotted for HIF-1 α, HIF-1 β, and β-actin. C, paired wild-type and Plk3−/− MEFs were treated with NiCl2 for indicated times. Conditioned media from MEFs with various treatments were collected and subjected to ELISA analysis for VEGF-A. The data were summarized from three independent experiments. *, significant statistical difference (P < 0.01). D, paired wild-type and Plk3−/− MEFs were treated with NiCl2 and/or MG132 for 24 h. Equal amounts of cell lysates were blotted for HIF-α and β-actin. Representative data from three independent experiments are shown.
Nickel ions are known to deplete cellular iron and ascorbic acid, resulting in an oxidative stress condition similar to hypoxia (25). Nickel also directly inhibits HIF prolyl hydroxylase (PHD; ref. 26), which otherwise “tags” HIF proteins for recognition by von Hippel-Lindau (VHL) ubiquitin E3 ligase for polyubiquitination and subsequent degradation by the proteasome (27). To determine whether Plk3 deficiency compromised the HIF-1α degradative pathway, paired Plk3−/− and wild-type MEFs were treated with NiCl2 for various times. Nickel ion treatment significantly enhanced HIF-1α, but not HIF-1β, levels in both types of MEFs; however, HIF-1α was induced to a much greater extent in Plk3−/− MEFs than that in wild-type MEFs (Fig. 3B). Because VEGF-A is a direct transcriptional target of HIF-1α, we also measured the level of VEGF-A in conditioned medium of paired MEFs treated with nickel ion. ELISA analysis revealed that VEGF-A levels were significantly higher in Plk3−/− MEF medium after 16 and 24 h of nickel treatment (Fig. 3C).
In hypoxia, HIF-1α is stabilized due to the inhibition of PHD that requires O2 (25). To determine whether protein stabilization was responsible for the increased HIF-1α levels in Plk3−/− MEFs, paired MEFs were treated with nickel ions and/or MG132, a specific proteasome inhibitor. MG132 alone stabilized HIF-1α to a much higher level in Plk3-null MEFs than in wild-type MEFs; in combination with nickel, the proteasome inhibitor superinduced HIF-1α and the magnitude of induction was much greater in Plk3−/− MEFs (Fig. 3D). These combined studies therefore strongly suggest that Plk3 deficiency renders cells hypersensitive to the induction of HIF-1α by hypoxia.
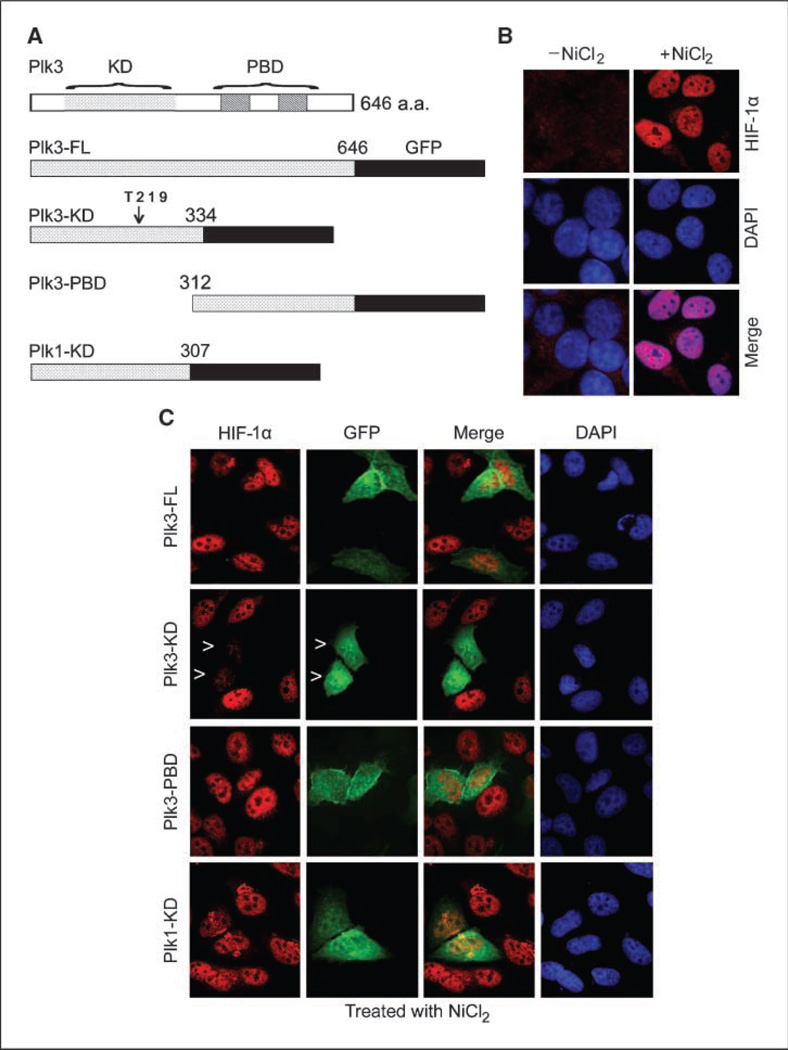
To further confirm that Plk3 is directly involved in regulating HIF-1α production, we made use of a series of human Plk3 and Plk1 expression constructs that have been described (19). Plk3 consists of a KD and a PBD. Plasmid constructs were made expressing KD, PBD, and FL Plk3 as GFP fusion proteins (Fig. 4A). The various expression constructs were transfected into HeLa cells for 24 h and then treated with nickel for 4 h. Indirect fluorescence microscopy revealed that, whereas little HIF-1α was detected in untreated HeLa cells, nickel treatment greatly stimulated the accumulation of this protein in the nucleus (Fig. 4B). Expression of Plk3-KD, but not Plk3-PBD, drastically suppressed the induction of nuclear HIF-1α by nickel; on the other hand, expression of Plk1-KD did not affect the induction of HIF-1α (Fig. 4C). Interestingly, ectopically expressed Plk3-FL did not have a noticeable effect on suppression of nuclear accumulation of HIF-1α (Fig. 4C). Expression of Plk3-KD, but not Plk3-PBD, also significantly suppressed the induction of HIF-1α by cobalt ions (Supplementary Fig. S2A and B). These data suggest that Plk3 plays a specific role in regulating the hypoxic response signaling pathway and that active Plk3 may be necessary for the destabilization and nuclear exclusion of HIF-1α.
Figure 4.
Ectopic expression of the Plk3-KD suppresses nuclear accumulation of HIF-1α in HeLa cells. A, schematic presentation of Plk3 domain structure and several Plk3 and Plk1 expression constructs. B, HeLa cells treated with NiCl2 for 4 h were stained with the HIF-1α antibody (red), DNA was stained with DAPI (blue). Representative cells are shown. C, HeLa cells transfected with various expression constructs for 24 h and then treated with NiCl2 for 4 h were stained with antibodies to HIF-1α (red) and GFP (green). DNA was stained with DAPI (blue). Representative cells are shown. Arrowheads, Plk3-KD–expressing cells with little accumulation of nuclear HIF-1α.
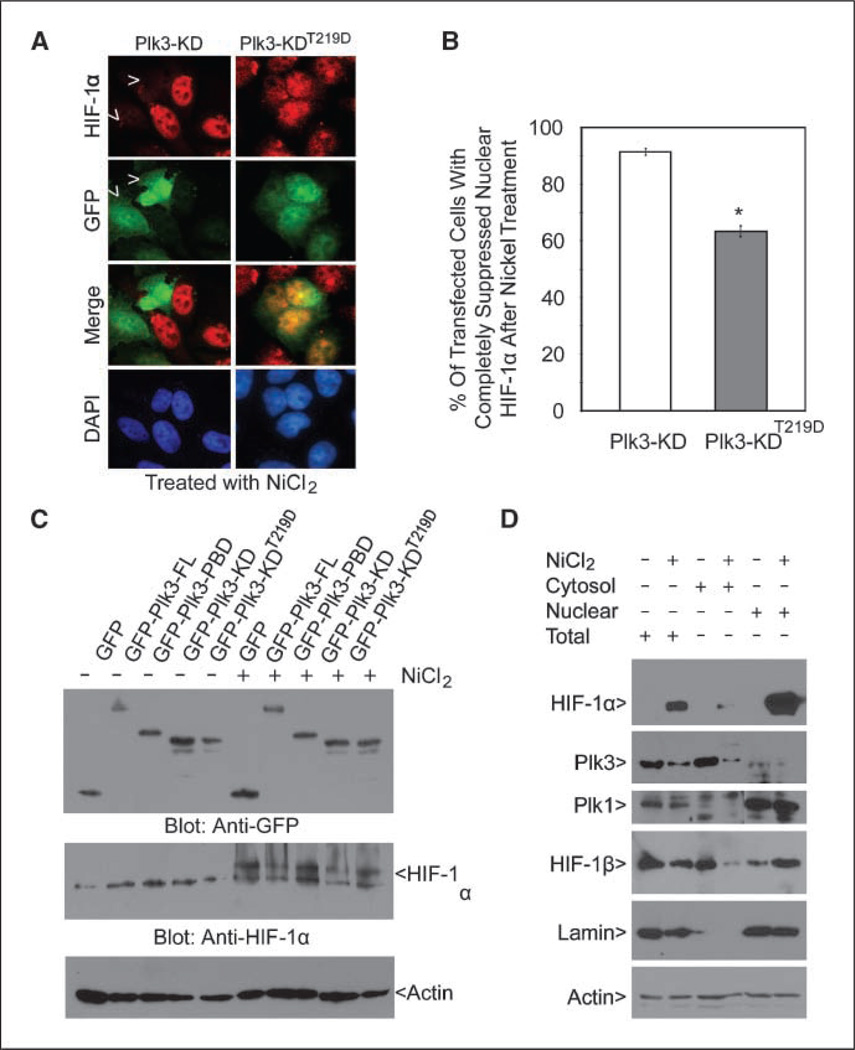
Expression of Plk3-KD greatly suppressed NiCl2- or CoCl2-mediated induction of HIF-1α, suggesting that the kinase activity may be required for negative regulation of this transcription factor. To test this possibility, we made a kinase-defective Plk3-KD by mutating a residue (Thr219) that is important for kinase activation. As expected, expression of this mutant construct significantly suppressed the accumulation of nuclear HIF-1α (Fig. 5A and B). On the other hand, we noticed that there existed a population of Plk3-KDT219D–expressing cells containing a significant amount of nuclear HIF-1α after nickel treatment (Fig. 5B). This is likely due to a basal level of the kinase activity associated with this construct (data not shown).
Figure 5.
HIF-1α expression is negatively correlated with Plk3 activity. A, HeLa cells transfected with GFP-Plk3-KD or GFP-Plk3-KDT219D expression constructs for 24 h and then treated with NiCl2 for 4 h were stained with antibodies to HIF-1α (red) and GFP (green). DNA was stained with DAPI (blue). Representative cells are shown. Arrows, Plk3-KD–expressing cells with suppressed accumulation of nuclear HIF-1α after NiCl2 treatment. B, HeLa cells transfected with GFP-Plk3-KD or GFP-Plk3-KDT219D expression constructs for 24 h and then treated with NiCl2 for 4 h were stained with antibodies to HIF-1α and GFP. The percent of transfected cells with completely suppressed nuclear HIF-1α after nickel ion treatment was summarized from three independent experiments. *, significant statistical difference (P < 0.01). C, HeLa cells were transfected with various GFP-Plk3 expression constructs as indicated for 24 h followed by treatment with NiCl2 for 4 h. Equal amounts of cell lysates were blotted for GFP, HIF-1α, and β-actin. D, HeLa cells were treated with or without NiCl2 for 24 h. Equal amounts of total cell lysates or cytoplasmic/nuclear fractions were blotted for HIF-1α, Plk3, Plk1, HIF-1β, lamin B, and β-actin. Lamin B, a nuclear protein, was examined to confirm effective subcellular fractionation. Each experiment was repeated at least thrice.
To further confirm the effect of Plk3 kinase activity on suppressing HIF-1α induction, we transfected HEK293 cells with various GFP-tagged Plk3 expression constructs for 24 h followed by treatment with nickel for 4 h. Equal amounts of cell lysates were blotted for HIF-1α. Blotting with the anti-GFP antibody revealed that transfected constructs were expressed with anticipated sizes at comparable levels (Fig. 5C). Transfection of Plk3-KD greatly reduced the level of HIF-1α compared with that of Plk3-PBD-transfected or GFP-transfected control; consistent with the notion that the kinase activity is required for the inhibition, transfection of Plk3-KD did not significantly suppress the level of HIF-1α induced by nickel (Fig. 5C;Supplementary Table S2). Interestingly, transfection of Plk3-FL construct also reduced the level of HIF-1α (Fig. 5C; supplementary Table S2), suggesting that FL Plk3 is capable of inhibiting the steady-state level of HIF-1α.
Because Plk3 is constitutively expressed, we asked whether hypoxic stress affected expression of endogenous Plk3. Immuno-blot analysis showed that Plk3, which was mostly cytosolic, was significantly down-regulated on treatment with NiCl2;on the other hand, expression of Plk1, which was mostly nuclear, was not affected by the nickel ion treatment (Fig. 5D). As expected, HIF-1α expression was induced and translocated into the nucleus after NiCl2 treatment (Fig. 5D). HIF-1β expression was not induced by nickel; however, it was translocated into the nucleus on hypoxic stress (Fig. 5D).
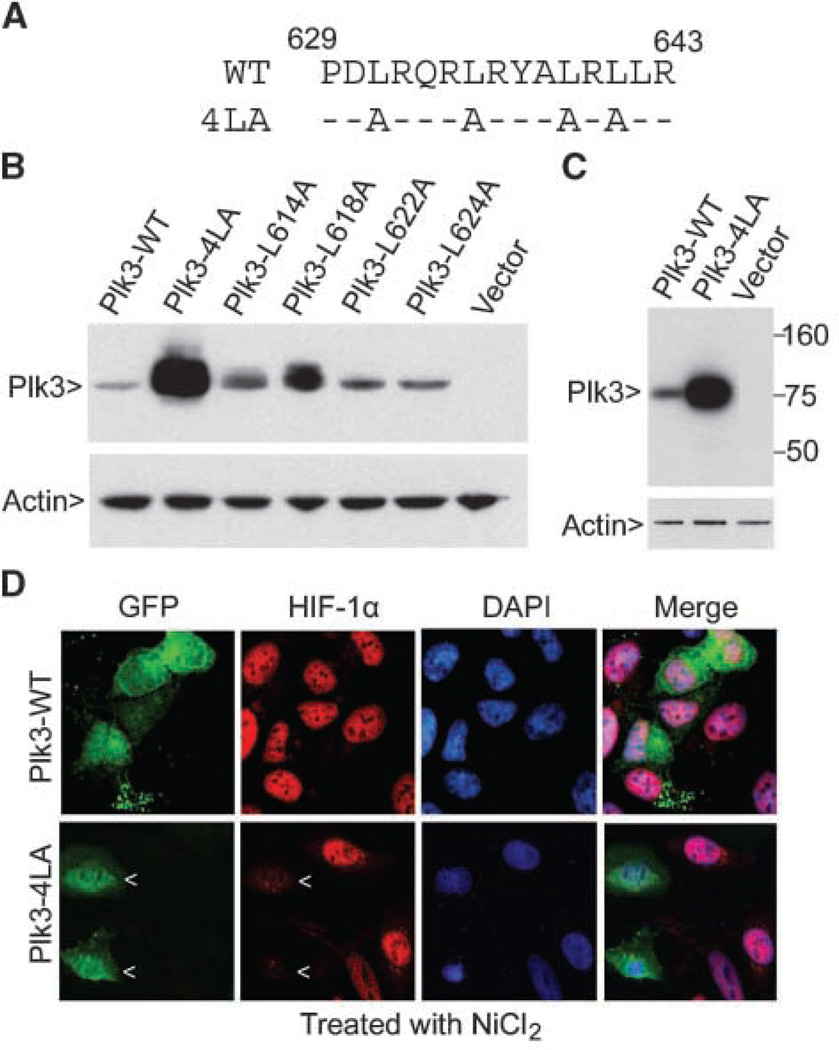
Activated Plk3 seems to undergo degradation via the ubiquitin-proteosome pathway (20). Through expressing various Plk3 mutants as well as wild-type Plk3, we found that mutations of a leucine-rich region in the Plk3 COOH-terminal tail greatly stabilized ectopically expressed Plk3 (Fig. 6A-C). To determine whether ectopic expression of a stable Plk3 protein affected HIF-1α accumulation after hypoxic stress, we transfected HeLa cells with Plk3 wild-type (Plk3-WT) and Plk3-4LA (Plk3 with four leucine mutations) expression constructs for 24 h before treatment with NiCl2 for 8 h. HIF-1α expression was greatly induced by nickel ion in cells transfected with the Plk3-WT construct; however, the induction of HIF-1α was significantly suppressed in cells transfected with the Plk3-4LA expression construct (Fig. 6D).
Figure 6.
Ectopic expression of a Plk3 COOH-terminal region mutant suppresses NiCl2-mediated nuclear accumulation of HIF-1α in HeLa cells. A, amino acid sequence of the relevant mouse Plk3 region (numbering based on the 648-amino acid Plk3 isoform). WT, wild-type sequence; 4LA, mutant sequence showing the positions of the four alanine substitutions in the 4LA protein. B, plasmid constructs expressing HA-tagged wild-type mouse Plk3 or the indicated Plk3 mutants were transfected into NIH3T3 cells for 24 h. Empty plasmid was also used for transfection. Equal amounts of cell lysates were blotted with anti-HA and anti-β-actin antibodies. C, plasmid constructs expressing HA-tagged wild-type mouse Plk3 or Plk3-4LA, or parental plasmid vector, were transfected into human HEK293 cells for 24 h. Equal amounts of cell lysates were blotted with the antibodies described above. D, HeLa cells transfected with the Plk3-WT or Plk3-4LA expression plasmid for 24 h and then treated with NiCl2 for 4 h were fixed and stained with antibodies to the HA tag (green) or HIF-1α (red). DNA was stained with DAPI (blue). Arrows, cells expressing the transfected HA-tagged Plk3 mutant protein. Representative cells are shown.
Given the newly identified role of Plk3 in response to hypoxic stress as well as the known regulators in the HIF-1α signaling pathway, we propose the following model for Plk3 function (Supplementary Fig. S3). Plk3 can potentially inhibit HIF-1α by physical interaction and direct phosphorylation. HIF-1 is a phosphoprotein (25), although the exact role of phosphorylation remains to be elucidated. PHD, which is inhibited under hypoxia or by treatment with nickel or cobalt ions, catalyzes prolyl hydroxylation of HIF-1α, promoting its association with VHL and subsequent ubiquitination. In theory, Plk3 could up-regulate the function of PHD and/or VHL, resulting in enhanced degradation of HIF-1α by the proteasomal pathway. On the other hand, it remains a possibility that Plk3 has unknown targets, which in turn modulate the activities of components in the HIF-1α signaling pathway.
Discussion
Our studies reveal a new role for Plk3 in controlling the stability of HIF-1α. By generating and analyzing Plk3-deficient mice, we show that Plk3 displays a tumor-suppressing function because ageing Plk3−/− mice develop tumors in various organs at an accelerated rate compared with their wild-type counterparts. By studying MEFs derived from Plk3−/− mice, we find that Plk3−/− MEFs are hypersensitive to the induction of HIF-1α and its transcriptional target VEGF-A under hypoxic conditions or by treatment with nickel ions. Through several independent cellular and molecular analyses, we have obtained additional lines of evidence that are consistent with a role of Plk3 in negative regulation of HIF-1α under hypoxic stresses: (a) ectopic expression of the Plk3-KD, but not its PBD or a Plk3 kinase-defective mutant, suppresses the nuclear accumulation of HIF-1α induced by nickel or cobalt ions;(b) hypoxia-induced HIF-1α expression is tightly associated with a significant down-regulation of Plk3 expression in HeLa cells; and (c) stabilization of Plk3 by mutating key residues in the PBD also suppresses HIF-1α accumulation induced by nickel ions. HIF-1α plays a key role in activating the “survival machinery”’ during malignancy. It is intriguing to observe that expression of Plk3-FL does not significantly suppress nuclear accumulation of HIF-1α in HeLa cells. This can be explained by the following possibilities. Compared with normal murine embryonic fibroblasts, Plk3 may function differently in HeLa cells, which are malignantly transformed. It is also possible that other members of the Plk family may interfere with the full function of Plk3. Alternatively, Plk3-KD may exhibit some off-target effects on other Plks in HeLa cells, leading to dramatic suppression of nuclear accumulation of HIF-1α during hypoxia.
At present, it remains unclear which component(s) of the HIF-1α pathway is the direct target of Plk3. Plk3 may directly regulate HIF-1α as the latter also undergoes phosphorylation during hypoxia. Supporting this, substrate recognition by E3 ubiquitin ligases and subsequent protein degradation are frequently phosphorylation dependent (28). Several HIF-1α kinases have been identified that modulate the stability or activation of this transcription factor during hypoxic responses. Mitogen-activated protein kinase (MAPK) is a protein kinase that positively stimulates HIF-1α in response to hypoxia and insulin-like growth factor-I (29). Analysis of MAPK-phosphorylated HIF-1α by mass spectroscopy reveals residues Ser641 and Ser643 as potential phosphorylation sites because mutations of these residues significantly reduce the phosphorylation of HIF-1α and decrease its transcriptional activity (30). In contrast, depletion of glycogen synthase kinase-3β (GSK-3β) induces HIF-1α, whereas overexpression of this kinase decreases the level of HIF-1α;GSK-3β–induced inhibition of HIF-1α is mediated through a proteasome-dependent mechanism (31). Thus, Plk3 seems to function in a manner similar to that of GSK-3β in promoting the degradation of HIF-1α.
Plk3 may directly affect the activity of components involved in regulating HIF-1α stability (refer to the model in Supplementary Fig. S3). In normoxia, hydroxylated HIF-1α interacts with VHL, a ubiquitin E3 ligase. VHL is a tumor suppressor gene product whose inactivation causes a familial cancer syndrome characterized by multiple primary renal cell carcinomas (32). Existing experimental evidence supports the possibility that Plk3 positively regulates the activity of VHL: (a) Plk3 also functions as a tumor suppressor, inactivation of which causes the development of common malignancies, including renal cancer (Fig. 2);(b) Plk3 is very responsive to oxidative stress and its kinase activity is rapidly activated on H2O2 treatment (11), suggestive of a role in oxygen homeostasis; and (c) VHL is necessary for cilia formation and maintenance. Specifically, a recent study showed that VHL is involved in directing the orientation of growing microtubules because loss of VHL function causes aberrant orientation of newly formed microtubules and inhibits ciliogenesis (33). Likewise, Plk3 regulates microtubule dynamics during the cell cycle. Ectopic expression of a kinasede-fective Plk3 mutant causes the aberrant formation of microtubule structures and dramatically changes the cell morphology (13).
Because both Plk3 and Plk4 are known to regulate the activity of p53 (12, 34), it is possible that Plk3 affects the stability and nuclear accumulation of HIF-1α through an indirect mechanism that involves p53. Under severe hypoxia, p53 protein is stabilized, which is correlated with S-phase arrest (35). Tumor hypoxia also induces and activates both HIF-1α and p53, which seem to have opposite functions. Whereas HIF-1α signaling promotes cellular adaptation to hypoxia, p53 mediates apoptosis induced by hypoxia (36). At the molecular level, p53 inhibits HIF-1α activity, which seems to be mediated through a direct interaction between p53 and the oxygen-dependent degradation domain of the HIF-1α subunit (37). The physical interaction between these two transcription factors could conceivably promote polyubiquitination of HIF-1α, resulting in its degradation. Supporting this, VHL is also associated with p53 and it increases p53 stability and activity by suppressing Mdm2-mediated ubiquitination and nuclear export of p53 and promoting acetylation by p300 (38). Because Plk3 interacts with and positively regulates p53 by phosphorylation in response to genotoxic stresses (11, 12), it is conceivable that it coordinates the activation of VHL and p53 and the inactivation of HIF-1α, which is essential for the initiation of hypoxic responses and cell growth arrest in normal cells.
In summary, we have identified Plk3 as a novel molecular component regulating the oxygen-sensing pathway. Given that Plk3 blocks cell proliferation and induces apoptosis and that HIF-1α promotes cell survival and neoplastic transformation, our current study underscores the importance of the oxygen-sensing pathway in suppressing tumorigenesis in vivo. Further studies on the mode of action of Plk3 will provide additional mechanistic insights that can guide the design of anticancer drugs tailored against the HIF-VHL pathway.
Supplementary Material
Acknowledgments
Grant support: NIH grants CA74229 (W. Dai) and HL67051 and HL39727 (J.A. Winkles).
We thank Dr. Steven Potter for the gene targeting vector, Drs. Ming Xu and Hong Sun for useful discussions, and Drs. Tongyi Liu, Yuqiang Fang, and Yang-Ming Yang for their involvement in mouse breeding and/or technical assistance.
Footnotes
Note: Supplementary data for this article are available at Cancer Research Online (http://cancerres.aacrjournals.org/).
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
References
- 1.Barr FA, Sillje HH, Nigg EA. Polo-like kinases and the orchestration of cell division. Nat Rev Mol Cell Biol. 2004;5:429–440. doi: 10.1038/nrm1401. [DOI] [PubMed] [Google Scholar]
- 2.Fenton B, Glover DM. A conserved mitotic kinase active at late anaphase-telophase in syncytial Drosophila embryos. Nature. 1993;363:637–640. doi: 10.1038/363637a0. [DOI] [PubMed] [Google Scholar]
- 3.Strebhardt K, Ullrich A. Targeting polo-like kinase 1 for cancer therapy. Nat Rev Cancer. 2006;6:321–330. doi: 10.1038/nrc1841. [DOI] [PubMed] [Google Scholar]
- 4.Xie S, Xie B, Lee MY, Dai W. Regulation of cell cycle checkpoints by polo-like kinases. Oncogene. 2005;24:277–286. doi: 10.1038/sj.onc.1208218. [DOI] [PubMed] [Google Scholar]
- 5.Donohue PJ, Alberts GF, Guo Y, Winkles JA. Identification by targeted differential display of an immediate early gene encoding a putative serine/threonine kinase. J Biol Chem. 1995;270:10351–10357. doi: 10.1074/jbc.270.17.10351. [DOI] [PubMed] [Google Scholar]
- 6.Fode C, Motro B, Yousefi S, Heffernan M, Dennis JW. Sak, a murine protein-serine/threonine kinase that is related to the Drosophila polo kinase and involved in cell proliferation. Proc Natl Acad Sci U S A. 1994;91:6388–6392. doi: 10.1073/pnas.91.14.6388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lee KS, Erikson RL. Plk is a functional homolog of Saccharomyces cerevisiae Cdc5, and elevated Plk activity induces multiple septation structures. Mol Cell Biol. 1997;17:3408–3417. doi: 10.1128/mcb.17.6.3408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Li B, Ouyang B, Pan H, et al. Prk, a cytokine-inducible human protein serine/threonine kinase whose expression appears to be down-regulated in lung carcinomas. J Biol Chem. 1996;271:19402–19408. doi: 10.1074/jbc.271.32.19402. [DOI] [PubMed] [Google Scholar]
- 9.Simmons DL, Neel BG, Stevens R, Evett G, Erikson RL. Identification of an early-growth-response gene encoding a novel putative protein kinase. Mol Cell Biol. 1992;12:4164–4169. doi: 10.1128/mcb.12.9.4164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Dai W. Polo-like kinases, an introduction. Oncogene. 2005;24:214–216. doi: 10.1038/sj.onc.1208270. [DOI] [PubMed] [Google Scholar]
- 11.Xie S, Wang Q, Wu H, et al. Reactive oxygen species-induced phosphorylation of p53 on serine 20 is mediated in part by polo-like kinase-3. J Biol Chem. 2001;276:36194–36199. doi: 10.1074/jbc.M104157200. [DOI] [PubMed] [Google Scholar]
- 12.Xie S, Wu H, Wang Q, et al. Plk3 functionally links DNA damage to cell cycle arrest and apoptosis at least in part via the p53 pathway. J Biol Chem. 2001;276:43305–43312. doi: 10.1074/jbc.M106050200. [DOI] [PubMed] [Google Scholar]
- 13.Wang Q, Xie S, Chen J, et al. Cell cycle arrest and apoptosis induced by human Polo-like kinase 3 is mediated through perturbation of microtubule integrity. Mol Cell Biol. 2002;22:3450–3459. doi: 10.1128/MCB.22.10.3450-3459.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eckerdt F, Yuan J, Strebhardt K. Polo-like kinases and oncogenesis. Oncogene. 2005;24:267–276. doi: 10.1038/sj.onc.1208273. [DOI] [PubMed] [Google Scholar]
- 15.Winkles JA, Alberts GF. Differential regulation of polo-like kinase 1, 2, 3, and 4 gene expression in mammalian cells and tissues. Oncogene. 2005;24:260–266. doi: 10.1038/sj.onc.1208219. [DOI] [PubMed] [Google Scholar]
- 16.Winkles JA, O’Connor ML, Friesel R. Altered regulation of platelet-derived growth factor A-chain and c-fos gene expression in senescent progeria fibroblasts. J Cell Physiol. 1990;144:313–325. doi: 10.1002/jcp.1041440218. [DOI] [PubMed] [Google Scholar]
- 17.Zambrowicz BP, Friedrich GA, Buxton EC, Lilleberg SL, Person C, Sands AT. Disruption and sequence identification of 2,000 genes in mouse embryonic stem cells. Nature. 1998;392:608–611. doi: 10.1038/33423. [DOI] [PubMed] [Google Scholar]
- 18.Yepes M, Brown SA, Moore EG, Smith EP, Lawrence DA, Winkles JA. A soluble Fn14-Fc decoy receptor reduces infarct volume in a murine model of cerebral ischemia. Am J Pathol. 2005;166:511–520. doi: 10.1016/S0002-9440(10)62273-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang N, Wang X, Jhanwar-Uniyal M, Darzynkiewicz Z, Dai W. Polo box domain of Plk3 functions as a centrosome localization signal, overexpression of which causes mitotic arrest, cytokinesis defects, and apoptosis. J Biol Chem. 2006;281:10577–10582. doi: 10.1074/jbc.M513156200. [DOI] [PubMed] [Google Scholar]
- 20.Alberts GF, Winkles JA. Murine FGF-inducible kinase is rapidly degraded via the nuclear ubiquitin-proteosome system when overexpressed in NIH 3T3 cells. Cell Cycle. 2004;3:678–684. [PubMed] [Google Scholar]
- 21.Ouyang B, Pan H, Lu L, et al. Human Prk is a conserved protein serine/threonine kinase involved in regulating M phase functions. J Biol Chem. 1997;272:28646–28651. doi: 10.1074/jbc.272.45.28646. [DOI] [PubMed] [Google Scholar]
- 22.Huang Y, Zhao Q, Zhou CX, et al. Antileukemic roles of human phospholipid scramblase 1 gene, evidence from inducible PLSCR1-expressing leukemic cells. Oncogene. 2006;25:6618–6627. doi: 10.1038/sj.onc.1209677. [DOI] [PubMed] [Google Scholar]
- 23.Ma S, Charron J, Erikson RL. Role of Plk2 (Snk) in mouse development and cell proliferation. Mol Cell Biol. 2003;23:6936–6943. doi: 10.1128/MCB.23.19.6936-6943.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ko MA, Rosario CO, Hudson JW, et al. Plk4 haploinsufficiency causes mitotic infidelity and carcinogenesis. Nat Genet. 2005;37:883–888. doi: 10.1038/ng1605. [DOI] [PubMed] [Google Scholar]
- 25.Ke Q, Costa M. Hypoxia-inducible factor-1 (HIF-1) Mol Pharmacol. 2006;70:1469–1480. doi: 10.1124/mol.106.027029. [DOI] [PubMed] [Google Scholar]
- 26.Davidson T, Chen H, Garrick MD, D’Angelo G, Costa M. Soluble nickel interferes with cellular iron homeostasis. Mol Cell Biochem. 2005;279:157–162. doi: 10.1007/s11010-005-8288-y. [DOI] [PubMed] [Google Scholar]
- 27.Ohh M. pVHL’s kryptonite: E2-EPF UCP. Cancer Cell. 2006;10:95–97. doi: 10.1016/j.ccr.2006.07.016. [DOI] [PubMed] [Google Scholar]
- 28.Gao M, Karin M. Regulating the regulators: control of protein ubiquitination and ubiquitin-like modifications by extracellular stimuli. Mol Cell. 2005;19:581–593. doi: 10.1016/j.molcel.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 29.Sutton KM, Hayat S, Chau NM, et al. Selective inhibition of MEK1/2 reveals a differential requirement for ERK1/2 signalling in the regulation of HIF-1 in response to hypoxia and IGF-1. Oncogene. 2007;26:3920–3929. doi: 10.1038/sj.onc.1210168. [DOI] [PubMed] [Google Scholar]
- 30.Mylonis I, Chachami G, Samiotaki M, et al. Identification of MAPK phosphorylation sites and their role in the localization and activity of hypoxiainducible factor-1α. J Biol Chem. 2006;281:33095–106. doi: 10.1074/jbc.M605058200. [DOI] [PubMed] [Google Scholar]
- 31.Flugel D, Gorlach A, Michiels C, Kietzmann T. Glycogen synthase kinase 3 phosphorylates hypoxia-inducible factor 1a and mediates its destabilization in a VHL-independent manner. Mol Cell Biol. 2007;27:3253–3265. doi: 10.1128/MCB.00015-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kaelin WG., Jr The von Hippel-Lindau tumor suppressor protein and clear cell renal carcinoma. Clin Cancer Res. 2007;13:680s–684s. doi: 10.1158/1078-0432.CCR-06-1865. [DOI] [PubMed] [Google Scholar]
- 33.Kuehn EW, Walz G, Benzing T. von Hippel-Lindau: a tumor suppressor links microtubules to ciliogenesis and cancer development. Cancer Res. 2007;67:4537–4540. doi: 10.1158/0008-5472.CAN-07-0391. [DOI] [PubMed] [Google Scholar]
- 34.Swallow CJ, Ko MA, Siddiqui NU, Hudson JW, Dennis JW. Sak/Plk4 and mitotic fidelity. Oncogene. 2005;24:306–312. doi: 10.1038/sj.onc.1208275. [DOI] [PubMed] [Google Scholar]
- 35.Hammond EM, Denko NC, Dorie MJ, Abraham RT, Giaccia AJ. Hypoxia links ATR and p53 through replication arrest. Mol Cell Biol. 2002;22:1834–1843. doi: 10.1128/MCB.22.6.1834-1843.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Fels DR, Koumenis C. HIF-1α and p53: the ODD couple? Trends Biochem Sci. 2005;30:426–429. doi: 10.1016/j.tibs.2005.06.009. [DOI] [PubMed] [Google Scholar]
- 37.Hansson LO, Friedler A, Freund S, Rudiger S, Fersht AR. Two sequence motifs from HIF-1α bind to the DNA-binding site of p53. Proc Natl Acad Sci U S A. 2002;99:10305–10309. doi: 10.1073/pnas.122347199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Roe JS, Kim H, Lee SM, Kim ST, Cho EJ, Youn HD. p53 stabilization and transactivation by a von Hippel-Lindau protein. Mol Cell. 2006;22:395–405. doi: 10.1016/j.molcel.2006.04.006. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.