Abstract
Host genetic variation is presently estimated to account for about one-fourth of the observed differences in control of HIV across infected individuals. Genome-wide association studies have confirmed that polymorphism within the HLA class I locus is the primary host genetic contributor to determining outcome after infection. Here we progress beyond the genetic associations alone to consider the functional explanations for these correlations. In this process, the complex and multidimensional effects of HLA molecules in viral disease become apparent.
Keywords: HLA-B, HLA-C 3′ UTR, genome-wide association study, cytotoxic T lymphocytes, microRNA
INTRODUCTION
When a blood test for HIV finally became available in April 1985, the world had its first glimpse of the rapid spread of this new epidemic of progressive immunodeficiency. Nowhere was this more apparent than in a cohort of gay men in the San Francisco area who had been recruited for a hepatitis B vaccine trial in the late 1970s. Based on retrospective analysis of samples collected throughout the trial, the actual timing of infection could be documented for all persons, and the results were alarming indeed. By 1982, 42% of the men in the trial had seroconverted to HIV (1). And based on subsequent progression to clinical AIDS in many of these persons, the general sense was that seropositivity was an indication of an inevitable future deterioration in immune function, the development of opportunistic infections, and subsequent death. Clinical follow-up largely supported this view, as did the experience in hospitals across the country, where new cases of AIDS were being identified at an alarming rate.
However, as the epidemic expanded, it became apparent that not everyone was progressing, at least by the one predictive parameter available at that time, CD4 cell count (2). More striking were the results of viral-load testing, which became widely available in the mid-1990s. In some persons known to be infected since early in the epidemic, levels remained below the limit of detection of increasingly sensitive assays. It is now recognized that ~1 in 300 persons who become HIV infected maintain spontaneous control of HIV to levels below 50 RNA copies/ml, the lower limit of detection by standard assays (3). And some cases of HIV infection have been documented without any evidence of clinical disease progression after more than three decades of follow-up. It slowly became apparent that some people were not only surviving this infection but continuing to thrive.
Multiple longitudinal studies have now provided ample evidence of dramatic differences in untreated disease outcome, ranging from the development of AIDS in less than a year to no evidence of progression decades later (4, 5). However, the reasons for these differences have remained unclear. With the advent of new human genome sequencing techniques that led to completion of the HapMap, a new approach to addressing a potential genetic basis for these differences became available (6-8).
Here we review the results of the International HIV Controllers Study, which was initiated in an attempt to define the contribution of host genetics to durable HIV control (5). Sites in HLA-B and HLA-C within the major histocompatibility complex (MHC) on chromosome 6 were highly associated with control. We analyze the results of this study and extend the interpretation of this and other studies in an attempt to define the involvement of those sites (9-13) and to provide insights into the nature of persistent HIV control.
ESTABLISHMENT OF THE INTERNATIONAL HIV CONTROLLERS STUDY
The International HIV Controllers Study was established to directly assess the genetic contribution to HIV control. The study examined host genetic variability in unprecedented numbers of persons able to spontaneously control HIV replication to <2,000 RNA copies/ml and compared these to persons with progressive infection. Cohort studies indicate that at this level of viral load, the chance of transmission is markedly diminished (14), and likewise at this level the chance of disease progression drops dramatically (4). This could be viewed as a possible model of successful vaccination: even if persons were to become infected, preventing subsequent disease progression and subsequent transmission would impede the spread of the epidemic.
Entry criteria into multicenter studies of HIV drug trials had amassed DNA samples from large numbers of persons with progressive disease, but HIV controllers were less likely to be involved in therapeutic clinical trials, and only a few research sites had assembled cohorts of HIV controllers (15-18). For the International HIV Controllers Study, existing cohorts were pooled, still falling short of the numbers needed for genetic studies. However, through a direct appeal to practitioners at major HIV treatment centers across the United States and around the globe, together with recruitment efforts through AIDS activist organizations and public announcements, >1,500 such subjects were successfully recruited through >200 collaborators, the majority of whom were physicians with large HIV practices.
SINGLE-NUCLEOTIDE POLYMORPHISMS AND HIV CONTROL
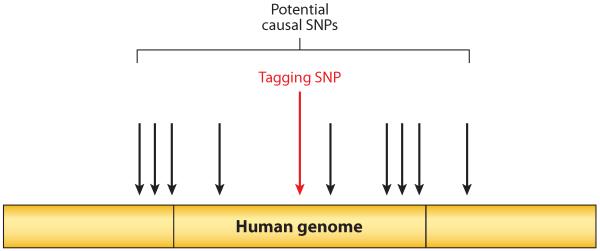
The initial goal of this study was to perform a genome-wide association study (GWAS) to define single-nucleotide polymorphisms (SNPs) associated with control or lack of control, making use of high-throughput technologies that allow rapid assessment of SNPs within the human genome. Automated systems enable more than a million SNPs to be readily analyzed on large cohorts. Because of linkage disequilibrium (LD), knowledge of one SNP (termed a tagging SNP) gives information about surrounding unmeasured SNPs (Figure 1), so that measurement of roughly one million SNPs yields information on the majority of variability in the three billion nucleotides of the human genome. Thus, one can take a nonbiased approach to defining SNPs that either influence disease outcome themselves or tag other SNPs that do. Such studies rely on defining associations between specific SNPs and a specific disease phenotype, and had been successfully applied to HIV in a landmark study of genetic determinants of HIV control after acute infection (10).
Figure 1.
GWAS analysis: individual “tagging SNPs” are measured by chip technology. Owing to linkage disequilibrium, the tagging SNP provides reliable information about surrounding SNPs, allowing the majority of variability in the human genome to be determined by measuring only one million of its three billion nucleotides. Abbreviations: GWAS, genome-wide association study; SNP, single-nucleotide polymorphism.
The initial International HIV Controllers Study included data on a total of 974 controllers specifically recruited to this study and compared these to 2,648 progressors obtained from AIDS Clinical Trials Group (ACTG) studies (5). In each person, 1,384,048 SNPs were assessed. Persons were stratified into ethnic groups, including those of European, Hispanic, and African-American ancestry, by applying a statistical technique called principal-components analysis. This computational approach defines the greatest variances in the data, which are due to ethnic differences, and allows one to group persons based on these parameters and thereby avoid potential confounding effects of ethnic genetic variability.
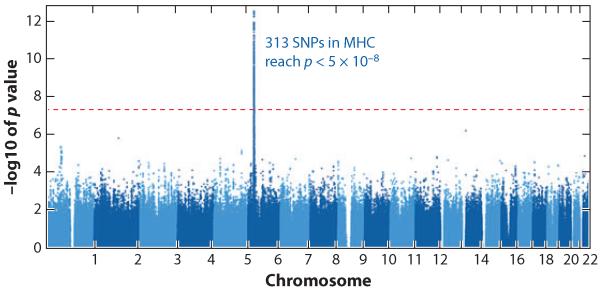
The results of this analysis were striking both in terms of numbers of SNPs and their location. In persons of European ancestry, a total of 313 SNPs were significantly different in the HIV controllers and progressors after correction for multiple comparisons, requiring a p-value of <5 × 10−8 for significance. All were within a three-megabase region concentrated around the HLA class I region on chromosome 6 (Figure 2). The HLA class I genes are the most polymorphic loci in the human genome. The proteins encoded by these genes are directly involved in immune function by binding and presenting a great variety of antigenic peptides to cytotoxic T lymphocytes (CTLs), and also by interacting with HLA receptors on the surface of other cell types (19, 20). There was no signal detected outside this region in chromosome 6, and no signal at all detected in any of the other 21 chromosomes examined.
Figure 2.
Manhattan plot of data derived from 974 HIV controllers and 2,648 progressors. Shown are p-values for 1,384,048 data points derived from comparing each measured single-nucleotide polymorphism (SNP) in HIV controllers versus progressors. 313 SNPs reach statistical significance in this population of European ancestry, appearing as a line, which is defined by p < 5 × 10−8 because of the need to correct for multiple comparisons. All significant SNPs lie within the major histocompatibility complex on chromosome 6.
Further analysis of these SNPs by stepwise regression analysis was performed in order to determine which were independently linked to HIV control. This was required because this region of the genome is characteristically associated with high levels of LD, where multiple SNPs are strongly correlated with one another. A total of four SNPs remained statistically significant after this analysis (Table 1). Two of these coincided with SNPs that had been associated with viral-load set point following acute infection (10, 12). SNP rs2395029 is a proxy for HLA-B*5701, already well known to be associated with a better outcome in HIV infection (16). A second SNP known to modulate viral set point after acute infection (10) was also significant: rs9264942 is a variant thought to be associated with HLA-C expression levels. In addition to these, two others reached independent statistical significance, one in a noncoding region near MICA and one implicated as a possible psoriasis candidate gene called PSORS1C3. However, different independently significant SNPs, also in the HLA class I region, were found in the African-American and Hispanic samples.
Table 1. Independent SNPs associated with HIV control.
| SNP(EA) | Location | Odds ratio (p-value) |
|---|---|---|
| rs9264942 | 35 kb upstream of HLA-C | 2.9 (2.8 × 10−35) |
| rs2395029 | Proxy for HLA-B*57:01 | 5.3 (9.7 × 10−26) |
| rs4418214 | SNP near MICA | 4.4 (1.4 × 10−34) |
| rs3131018 | In PSORS1C3 | 2.1 (4.2 × 10−16) |
This study also revealed that none of the previously reported genetic associations outside the MHC was significant in this population rigorously controlled for ethnicity. Limiting the analysis to variants outside the MHC that have been linked to disease progression in the literature, which required a more liberal correction for multiple comparisons since fewer were performed, revealed only variants in the CCR5-CCR2 locus as statistically significant, and the associations were much weaker than those found within the MHC (5). The four SNPs in the MHC explained 19% of the variability in viral load in HIV-infected persons, and combined with the CCR5-CCR2 variants explain 23% of the genetic variance in host control. Thus, there may be genetic factors that are not detectable by GWAS, at least using the current cohort size, that impact viral control, and there are almost certainly other nongenetic factors that modulate disease. This is consistent with the observation that the majority of persons with protective HLA alleles, including HLA-B*57 and -B*27, actually have progressive infection (21), indicating that other factors impact outcome.
LIMITATIONS OF GWAS
These GWAS results are consistent with previous studies with variable definitions of HIV control (10-13, 22) and show a relationship between MHC and HIV disease outcome. Variation within the MHC, and particularly within the HLA genes embedded in the MHC, associates with a remarkable plethora of different human diseases, as determined by other disease-specific GWAS (Table 2), and most of these associated variants map closely to specific HLA class I and II genes, implicating these genes as somehow causal in the disease process. And a large number of the disease associations identified through GWAS, including those for HIV disease, had already been identified prior to the development of genome-wide scans through direct genotyping of the HLA class I and II genes.
Table 2. Diseases and traits studied by genome-wide association studies implicating the major histocompatibility complexa.
| Nasopharyngeal carcinoma |
| Multiple sclerosis |
| Rheumatoid arthritis |
| Drug-induced liver injury (amoxicillin–clavulanate) |
| Vitiligo |
| HIV-1 control |
| Follicular lymphoma |
| Ulcerative colitis |
| Psoriatic arthritis |
| Psoriasis |
| Ankylosing spondylitis |
| Hepatocellular carcinoma |
| Drug-induced liver injury (flucloxacillin) |
| AIDS progression |
| Neonatal lupus |
| Lung adenocarcinoma |
| Lung cancer |
| Systemic lupus erythematosus |
| Age-related macular degeneration |
| Schizophrenia |
| Nephropathy |
| Lumiracoxib-related liver injury |
| Arthritis (juvenile idiopathic) |
| Knee osteoarthritis |
| Type 1 diabetes |
| Parkinson’s disease |
| Hodgkin’s lymphoma |
| Inflammatory bowel disease |
| Chronic lymphocytic leukemia |
| Immunoglobulin A deficiency |
| Response to interferon beta therapy |
| Nephropathy (idiopathic membranous) |
| Celiac disease |
| Asthma |
| Primary biliary cirrhosis |
| Systemic sclerosis |
| Alopecia areata |
| Hepatitis B |
| Leprosy |
| Adverse response to carbamapezine |
| Hematological and biochemical traits |
| CD4:CD8 lymphocyte ratio |
| Pulmonary function |
Data in this table were collected from http://www.genome.gov/26525384.
A number of important questions remain based on the GWAS results. Are the SNPs themselves causal, or do they tag something causal? Do the SNPs tag something linked to HLA, or are they simply tagging the HLA alleles? The SNPs differ by ethnicity; does this mean that the mechanisms of control are different? If HLA is causal, what about HLA confers this effect? Or does this represent LD with a non-HLA determinant? To what extent is the peptide-binding groove involved, versus other nonbinding variants? Are these SNPs tagging something that is functionally linked to control? Thus although the GWAS results implicated the HLA region as the key genetic determinant, the GWAS findings alone generated many new unanswered questions.
BEYOND GWAS: FROM TAGGING SNPs TO HLA-B AND -C
Because the major signals from HIV GWAS have been confined to SNPs in the MHC region, and specifically HLA-B and HLA-C, HLA sequence analysis should resolve at least some of the above questions regarding the cause of the marked differences. HLA typing in the International HIV Controllers Study cohort was performed by a novel imputation method, derived from large data sets in which both SNP and HLA data were collected (23). These data allowed analysis of every amino acid in the region defined by the GWAS to be important. After correction for multiple comparisons, HLA alleles associated with both control and lack of control were identified (5). Importantly, stepwise regression analysis revealed that HLA alone, and not the SNPs, was linked to control. This settled one of the major questions from earlier studies, which had left unresolved whether the GWAS was implicating HLA or something linked to HLA. Of the alleles defined, several are associated with protection (B*57:01, B*27:05, B*14/Cw*08:02, B*52, A*25) and two others with susceptibility (B*35 and B*07) in Europeans. Importantly, these associations with HIV control had in large part already been defined in previous reports (16, 24-29).
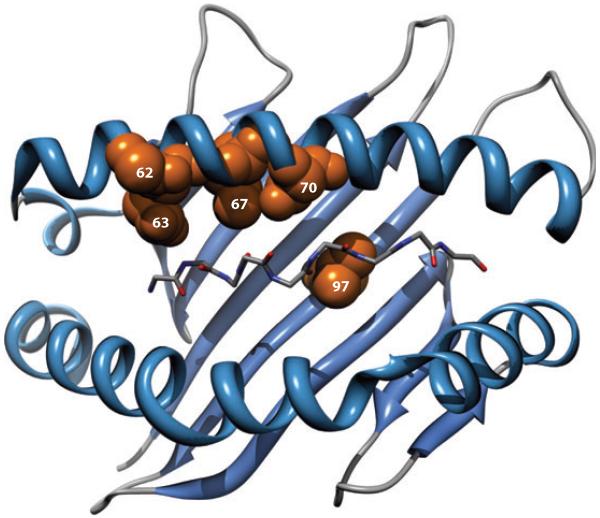
The above analysis refined the SNP associations by showing that they are due to an HLA signal. But knowledge of the HLA types of each person, and by inference the amino acid sequence in each person, allowed further extension of this analysis to determine the significance of individual amino acids in host genetic control. This revealed that the major genetic associations, after correction for multiple comparisons and replication in a second cohort, were due to polymorphisms in amino acids in the HLA-B peptide-binding groove, all of which line the peptide-binding groove (Figure 3), as well as an amino acid polymorphism at position 304 in the transmembrane domain of HLA-C. The most significant amino acid positions associated with control are positions 67, 70, and 97 in HLA-B, which are all involved in binding peptides within the peptide-binding groove and are in strong LD with the other variants in the peptide-binding groove. Positions 62 and 63 in the HLA-B peptide-binding groove were also found independently significant. Thus, this GWAS analysis extended previous GWAS, and previous population studies that linked specific HLA alleles to control and progression, by demonstrating that the peptide-binding groove is the major genetic determinant of HIV control and is what is being tagged by the SNPs.
Figure 3.
Three-dimensional ribbon representation of the HLA-B protein showing amino acid positions 62, 63, 67, 70, and 97 lining the peptide-binding groove. The peptide backbone of the epitope is also displayed. Adapted from Reference 5 with permission.
INTERPRETING THE HLA-B ASSOCIATIONS
The effects of balancing selection at the HLA class I loci are manifest in the high degree of polymorphism observed in this region (30), particularly in the nucleotides encoding the peptide-binding region (31). The amino acid sequences of pockets within this region determine the specificity of a given HLA class I allotype for the peptide bound within the groove, and it is the combination of >30 amino acid sites, most of which are polymorphic, forming six pockets in the groove, that provides such specificity (32).
The results showing that these HLA-B amino acids are more strongly associated with HIV control than any single HLA, including HLA-B*57, implicate the peptide-binding groove as the major factor influencing outcome and imply that the nature of peptide presentation, heavily affected by these amino acids and others, is key in terms of control. Prior population studies indicated an association between HLA-B alleles and control (28), but they did not define whether this was due to LD with something outside the HLA or whether this might be due to killer cell immunoglobulin-like receptors (KIR) or another receptor associated with HLA class I. Likewise, GWAS analyses have indicated specific SNPs associated with control but not the actual genetic determinants. These data thus extend prior studies by showing that the dominant impact of HLA on disease outcome is due to amino acid residues in the peptide-binding region (33).
Of the amino acids identified, position 97 in HLA-B was the most significant, and much more significant than any single classical HLA allele, including B*57:01. Positions 67 and 70 also showed very strong associations with level of HIV control. However, it remains to be determined whether positions such as 67, 70, and 97 are showing up with the most significance because of particular importance in peptide binding leading to induction of particularly effective T cell responses, or through some other mechanism. Each of these three positions resides in the peptide-binding groove. Position 97 forms part of the C and E pockets, and 67 and 70 form part of the B pocket, which are involved in anchoring the bound peptide into the groove. Other amino acids within the groove approached statistical significance in this relatively small cohort by genetic-association-study standards. Position 97 is among the most polymorphic of all positions in the peptide-binding groove and is also a position where variation is predicted to have a large impact on the conformation of the peptide–HLA complex (33), suggesting that the tertiary structure of this complex may be critical for induction of effective immune responses.
The data indicate that the nature of the peptide–HLA-B interaction and an independent influence of HLA-C are the major genetic determinants of HIV control. Important to note is that the entire HLA-B signal points to the peptide-binding groove, but the data should not be interpreted to mean that the amino acids that reached significance are the only important ones. Approximately 30 amino acids make peptide contact; it is likely more are involved and might reach significance with greater numbers of subjects. However, three amino acids at positions 97, 67, and 70 showed the most significant association with disease outcome. Different amino acids at these positions (position 97 has six, position 70 has four, and position 67 has five different amino acids across all HLA-B alleles) mark specific risk and protective HLA-B alleles in terms of HIV control. Permutation analyses where amino acid sequences were randomly shuffled indicated that positions 67, 70, and 97 are very unlikely to show such an effect by chance alone. Nevertheless, this type of analysis must be interpreted with caution because there are many sites within the groove that contribute to peptide binding and the LD among these variable sites is strong, such that no single polymorphic amino acid in the groove can be considered as an independent entity with regard to peptide binding.
A simple explanation for the finding that the nature of the peptide–HLA-B interaction is influential is that the specific peptides presented influence HIV control. The reason for the strong associations between these positions and control of HIV could be that multiple alleles marked by certain amino acids at these sites bind peptides that lead to strong control of HIV viral load, perhaps due to high sequence conservation preventing immune escape or due to targeting of vulnerable regions of the virus where mutations lead to fitness loss. Likewise, multiple alleles marked by alternative amino acids at these same sites could bind HIV peptides that lead to very poor control. Alternatively, it may be that the precise conformation of the peptide within the groove influences the efficacy of the CTLs that target these epitopes, perhaps due to differences in T cell receptor engagement or triggering. The individual amino acids are not causative on their own. What is causative is likely their overall impact on shaping the peptide-binding groove and, like many other polymorphic sites within the groove, determining which peptide will bind that particular HLA allotype, as well as the subsequent interaction with TCR.
Position 97, the most significant in this analysis, may have one of the strongest impacts on the conformation of the groove (33), and its extreme polymorphism (six amino acids in Europeans) suggests that this position may be of particular importance in generating the diverse repertoire of peptides that bind the various HLA-B allotypes. Still, this is difficult to test definitively because there is no set of HLA-B allotypes that are identical at all positions apart from the six amino acids known to occupy position 97. The impact of this position is suggested by the contrast between the protective allele B*5801 and the risk allele B*5802, which differ only at positions 94, 95, and 97 yet are associated with opposite extremes of outcome (34). Clearly, many additional amino acids lining the groove are involved in interactions with the viral peptide to a greater or lesser extent, and indeed, as mentioned, a number of other amino acids within the peptide-binding groove approach statistical significance. Although statistical analysis shows that no other amino acid sites are nearly as significant as 67, 70, and 97, functional data are needed to answer definitively whether and why these three positions are more influential than others in determining the peptide repertoire or HLA-peptide interaction with effector cells.
An alternative explanation for the exceptionally strong association between single amino acids and HIV control involves a novel mechanism of HLA function that is dependent on a single polymorphic amino acid position such as position 97. One well-defined example of such a situation is the dimorphic position 80 of the HLA-C molecule: a lysine at this position promotes interaction with the natural killer cell receptor KIR2DL1, but asparagine at this position does not allow the interaction (35). Position 97 is among the most polymorphic amino acid sites within the HLA-B molecule. Each of the HLA-B alleles shown to associate significantly with HIV control or lack of control has distinct amino acids at position 97, some with very different properties: B*57, valine, nonpolar; B*27:05, asparagine, polar/uncharged; B*14, tryptophan, aromatic; B*52, threonine, polar/uncharged; B*07, serine, polar/uncharged; B*35, arginine, positively charged. If position 97 were itself causative in determining HIV control through a previously undiscovered role in HLA-B function that is distinct from its role in the nature of peptide binding, then amino acids with similar properties might be expected to distinguish the protective alleles from the risk alleles. However, the protective alleles are marked by amino acids that range from polar to nonpolar to aromatic in their properties, and the two risk alleles are either polar or positively charged. It is difficult, therefore, to conceive of a novel mechanism of HLA-B function against HIV that is dependent on variation at position 97 alone, or even in some combination with only one or two other positions, other than through an impact on peptide binding. Rather, it seems likely that positions 97, 70, and 67 contribute to conferring significant protection or susceptibility due to the distinct sets of HIV peptides presented or the nature of their presentation, but each of these positions, along with many others, participates in determining which peptides will bind.
INTERPRETING THE HLA-C ASSOCIATION: BEYOND GWAS TO MICRO-RNA
In addition to the HLA-B associations, the other major genetic association identified in studies of genetic control of HIV is a polymorphism in HLA-C. The HLA-C locus has relatively limited polymorphism (36), lower expression on the cell surface (37, 38), and more extensive ligand-receptor interactions with KIR (39) than the other two classical class I loci, HLA-B and HLA-A. In large part, evidence to directly implicate HLA-C variation as a key determinant in the level of control of HIV has been lacking, whereas the importance of the HLA-B locus has been well appreciated (28). HLA-C molecules, however, have the unique advantage of restricting HIV epitopes without succumbing to HIV nef-mediated downregulation associated with HLA-A and HLA-B (40), which results in lack of recognition of infected cells by CTLs restricted by these alleles (41). Relative to HLA-A and -B, properly folded HLA-C molecules are expressed at a low level on the cell surface (37, 38, 42-44), partly because of their poor assembly with β2 microglobulin (42, 44) and retention in the endoplasmic reticulum where they are degraded to some extent (43). The cytoplasmic tail of HLA-C contains an internalization and lysosomal targeting signal that further regulates its surface expression (45). Given the many distinctions between HLA-C and the two other classical class I loci, it would not be surprising if the mechanisms by which HLA-C provides antiviral protection were distinct from those conferred by HLA-B.
The genetic association linking HLA-C to HIV control in the GWAS is due to a SNP located 35 kb upstream of the HLA-C gene (rs9264942), termed −35, which was first identified in viral set-point control following acute infection (10) and subsequently confirmed using a different definition of phenotype of HIV control in the International HIV Controllers Study (46). In both studies, −35C associated with protection and −35T associated with susceptibility to the outcomes tested. The −35 genotype also correlated with levels of HLA-C mRNA transcripts (47) and cell surface expression (46). These data suggest that high levels of HLA-C expression might confer an advantage against the virus, a potentially novel effect of an HLA gene that does not involve coding-region variation.
Understanding how the −35 SNP might be influencing HLA-C expression has been an important challenge. This SNP is not located in any canonical transcription factor binding site that might alter expression levels, and there are no polymorphisms in the cis-acting regulatory elements in the promoter region of HLA-C alleles (48) that are in LD with the −35 SNP or that show better association with viral-load control (46). Thus, the −35 SNP is probably not the causal variant for differential HLA-C expression but rather may be marking by LD another polymorphism that directly affects levels of HLA-C (46). Furthermore, Corrah et al. (49) showed that in their sample, differences in HLA-C cell surface expression levels as a function of −35 genotypes could be attributed to the low expression of the HLA-C*07 lineage of subtypes, which are located on a −35T haplotype. They concluded that the associations between −35 genotype and HIV outcomes reported previously (10, 46) were due to specific HLA-B and HLA-C alleles that are in significant LD with the −35 variant. The multiple studies addressing the effects of −35 genotype raise two questions: Is the -35 genotype marking through LD another variant(s) that directly affects expression levels of HLA-C, and is the association of −35 with HIV control attributable to levels of HLA-C expression or particular HLA-B and -C alleles that are in LD with −35?
A recent scan of variation in the 3′ untranslated region (UTR) of HLA-C identified quite extensive sequence polymorphism (7), including that at a binding site for the microRNA miR-148a (9) (Figure 4). MicroRNAs are a class of endogenous non-protein-coding RNAs composed of ~22 nucleotides that are estimated to regulate at least 30% of all genes in animals (50) by binding to specific sites in the 3′ UTR of protein-coding mRNAs, resulting in post-transcriptional destabilization of target mRNAs and, to a lesser extent, translational repression (51-54). A single miRNA may regulate hundreds of genes (50, 55-57), and their function has broad, diverse effects on normal cell function (58-60) and human disease (61-63).
Figure 4.
Alignment of full-length 3′ untranslated region (UTR) sequences of the common HLA-C alleles. Identical nucleotides are shown as dots, and deletions are indicated by hyphens. The miR-148 binding site is located within the blue boxed segment, where those alleles with an insertion at position 263 bind miR-148a and are downregulated, but those with a deletion at this position escape microRNA binding and downregulation.
Variation in the miR-148a binding site in the HLA-C 3′ UTR includes an insertion/deletion polymorphism termed 263I/D, which is in strong LD with the −35 variant in Caucasians. This variant was shown to affect levels of expression of a reporter construct, as well as endogenous HLA-C cell surface levels, and accounts in large part for the varied levels of HLA-C expression on the cell surface that was reported previously to associate with the −35 variant (9). These data add another layer of diversity to the HLA-C locus beyond its codingregion variation in that polymorphism in the 3′ UTR significantly affects differential expression of the various HLA-C allotypes, which may be important in disease pathogenesis.
Although miR-148a directly affects the level of HLA-C expression, it is unlikely to account completely for the varied expression of HLA-C. Previous data have shown a continuous gradient in expression levels of different HLA-C allotypes (46) rather than the bimodal expression pattern that would be expected if miR-148a regulation were the sole mechanism determining expression levels. Therefore, additional cis-acting factors may also affect HLA-C expression in an allotype-specific manner, or trans-acting factors unlinked to the HLA-C locus might affect expression levels in a manner that is independent of HLA-C allotype, leading to some degree of variation in expression levels of a given HLA-C allotype. Nevertheless, variation in the miR-148a-binding site, which is in very strong LD with −35, probably accounts for the majority of the previous association between −35 and HLA-C expression. Given the strong LD between −35 and 263D/I in the miR-148a-binding site of HLA-C, it was not surprising that the latter associated significantly with control of HIV, just as −35 does. The question remains whether levels of HLA-C have a direct influence on control of HIV or whether the effect observed can be attributed completely to individual alleles of HLA-B and HLA-C, such as B*57, B*27, and C*07, as suggested previously (49). To address this possibility from a genetic standpoint, all HLA class I alleles with frequencies of 5% or greater (n = 63), along with the 3′ UTR variant, were considered in a stepwise multivariate analysis. Only six of the 64 variables remained in the model and showed independent effects: B*57:01, B*57:03, B*58:01, B*27:05, C*14:01, and 263D/I. These data suggest that expression levels of HLA-C, as determined by miR-148a binding, have an effect on HIV control that is independent of individual HLA alleles, but functional data will be necessary to address this question definitively. It may prove difficult to identify the functional assays necessary to address the putative mechanism appropriately, as it could involve T cells, natural killer cells, dendritic cells, or other cell types that recognize HLA-C on an infected target cell.
CONCLUSIONS AND REMAINING QUESTIONS
The importance of the HLA class I loci in determining outcome in HIV infection has been known since the first HIV cohorts were genotyped and long before the agnostic genomewide scans were developed and employed. GWAS have added important findings including the primary importance of chromosome 6, and the HLA class I region in particular, in determining HIV outcome relative to the rest of the genome. Through more detailed sequencing of the HLA region, we have now moved beyond the GWAS results to show the central importance of the viral peptide–HLA-B interaction in disease outcome and to show that the levels of HLA class I molecules may also participate in antiviral activity.
Understanding the role of specific positions within the peptide-binding groove of HLA-B, as well as the impact of HLA expression levels, on HIV control clearly requires further investigation. It will be important to determine whether sites such as 67, 70, and 97 have functional characteristics that go beyond their role in peptide binding, but this seems unlikely. The association with HIV outcome may occur because these amino acid positions are particularly important along with others either by determining which peptide sits in the groove or by affecting the conformation of the peptide–HLA complex resulting in altered CD8 T cell function. One could also argue that position 97, for example, is no more important than any other polymorphic site lining the peptide-binding region but is simply a good marker of protective/risk alleles, in part due to its high level of polymorphism. Actual functional studies will be required to further define the mechanistic basis for these observations. Additional functional data will also be essential to definitively establish a role of high HLA-C expression levels in successful defense against HIV, and the genetic data call for a thorough investigation of differential expression of allotypes at all other HLA class I and II loci and the role their expression levels may have on human disease in general.
Some GWAS have identified variants outside of the MHC that may affect outcome in HIV disease, but unlike the HLA class I–region variants, none of these show a consistent effect across GWAS reports, and most failed to replicate in the International HIV Controllers Study, which was corrected for population differences. Eventually, it will be necessary to consider synergistic effects of variants throughout the genome, which may explain some of the associations outside of the MHC in particular GWAS. It is also necessary to consider epistatic effects of HLA variation with other loci, especially within the KIR genes, which encode highly polymorphic molecules that bind HLA class I and thereby regulate natural killer cell and CTL activity.
Investigations of the role of host genetic polymorphisms in HIV disease have generated a plethora of additional questions, but they have also refined our questions in a manner that moves us a bit closer to an educated, fact-based truth and a bit further from empirical trawling.
ACKNOWLEDGMENTS
The International HIV Controllers Study was initially made possible with a grant from the Mark and Lisa Schwartz Foundation and extended with support from the Bill and Melinda Gates Foundation. This project has been funded in part with federal funds from the National Institute for Allergy and Infectious Disease (AI30914) to B.D.W. and from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E, to M.C. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. We thank Drs. Arman Bashirova, Smita Kulkarni, Richard Apps, and Paul deBakker for helpful discussions.
Footnotes
DISCLOSURE STATEMENT
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
LITERATURE CITED
- 1.van Griensven GJ, Hessol NA, Koblin BA, et al. Epidemiology of human immunodeficiency virus type 1 infection among homosexual men participating in hepatitis B vaccine trials in Amsterdam, New York City, and San Francisco, 1978-1990. Am. J. Epidemiol. 1993;137:909–15. doi: 10.1093/oxfordjournals.aje.a116752. [DOI] [PubMed] [Google Scholar]
- 2.Buchbinder SP, Katz MH, Hessol NA, et al. Long-term HIV-1 infection without immunologic progression. AIDS. 1994;8:1123–28. doi: 10.1097/00002030-199408000-00014. [DOI] [PubMed] [Google Scholar]
- 3.Deeks SG, Walker BD. Human immunodeficiency virus controllers: mechanisms of durable virus control in the absence of antiretroviral therapy. Immunity. 2007;27:406–16. doi: 10.1016/j.immuni.2007.08.010. [DOI] [PubMed] [Google Scholar]
- 4.Lyles RH, Munoz A, Yamashita TE, et al. Natural history of human immunodeficiency virus type 1 viremia after seroconversion and proximal to AIDS in a large cohort of homosexual men. Multicenter AIDS Cohort Study. J. Infect. Dis. 2000;181:872–80. doi: 10.1086/315339. [DOI] [PubMed] [Google Scholar]
- 5.Pereyra F, Jia X, McLaren PJ, et al. The major genetic determinants of HIV-1 control affect HLA class I peptide presentation. Science. 2010;330:1551–57. doi: 10.1126/science.1195271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lander ES, Linton LM, Birren B, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. doi: 10.1038/35057062. [DOI] [PubMed] [Google Scholar]
- 7.Sabeti PC, Varilly P, Fry B, et al. Genome-wide detection and characterization of positive selection in human populations. Nature. 2007;449:913–18. doi: 10.1038/nature06250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.The International HapMap Project. Nature. 2003;426:789–96. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 9.Kulkarni S, Savan R, Qi Y, et al. Differential microRNA regulation of HLA-C expression and its association with HIV control. Nature. 2011;472:495–98. doi: 10.1038/nature09914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fellay J, Shianna KV, Ge D, et al. A whole-genome association study of major determinants for host control of HIV-1. Science. 2007;317:944–47. doi: 10.1126/science.1143767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fellay J, Ge D, Shianna KV, et al. Common genetic variation and the control of HIV-1 in humans. PLoS Genet. 2009;5:e1000791. doi: 10.1371/journal.pgen.1000791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dalmasso C, Carpentier W, Meyer L, et al. Distinct genetic loci control plasma HIV-RNA and cellular HIV-DNA levels in HIV-1 infection: the ANRS Genome Wide Association 01 study. PloS one. 2008;3:e3907. doi: 10.1371/journal.pone.0003907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pelak K, Goldstein DB, Walley NM, et al. Host determinants of HIV-1 control in African Americans. J. Infect. Dis. 2010;201:1141–49. doi: 10.1086/651382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Quinn TC, Wawer MJ, Sewankambo N, et al. Viral load and heterosexual transmission of human immunodeficiency virus type 1. Rakai Project Study Group. N. Engl. J. Med. 2000;342:921–29. doi: 10.1056/NEJM200003303421303. [DOI] [PubMed] [Google Scholar]
- 15.Harrer T, Harrer E, Kalams SA, et al. Cytotoxic T lymphocytes in asymptomatic long-term nonprogressing HIV-1 infection. Breadth and specificity of the response and relation to in vivo viral quasispecies in a person with prolonged infection and low viral load. J. Immunol. 1996;156:2616–23. [PubMed] [Google Scholar]
- 16.Migueles SA, Sabbaghian MS, Shupert WL, et al. HLA B*5701 is highly associated with restriction of virus replication in a subgroup of HIV-infected long term nonprogressors. Proc. Natl. Acad. Sci. USA. 2000;97:2709–14. doi: 10.1073/pnas.050567397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Emu B, Sinclair E, Favre D, et al. Phenotypic, functional, and kinetic parameters associated with apparent T-cell control of human immunodeficiency virus replication in individuals with and without antiretroviral treatment. J. Virol. 2005;79:14169–78. doi: 10.1128/JVI.79.22.14169-14178.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Navis M, Schellens I, van Baarle D, et al. Viral replication capacity as a correlate of HLA B57/B5801-associated nonprogressive HIV-1 infection. J. Immunol. 2007;179:3133–43. doi: 10.4049/jimmunol.179.5.3133. [DOI] [PubMed] [Google Scholar]
- 19.Martin MP, Gao X, Lee JH, et al. Epistatic interaction between KIR3DS1 and HLA-B delays the progression to AIDS. Nat. Genet. 2002;31:429–34. doi: 10.1038/ng934. [DOI] [PubMed] [Google Scholar]
- 20.Lichterfeld M, Kavanagh DG, Williams KL, et al. A viral CTL escape mutation leading to immunoglobulin-like transcript 4-mediated functional inhibition of myelomonocytic cells. J. Exp. Med. 2007;204:2813–24. doi: 10.1084/jem.20061865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pereyra F, Addo MM, Kaufmann DE, et al. Genetic and immunologic heterogeneity among persons who control HIV infection in the absence of therapy. J. Infect. Dis. 2008;197:563–71. doi: 10.1086/526786. [DOI] [PubMed] [Google Scholar]
- 22.Limou S, Le Clerc S, Coulonges CD, et al. Genomewide association study of an AIDS nonprogression cohort emphasizes the role played by HLA genes (ANRS Genomewide Association Study 02) J. Infect. Dis. 2009;199:419–26. doi: 10.1086/596067. [DOI] [PubMed] [Google Scholar]
- 23.Brown WM, Pierce J, Hilner JE, et al. Overview of the MHC fine mapping data. Diab. Obes. Metab. 2009;11(Suppl. 1):2–7. doi: 10.1111/j.1463-1326.2008.00997.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carrington M, Nelson GW, Martin MP, et al. HLA and HIV-1: heterozygote advantage and B*35-Cw*04 disadvantage. Science. 1999;283:1748–52. doi: 10.1126/science.283.5408.1748. [DOI] [PubMed] [Google Scholar]
- 25.Carrington M, O’Brien SJ. The influence of HLA genotype on AIDS. Annu. Rev. Med. 2003;54:535–51. doi: 10.1146/annurev.med.54.101601.152346. [DOI] [PubMed] [Google Scholar]
- 26.Costello C, Tang J, Rivers C, et al. HLA-B*5703 independently associated with slower HIV-1 disease progression in Rwandan women. AIDS. 1999;13:1990–91. doi: 10.1097/00002030-199910010-00031. [DOI] [PubMed] [Google Scholar]
- 27.Flores-Villanueva PO, Yunis EJ, Delgado JC, et al. Control of HIV-1 viremia and protection from AIDS are associated with HLA-Bw4 homozygosity. Proc. Natl. Acad. Sci. USA. 2001;98:5140–45. doi: 10.1073/pnas.071548198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kiepiela P, Leslie AJ, Honeyborne I, et al. Dominant influence of HLA-B in mediating the potential co-evolution of HIV and HLA. Nature. 2004;432:769–75. doi: 10.1038/nature03113. [DOI] [PubMed] [Google Scholar]
- 29.Klein MR, Keet IP, D’Amaro J, et al. Associations between HLA frequencies and pathogenic features of human immunodeficiency virus type 1 infection in seroconverters from the Amsterdam cohort of homosexual men. J. Infect. Dis. 1994;169:1244–49. doi: 10.1093/infdis/169.6.1244. [DOI] [PubMed] [Google Scholar]
- 30.Klein J, Satta Y, O’Huigin C, Takahata N. The molecular descent of the major histocompatibility complex. Annu. Rev. Immunol. 1993;11:269–95. doi: 10.1146/annurev.iy.11.040193.001413. [DOI] [PubMed] [Google Scholar]
- 31.Hughes AL, Nei M. Pattern of nucleotide substitution at major histocompatibility complex class I loci reveals overdominant selection. Nature. 1988;335:167–70. doi: 10.1038/335167a0. [DOI] [PubMed] [Google Scholar]
- 32.Bjorkman PJ, Saper MA, Samraoui B, et al. The foreign antigen binding site and T cell recognition regions of class I histocompatibility antigens. Nature. 1987;329:512–18. doi: 10.1038/329512a0. [DOI] [PubMed] [Google Scholar]
- 33.Fagerberg T, Cerottini JC, Michielin O. Structural prediction of peptides bound to MHC class I. J. Mol. Biol. 2006;356:521–46. doi: 10.1016/j.jmb.2005.11.059. [DOI] [PubMed] [Google Scholar]
- 34.Ngumbela KC, Day CL, Mncube Z, et al. Targeting of a CD8 T cell env epitope presented by HLA-B*5802 is associated with markers of HIV disease progression and lack of selection pressure. AIDS Res. Hum. Retrovir. 2008;24:72–82. doi: 10.1089/aid.2007.0124. [DOI] [PubMed] [Google Scholar]
- 35.Biassoni R, Falco M, Cambiaggi A, et al. Amino acid substitutions can influence the natural killer (NK)-mediated recognition of HLA-C molecules. Role of serine-77 and lysine-80 in the target cell protection from lysis mediated by “group 2” or “group 1” NK clones. J. Exp. Med. 1995;182:605–9. doi: 10.1084/jem.182.2.605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zemmour J, Parham P. Distinctive polymorphism at the HLA-C locus: implications for the expression of HLA-C. J. Exp. Med. 1992;176:937–50. doi: 10.1084/jem.176.4.937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.McCutcheon JA, Gumperz J, Smith KD, et al. Low HLA-C expression at cell surfaces correlates with increased turnover of heavy chain mRNA. J. Exp. Med. 1995;181:2085–95. doi: 10.1084/jem.181.6.2085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Snary D, Barnstable CJ, Bodmer WF, Crumpton MJ. Molecular structure of human histocompatibility antigens: the HLA-C series. Eur. J. Immunol. 1977;7:580–85. doi: 10.1002/eji.1830070816. [DOI] [PubMed] [Google Scholar]
- 39.Bashirova AA, Martin MP, McVicar DW, Carrington M. The killer immunoglobulin-like receptor gene cluster: tuning the genome for defense. Annu. Rev. Genom. Hum. Genet. 2006;7:277–300. doi: 10.1146/annurev.genom.7.080505.115726. [DOI] [PubMed] [Google Scholar]
- 40.Cohen GB, Gandhi RT, Davis DM, Mandelboim O, Chen BK, et al. The selective downregulation of class I major histocompatibility complex proteins by HIV-1 protects HIV-infected cells from NK cells. Immunity. 1999;10:661–71. doi: 10.1016/s1074-7613(00)80065-5. [DOI] [PubMed] [Google Scholar]
- 41.Yang OO, Nguyen PT, Kalams SA, Dorfman T, Gottlinger HG, et al. Nef-mediated resistance of human immunodeficiency virus type 1 to antiviral cytotoxic T lymphocytes. Journal of virology. 2002;76:1626–31. doi: 10.1128/JVI.76.4.1626-1631.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Neefjes JJ, Ploegh HL. Allele and locus-specific differences in cell surface expression and the association of HLA class I heavy chain with beta 2-microglobulin: differential effects of inhibition of glycosylation on class I subunit association. Eur. J. Immunol. 1988;18:801–10. doi: 10.1002/eji.1830180522. [DOI] [PubMed] [Google Scholar]
- 43.Neisig A, Melief CJ, Neefjes J. Reduced cell surface expression of HLA-C molecules correlates with restricted peptide binding and stable TAP interaction. J Immunol. 1998;160:171–9. [PubMed] [Google Scholar]
- 44.Setini A, Beretta A, De Santis C, Meneveri R, Martayan A, et al. Distinctive features of the alpha 1-domain alpha helix of HLA-C heavy chains free of beta 2-microglobulin. Hum. Immunol. 1996;46:69–81. doi: 10.1016/0198-8859(96)00011-0. [DOI] [PubMed] [Google Scholar]
- 45.Schaefer MR, Williams M, Kulpa DA, Blakely PK, Yaffee AQ, Collins KL. A novel trafficking signal within the HLA-C cytoplasmic tail allows regulated expression upon differentiation of macrophages. J Immunol. 2008;180:7804–17. doi: 10.4049/jimmunol.180.12.7804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Thomas R, Apps R, Qi Y, Gao X, Male V, et al. HLA-C cell surface expression and control of HIV/AIDS correlate with a variant upstream of HLA-C. Nat. Genet. 2009;41:1290–4. doi: 10.1038/ng.486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Stranger BE, Forrest MS, Clark AG, Minichiello MJ, Deutsch S, et al. Genome-wide associations of gene expression variation in humans. PLoS Genet. 2005;1 doi: 10.1371/journal.pgen.0010078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Yao Z, Volgger A, Scholz S, Albert E. Polymorphism of the HLA-C promotor region. Immunogenetics. 1997;45:428–31. doi: 10.1007/s002510050225. [DOI] [PubMed] [Google Scholar]
- 49.Corrah TW, Goonetilleke N, Kopycinski J, Deeks SG, Cohen MS, et al. Reappraisal of the relationship between the HIV-1-protective single-nucleotide polymorphism 35 kilobases upstream of the HLA-C gene and surface HLA-C expression. J Virol. 2011;85:3367–74. doi: 10.1128/JVI.02276-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 51.Giraldez AJ, Mishima Y, Rihel J, Grocock RJ, Van Dongen S, et al. Zebrafish MiR-430 promotes deadenylation and clearance of maternal mRNAs. Science. 2006;312:75–9. doi: 10.1126/science.1122689. [DOI] [PubMed] [Google Scholar]
- 52.Lim LP, Lau NC, Garrett-Engele P, Grimson A, Schelter JM, et al. Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature. 2005;433:769–73. doi: 10.1038/nature03315. [DOI] [PubMed] [Google Scholar]
- 53.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc. Natl. Acad. Sci. USA. 2006;103:4034–9. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Guo H, Ingolia NT, Weissman JS, Bartel DP. Mammalian microRNAs predominantly act to decrease target mRNA levels. Nature. 2010;466:835–40. doi: 10.1038/nature09267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Engels BM, Hutvagner G. Principles and effects of microRNA-mediated post-transcriptional gene regulation. Oncogene. 2006;25:6163–9. doi: 10.1038/sj.onc.1209909. [DOI] [PubMed] [Google Scholar]
- 56.Griffiths-Jones S. The MicroRNA Registry. Nucleic Acids Res. 2004;32:D109–11. doi: 10.1093/nar/gkh023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Miranda KC, Huynh T, Tay Y, Ang YS, Tam WL, et al. A pattern-based method for the identification of MicroRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–17. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 58.Ambros V. The functions of animal microRNAs. Nature. 2004;431:350–5. doi: 10.1038/nature02871. [DOI] [PubMed] [Google Scholar]
- 59.Baltimore D, Boldin MP, O’Connell RM, Rao DS, Taganov KD. MicroRNAs: new regulators of immune cell development and function. Nat Immunol. 2008;9:839–45. doi: 10.1038/ni.f.209. [DOI] [PubMed] [Google Scholar]
- 60.Neilson JR, Zheng GX, Burge CB, Sharp PA. Dynamic regulation of miRNA expression in ordered stages of cellular development. Genes Dev. 2007;21:578–89. doi: 10.1101/gad.1522907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Dai R, Ahmed SA. MicroRNA, a new paradigm for understanding immunoregulation, inflammation, and autoimmune diseases. Transl Res. 2011;157:163–79. doi: 10.1016/j.trsl.2011.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Di Leva G, Croce CM. Roles of small RNAs in tumor formation. Trends Mol Med. 2010;16:257–67. doi: 10.1016/j.molmed.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ryan BM, Robles AI, Harris CC. Genetic variation in microRNA networks: the implications for cancer research. Nat Rev Cancer. 2010;10:389–402. doi: 10.1038/nrc2867. [DOI] [PMC free article] [PubMed] [Google Scholar]