Abstract
Accurately identifying accessible sites in RNA is a critical prerequisite for optimising the cleavage efficiency of hammerhead ribozymes and other small nucleozymes. Here we describe a simple RNase H-based procedure to rapidly identify hammerhead ribozyme-accessible sites in gene length RNAs. Twelve semi-randomised RNA–DNA–RNA chimeric oligonucleotide probes, known as ‘gapmers’, were used to direct RNase H cleavage of transcripts with the specificity expected for hammerhead ribozymes, i.e. after NUH sites (where H is A, C or U). Cleavage sites were identified simply by the mobility of RNase H cleavage products relative to RNA markers in denaturing polyacrylamide gels. Sites were identified in transcripts encoding human interleukin-2 and platelet-derived growth factor. Thirteen minimised hammerhead ribozymes, miniribozymes (Mrz), were synthesised and in vitro cleavage efficiency (37°C, pH 7.6 and 1 mM MgCl2) at each site was analysed. Of the 13 Mrz, five were highly effective, demonstrating good initial rate constants and extents of cleavage. The speed and accuracy of this method commends its use in screening for hammerhead-accessible sites.
INTRODUCTION
Hammerhead ribozymes can act as sequence-specific ribonucleases to regulate gene expression and may have a variety of investigative and therapeutic applications (1,2). Conventional hammerhead ribozymes cleave after NUH sites, where N is any nucleotide and H is any nucleotide except guanosine (3–6). A randomly chosen transcript will therefore contain a hammerhead target site, on average, once in every 5.3 nt. However, RNA folds into complex structures that can prevent ribozymes or antisense molecules from binding and cleaving. Target sites that are obscured and blocked in this manner are described as inaccessible. The location of accessible sites is generally not known a priori. Given the gradual realisation over the last 10 years or so of the importance of target site selection to the efficiency of antisense and ribozyme strategies, it is not surprising that considerable effort has been expended trying to find a workable solution to this problem. Early studies tended to randomly select target sites (7,8) or try to make educated guesses, e.g. near the initiation codon, splice sites or the polyadenylation signal (9). Analysis of the effectiveness of these strategies is complicated by the perception that many unsuccessful studies remain unpublished (10). A number of studies (11,12) have used RNA folding algorithms to identify sites that were not occluded by secondary structure, however, target sites predicted to lie in single-stranded regions using RNA folding programs (13,14) did not necessarily correlate with experimental accessibility (11). Even when well-characterised RNA structures are considered, the relationship between accessibility and structure is both subtle and complex (15). Structure-dependent nucleases have been used to find accessible regions, but the correlation between apparently single-stranded regions and activity of ribozymes targeting those regions was not great (11). Trial and error methods have been successful in identifying effective molecules, but this approach requires the user to construct and test up to hundreds of separate oligonucleotides (16,17). Ribozymes with the substrate-binding regions randomised have been used to identify accessible sites both in vitro and in vivo. Some successes have been reported with these strategies (18–21), but the level of cleavage at any given site is often very low and distinguishing genuine cleavage sites from random degradation can be difficult. One way of reducing the level of degeneracy and the consequent low level of cleavage is to target only those sequences that exist within the target RNA. This can be achieved by inserting the hammerhead core sequence into segments of the appropriate antisense nucleic acid (22,23). In this way the complexity of, say, a ribozyme with two randomised arms of 8 nt each (which has a complexity of 4 × 109 members) is reduced to a complexity approximately equal to the length of the target RNA (e.g. 103 members). Again, this approach is technically difficult and is not widely used. Randomised hairpin ribozyme libraries have been used in vivo with phenotypic selection (20); in that communication a novel gene was discovered. Because this method when used in vivo targets all expressed genes, it may be very difficult to interpret any results obtained. A common feature of the randomised ribozyme approaches is that they are technically difficult to perform and replicate. Thus, despite the conceptual attractiveness of such approaches, they are not widely used. Therefore, identifying optimal target sites continues to remain a major obstacle to achieving both gene-suppressive ribozyme and antisense activity (24,25).
RNase H is a ubiquitous ribonuclease that cleaves the RNA portion of RNA–DNA duplexes. It has been used in conjunction with both defined sequence (26,27) and randomised oligonucleotides (15,16,28,29). Modifications to this basic approach have included ‘gapmer oligonucleotides’ (30), where windows or ‘gaps’ of DNA are inserted into an otherwise normal or modified RNA oligonucleotide. This modification increases oligonucleotide stability in vivo and the avidity of the interaction of the probe with the target, so that shorter probes can be used effectively. The performance of degenerate probes in such applications is often improved by specifying one or more nucleotides (29). This paper describes a series of experiments in which 3 nt were specified in the centre of a series of 12 RNA–DNA–RNA gapmers. The conserved nucleotides were constrained to only bind to the hammerhead target sites (NUH). Compared with other strategies, the reduction in complexity of the probes was expected to increase the sensitivity and simplify identification of cleavage sites.
MATERIALS AND METHODS
Preparation of oligonucleotides
Oligonucleotides were synthesised on a Perkin Elmer Applied Biosystems 394 DNA synthesiser (Foster City, CA). PE Applied Biosystems and Glen Research (Sterling, VA) supplied DNA and RNA phosphoramidite monomers, respectively. Phosphorothioate linkages were generated using Beaucage reagent (Glen Research). PE Applied Biosystems supplied other ancillary reagents. All oligonucleotides contained a 3′-deoxyribonucleotide for convenience of synthesis. Randomised 3′-nucleotides were prepared from equimolar mixtures of dG-, dA-, dT- and dC-derivatised polystyrene resin. Oligonucleotides, synthesised at 40 nM scale, were deprotected in 15 M NH4OH/ethanol (3:1) at 55°C for 8–10 h, prior to drying in a vacuum centrifuge. Silyl protecting groups were removed in 100 µl of triethylamine trihydrofluoride (Aldrich, Milwaukee, WI) for 24 h at room temperature prior to precipitation of the oligonucleotide by addition of 1 ml of 1-butanol. Oligonucleotides longer than 11mers were gel purified using 15% denaturing (7 M urea) polyacrylamide gel electrophoresis (PAGE). Nucleic acids were collected by NaOAc/ethanol precipitation and centrifugation. The homogeneity of all syntheses were verified by 5′-end-labelling an aliquot using T4 polynucleotide kinase (Roche Molecular Biochemicals, Germany) and [γ-32P]ATP (Geneworks, Australia), fractionating products by PAGE followed by ImageQuant (Molecular Dynamics, Sunnyvale CA) analysis.
Gapmer libraries
Twelve gapmer libraries, each targeting a specific NUH sequence, were designed. These gapmer libraries were 11 nt in length and of the general form 5′-NNNN(ndax)NpsNpsn-3′, where the bracketed nucleotides represent 1 of 12 separate DNA cassettes. Upper case letters represent ribonucleotides, lower case represent 2′ deoxyribonucleotides, d and x represent specific deoxyribonucleotides g, a or t and g, a, t or c, respectively, n and N represent randomised deoxyribonucleotides and ribonucleotides, respectively, and ps represents a phosphorothioate linkage. The sequences of the individual gapmers is given in Table 1.
Table 1. Sequence of NUH targeting gapmers.
| Target set |
NUH target |
Gapmer sequence (5′→3′) |
| GUH set | GUC | NNNNngacNpsNpsn |
| GUU | NNNNnaacNpsNpsn | |
| GUA | NNNNntacNpsNpsn | |
| AUH set | AUC | NNNNngatNpsNpsn |
| AUU | NNNNnaatNpsNpsn | |
| AUA | NNNNntatNpsNpsn | |
| UUH set | UUC | NNNNngaaNpsNpsn |
| UUU | NNNNnaaaNpsNpsn | |
| UUA | NNNNntaaNpsNpsn | |
| CUH set | CUC | NNNNngagNpsNpsn |
| CUU | NNNNnaagNpsNpsn | |
| CUA | NNNNntagNpsNpsn |
Upper and lower case letters represent RNA and DNA nucleotides, respectively. ps indicates a phosphorothioate linkage intended to impart increased nuclease resistance. Within any set of gapmer libraries the DNA cassette varies by only one defined nucleotide.
Preparation of RNAs and size markers
A full-length interleukin-2 (IL-2) template DNA was prepared by PCR using conventional methods with primers 5′-CCG GAA TTC ATC ACT CTC TTT AAT CAC TAC TCA CAG TAA CCT CAA CTC CTG CCA CAA TGT-3′ and 5′-CGC GGA TCC TTT TTA TAT TTA TCA-3′ and the partial length IL-2 cDNA clone pTCGF-11 (31) as template. The resultant PCR product was digested with EcoRI and BamHI (sites represented by underlined nucleotides in the primers) and ligated into similarly digested pBluescribe (Stratagene, La Jolla, CA), creating plasmid pFL5/3IL-2. Digestion of pFL5/3IL-2 with BamHI followed by T7 RNA polymerase-directed transcription generated an 811 nt IL-2 sense transcript.
A platelet-derived growth factor (PDGF)-longA cDNA was subcloned from pmetPDGF-LA (32) into pBluescribe to create pBSPDGF-LA. That plasmid was digested with EcoRI, permitting T3 RNA polymerase-directed run-off transcription of a 707 nt PDGF sense transcript. Transcripts were prepared using Ampliscribe transcription kits (Epicentre Technologies). Internally labelled transcripts were prepared by decreasing the UTP in the transcription reaction to ∼1.5 mM and adding 50 µCi (3000 Ci/mmol) [α-32P]UTP (Amersham).
RNA size markers for determining the size of RNase H cleavage products included internally labelled hammerhead cleavage products and short run-off transcripts. The sizes of Mrz cleavage products are given with the NUH target site (equals the length of the 5′-product) in plain type and the corresponding 3′-product in parenthesised italic. The IL-2 transcript was cleaved with four miniribozymes (Mrz) and these cleavage sites and products were GUC-80(731), AUU-425(386), GUU-605(206) and AUU-616(195). These Mrz had the following sequences: GUC-80, CAAUGCAACUGAUGAGUUUUCGAAACAGGAGt; AUU-425, CACACAUGCUGAUGAGUUUUCGAAUGUUGUt; GUU-605, AGGUAGCACUGAUGAGUUUUCGAAACCAUACa; AUU-616, AUAGUUACCUGAUGAGUUUUCGAAAUAGGUAg Underlined sequences represent the substrate-binding arms of the Mrz. Run-off transcription initiated at the T7 promoter of XbaI and StuI-digested pFL5/3IL-2 yielded 291 and 553 nt markers, respectively. The PDGF transcript was cleaved with three Mrz and their cleavage sites and resultant cleavage products were CUU-40(667), CUU-204(503) and GUC-625(82). These Mrz had the following sequences: CUU-40, GGCAAGCCCUGAUGAGUUUUCGAAAGGUCCUc; CUU-204, GGUGUCCACUGAUGAGUUUUCGAAAGAAUCCt; GUC-625, UUUUACCUCUGAUGAGUUUUCGAAACUCCCUa. Run-off transcription initiated at the T3 promoter of AccI-digested pBS-PDGF-LA produced a 357 nt marker.
3′-End-labelling of in vitro transcripts
Labelling the 3′-termini of transcripts with [α-32P]cordycepin was found to be a rapid method for end-labelling to reproducibly high specific activity. The buffer contained 200 mM Tris–HCl (pH 7.6), 10 mM MgCl2, 12.5 mM MnCl2, 1.25 M NaCl and 250 µg/ml nuclease-free BSA (33). Transcript was heated in water at 80°C for 1 min and snap cooled on ice. 3′-OH termini were labelled at 37°C for 45 min in a reaction containing 3′-end-labelling buffer, 5 µl (50 µCi) [α-32P]cordycepin (5000 Ci/mmol; NEN, Boston, MA), 15 U yeast poly(A) polymerase (Pharmacia) and 50 pmol transcript. The reaction was terminated and the labelled transcript precipitated by addition of 1 vol of 7.5 M ammonium acetate, cooling to –20°C for ∼1 h, then centrifugation at 14 000 g for 15 min. Pelleted RNA was washed twice in 70% ethanol and redissolved in water to 1 pmol/µl. 3′-End-labelled transcripts were probed (typically) within 24 h of being prepared.
RNase H probing assay
Reaction mixes were assembled on ice and contained 1 pmol 3′-end-labelled transcript, 50 mM Tris–HCl (pH 7.6), 1 U Escherichia coli RNase H (Roche Molecular Biochemicals) and 10 mM MgCl2 in a total of 4 µl. Combined transcript, RNase H and reaction buffer were warmed to 37°C for 1 min whilst an aliquot of gapmer was denatured at 75°C for 1 min. Cleavage was initiated by addition of 100 pmol (1 µl) denatured (75°C) probe to the prewarmed (37°C) 4 µl mixture. This mixture was incubated at 37°C for 15 min before quenching in 5 µl of loading buffer containing 10 mM EDTA and 95% deionised formamide. Fifteen minutes was chosen because time course studies indicated rapid and complete RNase H cleavage activity within this time (results not shown). An aliquot of quenched probing reaction (typically 3 µl) was heated to 80°C for 1 min and fractionated by denaturing (7 M urea) PAGE (3.5 or 4%) in a 60 cm gel. Gels were dried and exposed to PhosphorImager plates and analysed using ImageQuant software (Molecular Dynamics). The mobility of RNase H cleavage products was determined relative to RNA size markers using Kodak 1D Image Analysis Software (Eastman Kodak, NY). As 3′-end-labelled cleavage products carry an extra nucleotide as a result of end-labelling, this nucleotide was subtracted from the size of the mapped cleavage product.
Ribozyme cleavage studies
Cleavage sites in transcripts are identified according to their distance from the 5′-end. A target site referred to as GUC-80 or AUC-293 is a GUC or AUC triplet 80 or 293 nt, respectively, from the 5′-end of the transcript. All ribozymes are Mrz in which helix II was replaced by a single G-C base pair linked by a four U loop (34). Mrz IL-2 GUA-39 is shown schematically in Figure 1. Their sequences were as follows. IL-2 Mrz: GUA-39, GUUGAGGUCUGAUGAGUUUUCGAAACUGUGAg; AUU-87, CUUAGUGCCUGAUGAGUUUUCGAAAUGCAAGa; CUA-124, ACUUGAAGCUGAUGAGUUUUCGAAAGGUGCAc; UUA-484, ACAAAAGGCUGAUGAGUUUUCGAAAAUCCAUc; UUU-489, CUUUGACACUGAUGAGUUUUCGAAAAGGUAAt; AUU-616, AUAGUUACCUGAUGAGUUUUCGAAAUAGGUAg. PDGF Mrz: CUU-40, GGCAAGCCCUGAUGAGUUUUCGAAAGGUCCUc; CUA-243, GCACAUGCUCUGAUGAGUUUUCGAAAGUGGCGUg; AUC-293, AGCUUCCUCCUGAUGAGUUUUCGAAAUGCUUCUc; GUC-354, GGUCGACCUCUGAUGAGUUUUCGAAACUCCGAGg; GUC-359, CGUGGGGUCCUGAUGAGUUUUCGAAACCUGACUc; GUC-370, AAGUUGGCGCUGAUGAGUUUUCGAAACGUGGGGt; GUC-530, CCUCACCUGCUGAUGAGUUUUCGAAACUUCUUUt. Sequences are shown 5′→3′, upper case letters are RNA, lower case are DNA and the underlined segments represent the substrate-binding arms. Mrz rather than standard hammerhead ribozymes were chosen for this analysis because, first, such hammerheads generally have increased ability to cleave transcripts (31) and, secondly, the presence of G(10.1)UUUUC(11.1) linkers (35) can impart enhanced cleavage activity, especially at 1 mM MgCl2 (36), a physiologically relevant Mg2+ ion concentration (37). Cleavage reactions were performed at 37°C using 10 µM Mrz and 0.1 µM IL-2 or PDGF transcript internally labelled with [α-32P]UTP in 50 mM Tris–HCl (pH 7.6). High concentrations of Mrz were used to reduce the likelihood that cleavage would be limited by ribozyme–substrate duplex stability. Prior heat denaturing or annealing of transcript and Mrz was not performed. Cleavage was initiated by addition of MgCl2 to 1 mM. Aliquots from cleavage reactions were quenched by addition of 2 vol of loading dye containing 95% formamide and 10 mM EDTA before transferring to ice. Cleavage reactions were followed for 360 min and products separated by denaturing PAGE prior to phosphorimaging and ImageQuant analysis. The amount of product formed as a percentage of the total product plus substrate is referred to as the per cent cleavage product. The cleavage reactions were largely biphasic, with an initial phase that fitted well to a first order process followed by a slower reaction that was approximated by a linear function. Details of the curve fitting and calculated constants are available as Supplementary Material.
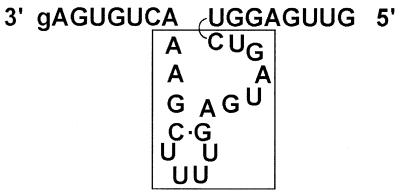
Figure 1.
Schematic representation of the Mrz used in this study. This example is of IL-2 GUA-39. The boxed section represents the conserved domain which is flanked by the two substrate-binding arms.
RESULTS
Gapmer design and function
Escherichia coli RNase H can be induced to cleave the RNA strand of an RNA–DNA heteroduplex when as few as four contiguous DNA nucleotides are hybridised to the RNA substrate (38). Under such circumstances RNase H cleaves the RNA strand between the third and fourth (counting 5′→3′) ribonucleotides, yielding 3′-hydroxyl and 5′-phosphate ends (38,39). A DNA tetramer is unlikely to form a stable duplex with any sequence of RNA at physiological temperatures. However, if that DNA tetramer is part of a longer largely RNA oligonucleotide a stable heteroduplex can form. In this study the DNA tetramer is contained within a chimeric 11mer. This length was selected as about the minimum required to generate sufficient duplex stability. The 3′-ends of the gapmers were protected with two phosphorothioate linkages as the same oligonucleotides were to be used in a further study that required 3′ exonuclease protection. Previous workers have described such chimeric oligonucleotides as ‘gapmers’ (30), referring to the ‘gap’ of DNA nucleotides within the otherwise RNA oligonucleotide. The novel feature of the gapmers described here is their targeting ability. Fixing the sequence of three of the nucleotides in the DNA window directs these gapmers to bind to sequences centred on specifically chosen complementary triplets. The hammerhead ribozyme cleaves after NUH sites. Therefore, we synthesised the 12 complementary populations of fixed-triplet gapmers with appropriately positioned DNA brackets to direct RNase H to cleave after NUH sites and thereby mimic the cleavage specificity of the hammerhead ribozyme.
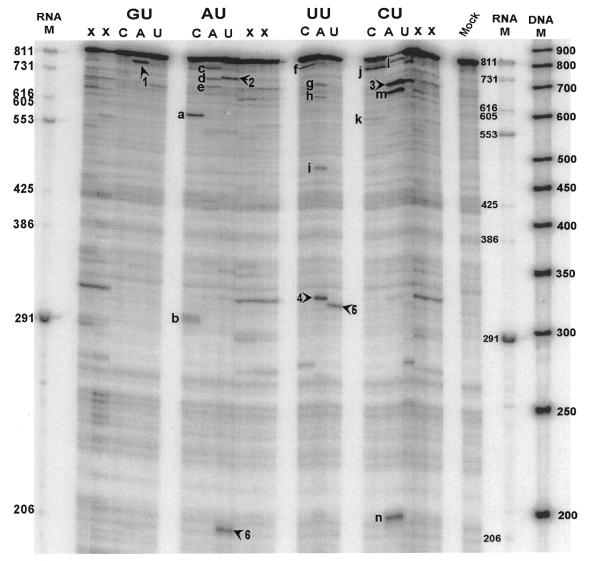
Mapped cleavage products were readily assigned to NUH triplets
Figure 2 shows that different gapmers facilitate cleavage by RNase H at different sites in the IL-2 transcript. We asked whether cleavage occurred at the expected positions, for example does the anti-GUA gapmer direct cleavage only after GUA sites? To answer this question the sites of cleavage were identified and compared with the sequence of the transcript. As transcripts were labelled at their 3′-ends, RNase H cleavage sites could be identified simply by determining the size of the cleavage products. Fragment lengths were determined by comparison with RNA size markers. Fragments were mapped back to the primary sequence (Table 2). Seventy-five per cent of the fragments mapped to within 7 nt of a complementary triplet. The remaining 25% were located within 6 nt of a triplet containing a single mismatch. An example is fragment c in Figure 2 (IL-2), in which an anti-AUA gapmer apparently cleaved the transcript at GUA-39 as there is no AUA site in the vicinity. This site is particularly accessible as strong cleavage was also observed at this site when the GUA-targeting gapmer was used (Fig. 2, band 1). A band (I) was also observed when the CUA gapmer was used. This indicates that some probe cross-hybridisation can occur when a site is particularly accessible. We were able to show that in all cases where cleavage occurred at a site of mismatch, but still at an NUH site, the correct gapmer to target the appropriate triplet sequence was also able to direct cleavage. In the case of band 5 in Figure 2 the assignment was confirmed by synthesising the specific Mrz, cleaving the target RNA with that Mrz and the semi-random gapmers and separating the products of these reactions on a single sequencing gel. Band 5 and the 3′-product of cleavage by UUU-489 co-migrated, confirming the assignment (data not shown). We concluded, therefore, that the majority of gapmer-mediated cleavage events occurred at cognate NUH sites.
Figure 2.
IL-2 transcript probed with RNase H in the presence of NUH-targeting gapmers and 6/8 and 9/9 arm length Mrz. Digests were fractionated by 3.5% denaturing PAGE. Mrz were synthesised against cleavage sites indicated by arrowheads and numbered 1–6. Lanes marked with an X are irrelevant. The assigned sites were: 1, GUA-39; 2, AUU-87; 3, CUA-124; 4, UUA-484; 5, UUU-489; 6, AUU-616. Bands indicated by letters immediately left of a cleavage product represent sites to which Mrz were not synthesised. Mock indicates equivalently treated transcript in the absence of probe. M indicates size markers.
Table 2. IL-2 and PDGF transcript mapped and assigned cleavage sites.
| Probe and product ID |
|
Mapped site |
Assigned sites |
| IL-2 | |||
| GUA | 1 | 36 | GUA-39 |
| AUU | 2 | 85 | AUU-87 |
| CUA | 3 | 124 | CUA-124 |
| UUA | 4 | 482 | UUA-484 |
| UUU | 5 | 490 | UUU-489 |
| AUU | 6 | 616 | AUU-616 |
| AUC | a | 213 | AUC-217 |
| AUC | b | 500 | AUC-501 |
| AUA | c | 39 | GUA-39 x |
| AUA | d | 87 | AUU-87 x |
| AUA | e | 120 | CUA-124 x |
| UUA | f | 28 | UUA-23 |
| UUA | g | 125 | CUA-124 x |
| UUA | h | 173 | UUA-168 |
| UUA | I | 339 | UUA-333 |
| CUC | j | 52 | CUC-49 |
| CUC | k | 235 | CUC-237 |
| CUA | l | 40 | GUA-39 x |
| CUA | m | 152 | CUA-153 |
| CUA | n | 611 | CUA-610 |
| PDGF | |||
| CUU | 1 | 43 | CUU-40 |
| CUA | 2 | 245 | CUA-243 |
| AUC | 3 | 294 | AUC-293 |
| GUC | 4 | 355 | GUC-354/359 |
| GUC | 5 | 366 | GUC-370 |
| GUC | 6 | 523 | GUC-530 |
| GUC | a | 309 | GUC-305 |
| GUC | b | 317 | GUC-314 |
| AUC | c | 145 | AUC-143 |
| AUC | d | 362 | ACC-363 x |
| AUU | e | 278 | AUU-275 |
| UUC | f | 362 | GUC-359 x |
| UUA | g | 335 | UUA-337 |
| UUA | h | 532 | UGA-537 x |
| CUC | I | 69 | CUC-74 |
| CUC | j | 163 | CUC-161 |
| CUC | k | 363 | CCC-364 x |
| CUC | l | 452 | CCC-455 x |
| CUA | m | 272 | CCA-273 x |
Bands labelled in Figures 2 and 3 were mapped for size based on their migration relative to RNA markers, then assigned to the closest cognate triplet. Assignments appended with an x in the last column had no perfectly matched triplet within the region of uncertainty of the assigned size of the band (±6 nt) and were assigned to the nearest triplet with a single mismatched.
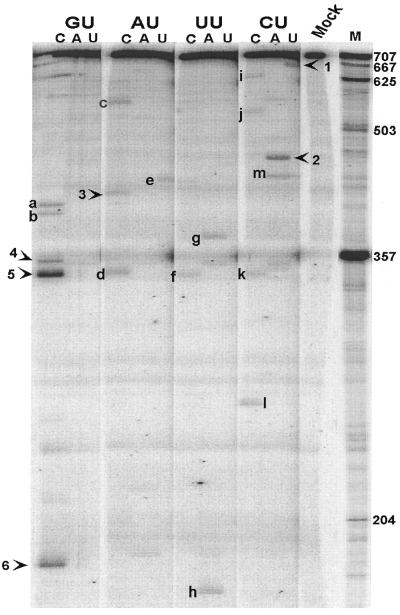
The PDGF transcript was similarly subjected to gapmer-directed RNase H digestion (Fig. 3). As observed for the IL-2 transcript, specific products were observed and 12 of the 19 products could be assigned to adjacent canonical triplet sequences (Table 2). In one case the estimated fragment size of 355 nt (band 4, Fig. 3) did not allow a definite assignment to either of two potential sites, GUC 354 and GUC 359. Like the IL-2 transcript products, a small but significant number (6/19) of bands were assigned to triplets containing a single mismatch. One of the bands in the PDGF experiment, band 1 in Figure 3, co-migrated with the 3′-product of cleavage by Mrz CUU-40 (data not shown), confirming its assignment.
Figure 3.
PDGF transcript probed with RNase H in the presence of NUH-targeting gapmers and fractionated by 4% PAGE. Mrz were synthesised against cleavage sites indicated by arrowheads and numbered 1–6. The assigned sites were: 1, CUU-40; 2, CUA-243; 3, AUC-293; 4, GUC-354,359; 5, GUC-370; 6, GUC-530. Bands indicated by letters immediately left of a cleavage product represent assigned products to which Mrz were not targeted. Mock indicates equivalently treated transcript in the absence of probe. M indicates size markers.
IL-2 and PDGF assigned sites were cleaved by specific Mrz in 1 mM MgCl2
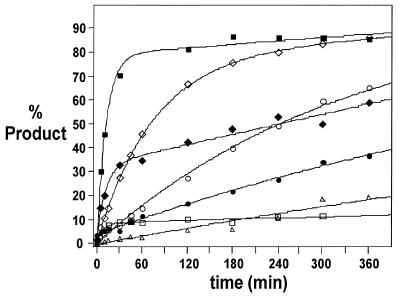
Specific Mrz were synthesised to 13 of the NUH sites assigned above. Six Mrz were directed against IL-2 sites and seven were targeted to PDGF sites (Table 2). These Mrz were tested for cleavage activity in vitro against the relevant transcripts at 37°C in 50 mM Tris buffer (pH 7.6) containing 1 mM MgCl2. Mrz and transcripts were not heat denatured or pre-annealed. Each Mrz cleaved its intended target at the expected position. Two sites in IL-2 transcript (AUU-87 and AUU-616) and three sites in PDGF (AUC-293, GUC-359 and GUC-370) were cleaved by their cognate Mrz with both high rate constants (kobs) and total extents of cleavage (%P360) (Figs 4 and 5).
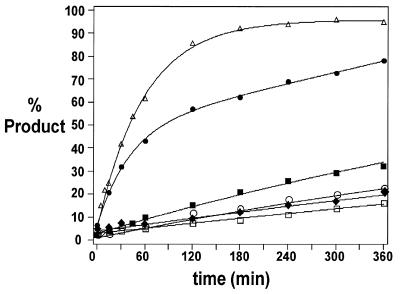
Figure 4.
Representative kinetics of in vitro cleavage of 811 nt IL-2 transcript by Mrz targeted to sites identified by gapmer-mediated RNase H cleavage assay. Sites: filled squares, GUA-39; open triangles, AUU-87; filled diamonds, CUA-124; open squares, UUA-484; open circles, UUU-489; filled circles, AUU-616. Cleavage conditions: 37°C, 50 mM Tris, pH 7.6, 1 mM MgCl2.
Figure 5.
Representative kinetics of in vitro cleavage of 707 nt PDGF transcript by Mrz targeted to sites identified by gapmer-mediated RNase H cleavage assay. Sites: open triangles, CUU-40; open diamonds, AUC-293; filled circles, CUA-243; open squares, GUC-354; filled squares, GUC-359; filled diamonds, GUC-370; open circles, GUC-530. Cleavage conditions: 37°C, 50 mM Tris, pH 7.6, 1 mM MgCl2.
DISCUSSION
Advantages of screening with RNase H and gapmers
This communication describes a simple procedure based on RNase H, transcript RNA (∼1013 copies) and NUH-targeting gapmers (6 × 1013 copies or 109 of each specificity) for identifying hammerhead ribozyme-accessible sites. This approach relies on the ability of E.coli RNase H to recognize a RNA–DNA heteroduplex of four contiguous base pairs. Under that condition RNase H cleaves the RNA strand in these heteroduplexes at a single internucleotide bond. We harnessed this ability in conjunction with chimeric oligonucleotide probes (gapmers) to direct RNase H cleavage with the specificity expected for hammerhead ribozymes. This was achieved by specifying the sequence of three DNA nucleotides in the gapmer probe so that the 12 potential NUH hammerhead cleavage triplets were targeted using 12 separate probe populations. The designs of the chimeric probes were such that the RNA transcript was cleaved on the 3′-side of the NUH triplet.
This approach permitted rapid and accurate identification of cleavage sites based simply on the mobility of the cleavage products and the sequence of the probe. We used separate probes complementary to the 12 NUH triplets. On average, targeting a random piece of RNA each probe should find a complementary RNA sequence once every 64 nt. Using sequencing gels and transcripts of the order of 700 nt in length it was possible to determine the size of cleavage fragments typically to within ∼6 nt. Thus, it was possible to unequivocally assign the majority of observed bands to specific NUH sites. In some cases, however, bands occurred in unexpected positions, i.e. where there was no corresponding Watson–Crick triplet sequence in that region. Such events are most likely explained by single base mismatches between one of the three defined nucleotides in the gapmer and the cleavage site. In support of this suggestion, in those cases where the probes existed, cleavage was also observed using the corresponding perfectly matched gapmer probe. In one case, in the PDGF experiment a band was observed that could not be defintively assigned to either of two closely spaced GUC sites. Therefore, Mrz were synthesised to target both sites. Despite being separated by only 5 nt, the differences in activity displayed by the Mrz targeting these two sites was striking. Mrz 359 cleaved >70% of the transcript in 30 min, whereas even after 5 h only ∼10% was cleaved by Mrz 354 (Fig. 5). This observation emphasises the highly exacting nature of RNA accessibility and the need to accurately identify potential target sites.
A major advantage of the targeted gapmer approach to mapping ribozyme targets is rapid and accurate identification of hammerhead recognition motifs without resorting to complex primer extension- (25,29) or RT–PCR-based analyses (18). The success of this approach relies on occupation of accessible sites by the NUH-targeting gapmers. This is achieved by virtue of a number of factors. First, defining the sequence of the three central nucleotides limits binding of separate gapmer libraries to a selective range of complementary sequences. Secondly, by constraining the randomised sequence in these libraries diversity is decreased. The reduction in probe sequence diversity permits a higher concentration (64-fold) of each targeting identity to be synthesised. This increases the likelihood that a given complementary site can be occupied and cleaved by RNase H. Finally, unlike unconstrained (fully randomised) oligonucleotide libraries (25,29), the definition of a central core of hybridising nucleotides minimises the potential for stable self-complementarity between individual members of the library.
A high proportion of identified sites are cleaved with good efficiency by their cognate Mrz
Mrzs were constructed to six of the 20 sites identified in IL-2 mRNA; of these two were very efficient, having both high rate constants and good extents of cleavage after 6 h (Fig. 4). Note that in these experiments the magnesium ion concentration was only 1 mM, 10-fold less than is typically used for in vitro experiments. We chose this concentration as it is close to the concentration of free magnesium ions found in mammalian cells (37). Likewise, for the PDGF mRNA we chose six fairly intense sites from the 19 identified in the RNase H screening procedure and synthesised seven Mrz to these sites (because one band could not be unequivocally assigned to either of two sites). Of these seven Mrz, three cleaved the target RNA very efficiently in vitro (Fig. 5). This is a very good hit rate and much more effective than would be expected by random selection of sites. Readily accessible sites in RNA have been estimated to occur about once in every 400 nt or, expressed in other terms, a typical success rate for screening antisense oligonucleotides is ∼1 in 20 to 1 in 40 (16,25,40). RNase H probing of human acetylcholinesterase mRNA with random dodecamers yielded five sites, only one of which was cleaved reasonably well by its cognate hammerhead ribozyme (11). The substrate half-life in the presence of a saturating concentration of ribozyme was 46 min (pH 7.5, 10 mM MgCl2, 37°C). The labile sites identified in this study (2/6 for IL-2 and 3/7 for PDGF) had early phase kinetics with half-lives between 7 and 60 min, despite the fact that the in vitro kinetics were performed in a 10-fold lower magnesium ion concentration than that used by Birikh et al. (11). In a study on α-lactalbumin mRNA (12) six hammerheads were targeted to sites considered accessible by RNA folding predictions. Of these, the most active hammerhead cleaved ∼50% of the transcribed RNA in 60 min (pH 8.0, 20 mM MgCl2, 37°C). In another study four ribozymes targeted to IL-6 mRNA were synthesised against all four GUC sites in the coding sequence and tested in vitro (41); the most active of these had a half-life of 170 min (pH 7.5, 20 mM MgCl2, 30°C) with saturating amounts of ribozyme, another was half as fast and two showed no cleavage at all. A recent report in which random armed hammerhead ribozymes were used to cleave the LTR mRNA of HIV identified two accessible NUH sites (42). The sites were cleaved by saturating concentrations of their cognate ribozymes with half-lives of ∼50 min (pH 7.5, 10 mM MgCl2, 37°C). In a study using randomised hairpin ribozymes (21) cleavage of pre-genomic RNA of hepatitis B virus was detected by primer extension and specific ribozymes made to 17 identified sites. No in vitro kinetics were reported, but ribozymes were expressed in cells from two different promoter systems and inhibition of HBV replication monitored. Of the 34 constructs, three inhibited replication by >50% and a further five by at least 40%. It is difficult to compare such disparate studies where the ribozyme design and Mg2+ ion concentrations used differ so dramatically, nevertheless these comparisons do demonstrate that the simple technique described here is of comparable efficacy to other in vitro methods. A recent publication described the in vivo selection of active hairpin ribozymes from a randomised population by expression in mammalian cells (20). That study identified a potential tumour suppressor gene by selection of cells that had gained the anchorage-independent growth phenotype. In vivo selection strategies have the advantage of eliminating uncertainty about the relevance of in vitro cleavage ability, but are much more technically demanding and, because they potentially target every expressed gene, may be very difficult to interpret.
In using this method to identify Mrz sites we make the assumption that a site that binds a simple oligonucleotide will support binding and cleavage by a Mrz. While this may be true in many cases, it is likely to be an over-simplification. When hammerheads assemble at an accessible site they form a complex junction of three helices. It is clear that this interaction is not perfectly modelled by simpler duplex-forming oligonucleotides. In addition, the kinetics of formation of RNase H–oligonucleotide–transcript trimolecular complexes is controlled by different factors to those that control the bimolecular hammerhead cleavage reaction. Consequently, it is not surprising that every potential hammerhead-cleavable site is not readily RNase H-cleavable and vice versa. In keeping with this, we found that one site in the IL-2 sequence, GUC-80, which is known to be accessible in vitro and in vivo (43), was not detected in this RNase H screen. In choosing likely Mrz sites we generally selected the most intense RNase H bands. Within this limited range of intensities the observed in vitro activity did not correlate with band intensity in the RNase H screen. Despite these limitations, it was encouraging to observe that all the NUH sites identified and tested were amenable to cleavage by Mrz and a high proportion of them were cleaved very efficiently. This indicates that there is general merit in screening for hammerhead-accessible sites by this simple, robust approach.
While this manuscript was in preparation a report of a similar method for the selection of accessible sites was published (44). In that study semi-random 13mer oligodeoxynucleotides (ODNs) were used as probes in conjunction with RNase H. The largely random probes possessed a central defined triplet complementary to one of the common hammerhead target sites, GUC, GUA, AUC or AUA. As we observed in this study, a relatively small number of specific cleavage events were observed for each specific ODN and their positions correlated with occurrence of the corresponding triplet in the mRNA sequence. Compared with this study, where a DNA quartet flanked by RNA (gapmer) directs RNase H to cleave at a single site in the RNA target, the cleavage directed by 13mer ODNs can occur anywhere in the complementary sequence, making target site identification potentially more difficult.
In summary, the potentiation of RNase H-mediated cleavage of transcripts by targeted gapmers is a simple and rapid method for identifying NUH sites that should be accessible to hammerhead ribozymes. We therefore commend this approach as a valuable aid for the rapid initial identification of such sites. Once appropriate sites have been identified, we have shown previously that cleavage efficiency can be markedly optimised (both in terms of initial rates and final extents of cleavage) by varying the length of the annealing arms (45,46) and, in the case of Mrz, the sequence of the linker region (36). Taken together, these approaches describe a robust pathway for designing high activity hammerhead ribozymes for the cleavage of mRNA.
SUPPLEMENTARY MATERIAL
Supplementary Material is available at NAR Online.
Acknowledgments
ACKNOWLEDGEMENT
We thank G.Brown (CSIRO Molecular Science) for the PDGF-LA-encoding plasmid.
References
- 1.Tanner N.K. (1999) Ribozymes: the characteristics and properties of catalytic RNAs. FEMS Microbiol. Rev., 23, 257–275. [DOI] [PubMed] [Google Scholar]
- 2.Sun L.Q., Cairns,M.J., Saravolac,E.G., Baker,A. and Gerlach,W.L. (2000) Catalytic nucleic acids: from lab to applications. Pharmacol. Rev., 52, 325–347. [PubMed] [Google Scholar]
- 3.Ruffner D.E., Stormo,G.D. and Uhlenbeck,O.C. (1990) Sequence requirements of the hammerhead RNA self-cleavage reaction. Biochemistry, 29, 10695–10702. [DOI] [PubMed] [Google Scholar]
- 4.Perriman R., Delves,A. and Gerlach,W.L. (1992) Extended target-site specificity for a hammerhead ribozyme. Gene, 113, 157–163. [DOI] [PubMed] [Google Scholar]
- 5.Shimayama T., Nishikawa,S. and Taira,K. (1995) Generality of the NUX rule: kinetic analysis of the results of systematic mutations in the trinucleotide at the cleavage site of hammerhead ribozymes. Biochemistry, 34, 3649–3654. [DOI] [PubMed] [Google Scholar]
- 6.Zoumadakis M. and Tabler,M. (1995) Comparative analysis of cleavage rates after systematic permutation of the NUX consensus target motif for hammerhead ribozymes. Nucleic Acids Res., 23, 1192–1196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cameron F.H. and Jennings,P.A. (1989) Specific gene supression by engineered ribozymes in monkey cells. Proc. Natl Acad. Sci. USA, 86, 9139–9144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sioud M. and Drlica,K. (1991) Prevention of human immunodeficiency virus type 1 integrase expression in Escherichia coli by a ribozyme. Proc. Natl Acad. Sci. USA, 88, 7303–7307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sun L.Q., Wang,L., Gerlach,W.L. and Symonds,G. (1995) Target sequence-specific inhibition of HIV-1 replication by ribozymes directed to tat RNA. Nucleic Acids Res., 23, 2909–2913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Krieg A.M. (1994) Instructions to authors. Antisense Res. Dev., 4(2). [DOI] [PubMed] [Google Scholar]
- 11.Birikh K., Berlin,Y., Soreq,H. and Eckstein,F. (1997) Probing accessible sites for ribozymes on human acetylcholinesterase RNA. RNA, 3, 429–437. [PMC free article] [PubMed] [Google Scholar]
- 12.l’Huillier P.J., Davis,S.R. and Bellamy,A.R. (1992) Cytoplasmic delivery of ribozymes leads to efficient reduction in α-lactalbumin mRNA levels in C1721 mouse cells. EMBO J., 11, 4411–4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zuker M., Jaeger,J. and Turner,D. (1991) A comparison of optimal and suboptimal RNA secondary structures predicted by free energy minimization with structures determined by phylogenetic comparison. Nucleic Acids Res., 19, 2707–2714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Matzura O. and Wennborg,A. (1996) RNAdraw: an integrated program for RNA secondary structure calculation and analysis under 32-bit Microsoft Windows. Comput. Appl. Biosci., 12, 247–249. [DOI] [PubMed] [Google Scholar]
- 15.Matveeva O., Felden,B., Audlin,S., Gesteland,R.F. and Atkins,J.F. (1997) A rapid in vitro method for obtaining RNA accessibility patterns for complementary DNA probes: correlation with an intracellular pattern and known RNA structures. Nucleic Acids Res., 25, 5010–5016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Monia B.P., Johnston,J.F., Geiger,T., Muller,M. and Fabbro,D. (1996) Antitumor activity of a phosphorothioate antisense oligodeoxynucleotide targeted against C-raf kinase. Nature Med., 21, 668–675. [DOI] [PubMed] [Google Scholar]
- 17.Peyman A., Helsberg,M., Kretzschmar,G., Mag,M., Grabley,S. and Uhlmann,E. (1995) Inhibition of viral growth by antisense oligonucleotides directed against the IE110 and the UL30 mRNA of herpes simplex virus type-1. Biol. Chem. Hoppe Seyler, 376, 195–198. [DOI] [PubMed] [Google Scholar]
- 18.Lieber A. and Strauss,M. (1995) Selection of efficient cleavage sites in target RNAs by using a ribozyme expression library. Mol. Cell. Biol., 15, 540–551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yu Q., Pecchia,D.B., Kingsley,S.L., Heckman,J.E. and Burke,J.M. (1998) Cleavage of highly structured viral RNA molecules by combinatorial libraries of hairpin ribozymes. The most effective ribozymes are not predicted by substrate selection rules. J. Biol. Chem., 273, 23524–23533. [DOI] [PubMed] [Google Scholar]
- 20.Welch P.J., Marcusson,E.G., Li,Q.-X., Beger,C., Kruger,M., Zhou,C., Leavitt,M., Wong-Staal,F. and Barber,J.R. (2000) Identification and validation of a gene involved in anchorage independent cell growth control using a library of randomised hairpin ribozymes. Genomics, 66, 274–283. [DOI] [PubMed] [Google Scholar]
- 21.zu Putlitz J., Yu,Q., Burke,J.M. and Wands,J.R. (1999) Combinatorial screening and intracellular antiviral activity of hairpin ribozymes directed against hepatitis B virus. J. Virol., 73, 5381–5387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pierce M.L. and Ruffner,D.E. (1998) Construction of a directed hammerhead ribozyme library: towards the identification of optimal target sites for antisense-mediated gene inhibition. Nucleic Acids Res., 26, 5093–5101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Tabler M. and Tsagris,M. (1991) Catalytic antisense RNAs produced by incorporating ribozyme cassettes into cDNA. Gene, 108, 175–183. [DOI] [PubMed] [Google Scholar]
- 24.Frank B.L. and Goodchild,J. (1997) Selection of accessible sites for ribozymes on large RNA transcripts. In Turner,P.C. (ed.), Methods in Molecular Biology, Vol. 74, Ribozyme Protocols. Humana Press, Totowa, NJ pp. 37–43. [DOI] [PubMed]
- 25.Ho S.P., Bao,Y., Lesher,T., Malhotra,R., Ma,L.Y., Fluharty,S.J. and Sakai,R.R. (1998) Mapping of RNA accessible sites for antisense experiments with oligonucleotide libraries. Nat. Biotechnol., 16, 59–63. [DOI] [PubMed] [Google Scholar]
- 26.Jarvis T.C., Wincott,F.E., Alby,L.J., McSwiggen,J.A., Beigelman,L., Gustofson,J., DiRenzo,A., Levy,K., Arthur,M., Matulic-Adamic,J., Karpeisky,A., Gonzalez,C., Woolf,T.M., Usman,N. and Stinchcomb,D.T. (1996) Optimizing the cell efficacy of synthetic ribozymes. Site selection and chemical modifications of ribozymes targeting the proto-oncogene c-myb. J. Biol. Chem., 271, 29107–29112. [DOI] [PubMed] [Google Scholar]
- 27.Scheer M. and Rossi,J.J. (1998) Rapid determination and quantitation of the accessibility to native RNAs by antisense oligodeoxynucleotides in murine cell extracts. Nucleic Acids Res., 26, 5079–5085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Lima W., Brown-Drivers,V., Fox,M., Hanecak,R. and Bruice,T.W. (1997) Combinatorial screening and rational optimization for hybridization to folded hepatitis C virus RNA for oligonucleotides with biological antisense activity. J. Biol. Chem., 272, 626–638. [PubMed] [Google Scholar]
- 29.Ho S.P., Britton,D.H., Stone,B.A., Behrens,D.L., Leffet,L.M., Hobbs,F.W., Miller,J.A. and Trainor,G.L. (1996) Potent antisense oligonucleotides to the human multidrug resistance-1 mRNA are rationally selected by mapping RNA-accessible sites with oligonucleotide libraries. Nucleic Acids Res., 24, 1901–1907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Monia B.P., Lesnik,E.A., Gonzalez,C., Lima,W.F., McGee,D., Guinosso,C.J., Kawasaki,A.M., Cook,P.D. and Freier,S.M. (1993) Evaluation of 2′-modified oligonucleotides containing 2′-deoxy gaps as anti-sense inhibitors of gene expression. J. Biol. Chem., 268, 14514–14522. [PubMed] [Google Scholar]
- 31.Clark S.C., Arya,S.K., Wong,S.F., Matsumoto,K.M., Kay,R.M., Kaufman,R.J., Brown,E.L., Shoemaker,C., Copeland,T., Oroszlan,S., Smith,K., Sarngadharan,M.G., Lindner,S.G. and Gallo,R.C. (1984) Human T-cell growth factor: partial amino acid sequence, cDNA cloning and organization and expression in normal and leukemic cells. Proc. Natl Acad. Sci. USA, 81, 2543–2547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kelly J.L., Sanchez,A., Brown,G.S., Chesterman,C.N. and Sleigh,M.J. (1993) Accumulation of PDGF B and cell-binding forms of PDGF A in the extracellular matrix. J. Cell Biol., 121, 1153–1163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lingner J. and Keller,W. (1993) 3′-End labeling of RNA with recombinant yeast poly(A) polymerase. Nucleic Acids Res., 21, 2917–2920. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hendry P., McCall,M.J. and Lockett,T.J. (1998) Small efficient hammerhead ribozymes. In Scanlon,K.J. (ed.), Methods in Molecular Medicine, Vol. 11, Therapeutic Applications of Ribozymes. Humana Press, Totowa, NJ, pp. 1–15.
- 35.Hertel K.J., Pardi,A., Uhlenbeck,O.C., Koizumi,M., Ohtsuka,E., Uesugi,S., Cedergren,R., Eckstein,F., Gerlach,W.L., Hodgson,R. and Symons,R.H. (1992) Numbering system for the hammerhead. Nucleic Acids Res., 20, 3252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Conaty J., Hendry,P. and Lockett,T. (1999) Selected classes of minimised hammerhead ribozyme have very high cleavage rates at low Mg2+ concentration. Nucleic Acids Res., 11, 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Flatman P.W. (1991) Mechanisms of magnesium transport. Annu. Rev. Physiol., 53, 259–271. [DOI] [PubMed] [Google Scholar]
- 38.Hogrefe H., Hogrefe,R., Walder,R. and Walder,J. (1990) Kinetic analysis of Escherichia coli RNase H using DNA-RNA-DNA/DNA substrates. J. Biol. Chem., 265, 5561–5566. [PubMed] [Google Scholar]
- 39.Crooke S.T., Lemonidis,K.M., Neilson,L., Griffey,R., Lesnik,E.A. and Monia,B.P. (1995) Kinetic characteristics of Escherichia coli RNase H1: cleavage of various antisense oligonucleotide–RNA duplexes. Biochem. J., 312, 599–608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Milner N., Mir,K. and Southern,E.M. (1997) Selecting effective antisense reagents on combinatorial oligonucleotide arrays. Nat. Biotechnol., 15, 537–541. [DOI] [PubMed] [Google Scholar]
- 41.Hendrix C., Anne,J., Joris,B., Van Aershot,A. and Herdewijn,P. (1996) Selection of hammerhead ribozymes for optimum cleavage of interleukin 6 mRNA. Biochem. J., 314, 655–661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bramlage B., Luzi,E. and Eckstein,F. (2000) HIV-1 LTR as a target for synthetic ribozyme-mediated inhibition of gene expression: site selection and inhibition in cell culture. Nucleic Acids Res., 28, 4059–4067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sioud M., Opstad,A., Hendry,P., Lockett,T.J., Jennings,P.A. and McCall,M.J. (1997) A minimised hammerhead ribozyme with activity against interleukin-2 in human cells. Biochem. Biophys. Res. Commun., 231, 397–402. [DOI] [PubMed] [Google Scholar]
- 44.Amarzguioui M., Brede,G., Babaie,E., Grotli,M., Sproat,B. and Prydz,H. (2000) Secondary structure prediction and in vitro accessibility of mRNA as tools in the selection of target sites for ribozymes. Nucleic Acids Res., 28, 4113–4124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McCall M.J., Hendry,P., Mir,A.A., Conaty,J., Brown,G. and Lockett,T.J. (2000) Small, efficient hammerhead ribozymes. In Walker,J.M. (ed.), Molecular Biotechnology, Vol. 14. Humana Press, Towota, NJ, pp. 5–17. [DOI] [PubMed]
- 46.Hendry P. and McCall,M. (1996) Unexpected anisotropy in substrate cleavage rates by asymmetric hammerhead ribozymes. Nucleic Acids Res., 24, 2679–2684. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.