Abstract
We genotyped 326 “frequently medicated” individuals of European-descent in Vanderbilt’s biorepository linked to de-identified electronic medical records, BioVU, on the ADME Core Panel to assess quality and performance of the assay. We compared quality control metrics and determined the extent of direct and indirect marker overlap between the ADME Core Panel and the Illumina Omni1-Quad. We found the quality of the ADME Core Panel data to be high, with exceptions in select copy number variants (CNVs) and markers in certain genes (notably CYP2D6). Most of the common variants on the ADME panel are genotyped by the Omni1, but absent rare variants and CNVs could not be accurately tagged by single markers. Finally, our frequently medicated study population did not convincingly differ in allele frequency from reference populations, suggesting that heterogeneous clinical samples (with respect to medications) follow similar allele frequency distributions in pharmacogenetics genes as their appropriate reference populations.
Keywords: ADME, pharmacogenomics, pharmacogenetics, BioVU, biorepository, CYP2D6, personalized medicine, precision medicine
Introduction
There is considerable inter-individual variation in the efficacy and risk of adverse events for many commonly prescribed medications. The inter-individual variation can be explained, in part, by genetic variation [1,2]. One vision of personalized or medicine is to use knowledge of a patient’s genetic profile prior to prescribing to maximize the likelihood of a beneficial outcome and minimize the risk of side effects. In response to this vision, patient genotypes for variants known to affect the efficacy of certain drugs (such as warfarin, clopidogrel, and tamoxifen) are being deposited into patients’ medical records in clinics to aid clinicians in formulating treatment plans [3].
The interest in using genetic variation to inform clinical care is driving a demand for the generation of high-quality genomic data in both research and clinical settings. However, many relevant loci in pharmacogenes are located in regions that are difficult to assay with conventional multiplexing methods used in arrays [4]. Moreover, unlike genome-wide association studies (GWAS), pharmacogenomic studies cannot rely on linkage disequilibrium to indirectly test or tag the relevant variation given the low coverage of pharmacogenes in general on these fixed-content GWAS products [4]. As an example, the cytochrome P450 (CYP) family of enzymes is responsible for 75% of phase I dependent drug metabolism, and variants in these genes have been associated with the outcomes of many drug responses [5]. Duplication events in the human genome have resulted in 57 functional genes in the CYP family and numerous pseudogenes [6]. However, many clinically important CYP genes often have high sequence similarity with pseudogenes and/or are located in repetitive gene clusters. Probes designed to genotype variants in these genes have been known to suffer from cross-hybridization problems and therefore have often been dropped during the development of genome-wide platforms [7]. Several pharmacogenetic platforms have been introduced to the market that target these variants directly and avoid cross-reactivity other homologous regions [7].
The first released multiplexed assay focused on pharmacogenomics was the Affymetrix DMET Panel, which has been used in multiple pharmacogenetic studies [8,9]. Unlike the DMET Panel, there are currently no published descriptions of the performance of two other marketed pharmacogenetic panels, Illumina’s ADME Core Panel and Sequenom’s iPLEX® ADME PGx. In contrast to the Affymetrix DMET, which covers 1,936 markers across 231 genes, the two newer panels specifically target the 184 markers in 32 genes that were identified by the PharmaADME group as the most important predictors for pharmacokinetic variability.
As part of an extensive institutional investment in personalized medicine, Vanderbilt University Medical Center (VUMC) has begun both a large-scale research effort and translational effort involving the Illumina ADME Core Panel. The research effort, known as the Vanderbilt Electronic Systems for Pharmacogenomic Assessment (VESPA), is the genotyping of almost 10,000 individuals for electronic medical record (EMR)-based pharmacogenomics studies. The translational effort is known as the Pharmacogenomic Resource for Enhanced Decisions in Care and Treatment (PREDICT) program, an effort to pre-emptively genotype VUMC patients using the Illumina ADME Core Panel in a Clinical Laboratory Improvement Amendments (CLIA)-certified environment as part of routine clinical care [3].
We present here an assessment of the performance of the ADME Core Panel in a sample of individuals with multiple prescribed medications identified in BioVU, the Vanderbilt biorepository linked to de-identified electronic medical records [10]. Our primary goal was to assess the ADME Core Panel’s content and quality with respect to pharmacogenomic research in clinical populations. We also considered the ADME Core Panel’s coverage of variants in comparison with other available pharmacogenetic genotyping methods. As a secondary analysis, while we expect a European-descent polypharmacy population to have similar allele frequencies to European-descent reference populations, we tested if our polypharmacy population within BioVU was enriched for pharmacokinetic functional variants compared with reference populations. Our data demonstrate that, as expected, the polypharmacy sample did not differ from reference frequencies and most of the data quality was high. However, the quality and utility of the variant content can vary dramatically, indicating that fixed-content panels are likely to be useful in only specific pharmacogenomic research or clinical settings.
Methods
Study Population
Our study population consisted of de-identified medical records from 326 “frequently medicated” individuals, which was defined as being prescribed warfarin or clopidogrel in addition to more than five drugs from the following classes: heparin, statins, immunosuppressives (sirolimus, tacrolimus, cyclosporine, and mycophenolate mofetil), tamoxifen, codeine, selective serotonin reuptake inhibitors (SSRIs), and anti-psychotics. These medications were chosen because at least one medication within the class has known pharmacogenomic interactions. All study samples were retrieved from BioVU, the Vanderbilt University biorepository linked to de-identified electronic medical records. BioVU as a resource, including its ethical, privacy, and other protections, has been described in detail elsewhere [10]. In brief, BioVU is composed of DNA is extracted and stored from blood collected from routine clinical testing that is scheduled to be discarded after a three-day waiting period. DNA samples are linked to a de-identified version of the individual’s electronic medical record, known as the Synthetic Derivative (SD), and can be accessed by investigators for research purposes after approval by the local internal review board and BioVU Review Committee. Records eligible for possible inclusion into BioVU include those with a laboratory blood draw, where the individual has signed the consent to treatment form, and has not indicated that they wish to opt-out. All records used in this study were coded as European American by administrative staff, which has been shown to be highly correlated in BioVU with genetic ancestry as determined by ancestry informative markers [11]. De-identified records in the SD that fit our “frequently medicated” definition were selected using the natural language processing system, MedEX. MedEX extracts medications from EMRs with at least one mention of a dose, route, frequency, or strength. A more detailed description of the software has been published elsewhere [12].
Genotyping
Illumina’s pharmacogenetic-targeted ADME Core panel is designed for the genotyping of 184 markers in 34 genes. The panel’s genotyping assay is a specific application of the Golden Gate assay technology [101]. The ADME Core Panel utilizes an additional primer extension step to avoid cross-reactivity with other homologous genes or regions that could interfere with probe hybridization. This initialization step is then followed by allele-specific primer extension and a ligation step.
Two-thirds of the 184 variants on the ADME Core Panel encode synonymous and non-synonymous amino acid changes whereas only 24 markers on the panel encode of non-coding variation (Table 1). Frameshift and splicing defects are encoded by 16 and 10 markers on the panel, respectively. A total of ten CNVs are targeted by the panel for the following genes: SULT1A1 (5 CNVs), CYP2A6, CYP2D6, GSTM1, GSTT1, and UGT2B17. Compared to the Pharmacogenetics Research Network’s (PGRN) Very Important Pharmacogenes (VIP) marker list of 135 variants in 46 genes, which includes variants that have been identified as having either in vitro or in vivo evidence of functional effects on drug response [13] the ADME Core Panel directly assays 25 of the 41 CYP variants (61%) and 18 of the 36 variants in other important pharmacogenes (50%).
Table 1.
Functional Distribution of ADME Core Panel Markers
| Category | Number of SNPs |
|---|---|
| Coding | 123 |
| Frameshift | 16 |
| CNVs | 10 |
| Non-Coding | 24 |
| Duplications | 1 |
| Splicing Defects | 10 |
| Monomorphic | 67 |
| 0.00 < MAF < 0.01 | 35 |
| 0.01 < MAF < 0.05 | 22 |
| 0.05 < MAF < 0.25 | 26 |
| 0.25 < MAF < 0.5 | 23 |
Abbreviations: copy number variants (CNVs) and minor allele frequency (MAF). MAFs are of non-CNV diallelic markers.
Genotyping for this study was conducted at the Vanderbilt University DNA Resources Core. ADME Core Panel genotype calling was performed with ADME Module Version 1.0.0.3. In addition to genotype calls by variant, the ADME Module software outputs star nomenclature gene results for each gene. Ninety-eight individuals were also genotyped on Illumina’s Human Omni1-Quad as part of other genotyping efforts. The Illumina HumanOmni1-Quad is a genome-wide BeadChip that targets over one million SNPs selected from all three HapMap phases, the 1000 Genomes Project, and previously confirmed genetic associations from the NHGRI GWAS catalog [14]. Genotype calling for the HumanOmni1-Quad was performed using Illumina’s Genome Studio Version 1.7.4. Genotyping runs of individual samples in the laboratory that did not produce data are referred to in this study as sample failures. We defined failed markers as those with a genotyping efficiency below 90%.
Statistical Methods
To assess coverage, linkage disequilibrium (LD) calculations for tagging ADME Core markers with HumanOmni1-Quad markers were performed with PLINK’s function for tagging with a specified marker list (v1.07) [15]. We restricted our search to a 250kb window around each marker. Concordance for overlapping samples genotyped with the ADME Core Panel and the HumanOmni1-Quad was calculated using PLATO [16].
Quality control measures for diallelic ADME SNPs and 70 SNPs from the HumanOmni1- Quad including genotyping efficiency, Hardy-Weinberg Equilibrium (HWE), minor allele frequency were calculated using PLINK. Tests of HWE for triallelic SNPs were calculated manually using Pearson’s chi-squared test. ADME Core Panel marker allele frequencies for comparison with the present study were abstracted from the primary literature, The International HapMap Project, 1000 Genomes Project, dbSNP, PharmGKB, the Environmental Genome Project, and SNP500Cancer [17-22]. All frequencies were selected from populations of European-descent, similar to the study population described here. Tests of association were calculated with the chi-squared test, and in the case of cell counts < 5, Fisher’s exact was used. All statistical analyses were performed with the statistical software package STATA 11.
Results
Demographics
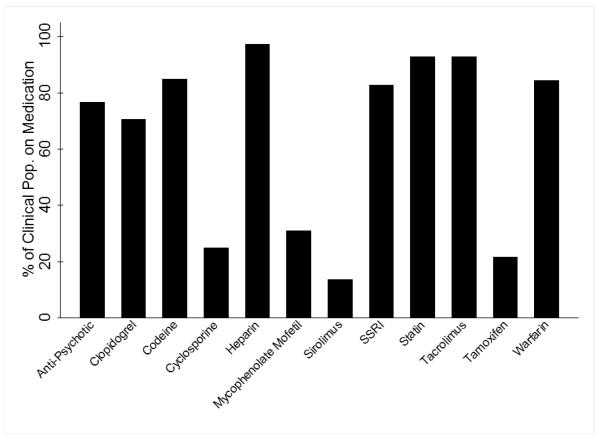
Table 2 presents the clinical characteristics of the study population. Three hundred and twenty six samples were genotyped on the ADME Core Panel for this assessment of the panel’s performance. Overall, there were more males than females in this cohort of frequently medicated individuals (55% versus 45%), and the average individual was overweight (body mass index or BMI >25 kg/m2; Table 2). The mean age of individuals at the time of their first prescription and final prescriptions was 57.0 ±12.9 and 64.8 ±12.6 years, respectively. The frequencies of drug classes prescribed are shown in Figure 1.
Table 2.
Study population characteristics (n=326)
| Variable | Mean / % | Standard Deviation (Min., Max.) |
|---|---|---|
| % European American | 100 | - |
| % Female | 45 | - |
| Age at First Drug, in years | 56.99 | 12.86 (24.33,84.94) |
| Age at Last Drug, in years | 63.73 | 12.64 (32.77,93.04) |
| BMI (kg/m2) | 29.67 | 6.18 (15.4,65.7) |
Abbreviations: body mass index (BMI).
Figure 1. Prescribed medications among the heavily medicated clinical population.
On the x-axis are the 12 classes of medications selected for our heavily medicated cohort definition. The y-axis is the proportion of our clinical population prescribed at least one medication of the given drug class.
Coverage
There are several options available for genotyping pharmacogenetic variants, ranging from single variant assays to genome-wide arrays, and each approach has its strengths and weaknesses with respect to assay performance and cost effectiveness (Table 3). Given that most datasets submitted to dbGaP have genome-wide SNP data [14,23], we assessed in our study sample whether these existing data adequately assess known pharmacogenetic variants. To do this, we characterized the linkage disequilibrium patterns in the immediate genomic regions of the ADME Core Panel variants and identified HumanOmni1-Quad SNPs that tag ADME Core Panel markers at three different linkage disequilibrium thresholds (r2): 1.0, 0.8, and 0.5. We found one SNP (CYP2A6 rs28399468) that can be indirectly tested or tagged at an r2 of 1.0 with a single marker (rs3212976) in populations of European-descent genotyped on the HumanOmni1-Quad. When the LD threshold was relaxed to 0.5, NAT2 rs1208 and SCLO1B3 rs7311358 could also be tagged by single markers (rs1802380 and rs4149117, respectively) targeted by the HumanOmni1-Quad.
Table 3.
Comparison of pharmacogenetic genotyping methods
| Method | Description | Cost per sample | Optimal Study Design |
Drawbacks |
|---|---|---|---|---|
| TaqMan | Accurately assays the genotype of a single nucleotide variant |
Low | Ideal for projects testing a small number of SNPs in a large population. |
Cost approaches that of GWAs platform for more than ~330 ‡ of SNPs. |
|
Genome-Wide
Fixed Content Platforms |
Current platforms range from 1-5 million common (MAF >1%) variants across the genome |
Mid | Holistic approach that targets most of the genome. Best candidate for pharmacodynamic studies, especially if the HLA* complex is a candidate† |
Overall highly accurate but drops in quality in repetitive genomic regions found around many CYP variants. |
|
ADME Core andother PGX* Panels |
Accurately targets 184-1,936§ variants in PGX genes |
Mid-High | Selective coverage of functional variants in VIPs*. Results of PGX studies have so far converged around pharmacokinetic genes covered here (with the exception of HLA). |
Large proportions of variants are too rare and lack statistical power in small to mid-range sized study designs. |
|
Targeted, Exome,
and Whole Genome Sequencing |
Selective sequencing of targeted genes/regions, all coding regions, or the whole genome |
High | Holistic approach to genome limited to coding regions. Most suitable for identifying the effect of burden of rare variants on drug response. |
Massive data storage requirements and unfamiliar analysis tools available may be prohibitive for some investigators. Little information in the literature on performance in VIP variants. |
Daly AK: Drug-induced liver injury: past, present and future. Pharmacogenomics. 11(5), 607-611 (2010)
Estimated prices for one sample as follows: $0.72 per SNP assayed with TaqMan assay and $240.00 per genome-wide genotyping platform.
1,936 variants captured with the Affymetrix DMET (drug metabolism enzymes and transporters)
Abbreviations: Pharmacogenetics/Pharmacogenomics (PGX), Very Important Pharmacogene (VIP), and Human Leukocyte Antigen (HLA)
We also compared the marker content on the ADME Core Panel to Affymetrix’s pharmacogenetic platform, the Drug Metabolizing Enzymes and Transporters (DMET) Plus, to identify overlapping and unique markers to the ADME Core Panel [24]. The DMET Plus interrogates 1,936 markers across 231 genes, with an emphasis on pharmacogenes. Of the 184 markers targeted by the ADME Core Panel, 159 overlap with DMET Plus. A total of 25 markers are unique to the ADME Core Panel. The ADME specific markers are 18 SNPs with rsIDs, five SULT1A1 CNVs, one multi SNP assay designed to probe the SLC22A1 M420del, and CYP2A6*1B.
Quality and Performance
We assessed basic quality control metrics on the 326 samples genotyped on the ADME Core panel. The vast majority of the samples (92%) were genotyped successfully using the ADME Core panel. A total of 27 samples failed and were not considered further in this analysis. Of the 184 markers targeted for genotyping on the ADME panel, four SNPs and three CNVs failed. The four failed SNPs include GSTM1 rs1065411, CYP2A6 rs28399447, CYP2A6*B1 and SLCO1B3 rs7311358. The three failed CNVs were in the following genes GSTM1, GSTT1, and UGT2B17. The total number of markers (excluding CNVs) with 100% and >99% genotyping efficiency were 84 and 114, respectively (Table 4). There were four CNVs with 100% (SULT1A1) genotyping efficiency and three between 90% and 99% (CYP2D6, CYP2A6, and SULT1A1).
Table 4. ADME Core Panel genotyping quality control statistics.
Genotyping efficiency (GE) was calculated for each of the 174 SNPs and 10 copy number variants (CNVs) targeted by the ADME custom assay in 299 samples that were successfully genotyped. Tests of Hardy Weinberg Equilibrium (HWE) were only performed for each of the SNPs.
| Category | SNPs (%) |
CNVs (%) |
|---|---|---|
| Monomorphic | 67 (38.50) |
7 (70.00) |
| GE < 90% | 4 (2.22) |
3 (30.00) |
| GE < 95% | 13 (7.47) |
4 (40.00) |
| GE > 99% | 115 (66.09) |
4 (40.00) |
| GE = 100% | 88 (50.57) |
4 (40.00) |
| HWE, p < 0.001 | 4 (2.29) |
- |
Four SNPs significantly departed from Hardy-Weinberg Equilibrium (HWE; p<0.001, Table 4): CYP2B6 rs8192709, GSTM1 rs1065411, CYP2D6 rs3892097, and the triallelic marker ABCB1 rs2032582. One of these HWE deviating markers, rs1065411, also had very low genotyping efficiency (49%). More than one-third of the 173 diallelic markers were monomorphic in our sample population (67; Table 1), and one-third (57) of the markers were rare (MAF ≤ 0.05; Table 1). The remaining markers (49; 28%) were common (MAF ≥ 0.05) in this study population. Table 5 displays the frequency of Clinical Pharmacogenetics Implementation Consortium (CPIC) “likely phenotypes” and star genotypes, as opposed to frequencies by individual rs number, in this sample population.
Table 5.
Frequency of CPIC “likely phenotypes” and star genotypes with published guidelines for drug dosing
| ADME Core Gene | CPIC* Categories | Frequency |
|---|---|---|
| CYP2D6 | Poor Metabolizer (PM) | 0.07 |
| Intermediate Metabolizer (IM) | 0.02 | |
| Extensive Metabolizer (EM) | 0.60 | |
| Ultrarapid Metbolizer (UM) | 0 | |
| No Call Undetermined ¶ |
0.21 0.10 |
|
| CYP2C9 | *1/*1 | 0.61 |
| *1/*2 | 0.20 | |
| *2/*2 | 0.02 | |
| *2/*3 | 0.03 | |
| *1/*3 | 0.12 | |
| No Call | 0.03 | |
| CYP2C19 | PM | 0.01 |
| IM | 0.20 | |
| EM | 0.42 | |
| UM | 0.28 | |
| *2/*17 | 0.07 | |
| No Call | 0.02 | |
| CYP3A5 ∥ | *1/*1 | 0.01 |
| *1/*3 | 0.11 | |
| *3/*3 | 0.88 | |
| TPMT | No/Low Activity | 0.00 |
| Intermediate Activity | 0.04 | |
| Normal Activity | 0.89 | |
| No Call | 0.06 | |
| SLCO1B1 | *5 | 0.26 |
| WT/*5 | 0.73 | |
| *5/*5 | 0.01 | |
| VKORC1 (−1639G>A) | GA | 0.36 |
| AA | 0.13 | |
| GG | 0.51 |
Clinical Pharmacogenetics Implementation Consortium (CPIC)
Dutch Working Group CYP3A5 categories
Samples called *4 HET *10 HET by ADME Module. The module uses the genotype of rs1065852 for calling the *10 haplotype. However this genotype does not distinguish *4 and *10 and these patients could either be *1/*4 (EM) or *4/*10 (IM).
Within this frequently medicated de-identified population genotyped on the ADME Core Panel, a subset of 98 individuals were genotyped on Illumina’s HumanOmni1-Quad platform for previous genetic association studies conducted in BioVU. More than one-third of markers (41%) of 70 diallelic ADME Core Panel markers overlap with the HumanOmni1-Quad. Out of the set of overlapping markers, the majority (59/70) had 100% concordance, and eight SNPs were >99% concordant.
As an supplemental step in our assessment of panel performance in this study, we compared the allele frequencies observed here with reference allele frequencies abstracted from multiple sources including the primary literature, The International HapMap Project, 1000 Genomes Project (Pilot 1 Low Coverage Panel), dbSNP, PharmGKB, the Environmental Genome Project, and SNP500Cancer [17-22]. Of the 173 diallelic, non-CNV markers targeted by the panel, we were able to abstract allele frequencies from European-descent populations for 109 (63%). Of the subset of markers that were without a reference frequency in the literature or public databases, all but two, SLC22A1 M420del and CYP2D6 rs35742686, were rare (MAF<0.01) or monomorphic in our sample population. These data suggest there is little population data on low frequency but functional pharmacogenetic variants. As might expected, the vast majority of marker allele frequencies in this frequently medicated sample from BioVU did not differ from allele frequencies previously reported for European-descent populations (Supplementary Table 1). In fact, after accounting for multiple testing, only one marker (CYP2D6 rs1080985) had a significantly different allele frequency in this BioVU sample compared with the 1000 Genomes Project CEU data: 0.11 versus 0.24, respectively (p=2.74 × 10−5; Supplementary Table 1).
Results of comparisons between this study sample and CPIC “likely phenotypes” frequencies [25-29](Table 5) were similar to that observed at the single variant level. The difference observed in frequency of CYP2D6 genotypes between CPIC and this study sample is likely due to the large proportion of individuals with missing calls for various markers, which in such cases were assigned “no call.” This underscores the difficulty in accurately assigning CYP2D6 genotypes even with the targeted ADME Panel. Overall, the small sample size and subsequent low power may have impacted our ability to detect small differences in frequencies of other alleles tested.
Discussion
We sought to assess the performance of the ADME Core Panel, a fixed content panel for pharmacogenomic research and clinical use, in a “frequently medicated” sample of individuals from BioVU, a biorepository of DNA samples linked to de-identified electronic medical records. These samples did not display any convincing evidence of having a different genomic profile of rare variants in pharmacogenetic genes compared with reference populations. The one significant SNP (CYP2D6 rs1080985) that differed between this study population and 1000 Genomes CEU samples may represent a true difference in frequency or may represent a sequencing error in the 1000 Genomes Pilot study due to the repetitive region of CYP2D6.
Our assessment of performance was based upon two major criteria: coverage and quality. Overall, the ADME Core Panel targets approximately one-third of the PGRN VIP marker list. Of the 184 markers targeted by the panel, the majority is coding or considered functional. Data from DNA samples extracted from frequently medicated individuals in BioVU suggest that the ADME Core Panel produced high quality and reproducible genotypes for the majority of variants targeted by the panel. CNVs targeted by the ADME Core Panel proportionally performed worse than SNPs in genotyping efficiency.
Overall, the quality of the data produced by the ADME Core Panel was high; however, there were variants with low quality data. In this study, several variants were out of HWE or had low quality of genotyping. Of note are two markers (rs1080985 and rs928286) in the highly polymorphic and difficult genotype gene CYP2D6. These two markers had lower than average call rates of 93.1% and 93.8%, respectively, suggesting that the panel’s assays are not completely optimized.
Limitations
A limitation of this study is the sample size. With only 299 individuals, a large proportion of markers targeted by the ADME Core Panel were monomorphic in this sample. However, this limitation also reflects a limitation of the ADME Core Panel. That is, more than one-third of the panel targets relatively rare variation based on European-descent populations. For a variant with a minor allele frequency of 1%, fewer than six heterozygotes would be expected in this study sample. Thirty-five of the 184 variants (~19%) targeted by the ADME Core Panel have a minor allele frequency <1% in European-descent populations (Table 1), and these will require thousands of samples genotyped to detect heterozygotes at an appreciable frequency for single SNP tests of association. Therefore, depending on the study design and study population, the panel’s content may decrease as the number of monomorphic markers increases.
ADME Core Panel for Genetics Research
Despite the potential decrease in content based on sample size and population, it is important to note that the ADME Core Panel targets important pharmacogenomic variants that are not tagged well or in LD with variants directly assayed by fixed-content GWAS arrays. While 70 non-CNVs were directly assayed by the Illumina HumanOmni1-Quad, only three of the remaining SNPs not directly assayed were in LD or tagged with a SNP genotyped directly on the array. Since the panel targets specifically markers that influence the inter-individual pharmacokinetics of drug metabolism, 84 of the 184 variants are functional variants in the CYP450 family of genes because of their ubiquitous role in the oxidation step of numerous medications. The remaining genes are Phase II enzymes and transporters, which catalyze addition modifications to drugs by catalyzing conjugation reactions and facilitate differential tissue distribution, respectively [30]. Many pharmacogenetic genes are redundant or lack endogenous substrates and consequently are presumed to be under less evolutionary pressure. This relaxed selection over human history has produced hypermorphic and highly deleterious alleles at relatively common frequencies. For instance, 7% of individuals of European-descent do not have a fully functional copy of the CYP2D6 gene, i.e. they are homozygotes or compound heterozygotes for loss of function alleles [31]. Thus, the variants that the panel targets are common functional variants encoding splicing mutations, non-synonymous SNPs, and more dramatic structural changes such as indels and CNVs. Compared with GWAS fixed-content arrays where the variants are mostly non-coding and functionality of significant markers can be difficult to interpret, markers from the ADME Core Panel in a pharmacogenetic association study have moderate to extensive biological data on their function.
Comment on Array-Based Pharmacogenomics
Perhaps the biggest limitation of the array-based approach for pharmacogenomics research and clinical implementation is the fact that only specific variants are being targeted in any one experiment or diagnostic order. As already mentioned, advances in sequencing technology now make it possible to generate complete variation data on an individual or patient for the whole genome, whole exome, and targeted regions in a cost-effective manner (Table 3). In recognition of this genomic evolution of technologies, Illumina recently announced that it will no longer be accepting orders for the Illumina BeadXpress, effectively discontinuing sales of mid-throughput genotyping instrumentation that process custom and fixed-content panels such as the ADME Core Panel. In its place will likely be targeted sequencing of individuals for research and patients for clinical diagnostics for genes and genomic regions important in drug therapy. While the generation of the data may be different compared with the arrays, the challenges of data quality and interpretation or implementation will be the same and will continue to be an active area of research and clinical oversight.
Future Perspective
We demonstrate that most of the data produced by the ADME Core Panel is high quality based on conventional quality control metrics. However, this as well as other fixed content panels have limitations on the number of types of variants targeted for pharmacogenomic research and clinical diagnostics. Targeted or whole genome/exome sequencing will likely remedy the content issue typical of genotyping panels and accelerate the understanding of the underlying genetic architecture that impacts responses to drug therapy in the clinic.
Supplementary Material
Executive Summary.
Background
The Illumina ADME Core Panel targets 184 functional and clinically interesting markers for pharmacogenomic studies.
Little is known about the quality or research utility of the panel compared with other fixed content assays.
Also, few data exist on the allele frequency of many of the variants targeted by the Illumina ADME Core Panel.
Methods
We used BioVU, the Vanderbilt University Medical Center biorepository linked to de-identified electronic medical records, to identify 326 individuals of European-descent that were prescribed warfarin or clopidogrel in addition to more than five drugs from the following classes: heparin, statins, immunosuppressives (sirolimus, tacrolimus, cyclosporine, and mycophenolate mofetil), tamoxifen, codeine, selective serotonin reuptake inhibitors (SSRIs), and anti-psychotics.
Individuals were genotyped using the Illumina ADME Core Panel. A subset of these individuals also was genotyped on the Illumina Human Omni1-Quad for previous genetic association studies.
Basic quality control metrics (call rates and Hardy-Weinberg Equilibrium) as well as concordance between the two arrays were calculated to assess genotyping quality of the Illumina ADME Core Panel.
Allele frequencies from public reference datasets and the literature were used to compare to this study sample of “frequently medicated” European-descent individuals.
Results
A proportion of the genetic variants targeted by the Illumina ADME Core Panel were neither targeted nor tagged by the fixed-content genome-wide association study (GWAS) assays such as the Illumina Human Omni1-Quad.
As expected, the Illumina ADME Core Panel performed well for diallelic variants in this modest sample of polypharmacy individuals.
A proportion of the Illumina ADME Core Panel variants were rare or monomorphic in this modest sample of European Americans, and no reference allele frequencies wre available in public databases or the literature.
All but one variant assayed by the Illumina ADME Core Panel was observed as a higher allele frequency in this polypharmacy sample compared with a reference population.
Discussion
The Illumina ADME Core panel produces high quality diallelic genotypes suitable for studies of pharmacogenomics, many of which are not available or tagged by fixed-content genome-wide association study arrays.
Acknowledgements
We would like to thank Ms. Allison R. Baker and Ms. Victoria M. Youngblood for their assistance in organizing a portion of this dataset. The Vanderbilt University Center for Human Genetics Research, Computational Genomics Core provided computational and/or analytical support for this work.
The dataset(s) used for the analyses described were obtained from Vanderbilt University Medical Center’s BioVU which is supported by institutional funding and by the Vanderbilt CTSA grant UL1 TR000445 from NCATS/NIH. This study was supported in part by RC2 GM092618.
Footnotes
Financial & Competing Interests Disclosure The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reference List
- 1.Crawford DC, Ritchie MD, Rieder MJ. Identifying the genotype behind the phenotype: a role model found in VKORC1 and its association with warfarin dosing. Pharmacogenomics. 2007;8(5):487–496. doi: 10.2217/14622416.8.5.487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wells QS, Delaney JT, Roden DM. Genetic determinants of response to cardiovascular drugs. Curr.Opin.Cardiol. 2012;27(3):253–261. doi: 10.1097/HCO.0b013e32835220e3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pulley JM, Denny JC, Peterson JF, et al. Operational Implementation of Prospective Genotyping for Personalized Medicine: The Design of the Vanderbilt PREDICT Project. Clin.Pharmacol.Ther. 2012;92(1):87–95. doi: 10.1038/clpt.2011.371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters EJ, McLeod HL. Ability of whole-genome SNP arrays to capture ’must have’ pharmacogenomic variants. Pharmacogenomics. 2008;9(11):1573–1577. doi: 10.2217/14622416.9.11.1573.** Discusses the Pharmacogenetics Research Network’s very important pharmacogenes list and their coverage of genome-wide platforms
- 5.Sim SC, Ingelman-Sundberg M. Pharmacogenomic biomarkers: new tools in current and future drug therapy. Trends Pharmacol.Sci. 2011;32(2):72–81. doi: 10.1016/j.tips.2010.11.008. [DOI] [PubMed] [Google Scholar]
- 6.Nelson DR, Zeldin DC, Hoffman SM, Maltais LJ, Wain HM, Nebert DW. Comparison of cytochrome P450 (CYP) genes from the mouse and human genomes, including nomenclature recommendations for genes, pseudogenes and alternative-splice variants. Pharmacogenetics. 2004;14(1):1–18. doi: 10.1097/00008571-200401000-00001. [DOI] [PubMed] [Google Scholar]
- 7.Dumaual C, Miao X, Daly TM, et al. Comprehensive assessment of metabolic enzyme and transporter genes using the Affymetrix Targeted Genotyping System. Pharmacogenomics. 2007;8(3):293–305. doi: 10.2217/14622416.8.3.293. [DOI] [PubMed] [Google Scholar]
- 8.Birdwell KA, Grady B, Choi L, et al. The use of a DNA biobank linked to electronic medical records to characterize pharmacogenomic predictors of tacrolimus dose requirement in kidney transplant recipients. Pharmacogenet.Genomics. 2012;22(1):32–42. doi: 10.1097/FPC.0b013e32834e1641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Mega JL, Close SL, Wiviott SD, et al. Cytochrome p-450 polymorphisms and response to clopidogrel. N.Engl.J.Med. 2009;360(4):354–362. doi: 10.1056/NEJMoa0809171. [DOI] [PubMed] [Google Scholar]
- 10.Roden DM, Pulley JM, Basford MA, et al. Development of a large-scale de-identified DNA biobank to enable personalized medicine. Clin.Pharmacol.Ther. 2008;84(3):362–369. doi: 10.1038/clpt.2008.89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dumitrescu L, Ritchie MD, Brown-Gentry K, et al. Assessing the accuracy of observer-reported ancestry in a biorepository linked to electronic medical records. Genet.Med. 2010;12(10):648–650. doi: 10.1097/GIM.0b013e3181efe2df.* Measured the concordance between administratively assigned race/ethinicity and genetic ancestry in Vanderbilt’s DNA Databank BioVU.
- 12.Xu H, Stenner SP, Doan S, Johnson KB, Waitman LR, Denny JC. MedEx: a medication information extraction system for clinical narratives. J.Am.Med.Inform.Assoc. 2010;17(1):19–24. doi: 10.1197/jamia.M3378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Giacomini KM, Brett CM, Altman RB, et al. The pharmacogenetics research network: from SNP discovery to clinical drug response. Clin.Pharmacol.Ther. 2007;81(3):328–345. doi: 10.1038/sj.clpt.6100087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hindorff LA, Sethupathy P, Junkins HA, et al. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc.Natl.Acad.Sci.U.S.A. 2009;106(23):9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am.J.Hum.Genet. 2007;81(3):559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Grady BJ, Torstenson E, Dudek SM, Giles J, Sexton D, Ritchie MD. Finding unique filter sets in plato: a precursor to efficient interaction analysis in gwas data. Pac.Symp.Biocomput. 2010:315–326. [PMC free article] [PubMed] [Google Scholar]
- 17.Packer BR, Yeager M, Burdett L, et al. SNP500Cancer: a public resource for sequence validation, assay development, and frequency analysis for genetic variation in candidate genes. Nucleic Acids Res. 2006;34(Database issue):D617–D621. doi: 10.1093/nar/gkj151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The International HapMap Project Nature. 2003;426(6968):789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 19.Hewett M, Oliver DE, Rubin DL, et al. PharmGKB: the Pharmacogenetics Knowledge Base. Nucleic Acids Res. 2002;30(1):163–165. doi: 10.1093/nar/30.1.163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Rieder MJ, Livingston RJ, Stanaway IB, Nickerson DA. The environmental genome project: reference polymorphisms for drug metabolism genes and genome-wide association studies. Drug Metab Rev. 2008;40(2):241–261. doi: 10.1080/03602530801952880. [DOI] [PubMed] [Google Scholar]
- 21.Sherry ST, Ward MH, Kholodov M, et al. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29(1):308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Nakamoto K, Kidd JR, Jenison RD, et al. Genotyping and haplotyping of CYP2C19 functional alleles on thin-film biosensor chips. Pharmacogenet.Genomics. 2007;17(2):103–114. doi: 10.1097/FPC.0b013e32801152c2. [DOI] [PubMed] [Google Scholar]
- 23.Mailman MD, Feolo M, Jin Y, et al. The NCBI dbGaP database of genotypes and phenotypes. Nat.Genet. 2007;39(10):1181–1186. doi: 10.1038/ng1007-1181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sissung TM, English BC, Venzon D, Figg WD, Deeken JF. Clinical pharmacology and pharmacogenetics in a genomics era: the DMET platform. Pharmacogenomics. 2010;11(1):89–103. doi: 10.2217/pgs.09.154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Crews KR, Gaedigk A, Dunnenberger HM, et al. Clinical Pharmacogenetics Implementation Consortium (CPIC) guidelines for codeine therapy in the context of cytochrome P450 2D6 (CYP2D6) genotype. Clin.Pharmacol.Ther. 2012;91(2):321–326. doi: 10.1038/clpt.2011.287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnson JA, Gong L, Whirl-Carrillo M, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for CYP2C9 and VKORC1 genotypes and warfarin dosing. Clin.Pharmacol.Ther. 2011;90(4):625–629. doi: 10.1038/clpt.2011.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Relling MV, Gardner EE, Sandborn WJ, et al. Clinical Pharmacogenetics Implementation Consortium Guidelines for Thiopurine Methyltransferase Genotype and Thiopurine Dosing: 2013 Update. Clin.Pharmacol.Ther. 2013 doi: 10.1038/clpt.2013.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Scott SA, Sangkuhl K, Gardner EE, et al. Clinical Pharmacogenetics Implementation Consortium guidelines for cytochrome P450-2C19 (CYP2C19) genotype and clopidogrel therapy. Clin.Pharmacol.Ther. 2011;90(2):328–332. doi: 10.1038/clpt.2011.132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wilke RA, Ramsey LB, Johnson SG, et al. The clinical pharmacogenomics implementation consortium: CPIC guideline for SLCO1B1 and simvastatin-induced myopathy. Clin.Pharmacol.Ther. 2012;92(1):112–117. doi: 10.1038/clpt.2012.57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilke RA, Reif DM, Moore JH. Combinatorial pharmacogenetics. Nat.Rev.Drug Discov. 2005;4(11):911–918. doi: 10.1038/nrd1874. [DOI] [PubMed] [Google Scholar]
- 31.Ingelman-Sundberg M. Genetic polymorphisms of cytochrome P450 2D6 (CYP2D6): clinical consequences, evolutionary aspects and functional diversity. Pharmacogenomics.J. 2005;5(1):6–13. doi: 10.1038/sj.tpj.6500285.* Interesting review of the clinical and evolutionary history of cytochrome P450 2D6.
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.