Abstract
Background
We explored use of a canine model of heart failure (HF) for pharmacogenomic discovery, specifically analyzing response to beta blockers (BB)
Methods
Dogs with HF that received BB (n=39) underwent genome-wide genotyping to test the association with changes in left ventricular (LV) volume and ejection fraction after treatment. Resulting candidate genes underwent RNA quantification in cardiac tissue from normal (n=5), placebo-HF (n=5), and BB-HF (n=7) dogs.
Results
Three markers met whole-genome significance for association with improved LV end-systolic volume after BB therapy (each p<5×10−7). RNA quantification of three candidate genes near these markers -- GUCA1B, RRAGD, and MRPS10 --revealed that gene expression levels in BB-HF dogs were between that of placebo-HF dogs and normal dogs.
Conclusion
Genome-wide pharmacogenomic screening in a canine model of HF suggests 3 novel BB response candidate loci. This approach is adaptable to discovering mechanisms of action for other drug therapies, and may be a useful strategy for identifying candidate genes for drug response in the pre-clinical setting.
Introduction
Heart failure (HF) continues to be an enormous public health problem despite the many advances in its pharmacotherapy over the past two decades (AHA, 2009). The availability of so many therapeutic options for HF makes optimization of therapy for an individual challenging due to the risks of polypharmacy (Francis and Young 2001; Tang and Francis, 2001) and because diversity of response to therapy between individuals or population subgroups is well recognized (van Campen et al., 1998). The expansion of human genomic information has led to exciting new opportunities for pharmacogenetics to improve the selection of drug therapy. Beta adrenergic antagonists (beta-blockers, BB) are an illustrative example where there have been numerous studies relating genetic sequence variants to a variety of BB effects (Johnson et al., 2003; Lanfear et al., 2005) including reverse-remodeling (Kaye et al., 2003; Terra et al., 2005) and survival in HF patients (Shin et al., 2007; Bristow et al., 2010).
Despite progress in discovering drug-response determinants, the candidate gene approach is limited to genes with a priori associations to the phenotype of interest, such as beta receptor variants with BBs. On the other hand, genome-wide association studies (GWAS) may be able to identify important genetic targets without preexisting knowledge or hypotheses. Advances in genotyping technology have made whole genome arrays more accessible, but despite this, pharmacogenomic progress remains costly and challenging due to the extensive genotyping needed for adequate genomic coverage in humans and the large sample size requirements due to the high number of multiple comparisons and the inherent “noise” in human clinical phenotypes. In addition, the number of studies required (to first detect and then validate an association), and the complexity of drug-response phenotypes, add to this difficulty. As a result almost all studies to date, save a few recent examples (Link et al., 2008), have taken a candidate gene approach. Specifically for BB, to our knowledge there are no published studies of genome-wide analysis of BB response in HF. If pharmacogenomics could take advantage of animal models of human disease, it may be able to accelerate the pace of discovery. Animal models have several advantages such as allowing for more uniform experimental conditions, study designs, and phenotype assessments that are not possible in humans (such as placebo-controlled trials of BB or routine myocardial sampling), as well as the potential advantage of increased efficiency of genetic mapping in some organisms. For example, an animal model would allow myocardial genotype:gene expression correlative experiments, but in contrast similar studies in humans would be extremely difficult to carry out due to ethical concerns (e.g., risks of myocardial biopsy in heart failure patients) and the inability to get a sufficient sample size to assess inter-individual variability by treatment assignment. For example, there are published studies examining gene expression pattern changes in humans with heart failure after BB therapy or cardiac resynchronization but the sample sizes in these studies are roughly 10–15 subjects and are all treated (i.e., no comparison between treated and untreated subjects).
Canine models offer many of these advantages; dogs display a number of diseases that are very similar to humans (Tsai et al., 2007), most known canine genes have human homologues (Lindblad-Toh et al., 2005), and the canine genome has long blocks of linkage disequilibrium which have enabled efficient mapping of genetic traits (Karlsson et al., 2007; Karlsson and Lindblad-Toh, 2008). Importantly, an established canine model of HF exists which has been tested extensively and is an excellent reflection of human HF (Kono et al., 1992; Sabbah et al., 1992), particularly in terms of response to drug therapies (Sabbah et al., 1991; Sabbah et al., 1994; Morita et al., 2002). Given this high-fidelity to human HF drug response and that the dog is a favorable model for genetic study, the canine HF model becomes an important setting to explore the possibility for pre-clinical pharmacogenomic study. Another advantage is the fact that there are existing data and samples from completed studies that could be leveraged. Thus, we sought to explore whether this canine model of HF could be used for pharmacogenomic discovery, focusing on response to BB as a test case.
Methods
Overview
This study utilized tissue from previously conducted studies of BB therapy in the canine model. Our group has conducted 4 previous experiments of BB therapy in dogs with heart failure, the preserved samples and data from which were used to complete the present studies. Samples from all BB treated animals (39 total), 5 control group animals (treated with placebo), as well as 5 normal dogs (i.e., no HF) were used in the described experiments.
Canine subjects
The studies were approved by the Institutional Review Board. The canine micro-embolic model of chronic HF has been previously described in detail (Morita et al., 2002). Briefly, HF is produced in dogs by multiple sequential intracoronary embolizations with polystyrene latex microspheres (~90 µm in diameter) that lead to loss of viable myocardium. These embolizations are performed weekly and the LV ejection fraction is repeatedly assessed until the goal level of LV dysfunction is reached, in this case an EF ≤35%. The model manifests many of the sequelae of HF in humans including profound systolic and diastolic left ventricular (LV) dysfunction, LV dilation and compensatory hypertrophy, increased LV filling pressures, increased systemic vascular resistance, and decreased cardiac output (Gengo et al., 1992; Sabbah et al., 1992; Sabbah et al., 1993). The HF cohort for this study is made up of 44 male dogs which had HF induced as above and then were treated with either BB (39 dogs, included several agents and doses, see below) or placebo (8 dogs). They were subsequently evaluated for response to treatment in terms of LV dimensions and ejection fraction after 3 months of treatment. In addition, samples from 5 dogs without HF were utilized in the gene-expression experiments (detailed below).
Beta blocker studies
There were 4 similar BB experiments from which the included animals were drawn. The time to achieve the goal EF (≤35%) varies from animal to animal, ranging from 2 to 8 weeks. Once the goal EF is achieved BB therapy was initiated after 2 weeks of recovery. The BB were initiated at half of the target dose for one week and then increased to the target dose which was continued for 3 months. The agent and goal dose in the 4 groups were as follows: 1) Metoprolol succinate 100 mg daily for 3 months (n=17); 2) Carvedilol 0.3 mg/kg Bid for 3 months (n=8); 3) Carvedilol 1 mg/kg Bid for 3 months (n=8); and 4) Atenolol 50 mg daily for 3 months (n=6). Outcomes assessed included the change in LV size and function from baseline (after inducing HF but before BB initiation) to the end of study period (3 months). This was achieved using cardiac catheterization with ventriculogram and the area-length method (Dodge et al., 1966) to assess LV end-systolic volume, end-diastolic volume, and ejection fraction. The dogs weighed between 20 and 25kg at baseline.
Genotyping and gene expression
DNA isolated from frozen tissue which had been stored at −70°C. genome-wide genotyping was accomplished using a commercially available canine genotyping array from Illumina® (CanineSNP20). The array utilizes the Infinium platform (Steemers et al., 2006) and tests ~22,000 validated polymorphisms across the canine genome, roughly equally spaced at an average distance of roughly 8kb. It has been validated in 10 different breeds and containing many breed informative markers. Markers meeting whole genome significance for association with LV reverse remodeling after BB treatment were further investigated, and 5 nearby genes were chosen for gene expression analysis. Canine myocardial samples were lysed and RNA was isolated according to standard techniques. Real time reverse transcriptase polymerase chain reaction (RT-PCR) was performed to quantify transcript abundance of genes of interest as well as a house keeper gene (GAPDH). The target gene RNA abundance was normalized to that of GAPDH. These ratio values were then divided by the ratio in the normal dog samples, to find a relative change in expression for heart failure dogs versus normal dogs.
Statistical analysis
The association of change in LV end-systolic volume (ESV), LV end-diastolic volume (EDV), and LV ejection fraction (EF) with genotype was tested using the ANOVA. Bonferonni correction was used to adjust for the 22,000 multiple comparisons, yielding a critical p value of 2.2 × 10−6
Results
Compared to placebo, BB treatment led to improvement in LV ESV and EF, regardless of BB regimen. Average changes in LV size and function (compared to baseline) during treatment were as follows: ESV −4.33 ml in BB treated dogs vs. +10.6 ml in placebo dogs (p<0.001); for EF it was +4.9 vs. −6.9 (p<0.001); and for EDV it was −1.6 ml vs. +8.5 ml (p<0.001). Among dogs receiving BB there was a substantial amount of variation in terms of improvement in LV size and function (Table 1). For example, the change in ESV over the treatment period ranged from −15 ml to +1 ml.
Table 1.
Changes in Ventricular Size and Function in HF Dogs Treated with BB (n=39)
| BB Group | Parameter | Mean Change | Std. Deviation | Range |
|---|---|---|---|---|
| Combined BB Group (n=39) | Δ Ejection Fraction (%) | 4.92 | 2.97 | −1, +11 |
| Δ End-Systolic Volume (ml) | −4.33 | 3.62 | −15, +1 | |
| Δ End-Diastolic Volume (ml) | −1.62 | 2.93 | −11, +7 | |
| Carvedilol 1mg/kg bid (n=8) | Δ Ejection Fraction (%) | 6.38 | 2.97 | +2, +11 |
| Δ End-Systolic Volume (ml) | −7.75 | 3.49 | −5, −15 | |
| Δ End-Diastolic Volume (ml) | −5.00 | 2.73 | −2, −11 | |
| Carvedilol 0.3mg/kg bid (n=8) | Δ Ejection Fraction (%) | 2.38 | 1.41 | 0, +5 |
| Δ End-Systolic Volume (ml) | −1.63 | 2.07 | −4, +1 | |
| Δ End-Diastolic Volume (ml) | 0.13 | 3.23 | −3, +7 | |
| Metoprolol Succinate 100mg daily (n=17) | Δ Ejection Fraction (%) | 6.65 | 1.73 | +3, +10 |
| Δ End-Systolic Volume (ml) | −5.18 | 2.98 | −15, −1 | |
| Δ End-Diastolic Volume (ml) | −1.47 | 1.91 | −5, 0 | |
| Atenolol 50mg daily (n=6) | Δ Ejection Fraction (%) | 1.50 | 2.07 | −1, 5 |
| Δ End-Systolic Volume (ml) | −1.00 | 1.26 | −3, 0 | |
| Δ End-Diastolic Volume (ml) | 0.17 | 1.33 | −1, +2 | |
| Placebo (n=8) | Δ Ejection Fraction (%) | −6.88 | 1.64 | −10, −5 |
| Δ End-Systolic Volume (ml) | 10.63 | 6.94 | +4, +24 | |
| Δ End-Diastolic Volume (ml) | 8.75 | 8.73 | 1, +25 |
Note: p<0.05 vs. baseline show in bold.
Association of beta blocker response with genotype
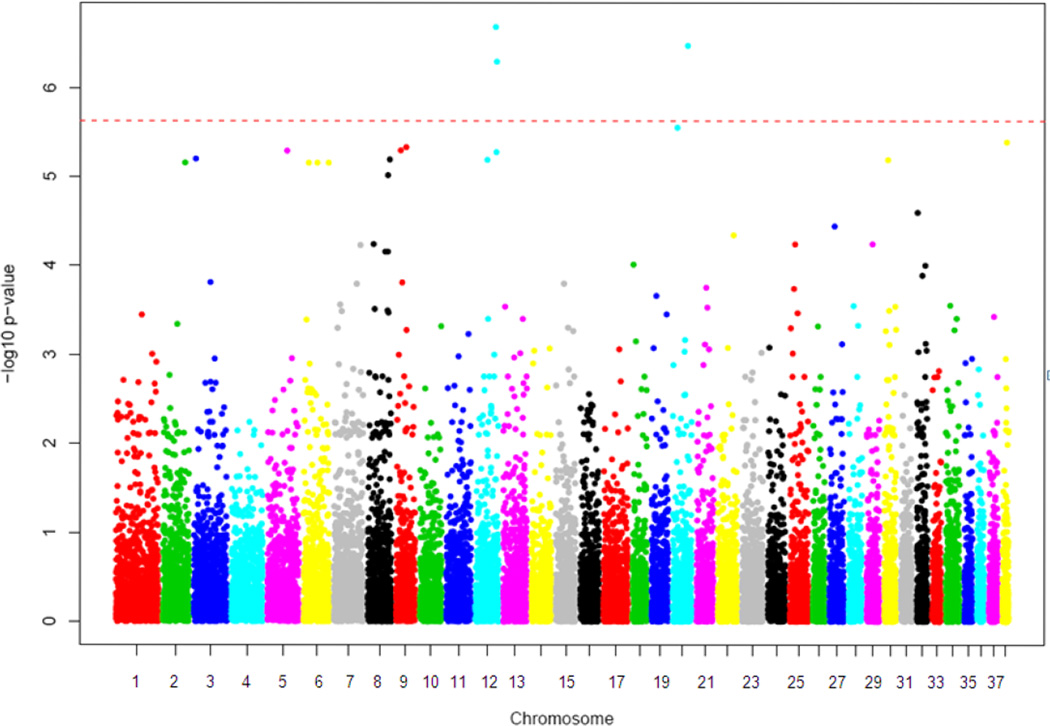
genome-wide markers were tested for association with changes in ESV, EDV, and EF after BB treatment. This revealed 3 markers meeting whole genome significance after Bonferonni correction for association with changes in ESV (Figure 1). There were several more markers in the near-significant range. No markers met genome-wide significance for change in EDV or EF. Upon interrogation of the locations of the statistically significant variants it was discovered that they were not nearby expected BB response elements (i.e., ADRB1, ADRB2) but instead were in genes/regions not previously associated with BB response. Further examination of the genes in these regions revealed that several had potential biologic plausibility in terms of effecting myocardial physiology or BB effects (summarized in Table 2).
Figure 1.
Manhattan plot of log(p) vs. chromosomal position of genetic markers. (Red dashed line indicates whole-genome significance level by Bonferonni correction.)
Table 2.
Genes in Region Near Markers That Reached Genome-wide Significance
| Variant ID | Chr. | p-value | Gene Symbol | Gene Name | Classification | Function |
|---|---|---|---|---|---|---|
| BICF2P138111 | 12 | 2.07E-07 | Predicted gene | Unknown | Unknown | Unknown |
| GUCA1A | Guanylate cyclase activator 1A | Calmodulin Superfamily | Regulator of guanylate cyclase activity | |||
| GUCA1B | Guanylate cyclase activator 1B | Calmodulin Superfamily | Regulator of guanylate cyclase activity | |||
| MRPS10 | Mitochondrial Ribosomal 28s protein S10 | Small subunit of mitoribosomes, mitochondrial protein synthesis | ||||
| BICF2P1313841 | 12 | 5.07E-07 | RRAGD | Ras-related GTP binding D | Ras Superfamily | Unknown |
| BICF2P351263 | 20 | 3.36E-07 | No known genes – lies within conserved intergenic region | |||
| near by (<1Mb) LMCD1 | Lim and cysteine rich domain protein 1 | transcriptional cofactor expressed in myocardium | involved in cell differentiation and cytoskeletal remodeling | |||
Gene expression analysis of genes indicated by genome-wide genotyping
Gene expression quantification using real time RT-PCR was attempted for each gene listed in Table 2 due to their proximity to an associated marker. Despite multiple attempts working probe sets were not able to be generated to interrogate Guanylate Cyclase Activator 1A (GUCA1A) and Lim and cysteine rich domain protein 1 (LMCD1). The remaining three genes of interest [GUCA1B, mitochondrial ribosomal 28s protein S10 (MRPS10), and ras-related GTP binding D (RRAGD)] successfully underwent further testing. We quantified RNA using myocardial tissue samples from 7 BB treated dogs (from high-dose carvedilol group), 5 placebo treated dogs, and 5 normal dogs (i.e., no HF). This was quantified via real-time RT-PCR, normalizing RNA abundance of the gene of interest to that of GAPDH. These ratio values were then divided by the ratio in the normal dog samples, to find a relative change in expression for heart failure animals (ΔΔCt method). The results revealed that each of the genes tested showed higher expression in placebo treated HF dogs compared to normal dogs, with roughly 3 to 4 fold greater expression (Table 3). This reached statistical significance for GUCA1B (p=0.02) and was borderline significant for MRPS10 (p=0.09). These expression differences appeared attenuated in HF dogs treated with carvedilol, which showed expression 2 to 3 fold that of normal dogs.
Table 3.
Candidate Gene Expression in BB Treated HF Dogs, Placebo Treated HF Dogs, and Normal Dogs for Novel Candidate Genes
| Gene | Normal (N=5) | Placebo (N=5) | Carvedilol (N=7) |
|---|---|---|---|
| Expression relative to Normal | |||
| GUCA1B (Guanylate cyclase activator 1B) |
1 | 3.7* | 2.8 |
| RRAGD (Ras related G protein D) |
1 | 2.95 | 2.75 |
| MRPS10 (Mitochondrial ribosomal 28s protein S10) |
1 | 3.8† | 2.6 |
Note: Shown as average relative expression compared to normal dog samples, using ΔΔCt method to normalize to simultaneous sample GAPDH expression
p<0.05 vs. Normal
p<0.01 vs. Normal
Discussion
In this study we were able to identify genetic markers associated with favorable remodeling in response to BB treatment in a canine model of HF. Several markers met genome-wide significance after Bonferonni correction, which tends to be conservative, and this was achieved with a relatively low number of animals (n=39). Subsequent gene expression analysis showed differences in transcript abundance between BB treated dogs, placebo treated HF dogs, and normal dogs which, while modest in magnitude, were consistent in direction with the hypothesis that these novel loci are relevant to BB response in HF.
To varying degrees, each of these three genes has some biologic plausibility. The function of RRAGD is not completely established, but it is a member of the ras-like GTPase superfamily of proteins which are known to regulate a wide range of intracellular processes. These include gene expression, cytoskeletal processes, translation, and cell division. Its specific function and role in HF are not established. The function of GUCA1B is better known, it being a recognized stimulator of guanylate cyclase in the retina. Whether it could have effects elsewhere is not known, though some guanylate cyclases are known to have important cardiovascular effects (GCA/NPRA) and GUCA1B appears to be expressed in heart, skeletal muscle, and kidney, though not at the level of the retinal expression. On the other hand, MRPS10 is known to code for the small subunit of mitochondrial ribosomes. Myocardial energetics is being increasingly recognized to play an important role in HF pathophysiology (Huss and Kelly, 2004; Qanud et al., 2008), and BB treatment is known to impact this (Wallhaus et al., 2001; Sabbah 2004). Thus, it would not be difficult to hypothesize that variability in how individuals’ mitochondria function may impact the development of HF or reverse-remodeling in response to BB therapy.
Our data are the first we are aware of demonstrating pharmacogenomic discovery in a large mammal disease model. Our model system has the distinct advantage that it is known to be an accurate predictor of drug response in human HF, across multiple drug classes and specifically for BBs. While pharmacogenetic and pharmacogenomic studies have been pursued in mice, the ability to use the canine model would be a significant advance because the findings may be more directly applicable to human disease and treatment, as evidenced by the high fidelity of this canine model to human heart failure. Furthermore, the potentially favorable genomic structure in dogs may allow genome-wide screening for pharmacogenetic interactions using a relatively small number of animals, thus increasing speed while decreasing cost of pharmacogenomic discovery. This may also allow subsequent human studies to utilize an expanded candidate gene approach (rather than whole genome), greatly accelerating the pace of progress while not limiting discovery to genes with a priori associations to the drug of interest. Finally, this is of high potential impact because the approach could be applied to any drug of interest or conceivably even medical devices.
Our study has limitations that should be noted. First, we did not study a large enough sample of placebo treated HF dogs to perform drug x genotype interactions. Despite this, predicting favorable response within treated animals (i.e., pre-post change) is an important end-point, and this study design has been often used in comparable human studies. Second, this analysis includes four individual BB studies that were previously carried out. While the drug and dose regimens were different, the heart failure induction, duration of treatment, and outcomes assessed were all identical, making it suitable to merge them in order to increase power. Conversely the individual study sizes were not large enough to allow evaluation of individual agents. Another concern is that the dogs were mongrels, so that population stratification could theoretically lead to false-positive associations. Confirmatory studies are needed to define this further. Finally, two of five candidates could not have RNA quantified so the relationship of gene expression to HF and BB response for these genes remains unknown.
Conclusion
We described the first use of a canine model of HF in a genome-wide search for genetic predictors of response to drug therapy, identifying novel candidate genes for BB response. Our data suggest that this approach may be useful, having the potential to yield significant results with a modest sample size in a pre-clinical research setting. This type of approach requires further scrutiny, but if proven useful, could greatly accelerate pharmacogenomic discovery.
Acknowledgment
This work was supported in part by research grants from the National Heart, Lung, and Blood Institute K23HL085124 (to D.E.L.) and PO1HL074237-06 (to H.N.S.).
Abbreviations
- BB
beta blocker
- EDV
end diastolic volume
- EF
ejection fraction
- ESV
end-systolic volume
- GWAS
genome-wide association studies
- HF
heart failure
- LV
left ventricular
Footnotes
Disclosure
The authors report no conflicts of interest.
Contributor Information
David E. Lanfear, Division of Cardiology, Department of Internal Medicine, Henry Ford Hospital, Detroit, Michigan 48202, USA
James J. Yang, Department of Public Health Sciences, Henry Ford Hospital, Detroit, Michigan 48202, USA
Sudhish Mishra, Division of Cardiology, Department of Internal Medicine, Henry Ford Hospital, Detroit, Michigan 48202, USA
Hani N. Sabbah, Division of Cardiology, Department of Internal Medicine, Henry Ford Hospital, Detroit, Michigan 48202, USA
References
- AHA. Heart Disease and Stroke Statistics - 2009 Update. Dallas, Texas, USA: American Heart Association; 2009. [Google Scholar]
- Bristow MR, Murphy GA, Krause-Steinrauf H, Anderson JL, Carlquist JF, Thaneemit-Chen S, Krishnan V, Abraham WT, Lowes BD, Port JD, Davis GW, Lazzeroni LC, Robertson AD, Lavori PW, Liggett SB. An alpha2C–adrenergic receptor polymorphism alters the norepinephrine-lowering effects and therapeutic response of the beta-blocker bucindolol in chronic heart failure. Circ Heart Fail. 2010;3(1):21–28. doi: 10.1161/CIRCHEARTFAILURE.109.885962. [DOI] [PubMed] [Google Scholar]
- Dodge HT, Sandler H, Baxley WA, Hawley RR. Usefulness and limitations of radiographic methods for determining left ventricular volume. Am J Cardiol. 1966;18(1):10–24. doi: 10.1016/0002-9149(66)90191-3. [DOI] [PubMed] [Google Scholar]
- Francis GS, Young JB. The looming polypharmacy crisis in the management of patients with heart failure. Potential solutions. Cardiol Clin. 2001;19(4):541–545. doi: 10.1016/s0733-8651(05)70241-1. [DOI] [PubMed] [Google Scholar]
- Gengo PJ, Sabbah HN, Steffen RP, Sharpe JK, Kono T, Stein PD, Goldstein S. Myocardial beta adrenoceptor and voltage sensitive calcium channel changes in a canine model of chronic heart failure. J Mol Cell Cardiol. 1992;24(11):1361–1369. doi: 10.1016/0022-2828(92)93100-x. [DOI] [PubMed] [Google Scholar]
- Huss JM, Kelly DP. Nuclear receptor signaling and cardiac energetics. Circ Res. 2004;95(6):568–578. doi: 10.1161/01.RES.0000141774.29937.e3. [DOI] [PubMed] [Google Scholar]
- Johnson JA, Zineh I, Puckett BJ, McGorray SP, Yarandi HN, Pauly DF. Beta 1-adrenergic receptor polymorphisms and antihypertensive response to metoprolol. Clin Pharmacol Ther. 2003;74(1):44–52. doi: 10.1016/S0009-9236(03)00068-7. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Baranowska I, Wade CM, Salmon Hillbertz NH, Zody MC, Anderson N, Biagi TM, Patterson N, Pielberg GR, Kulbokas EJ, 3rd, Comstock KE, Keller ET, Mesirov JP, von Euler H, Kampe O, Hedhammar A, Lander ES, Andersson G, Andersson L, Lindblad-Toh K. Efficient mapping of mendelian traits in dogs through genome-wide association. Nat Genet. 2007;39(11):1321–1328. doi: 10.1038/ng.2007.10. [DOI] [PubMed] [Google Scholar]
- Karlsson EK, Lindblad-Toh K. Leader of the pack: gene mapping in dogs and other model organisms. Nat Rev Genet. 2008;9(9):713–725. doi: 10.1038/nrg2382. [DOI] [PubMed] [Google Scholar]
- Kaye DM, Smirk B, Williams C, Jennings G, Esler M, Holst D. Beta-adrenoceptor genotype influences the response to carvedilol in patients with congestive heart failure. Pharmacogenetics. 2003;13(7):379–382. doi: 10.1097/00008571-200307000-00002. [DOI] [PubMed] [Google Scholar]
- Kono T, Sabbah HN, Rosman H, Alam M, Stein PD, Goldstein S. Left atrial contribution to ventricular filling during the course of evolving heart failure. Circulation. 1992;86(4):1317–1322. doi: 10.1161/01.cir.86.4.1317. [DOI] [PubMed] [Google Scholar]
- Lanfear DE, Jones PG, Marsh S, Cresci S, McLeod HL, Spertus JA. Beta2-adrenergic receptor genotype and survival among patients receiving beta-blocker therapy after an acute coronary syndrome. JAMA. 2005;294(12):1526–1533. doi: 10.1001/jama.294.12.1526. [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K, Wade CM, Mikkelsen TS, Karlsson EK, Jaffe DB, Kamal M, Clamp M, Chang JL, Kulbokas EJ, 3rd, Zody MC, Mauceli E, Xie X, Breen M, Wayne RK, Ostrander EA, Ponting CP, Galibert F, Smith DR, DeJong PJ, Kirkness E, et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature. 2005;438(7069):803–819. doi: 10.1038/nature04338. [DOI] [PubMed] [Google Scholar]
- Link E, Parish S, Armitage J, Bowman L, Heath S, Matsuda F, Gut I, Lathrop M, Collins R. SLCO1B1 variants and statin-induced myopathy — a genomewide study. N Engl J Med. 2008;359(8):789–799. doi: 10.1056/NEJMoa0801936. [DOI] [PubMed] [Google Scholar]
- Morita H, Suzuki G, Mishima T, Chaudhry PA, Anagnostopoulos PV, Tanhehco EJ, Sharov VG, Goldstein S, Sabbah HN. Effects of long-term monotherapy with meto-prolol CR/XL on the progression of left ventricular dysfunction and remodeling in dogs with chronic heart failure. Cardiovasc Drugs Ther. 2002;16(5):443–449. doi: 10.1023/a:1022142620189. [DOI] [PubMed] [Google Scholar]
- Qanud K, Mamdani M, Pepe M, Khairallah RJ, Gravel J, Lei B, Gupte SA, Sharov VG, Sabbah HN, Stanley WC, Recchia FA. Reverse changes in cardiac substrate oxidation in dogs recovering from heart failure. Am J Physiol Heart Circ Physiol. 2008;295(5):H2098–H2105. doi: 10.1152/ajpheart.00471.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabbah HN. Biologic rationale for the use of beta-blockers in the treatment of heart failure. Heart Fail Rev. 2004;9(2):91–97. doi: 10.1023/B:HREV.0000046363.59374.23. [DOI] [PubMed] [Google Scholar]
- Sabbah HN, Goldberg AD, Schoels W, Kono T, Webb C, Brachmann J, Goldstein S. Spontaneous and inducible ventricular arrhythmias in a canine model of chronic heart failure: relation to haemodynamics and sympathoadrenergic activation. Eur Heart J. 1992;13(11):1562–1572. doi: 10.1093/oxfordjournals.eurheartj.a060102. [DOI] [PubMed] [Google Scholar]
- Sabbah HN, Kono T, Stein PD, Mancini GB, Goldstein S. Left ventricular shape changes during the course of evolving heart failure. Am J Physiol. 1992;263(1 Pt 2):H266–H270. doi: 10.1152/ajpheart.1992.263.1.H266. [DOI] [PubMed] [Google Scholar]
- Sabbah HN, Levine TB, Gheorghiade M, Kono T, Goldstein S. Hemodynamic response of a canine model of chronic heart failure to intravenous dobutamine, nitroprusside, enalaprilat, and digoxin. Cardiovasc Drugs Ther. 1993;7(3):349–356. doi: 10.1007/BF00880158. [DOI] [PubMed] [Google Scholar]
- Sabbah HN, Shimoyama H, Kono T, Gupta RC, Sharov VG, Scicli G, Levine TB, Goldstein S. Effects of long-term monotherapy with enalapril, metoprolol, and digoxin on the progression of left ventricular dysfunction and dilation in dogs with reduced ejection fraction. Circulation. 1994;89(6):2852–2859. doi: 10.1161/01.cir.89.6.2852. [DOI] [PubMed] [Google Scholar]
- Sabbah HN, Stein PD, Kono T, Gheorghiade M, Levine TB, Jafri S, Hawkins ET, Goldstein S. A canine model of chronic heart failure produced by multiple sequential coronary microembolizations. Am J Physiol. 1991;260(4 Pt 2):H1379–H1384. doi: 10.1152/ajpheart.1991.260.4.H1379. [DOI] [PubMed] [Google Scholar]
- Shin J, Lobmeyer MT, Gong Y, Zineh I, Langaee TY, Yarandi H, Schofield RS, Aranda JM, Jr, Hill JA, Pauly DF, Johnson JA. Relation of beta(2)-adrenoceptor haplotype to risk of death and heart transplantation in patients with heart failure. Am J Cardiol. 2007;99(2):250–255. doi: 10.1016/j.amjcard.2006.08.020. [DOI] [PubMed] [Google Scholar]
- Steemers FJ, Chang W, Lee G, Barker DL, Shen R, Gunderson KL. Whole-genome genotyping with the single-base extension assay. Nat Methods. 2006;3:31–33. doi: 10.1038/nmeth842. [DOI] [PubMed] [Google Scholar]
- Tang WH, Francis GS. Polypharmacy of heart failure. Creating a rational pharmacotherapeutic protocol. Cardiol Clin. 2001;19(4):583–596. doi: 10.1016/s0733-8651(05)70245-9. viii. [DOI] [PubMed] [Google Scholar]
- Terra SG, Hamilton KK, Pauly DF, Lee CR, Patterson JH, Adams KF, Schofield RS, Belgado BS, Hill JA, Aranda JM, Yarandi HN, Johnson JA. Beta1-adrenergic receptor polymorphisms and left ventricular remodeling changes in response to beta-blocker therapy. Pharmacogenet Genomics. 2005;15(4):227–234. doi: 10.1097/01213011-200504000-00006. [DOI] [PubMed] [Google Scholar]
- Tsai KL, Clark LA, Murphy KE. Understanding hereditary diseases using the dog and human as companion model systems. Mamm Genome. 2007;18(6–7):444–451. doi: 10.1007/s00335-007-9037-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Campen LC, Visser FC, Visser CA. Ejection fraction improvement by beta-blocker treatment in patients with heart failure: an analysis of studies published in the literature. J Cardiovasc Pharmacol. 1998;32(Suppl 1):S31–S35. doi: 10.1097/00005344-199800003-00006. [DOI] [PubMed] [Google Scholar]
- Wallhaus TR, Taylor M, DeGrado TR, Russell DC, Stanko P, Nickles RJ, Stone CK. Myocardial free fatty acid and glucose use after carvedilol treatment in patients with congestive heart failure. Circulation. 2001;103(20):2441–2446. doi: 10.1161/01.cir.103.20.2441. [DOI] [PubMed] [Google Scholar]