Abstract
Genome wide association studies revealed that variation in the Melatonin Receptor 1B gene (MTNR1B) is associated with insulin and glucose concentrations. Here we show that the risk genotype of this SNP predicts future type 2 diabetes (T2D) in two large prospective studies. Specifically, the risk genotype was associated with impairment of early insulin response to both oral and intravenous glucose and with faster deterioration of insulin secretion over time. We also show that the Melatonin Receptor 1B mRNA is expressed in human islets, and immunocytochemistry confirms that it is primarily localized in β-cells in islets. Non-diabetic individuals carrying the risk allele and patients with T2D showed increased expression of the receptor in islets. Insulin release from clonal β-cells in response to glucose was inhibited in the presence of melatonin. These data suggest that the circulating hormone melatonin, which is predominantly released from the pineal gland in the brain, is involved in the pathogenesis of T2D. Given the increased expression of Melatonin Receptor 1B in individuals at risk of T2D, the pathogenic effects are likely exerted via a direct inhibitory effect on β-cells. In view of these results, blocking the melatonin ligand-receptor system could be a therapeutic avenue in T2D.
Type 2 diabetes (T2D) incidence and prevalence are increasing at an alarming rate worldwide. It is well established that T2D is multifactorial and that multiple genes and environmental and behavioral factors combine to cause the disease. The recent genome-wide association studies (GWAS) have provided new insights into the nature of these genetic factors1–5. Many of the T2D-associated variants identified in these studies appear to influence the capacity of β-cells to cope with increased insulin demands imposed by insulin resistance. One of the GWAS (Diabetes Genetics Inititative; DGI) also provided information on association with 18 quantitative traits (www.broad.mit.edu/diabetes)1, including measures of insulin secretion and action. One of the strongest signals for glucose-stimulated insulin secretion in the DGI scan emanated from a SNP (rs10830963) in the melatonin receptor gene (MTNR1B) on chromosome 11 (P=7×10−4, rank order 595). Given that the melatonin pathway had previously been suggested to be involved in pathogenesis of T2D made the MTNR1B gene a prime candidate gene for T2D. This SNP was also strongly associated (P=3.2 × 10−50) with elevated fasting glucose concentrations in a meta-analysis of the recent GWAS of T2D 6.
Melatonin is a circulating hormone predominantly secreted from the pineal gland, although other endocrine cell systems may also synthesize and release this hormone7, which then could exert hitherto unknown autocrine and paracrine effects8. Melatonin is an indoleamine formed from tryptophan via acetylation and subsequent methylation of the neurotransmitter serotonin. It has primarily been implicated in the regulation of circadian rhythms and circulating levels of the hormone are high during night and drop during daylight7. In fact, it has been proposed that melatonin could be involved in a circadian lowering of nocturnal insulin levels9. Effects of melatonin are mediated by two distinct receptors, MTNR1A and MTNR1B10, which are members of the G-protein coupled receptor family, specifically inhibitory G-proteins (Gi). Both receptors have been found to be expressed in human and rodent islets11, with MTNR1A predominating, especially in glucagon-producing α-cells12. There is some evidence that melatonin may exert an effect on insulin secretion, in that acute effects exerted by cAMP-elevating agents are inhibited by melatonin, while prolonged effects of the hormone may be stimulatory7. Here we provide novel evidence that the common variant rs10830963 in the MTNR1B gene or variant(s) in linkage disequilibrium with it increases risk of future T2D by causing impaired early insulin secretion. Further we present functional data that suggest a potential role of the melatonin system, in particular the MTNR1B receptor for regulation of glucose homeostasis in man.
A variant in the MTNR1B gene increases future risk of T2D and is associated with increased fasting glucose levels
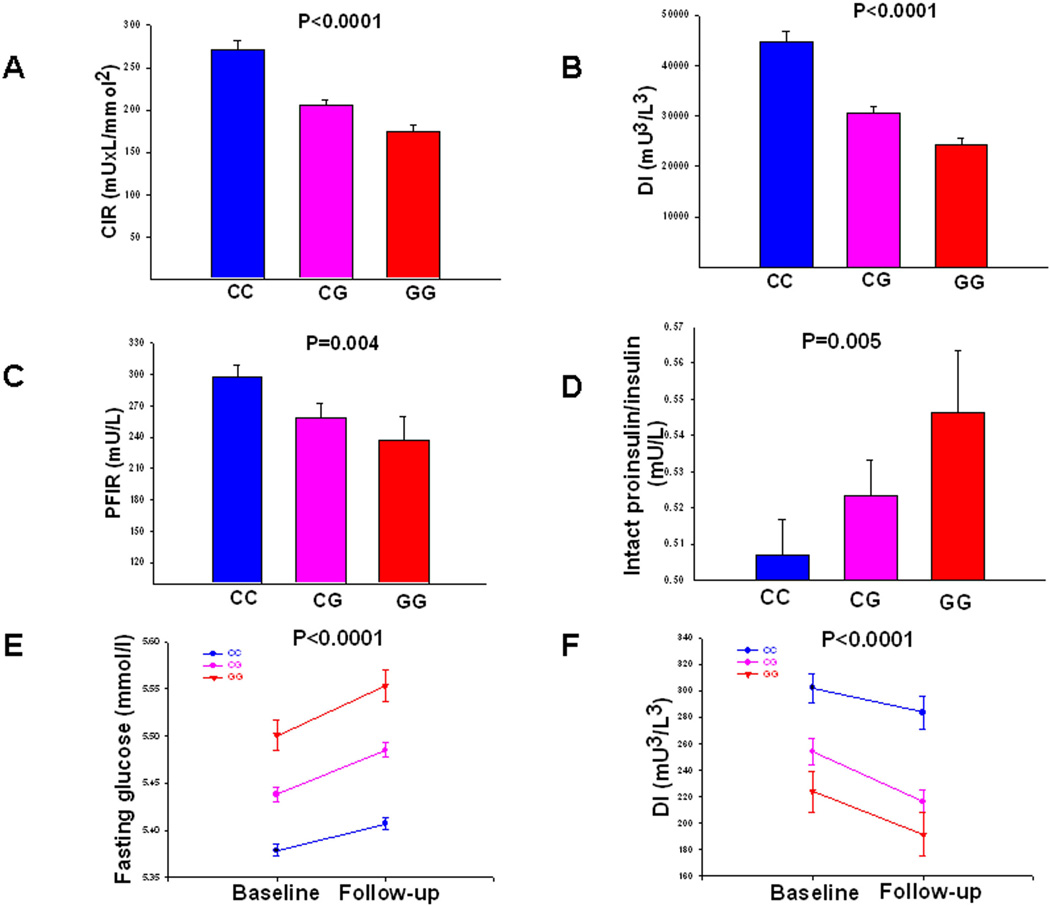
First, we studied whether the MTNR1B rs10830963 SNP predicts future T2D in 16,061 Swedish (from the Malmoe Preventive Project, MPP) and 2,770 Finnish (from the Botnia study) subjects, 2,201 (2063/138) of whom developed diabetes during 400,000 follow-up years (Table 1). The frequency of the risk G-allele of SNP rs10830963 was higher in individuals from the MPP study who converted to T2D compared to non-converters (30.2% vs 28.0%, P=0.002). This yielded a modestly increased risk of 1.12 (95%CI 1.04–1.20, P=0.002). There was no significant difference between converters and non-converters in the Botnia study, but here only 138 individuals developed T2D during a 7 year follow-up period (31.0% vs 29.3%; OR 1.09, 95%CI 0.82–1.43, P=0.56). In the combined analysis of the two cohorts, the risk allele was associated with a 1.11-fold increased risk of future T2D (95% CI 1.03–1.18, P=0.004). This relatively modest risk for future T2D probably explains why this SNP was not identified as being associated with T2D in previous GWAS (OR 1.12 (95% CI 1.04– 1.20), P=0.003 in DIAGRAM). However, the effect on glucose levels seems much stronger; in non-diabetic individuals from the MPP study, risk G-allele carriers displayed a higher fasting plasma glucose concentration at baseline (CC: 5.38±0.54 mmol/l, CG: 5.44±0.55 mmol/l, GG 5.50±0.55 mmol/l, P=3×10−19), which remained elevated throughout the 25-year follow-up period (CC: 5.41±0.54 mmol/l, CG: 5.49±0.54 mmol/l, GG 5.55±0.54 mmol/l, P=2×10−31) (Figure 1E).
Table 1.
Samples used in this study.
| Study | N (with diabetes) | Geographic Origin |
Age (yrs) | BMI (kg/m2) |
|---|---|---|---|---|
| Malmö Preventive Project (MPP) |
16,061 (2,063) | Sweden | 45.5 (6.9) | 24.3 (3.3) |
| Botnia PPP | 3,300 | Finland | 48.5 (15.9) | 26.1 (4.2) |
| Botnia prospective cohort | 2,770 (138) | Finland | 44.9 (14.2) | 25.6 (4.1) |
| Helsinki Birth Cohort | 1,600 | Finland | 61. 6 (3.0) | 27.1 (4.3) |
| FUSION | 522 | Finland | 39.1 (12.2) | 26.0 (6.4) |
| METSIM | 4,369 | Finland | 59.3 (2.8) | 26.9 (3.8) |
Data are mean (SD).
Figure 1. Insulin secretion according to different MTNR1Brs10830963 genotypes.
(A) Corrected early insulin response to glucose (CIR) during OGTT (Botnia PPP cohort; N=3,300), (B) Disposition index (DI) represents early insulin response to glucose corrected for insulin sensitivity by the Matsuda index (CIR × ISI, Botnia PPP cohort; N=3,300). (C) Insulin secretion measured as first phase insulin response during an IVGTT (Botnia cohort; N=505). (D) Intact proinsulin-to-insulin ratio in the fasting state (Helsinki Birth Cohort, N=1,600). (F) Change in fasting plasma glucose concentrations during 24-year follow-up in non-diabetic subjects (Malmoe study, N=13,674) (E) Change in insulin secretion (disposition index) over time in non-diabetic subjects (Botnia prospective cohort, N=2,444). Bars represent mean ± SEM. Blue lines represent non-risk and red lines risk genotype carriers of rs10830963 in MTNR1B.
A variant in the MTNR1B gene is associated with impaired early insulin response to glucose
Next, we examined insulin secretion in 3,300 non-diabetic participants from the population-based Botnia PPP study. We observed a dose-dependent decrease (corrected early insulin response to glucose (CIR), beta −0.170±0.021, P=5×10−16 and disposition index (DI), −0.241±0.022, P=1×10−26) with increasing number of G-alleles of rs10830963 (Table 2, Figure 1A and B). These findings were replicated in the METabolic Syndrome In Men (METSIM) study where both CIR (beta −0.143±0.022, P=1×10−10) and DI (beta − 0.128±0.022, P=9×10−9) were associated with rs10830963 in 4,257 subjects.
Table 2.
Effect of the MTNR1B rs10830963 on insulin secretion in the studied cohorts.
| Study | Phenotype | Genotypes | Additive model |
|||||
|---|---|---|---|---|---|---|---|---|
| CC | CG | GG | RA | BETA | SE | P Value | ||
|
DGI WGAS (OGTT n=1,020) |
Age (yrs) | 59±10 | 59±10 | 58±10 | - | 0.74 | ||
| BMI (kg/m2) | 26.5±3.6 | 26.7±4.0 | 27.3±3.7 | - | 0.14 | |||
| Fasting P-glucose (mmol/l) | 5.28±0.53 | 5.32±0.52 | 5.38±0.60 | 0.31 | 0.045 | 0.022 | 0.039 | |
| CIR (mUxL/mmol2) | 180±360 | 165±1912 | 144±163 | −0.166 | 0.048 | 7×10−4 | ||
| DI (mU3/L3) | 24036±29445 | 20285±27763 | 16555±22974 | −0.173 | 0.046 | 2×10−4 | ||
|
Botnia PPP (OGTT n=3,300) |
CC | CG | GG | |||||
| Age (yrs) | 48.3±16.0 | 48.5±15.9 | 49.6±15.9 | - | 0.38 | |||
| BMI (kg/m2) | 26.12−4.21 | 26.22−4.22 | 26.19−3.82 | - | 0.79 | |||
| Fasting P-glucose (mmol/l) | 5.06±0.54 | 5.25±0.55 | 5.28±0.55 | 0.30 | 0.134 | 0.014 | 2×10−22 | |
| CIR (mUxL/mmol2) | 271±415 | 205±245 | 175±134 | −0.170 | 0.021 | 5×10−16 | ||
| DI (mU3/L3) | 44631±87537 | 30499±49947 | 24316±21582 | −0.241 | 0.022 | 1×10−26 | ||
|
Botnia prospective (OGTT n=2,444) |
Baseline | CC | CG | GG | ||||
| Age (yrs) | 45.8±13.2 | 45.1±13.8 | 45.6±14.2 | - | 0.52 | |||
| BMI (kg/m2) | 25.5±4.1 | 25.7±3.7 | 25.7±3.8 | - | 0.48 | |||
| Fasting P-glucose (mmol/l) | 5.47±0.57 | 5.55±0.57 | 5.64±0.54 | 0.29 | 0.081 | 0.019 | 1×105 | |
| CIR (mUxL/mmol2) | 176±183 | 150±164 | 129±137 | −0.160 | 0.026 | 6×10−10 | ||
| DI (mU3/L3) | 302±361 | 254±306 | 224±212 | −0.171 | 0.026 | 9×10−11 | ||
| Follow-up | CC | CG | GG | |||||
| Age (yrs) | 53.8±13.8 | 52.7±14.3 | 53.3±14.9 | - | 0.25 | |||
| BMI (kg/m2) | 26.5±4.1 | 26.7±4.2 | 26.7±4.2 | - | 0.41 | |||
| Fasting P-glucose (mmol/l) | 5.25±0.56 | 5.34±0.56 | 5.41±0.61 | 0.086 | 0.019 | 5×10−6 | ||
| CIR (mUxL/mmol2) | 234±238 | 188±192 | 145±125 | −0.188 | 0.026 | 1×10−12 | ||
| DI (mU3/L3) | 284±429 | 217±259 | 191±221 | −0.179 | 0.029 | 8×10−10 | ||
|
Helsinki Birth Cohort (OGTT n=1,600) |
CC | CG | GG | |||||
| AGE (yrs) | 61.6±3.0 | 61.5±3.0 | 61.6±3.1 | - | - | 0.96 | ||
| BMI (kg/m2) | 27.0±4.2 | 27.2±4.4 | 27.1±4.2 | - | - | 0.53 | ||
| Fasting P-glucose (mmol/l) | 5.41±0.55 | 5.55±0.56 | 5.59±0.53 | 0.34 | 0.096 | 0.019 | 3×10−7 | |
| CIR (mUxL/mmol2) | 209±196 | 175±150 | 177±188 | −0.109 | 0.027 | 5×10−5 | ||
| DI (mU3/L3) | 19646±21504 | 15552±15063 | 15699±17881 | −0.122 | 0.027 | 8×10−6 | ||
| Intact proinsulin/insulin | 0.51±0.26 | 0.52±0.26 | 0.55±0.24 | 0.024 | 0.009 | 0.005 | ||
|
METSIM (n=4,257) |
Age (yrs) | 59.3±5.8 | 59.4±5.8 | 59.1±5.7 | 0.36 | - | - | - |
| BMI (kg/m2) | 26.9±3.9 | 26.9±3.7 | 26.5±3.7 | −0.058 | 0.020 | 4.3×10−3 | ||
| Fasting P-glucose (mmol/L) | 5.6±0.5 | 5.7±0.5 | 5.8±0.5 | 0.165 | 0.022 | 9.4×10−14 | ||
| CIR (mU×L/mmol2) | 196±212 | 168±165 | 152±143 | −0.143 | 0.022 | 1.3×10−10 | ||
| DI (mU3/L3) | 21554±28426 | 17878±18235 | 16798±16461 | −0.128 | 0.022 | 9.8×10−9 | ||
|
Botnia (IVGTT n=505) FUSION (FSIGT n=522) |
CC | CG | GG | |||||
| FPIR | 297±195 | 259±194 | 237±139 | 0.27 | −0.065 | 0.023 | 0.004 | |
| AIR (pM×8 min) | 2632±1731 | 2064±1468 | 1554±1092 | 0.35 | −0.316 | 0.067 | 2×10−6 | |
Data are means ± SD. CIR=corrected early insulin response to glucose during OGTT. DI=disposition index. FPIR=first phase insulin response during IVGTT. AIR = acute insulin response during frequently-sampled intravenous glucose tolerance test (FSIGT). RA= risk allele.
In the Botnia prospective study, 2,444 non-diabetic carriers of the G-allele showed lower insulin secretion at baseline (CIR, −0.160±0.026, P=6×10−10 and DI, − 0.171±0.026, P=9×10−11), which was maintained lower throughout the 7-year follow-up period (CIR, −0.188±0.026, P=1×10−12 and DI, −0.179±0.029, P=8×10−10) (Figure 1F). Further, the G allele of rs10830963 was also associated with impaired insulin secretion during an intravenous glucose tolerance test in 505 non-diabetic individuals from the Botnia study (FPIR, −0.065±0.023, P=0.004) (Figure 1C). The G allele of rs10830963 was also associated with reduced acute insulin response to glucose (AIR; P=2.2×10−6 and DI, P=5.0×10−3) in 522 non-diabetic individuals from the FUSION study13 (Table 2).
Finally, we examined whether the SNP would influence proinsulin processing as reflected in the ratio between proinsulin and insulin in 1,600 non-diabetic participants of the Helsinki Birth Cohort Study14. Also here, carriers of the MTNR1B risk genotype had impaired early insulin response to oral glucose (CIR, −0.109±0.027, P=5×10−5 and DI, − 0.122±0.027, P=8×10−6) (Table 2). In addition, risk allele carriers had an elevated intact proinsulin-to-insulin ratio (P=0.005) (Table 2, Figure 1D). However, an increased proinsulin-to-insulin ratio does not a priori imply a specific defect in proinsulin processing as proinsulin concentrations rise under most conditions of stressed β-cells.
The melatonin 1 B receptor (MTNR1B) is expressed in human islets and in β-cells
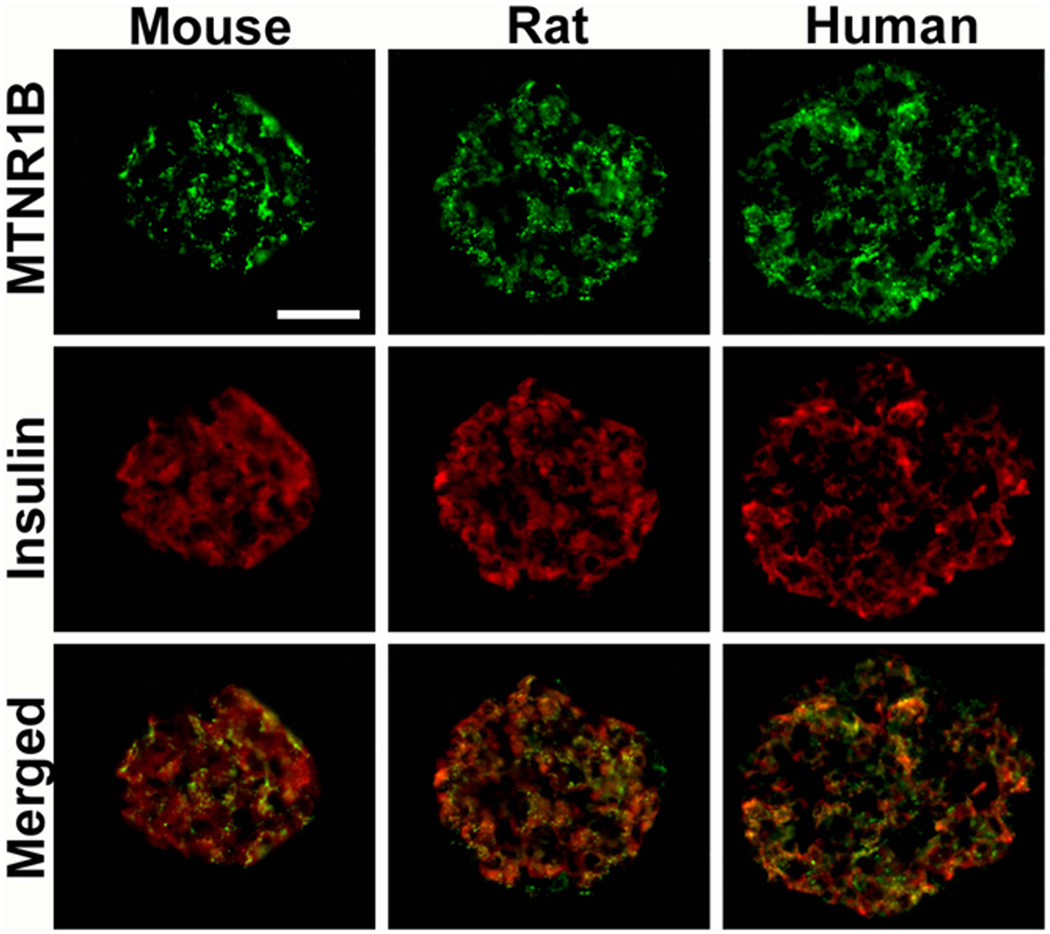
Using quantitative RT-PCR (Taqman®), we observed that both MTNR1A and MTNR1B were expressed in human islets as well as in clonal β-cells. In contrast to previous findings11,12, both receptors were expressed at near equal level in human islets. Moreover, islet expression of MTNR1B was confirmed by immunocytochemistry (Figure 2). Again, in contrast to a previous report, where single cell PCR identified MTNR1A mRNA primarily in α-cells12, we observed expression of MTNR1B predominantly in β-cells in both human and rodent islets (Figure 2). Also MTNR1A was observed in islets; its expression was less abundant and appeared to be restricted to a population of peripherally located β-cells in human, mouse, and rat islets.
Figure 2. Co-localization of MTNR1B expression with insulin in mouse, rat and human pancreatic islets.
Scale bar = 50um.
Expression of the MTNR1B gene is increased in carriers of the risk genotype of MTNR1B SNP and correlates negatively with insulin secretion
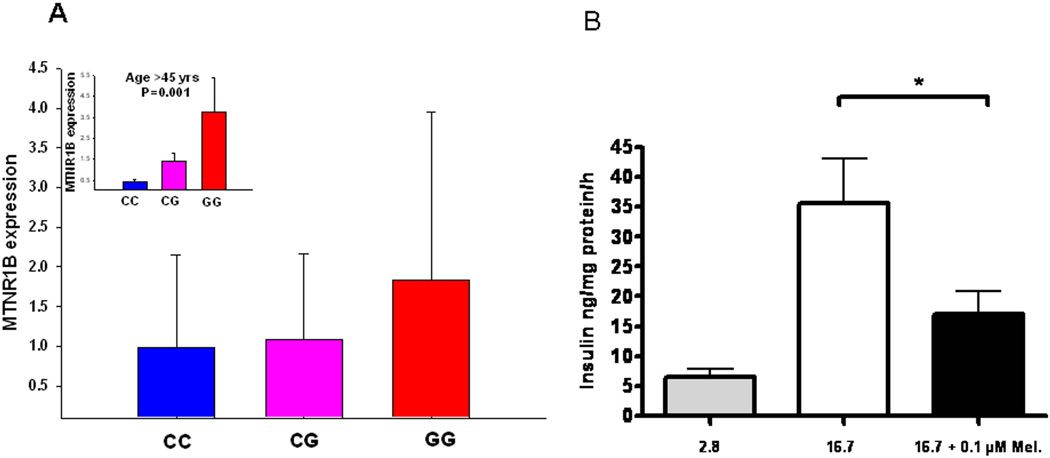
Next, we analyzed whether islet expression of MTNR1B, which we now had established in β-cells, correlated with presence of the risk G-allele rs10830963 in the MTNR1B gene as well as T2D. To this end, we used both quantitative RT-PCR and microarray. Using RT-PCR, we found that individuals carrying the G-allele showed higher expression of MTNR1B as compared with carriers of the C allele (age-adjusted P=0.01, Figure 3A). Notably, this effect was almost exclusively seen in individuals older than 45 years (P=0.001, Figure 2A insert). The microarray experiments (Affymetrix HU 133) were performed on islets isolated from 4 non-diabetic and 4 T2D islet donors15. There was a trend towards higher expression of MTNR1B in T2D than in non-diabetic islets (P =0.20, Supplementary Figure 1A) and expression correlated inversely with glucose-stimulated insulin secretion (Supplementary Figure 1B).
Figure 3. Expression of MTNR1B in human pancreatic islets.
(A) The MTNR1B mRNA levels were higher in risk GG genotype carriers (total n=51, CC=21, CG=25, GG=5; non-adjusted P=0.25, age-adjusted P=0.01). The insert graph shows expression of the MTNR1B mRNA levels in the individuals above mean age of 45 years (total n=25, CC=10, CG=13, GG=2; P=0.001): The MTNR1B mRNA levels were higher in risk GG genotype carriers. (B) Insulin secretion in INS-1 832/13 clonal β-cells in response to stimulation with 2.8 mM (grey bar) and 16.7 mM glucose (white bar) in with the presence and absence of 0.1µM melatonin (black bar). Individual experiments were performed in triplicate (n = 7, * p < 0.037). Bars represent mean ± SEM.
Melatonin impairs insulin secretion in clonal β-cells
To determine the effects of melatonin on insulin secretion, clonal β-cells (832/13) were acutely incubated at low and high glucose concentrations in the presence of 0.1 µM melatonin. Addition of melatonin exerted a clear inhibitory effect on insulin secretion provoked by glucose (Figure 3B). The present findings provide strong support for a role of melatonin and its receptor MTNR1B in the pathogenesis of T2D. A common variant in the MTNR1B receptor was associated with an increase in fasting glucose over time and predicted future T2D, most likely through impairment of insulin secretion from the pancreatic β-cell function7. Notably, this effect became more pronounced with increasing age most likely as a consequence of the increased demands imposed by increased age-related insulin resistance. This effect can be understood in light of what is known about the function of melatonin in islets based on previous studies as well as our present results. The MTNR1B is coupled to an inhibitory G-protein10. Activation of MTNR1B by melatonin would therefore block activation of adenylate cyclase, which is the predominant mode of action for incretin hormones, such as GLP-1 and gastric inhibitory polypeptide (GIP), both of which raise intracellular cAMP. There is also evidence supporting that glucose stimulation of the β-cell by itself leads to a rise in intracellular cAMP. Indeed, it has previously been observed that addition of melatonin blocks cAMP formation in β-cells16. Here, we confirmed previous observations, although discrepant results have been reported12, that melatonin acutely blocks glucose-induced insulin secretion7. Thus, in a situation where expression of MTNR1B is increased, it could be anticipated that cellular cAMP levels will be lower. Hence, the potentiating effect that this nucleotide exerts on insulin secretion, via both protein kinase A-dependent and -independent mechanisms, would be diminished, leading to impaired insulin secretion. This potential pathogenic situation would be further aggravated if melatonin levels are elevated. In fact, this appears to be the case: studies have reported that the circadian rhythm in melatonin secretion is perturbed in T2D17. It has been suggested that secretion of the hormone is elevated during the day, when it normally should be low, which could lead to reduced insulin secretion.
There are therapeutic implications of our findings. First, if melatonin plays a negative role in the development of T2D, antagonists of the receptors targeted to β-cells could be of utility. Second, patients with the risk profile conferred by the MTNR1B rs10830963 SNP may be less responsive to treatment with GLP-1 analogs as well as inhibitors of GLP-1 degradation (DPP-IV inhibitors). Identifying these individuals may allow tailoring of a more precise therapy in T2D.
In conclusion, our findings lend support to earlier reports of a role of the melatonin system for islet function but also provide novel insights into the mechanisms by which the system may play a role in the pathogenesis of T2D. Interfering with its action may be a new therapeutic avenue in T2D.
METHODS
Study populations
In the Malmoe Preventive Project (MPP), 33,346 Swedish subjects (22,444 men and 10,902 women; mean age 49 years, 24.5% with impaired fasting (IFG) and/or impaired glucose tolerance (IGT) from the city of Malmoe in southern Sweden participated in a health screening during 1974–99218. All individuals underwent a physical examination and blood was drawn for measurements of fasting blood glucose and lipid concentrations. In addition, 18,900 consecutively enrolled persons also had an oral glucose tolerance test (OGTT). Information on lifestyle factors and medical history was obtained by questionnaire. Of individuals participating in the initial screening 4,931 are deceased and 551 are lost from follow-up. 25,000 of the eligible individuals were invited to a re-screening visit during 2002–2006, which included a physical examination and fasting blood samples for measurements of plasma glucose and lipids. Of the invited subjects, 17,284 persons participated in the re-screening. Of them 1,223 were excluded because of lacking information or DNA (or T2D at baseline)19. Thereby, 16,061 non-diabetic subjects, 2,063 of whom developed T2D, were included in the current analyses. Diagnosis of diabetes was confirmed from patient records or based upon a fasting plasma glucose concentration greater than 7.0 mmol/l.
The Botnia study started in 1990 at the West coast of Finland aiming at identification of genes increasing susceptibility to T2D in members from families with T2D. The prospective part included 2,770 non-diabetic family members and/or their spouses (1,263 men and 1,507 women, mean age 45 years), 138 of whom developed T2D during a 7.7 year (median) follow-up period19–21. All subjects were given information about exercise and healthy diet and exposed at 2–3 years intervals to a new OGTT.
Prevalence, Prediction and Prevention of T2D (PPP Botnia) study is a population based study in the Botnia region which included approximately 10% of the population aged 18– 74 years (mean age 51 ± 17 years.) Diagnosis of diabetes was confirmed from patient records or based upon a fasting plasma glucose concentration greater than 7.0 mmol/l and/or 2 hr glucose greater than 11.1 mmol/l. 2,328 non-diabetic individuals also had serum insulin concentrations measured at baseline and during follow-up.
The Finland-United States Investigation of Non-insulin-dependent Diabetes Mellitus Genetics (FUSION) study has been described in detail2,13. For this study 578 non-diabetic spouses or offspring were included in the study of insulin response to intravenous glucose using a tolbutamide-modified frequently-sampled intravenous glucose tolerance tests (FSIGTs)22,23, and analyzed by the Minimal Model method24 to derive quantitative measures of insulin sensitivity (SI) and glucose effectiveness (SG). Insulin secretion was assessed as the acute insulin response to glucose (AIR) as described by Ward et al. and beta-cell function was assessed using the disposition index (DI=SI×AIR)25.
Detailed information on the Helsinki Birth Cohort Study (HBCS) has been previously described. In the present study, 1,600 non-diabetic subjects (698 men and 902 women, mean age 62 ± 3 years) were included14. In 2001–2004 all subjects participated in a clinical examination, including a standard 75 g OGTT. Intact proinsulin concentration was measured at 0 min and the fasting proinsulin/insulin ratio (PI/I) was calculated.
The METabolic Syndrome In Men (METSIM) study includes 650–70 years old men, , randomly selected from the population of the town of Kuopio, Eastern Finland, Finland (population 95,000). The present analysis is based on the first 4,386 non-diabetic subjects examined for METSIM with available OGTT data. Samples for the OGTT were obtained at fasting, and at 30 and 120 minutes post-load. The CIR and ISI were calculated from OGTT glucose and insulin data as described below. All participants gave informed consent for the studies and the local ethics committees approved the protocols.
Measurements
Weight, height, and waist and hip circumferences were measured as previously reported18,19. In the MPP cohort at baseline, blood samples were drawn at 0, 40 and 120 minutes of the 75 g OGTT for measurements of blood glucose and serum insulin concentrations, while fasting samples were drawn at the follow-up visit for measurement of plasma glucose and lipid concentrations using standard techniques. In the Botnia study, blood samples were drawn at −10, 0, 30, 60, and 120 minutes of the OGTT. Insulin sensitivity index (ISI) from the OGTT was calculated as 10,000/√[(fasting plasma glucose x fasting plasma insulin)(mean OGTTglucose x mean OGTTinsulin)]26. The basal insulin resistance index (HOMA) was calculated from fasting insulin and glucose concentrations (http://www.dtu.ox.ac.uk). β-cell function was assessed as corrected incremental insulin response during OGTT (CIR= (100 x insulin at 30 min or 40 min in MPP)/(glucose at 30 min or 40 min in MPP) x (glucose 30 min or 40 min in MPP −3.89))27 or as disposition index, i.e. insulin secretion adjusted for insulin sensitivity (CIR x ISI).
Plasma glucose was measured by hexokinase (MPP, FUSION), glucose oxidase (Botnia, FUSION, METSIM) methods. Plasma insulin concentrations were measured by an ELISA assay (Dako, Cambridgeshire; Botnia study), by a local radioimmunoassay (MPP), by radioimmunoassay using dextran-charcoal separation (FUSION), or by a commercial double-antibody solid-phase radioimmunoassay (METSIM).
Genotyping
In the DGI and FUSION GWAS genotyping was performed using Affymetrix 500K chip array1 and Illumina HumanHap300 BeadChip Version 1.02. In the FUSION and METSIM studies, SNP rs10830963 was genotyped by Sequenom iPlex gold SBE (Sequenom, San Diego, CA); in all other replication studies rs10830963 was genotyped by an allelic discrimination assay-by-design method on ABI 7900 (Applied Biosystems). Genotypes were in Hardy-Weinberg equilibrium. In MPP and Botnia, we obtained an average genotyping success rate of >95% and the concordance rate was 98.7%, using two different methods (allelic discrimination on ABI7900 and Affymetrix). Replication genotyping for FUSION and METSIM studies was performed using Sequenom iPlex gold SBE (Sequenom, San Diego, CA).
Immunocytochemistry
For histochemical analysis pancreatic specimens were dissected, fixed overnight in Stefanini's solution (2% paraformaldehyde and 0.2% picric acid in 0.1 M phosphate buffered saline, pH 7.2), rinsed thoroughly in Tyrode solution containing 10% sucrose, and frozen on dry ice. Sections (10 mm thickness) were cut and thaw-mounted on slides. Antibodies were diluted in phosphate buffered saline (PBS) (pH 7.2) containing 0.25% bovine serum albumin and 0.25% Triton X-100. Sections were incubated with primary antibodies (goat anti- Melatonin receptor 1B; code sc-13177; dilution 1:400, Santa Cruz Biotech. Inc., CA, goat anti- Melatonin receptor 1A; code sc-13186, dilution 1:400, Santa Cruz, and guinea pig anti-proinsulin; code 9003; dilution 1:2560; EuroDiagnostica, Malmoe, Sweden) overnight at 4° C in moisturizing chambers. The sections were rinsed in PBS with Triton X-100 for 2 × 10 min. Thereafter secondary antibodies with specificity for rabbit- or guinea pig- IgG, and coupled to either fluorescein isothiocyanate (FITC) or Texas-Red (Jackson, West Grove, PA), were applied on the sections. Incubation was for 1h at room temperature in moisturizing chambers. The sections were again rinsed in PBS with Triton X-100 for 2 × 10 min and then mounted in PBS:glycerol, 1:1. The specificity of immunostaining was tested using primary antisera pre-absorbed with homologus antigen (100 mg of peptide per ml antiserum at working dilution). Immunofluorescence was examined in an epifluorescence microscope (Olympus, BX60). By changing filters the location of the different secondary antibodies in double staining was determined. Images were captured with a digital camera (Nikon DS-2Mv)28.
Gene expression using real-time PCR
Total RNA was isolated with the AllPrep DNA/RNA Mini Kit (Qiagen GmbH, Hilden, Germany) (LUDC Human Tissue facility); RNeasy protect mini kit (Qiagen, Santa Clara, CA, USA) as previously described15 (Italy); Trizol (Invitrogen, Carlsbad, CA, USA) and further purified using RNeasy mini kit (Qiagen, Santa Clara, CA, USA) (NIH). RNA quantity was determined by evaluating the absorbance at 260 and 280 nm in a Perkin-Elmer spectrophotometer (Waltham, MA, USA), and quality was assessed by running samples on Agilent 2100 Bioanalyzer (Agilent Technologies, Santa Clara,CA, USA) (Italy). cDNA was synthesized from 0.4µg total RNA using RevertAid™ First Strand cDNA Synthesis Kit (Fermentas Life Sciences, Burlington, Canada) (LUDC Human Tissue facility); 0.5µg total RNA using the High Capacity RNA-to-cDNA Kit (Applied Biosystems, Foster City CA, USA) (NIH) ; 1 µg total RNA using iScript cDNA synthesis kit (Biorad Laboratories, Hercules, CA,USA) (Italy). TaqMan® gene expression assays were purchased from Applied Biosystems (ABI; Foster City, California, USA) for the various target genes: Hs00173794_m1 directed against human melatonin receptor 1B (MTNR1B) (LUDC Human Tissue facility; Italy, NIH) and human hypoxanthine-guanine phosphoribosyl transferase (HPRT) (LUDC Human Tissue facility, NIH); cyclophyllin (Italy) , which served as endogenous control gene. Q-PCR reactions were performed on the ABI 7900HT (Applied Biosystems) (LUDC Human Tissue facility, NIH) by mixing 2x TaqMan® Universal Master Mix, 20x TaqMan® Gene Expression Assays, nuclease free water and cDNA for a final reaction volume of 10µl (LUDC Human Tissue facility); as described earlier29 (Italy) . The relative quantity of MTNR1B mRNA was calculated using the comparative CT – method (LUDC Human Tissue facility, NIH). All experiments were performed in triplicate.
For microarray experiments, 100 ng total RNA was subjected to two rounds of amplification (GeneChip Two-Cycle Kit, Affymetrix, Santa Clara, CA, USA) and biotinylated RNA was generated using GeneChip IVT Labeling Kit (Affymetrix). RNA products were fragmented and hybridised to GeneChip Human HG U 133A Array (Affymetrix). The array data were normalised and analysed using DNA-Chip Analyzer (dChip) software (available from: http://biosun1.harvard.edu/complab/dchip/, last accessed in January 2008) that assesses the standard errors for the expression indexes and calculates confidence intervals for fold changes (Italy, NIH).
Effect of melatonin on insulin secretion
To determine the effects of melatonin on insulin secretion, the clonal β-cells from the line 832/13 was incubated with 0.1 µM melatonin for 1h. Then, released insulin into the buffer was determined by radioimmunoassay.
Statistical analyses
Differences in expression levels were tested by analysis of variance or non-parametric Mann-Whitney tests. The odds ratios for risk of developing T2D were calculated using logistic regression analyses adjusted for age at participation and time to last follow-up, body mass index and gender. Multivariate linear regression analyses were used to test genotype-phenotype correlations adjusted for age, gender, body mass index (apart from body mass index) and pedigree. Non-normally distributed variables were log-transformed before analysis. Analysis of FUSION FSIGT and METSIM OGTT data was carried out using a regression framework in which regression coefficients were estimated in the context of a variance component model to account for relatedness among individuals30. Trait values for both studies were adjusted for age and age2. For FUSION data sex was included as an additional covariate. Analyses were carried out in nondiabetic individuals excluding those known to be taking medications that directly affect glucose or insulin concentrations. Covariate-adjusted trait values were transformed to approximate univariate normality by applying an inverse normal scores transformation; the scores were ranked, ranks were transformed into quantiles, and quantiles were converted to normal deviates.
All statistical analyses were performed using SPSS version 14.0, PLINK (http://pngu.mgh.harvard.edu/~purcell/plink/), Stata (StataCorp, College Station, Texas, United States), or MERLIN30.
Supplementary Material
Acknowledgements
The DGI study was supported by a grant from Novartis.
Studies in Malmoe were supported by grants from the Swedish Research Council, including a Linné grant (No.31475113580), the Diabetes Programme at the Lund University, the Påhlsson Foundation, the Heart and Lung Foundation, the Wallenberg Foundation, the Swedish Diabetes Research Society, the Crafoord Foundation, Swedish Medical Society, Swedish Royal Physiographic Society, a Nordic Centre of Excellence Grant in Disease Genetics, the Finnish Diabetes Research Society, the Sigrid Juselius Foundation, Folkhälsan Research Foundation, Novo Nordisk Foundation, ENGAGE and EFSD. Studies in human islets were supported in part by the Italian Ministry of University and Research (PRIN 2007–2008) and the European Community (LSHM-CT-2006-518153)
Pancreatic islets at NIH were obtained through the ICR Basic Science Islet Distribution Program (City of Hope Hospital, Joslin Diabetes Center, Northwestern U.SCIC, UAD, U. Ill., U. Miami, UMN, U. Penn., U. Wisc., and Wash. U.); the JDRF Islet Resources (Wash. U.) and the National Disease Resource Interchange (NDRI).
The FUSION study would like to thank the many research volunteers who generously participated in the various studies represented in FUSION. We also thank Amy J. Swift, Mario Morken, Peter S. Chines, and Narisu Narisu for genotying and informatics support. Support for FUSION was provided by the following: NIH Grant DK062370 (M.B.), American Diabetes Association Research Grant 1-05-RA-140 (R.M.W.), DK072193 (Karen L. Mohlke), and National Human Genome Research Institute intramural project number 1 Z01 HG000024 (Francis S. Collins). The METSIM study was supported by Academy of Finland grant 124243 (M.L.).
Footnotes
Author contributions
VL: DGI GWAS, data analysis, and drafted the report.
CN, MRE: in vitro expression experiments and analysis, and drafted the report.
NW: immunocytochemistry.
AJ: genotyping and data analysis.
PS: in vitro expression experiments.
MB: microarray and human islets experiments.
RS: DGI GWAS analysis.
MF: in vitro physiology.
NP: genotyping.
BI, TT: phenotyping in the Botnia study.
PN: phenotyping in the Malmoe study.
JK: data analysis in METSIM study.
JT: phenotyping in the FUSION study.
MB: PI of the FUSION study.
DA: PI of the DGI study.
FS: immunocytochemistry.
JGE: phenotyping in the Helsinki Birth Cohort Study.
AUJ: FUSION GWAS and data analysis.
ML: PI of the METSIM study.
PM: microarray and human islets experiments.
RMW: FUSION GWAS analysis.
HM: design and supervision of in vitro study experiments, and drafted the report.
LG: designed and supervised all parts of the study, and drafted the report.
All researchers took part in the revision of the report and approved the final version.
References
- 1.Diabetes Genetics Initiative of Broad Institute of Harvard and MIT, L.U., and Novartis Institutes of BioMedical Research, et al. Genome-wide association analysis identifies loci for type 2 diabetes and triglyceride levels. Science. 2007;316:1331–1336. doi: 10.1126/science.1142358. [DOI] [PubMed] [Google Scholar]
- 2.Scott LJ, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sladek R, et al. A genome-wide association study identifies novel risk loci for type 2 diabetes. Nature. 2007;445:881–885. doi: 10.1038/nature05616. [DOI] [PubMed] [Google Scholar]
- 4.Zeggini E, et al. Meta-analysis of genome-wide association data and large-scale replication identifies additional susceptibility loci for type 2 diabetes. Nat Genet. 2008;40:638–645. doi: 10.1038/ng.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zeggini E, et al. Replication of genome-wide association signals in UK samples reveals risk loci for type 2 diabetes. Science. 2007;316:1336–1341. doi: 10.1126/science.1142364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Prokopenko I, Langenberg C, Florez JC, Saxena R, Soranzo N, et al. Variants in the melatonin receptor 1B gene (MTNR1B) influence fasting glucose levels and risk of type 2 diabetes. Nature Genetics. 2008 Submitted. [Google Scholar]
- 7.Peschke E. Melatonin, endocrine pancreas and diabetes. J Pineal Res. 2008;44:26–40. doi: 10.1111/j.1600-079X.2007.00519.x. [DOI] [PubMed] [Google Scholar]
- 8.Kvetnoy IM. Extrapineal melatonin: location and role within diffuse neuroendocrine system. Histochem J. 1999;31:1–12. doi: 10.1023/a:1003431122334. [DOI] [PubMed] [Google Scholar]
- 9.Boden G, Ruiz J, Urbain JL, Chen X. Evidence for a circadian rhythm of insulin secretion. Am J Physiol. 1996;271:E246–E252. doi: 10.1152/ajpendo.1996.271.2.E246. [DOI] [PubMed] [Google Scholar]
- 10.Pandi-Perumal SR, et al. Physiological effects of melatonin: Role of melatonin receptors and signal transduction pathways. Prog Neurobiol. 2008 doi: 10.1016/j.pneurobio.2008.04.001. [DOI] [PubMed] [Google Scholar]
- 11.Muhlbauer E, Peschke E. Evidence for the expression of both the MT1- and in addition, the MT2-melatonin receptor, in the rat pancreas, islet and beta-cell. J Pineal Res. 2007;42:105–106. doi: 10.1111/j.1600-079X.2006.00399.x. [DOI] [PubMed] [Google Scholar]
- 12.Ramracheya RD, et al. Function and expression of melatonin receptors on human pancreatic islets. J Pineal Res. 2008;44:273–279. doi: 10.1111/j.1600-079X.2007.00523.x. [DOI] [PubMed] [Google Scholar]
- 13.Valle T, et al. Mapping genes for NIDDM Design of the Finland-United States Investigation of NIDDM Genetics (FUSION) Study. Diabetes Care. 1998;21:949–958. doi: 10.2337/diacare.21.6.949. [DOI] [PubMed] [Google Scholar]
- 14.Eriksson JG, Osmond C, Kajantie E, Forsen TJ, Barker DJ. Patterns of growth among children who later develop type 2 diabetes or its risk factors. Diabetologia. 2006;49:2853–2858. doi: 10.1007/s00125-006-0459-1. [DOI] [PubMed] [Google Scholar]
- 15.Marselli L, et al. Gene expression of purified beta-cell tissue obtained from human pancreas with laser capture microdissection. J Clin Endocrinol Metab. 2008;93:1046–1053. doi: 10.1210/jc.2007-0931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Peschke E, Bach AG, Muhlbauer E. Parallel signaling pathways of melatonin in the pancreatic beta-cell. J Pineal Res. 2006;40:184–191. doi: 10.1111/j.1600-079X.2005.00297.x. [DOI] [PubMed] [Google Scholar]
- 17.Peschke E, et al. Melatonin and type 2 diabetes - a possible link? J Pineal Res. 2007;42:350–358. doi: 10.1111/j.1600-079X.2007.00426.x. [DOI] [PubMed] [Google Scholar]
- 18.Berglund G, et al. Long-term outcome of the Malmo preventive project: mortality and cardiovascular morbidity. J Intern Med. 2000;247:19–29. doi: 10.1046/j.1365-2796.2000.00568.x. [DOI] [PubMed] [Google Scholar]
- 19.Lyssenko V, et al. Clinical Risk Factors, DNA Variants, and the Development of Type 2 Diabetes. NEJM. 2008;359 doi: 10.1056/NEJMoa0801869. [DOI] [PubMed] [Google Scholar]
- 20.Lyssenko V, et al. Genetic prediction of future type 2 diabetes. PLoS Med. 2005;2:e345. doi: 10.1371/journal.pmed.0020345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lyssenko V, et al. Predictors of and longitudinal changes in insulin sensitivity and secretion preceding onset of type 2 diabetes. Diabetes. 2005;54:166–174. doi: 10.2337/diabetes.54.1.166. [DOI] [PubMed] [Google Scholar]
- 22.Steil GM, Volund A, Kahn SE, Bergman RN. Reduced sample number for calculation of insulin sensitivity and glucose effectiveness from the minimal model. Suitability for use in population studies. Diabetes. 1993;42:250–256. doi: 10.2337/diab.42.2.250. [DOI] [PubMed] [Google Scholar]
- 23.Yang YJ, Youn JH, Bergman RN. Modified protocols improve insulin sensitivity estimation using the minimal model. Am J Physiol. 1987;253:E595–E602. doi: 10.1152/ajpendo.1987.253.6.E595. [DOI] [PubMed] [Google Scholar]
- 24.Bergman RN, Ider YZ, Bowden CR, Cobelli C. Quantitative estimation of insulin sensitivity. Am J Physiol. 1979;236:E667–E677. doi: 10.1152/ajpendo.1979.236.6.E667. [DOI] [PubMed] [Google Scholar]
- 25.Ward WK, Bolgiano DC, McKnight B, Halter JB, Porte D., Jr. Diminished B cell secretory capacity in patients with noninsulin-dependent diabetes mellitus. J Clin Invest. 1984;74:1318–1328. doi: 10.1172/JCI111542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuda M, DeFronzo RA. Insulin sensitivity indices obtained from oral glucose tolerance testing: comparison with the euglycemic insulin clamp. Diabetes Care. 1999;22:1462–1470. doi: 10.2337/diacare.22.9.1462. [DOI] [PubMed] [Google Scholar]
- 27.Hanson RL, et al. Evaluation of simple indices of insulin sensitivity and insulin secretion for use in epidemiologic studies. Am J Epidemiol. 2000;151:190–198. doi: 10.1093/oxfordjournals.aje.a010187. [DOI] [PubMed] [Google Scholar]
- 28.Wierup N, Bjorkqvist M, Kuhar MJ, Mulder H, Sundler F. CART regulates islet hormone secretion and is expressed in the beta-cells of type 2 diabetic rats. Diabetes. 2006;55:305–311. doi: 10.2337/diabetes.55.02.06.db04-1383. [DOI] [PubMed] [Google Scholar]
- 29.Del Guerra S, et al. Functional and molecular defects of pancreatic islets in human type 2 diabetes. Diabetes. 2005;54:727–735. doi: 10.2337/diabetes.54.3.727. [DOI] [PubMed] [Google Scholar]
- 30.Chen WM, Abecasis GR. Family-based association tests for genomewide association scans. Am J Hum Genet. 2007;81:913–926. doi: 10.1086/521580. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.