Abstract
Evolution balances DNA replication speed and accuracy to optimize replicative fitness and genetic stability. There is no selective pressure to improve DNA replication fidelity beyond the background mutation rate from other sources, such as DNA damage. However, DNA polymerases remain amenable to amino-acid substitutions that lower intrinsic error rates. Here, we review these ‘antimutagenic’ changes in DNA polymerases and discuss what they reveal about mechanisms of replication fidelity. Pioneering studies with bacteriophage T4 DNA polymerase (T4 Pol) established the paradigm that antimutator amino-acid substitutions reduce replication errors by increasing proofreading efficiency at the expense of polymerase processivity. The discoveries of antimutator substitutions in proofreading-deficient ‘mutator’ derivatives of bacterial Pols I and III and yeast Pol δ suggest there must be additional antimutagenic mechanisms. Remarkably, many of the affected amino-acid positions from Pol I, Pol III, and Pol δ are similar to the original T4 Pol substitutions. The locations of antimutator substitutions within DNA polymerase structures suggest that they may increase nucleotide selectivity and/or promote dissociation of primer termini from polymerases poised for misincorporation, leading to expulsion of incorrect nucleotides. If misincorporation occurs, enhanced primer dissociation from polymerase domains may improve proofreading in cis by an intrinsic exonuclease or in trans by alternate cellular proofreading activities. Together, these studies reveal that natural selection can readily restore replication error rates to sustainable levels following an adaptive mutator phenotype.
Keywords: DNA replication fidelity, mutator, genetic adaptation, protein structure/function, genetic instability, cancer
INTRODUCTION
Genetic information must be accurately duplicated during cell division to ensure that daughter cells retain essential genes and the beneficial traits accrued through evolution. For this reason, organisms invest in an array of conserved mechanisms that promote genetic continuity (Friedberg et al., 2006). DNA polymerase errors during cell division represent the largest potential source of genetic instability (Preston et al., 2010). To minimize spontaneous mutation, these errors are corrected by exonucleolytic proofreading and post-replicative mismatch repair (MMR) (Iyer et al., 2006; McCulloch and Kunkel, 2008; Reha-Krantz, 2010), thereby lowering the contribution of replication errors to the background mutation rate derived from other sources (Quah et al., 1980; Morrison et al., 1993; Roche et al., 1994; Datta et al., 2000). Defects in polymerase proofreading or MMR, as well as other DNA repair enzymes, elevate spontaneous mutation rate, producing ‘mutator’ phenotypes (Friedberg et al., 2006).
While deleterious to long-term fitness, short-term genetic instability provides variation upon which natural selection can act (Sturtevant, 1937; Drake et al., 1998; Sniegowski et al., 2000; Giraud et al., 2001b; de Visser, 2002; Elena and Lenski, 2003; Denamur and Matic, 2006). Mutator phenotypes due to defects in MMR or polymerase proofreading promote somatic evolution of cancer cells in mice and humans (Loeb et al., 1974; Fishel et al., 1993; Parsons et al., 1993; de Wind et al., 1995; Reitmair et al., 1996; Goldsby et al., 2001; Goldsby et al., 2002; Albertson and Preston, 2006; Loeb et al., 2008; Daee et al., 2010). Similarly, microbial mutators arise spontaneously when populations are subjected to repeated rounds of selection (Chao and Cox, 1983; Mao et al., 1997; Sniegowski et al., 1997; Giraud et al., 2001a; Nilsson et al., 2004). Interestingly, following adaptation, bacterial clones with lower mutation rates evolve from mutator lineages, suggesting that there is selection pressure for genetic suppressors that restore DNA replication fidelity (Trobner and Piechocki, 1984; Schaaper and Cornacchio, 1992; Fijalkowska and Schaaper, 1995; Giraud et al., 2001a). Understanding the nature of these ‘antimutagenic’ changes may offer new insight into DNA replication fidelity and the evolution of genetic stability.
This review focuses on DNA polymerase variants that impart an antimutator phenotype due to amino-acid substitutions that increase DNA replication fidelity. To provide a framework for understanding the mechanisms of antimutagenesis, we begin our review with an exploration of the determinants of DNA replication fidelity. Several excellent in-depth reviews of DNA replication fidelity offer additional background information that readers may find useful (Pavlov et al., 2006b; Kunkel, 2009; Johnson, 2010; Reha-Krantz, 2010). We continue by describing the origins of the four main antimutator polymerase collections, focusing on details that are pertinent for understanding the influence of antimutator substitutions on DNA polymerase fidelity. We discuss possible antimutator mechanisms in light of X-ray crystallographic structures of relevant DNA polymerase replication complexes. Finally, we consider the relevance of these findings for DNA polymerase evolution and plasticity in the natural world.
DETERMINANTS OF DNA REPLICATION FIDELITY
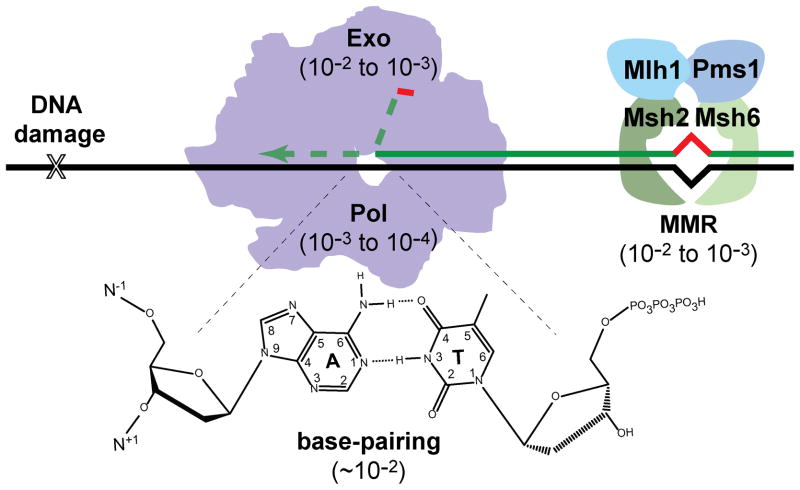
DNA replication is the sequential joining of nucleotides to the 3′ end of a DNA primer sequence annealed to a complementary DNA template. Cells accomplish this task with astonishing precision (10−9 – 10−10 mutations per base-pair synthesized), providing evidence of the strong evolutionary pressure to preserve genetic information. No single process accounts for this accuracy. Polymerases restrict mutation through a combination of nucleotide selection, exonucleolytic proofreading of misinsertions, and post-replicative mismatch repair (MMR) (Figure 1).
Figure 1.
Determinants of DNA replication fidelity. Accurate DNA synthesis is derived from stability differences between correct and incorrect base-pairs (base-pairing), polymerase selectivity (Pol), 3′→5′ exonucleolytic proofreading (Exo), and post-replicative mismatch repair (MMR). Reductions in mutation rate attributed to each mechanism are shown in parentheses. For proofreading, the primer strand partitions to a separate Exo active site, either integral to the polymerase or associated with the holoenzyme. In yeast, the MMR recognition complex is composed of MutS homologues (Msh) 2 and 6 (or a related complex, Msh2 and 3), which recruits an effector complex composed of MutL homologue (Mlh) 1 and post-meiotic segregation (PMS) 1 (both MutL homologues). The background mutation rate may also be influenced by damage to nucleotide pools or template DNA (X). A color version of this figure is available online.
Nucleotide Selection
DNA polymerases perform a variety of roles in diverse organisms, and based on homology, are grouped into families A, B, C, D, X, Y and RT [reviewed in (Pavlov et al., 2006b; Kunkel, 2009)]. In bacteria, C-family polymerases perform the bulk of chromosomal DNA synthesis, while B-family polymerases assume this role in eukaryotes (Pavlov et al., 2006b; Kunkel, 2009). In archaea, both B- and D-family polymerases appear to be required (Henneke et al., 2005; Barry and Bell, 2006; Li et al., 2010b). These main replicative DNA polymerases require speed and accuracy to efficiently and faithfully replicate large genomes. Other specialized DNA polymerases copy only short stretches of DNA during DNA repair or translesion synthesis (TLS) and thus do not require the same level of speed and accuracy as the replicative enzymes. TLS polymerases of the error-prone Y family are deployed to bypass replication-blocking lesions (Pavlov et al., 2006b; Kunkel, 2009), while reverse transcriptases (RT) catalyze both RNA- and DNA-dependent DNA synthesis of retroviral genomes (Sarafianos et al., 2009).
Amino-acid sequence alignments and polymerase crystal structures indicate that A, B, D, Y and RT families share a common core structure resembling a cupped right hand (Beese et al., 1993; Eom et al., 1996; Wang et al., 1997; Ding et al., 1998; Huang et al., 1998a; Li et al., 1998; Franklin et al., 2001; Ling et al., 2001). The palm contains the catalytic residues, the thumb positions the double-stranded DNA in the active site, and the highly mobile fingers domain closes over deoxyribonucleoside-triphosphates (dNTPs) during nucleotide selection. C- and X-family polymerases are ‘left-handed’ based on the direction of DNA synthesis relative to the thumb and fingers domains, which also bind DNA and dNTPs, respectively, despite being structurally different than their right-handed counterparts (Sawaya et al., 1997; Wing et al., 2008).
With each elongation step, highly accurate DNA polymerases distinguish correct from incorrect dNTPs with an average error rate of 10−4 to 10−5 misinsertions per base-pair synthesized (Kunkel, 2009). Hydrogen bonds (H-bonds) between cognate base pairs were initially assumed to make the largest contribution to nucleotide selection (Watson and Crick, 1953a; Watson and Crick, 1953b). However, free energy differences of correct and incorrect base pairs in solution only account for a fraction (10−2) of the observed specificity (Mildvan, 1974), because favorable gains in enthalpy due to correct base-pair formation are offset by unfavorable decreases in entropy of the solvent (Petruska and Goodman, 1995). Crystal structures of replication complexes reveal that high-fidelity DNA polymerases form tight nucleotide binding pockets that limit solvent access and select incipient base pairs that specifically conform to Watson-Crick geometry (Beese et al., 1993; Eom et al., 1996; Sawaya et al., 1997; Doublie et al., 1998; Kiefer et al., 1998; Li et al., 1998; Franklin et al., 2001; Wing et al., 2008; Swan et al., 2009).
Watson-Crick base pairs share pseudo-dyad symmetry, similar distances between the C1′ atoms of the sugars, and equivalent glycosyl bond angles (Saenger, 1984). In addition, Watson-Crick base pairs have minor groove H-bond acceptors (O2 of pyrimidines and N-3 of purines, Figure 1) in essentially identical positions (Saenger, 1984). While some mispairs form H-bond interactions of similar strength to Watson-Crick base pairs, their overall geometry differs dramatically. Thus, in theory, mispairs could be excluded on the basis of shape or absence of appropriately positioned minor-groove contacts (Echols and Goodman, 1991; Beard and Wilson, 2003). Kool and colleagues found that close structural mimics of thymidine and adenine that lack H-bond donors or acceptors are more easily incorporated by DNA polymerases than noncognate nucleotides, suggesting that the shape of the incipient base pair plays a role in selection (Moran et al., 1997; Morales and Kool, 1998; Morales and Kool, 2000; Potapova et al., 2006; Lee et al., 2008a). However, both A- and B-family polymerases also incorporate nucleotide analogues that do not conform to Watson-Crick geometry (Chiaramonte et al., 2003; Kincaid et al., 2005; Matsuda et al., 2006; Leconte et al., 2008). B-family polymerases, for instance, readily incorporate benzimidazole opposite all four natural nucleotides (Chiaramonte et al., 2003; Kincaid et al., 2005). Benzimidazole resembles adenine and guanine except that it lacks all heterocyclic amines (N-1, N-3, N-7 and N-9, Figure 1) and is bereft of extracyclic chemical groups (N2, N6 and O6). While incorporation of benzimidazole opposite T or C would satisfy Watson-Crick geometry, incorporation opposite A or G would clearly not. Systematic addition of substituents found in purines to benzimidazole reveals that they increase discrimination. N-1, N-3, and N6 help B-family polymerases exclude incorrect nucleotides, while groups involved in H-bonding (N-1, N2, and N6) contribute positively to the selection of correct nucleotides (Beckman et al., 2007; Cavanaugh et al., 2009). A-family polymerases, which do not readily incorporate benzimidazole, utilize N-1, N-3, N2, and N6 for both positive and negative selection (Chiaramonte et al., 2003; Kincaid et al., 2005; Trostler et al., 2009). Thus, minor-groove interactions appear to position nucleotides for optimal shape discrimination, while Watson-Crick H-bonding groups may contribute to negative discrimination through formation of non-standard base pairs or through solvent interactions that increase the effective size of the nucleotide (Kool, 2002).
Recent observations with the A-family polymerase T7 Pol suggests that, for some enzymes, negative discrimination of nucleotides is more than just passive steric exclusion (Tsai and Johnson, 2006; Johnson, 2010). The fingers domain resides in an open conformation in the absence of dNTP (Eom et al., 1996; Wang et al., 1997; Bailey et al., 2006). Upon dNTP binding the fingers close, cradling the new base pair in a binding pocket and coordinating the triphosphate through conserved basic residues (Sawaya et al., 1997; Doublie et al., 1998; Kiefer et al., 1998; Franklin et al., 2001; Wing et al., 2008). Initial kinetic experiments revealed only a small reduction in reaction rate with α-phosphothioate dNTP analogues, suggesting that a slow conformational change prior to phosphoryl-transfer amplifies binding differences between correct and incorrect nucleotides [reviewed in (Echols and Goodman, 1991; Joyce and Benkovic, 2004)]. Crystallographic evidence for movement of the fingers domain presented an ideal mechanism for this proposed isomerization (Joyce and Benkovic, 2004). However, fluorescence resonance energy transfer (FRET) measurements with Thermus aquaticus (Taq) Pol I indicate that closing of the fingers domain is much faster than catalysis (Rothwell et al., 2005). Further evidence for rapid closing was obtained by Tsai and Johnson who introduced a fluorescent label into the fingers domain of T7 Pol near an arginine involved in coordination with the triphosphate (Tsai and Johnson, 2006). Fluorescence corresponding to the open conformation rapidly decreased upon addition of correct dNTPs and unexpectedly increased with incorrect dNTPs. Although the structural reasons underlying the changes in fluorescence are unclear, they allowed Tsai and Johnson to monitor how rapidly the polymerase reverts to the open conformation following correct or incorrect dNTP binding. Reversal of the conformational change and dNTP release was 260-times slower for correct nucleotides than for incorrect nucleotides. Thus, T7 Pol appears to harness the binding energy of incorrect dNTPs to adopt a conformation that actively promotes dissociation (Tsai and Johnson, 2006).
The studies of T7 Pol also show that the rate of phosphoryl transfer differs between correct and incorrect nucleotides (Tsai and Johnson, 2006). All DNA polymerases utilize two divalent metal ions that are coordinated by highly conserved carboxylate residues to catalyze nucleotidyl transfer (Steitz et al., 1994; Beard and Wilson, 2003). With correct geometry, the first metal ion (Metal A) activates the 3′-OH of the primer terminus for in-line nucleophilic attack on the α-phosphate of the incoming dNTP. Metal A and a second metal (Metal B) stabilize the developing negative charge on the pyrophosphate leaving group (Steitz et al., 1994). Crystal structures of some DNA polymerases, such as RB69 Pol and yeast Pol δ, reveal a third metal ion (Metal C) that may further stabilize the leaving group (Franklin et al., 2001; Swan et al., 2009). Proper alignment of the dNTP and stabilization of the leaving group are required for efficient chemistry (Beard and Wilson, 2003). In the T7 Pol studies, phosphoryl transfer was 100-times faster for correct compared to incorrect base pairs, suggesting that the incorrect nucleotides are misaligned in the active site, slowing phosphoryl transfer (Tsai and Johnson, 2006). Thus, T7 Pol increases base-pair selectivity some 104-fold above that attributed to free-energy differences in solution (10−2) by two mechanisms: preferential stabilization and faster phosphoryl transfer of correct dNTPs (Tsai and Johnson, 2006; Johnson, 2010).
Proofreading and Mismatch Repair
Despite the above mechanisms to ensure accurate nucleotide selection, DNA polymerases make hundreds to thousands of mistakes every cell cycle, depending on the size of the genome. DNA sequence context influences the distribution of errors, creating ‘hot-spots’ in the mutation spectra. Intra- or inter-strand base-base stacking interactions in some contexts may stabilize mispairs (Echols and Goodman, 1991). Other mutational hotspots result from sequences where the primer and template easily misalign, causing frameshifts, insertions, deletions or, if transient, base-base mismatches (Bebenek and Kunkel, 2000). To correct replication errors, many DNA polymerases have an exonucleolytic proofreading activity [reviewed in (Reha-Krantz, 2010)]. Kinetic studies indicate that mispaired primer termini reduce polymerization of added nucleotides [reviewed in (Echols and Goodman, 1991; Goodman et al., 1993; Bebenek and Kunkel, 1995)]. Structural studies of an A-family polymerase with mispairs in any one of the first four positions of the upstream primer•template duplex, reveal that mispairs distort the active site by multiple mechanisms that slow catalysis (Johnson and Beese, 2004). Polymerase pausing permits the melting of double-stranded DNA and partitioning of primer termini to exonucleases for editing. These ‘proofreading’ exonucleases can be integral to the polymerase (as in many A- and B-family polymerases) or encoded as separate polypeptides that tightly associate with the polymerase (as in C-family polymerases) (Johnson and O’Donnell, 2005). Amino-acid substitutions that inactivate exonuclease activity confer a mutator phenotype characterized predominantly by increased base-substitution errors and frameshift mutations (Simon et al., 1991; Morrison et al., 1993; Reha-Krantz and Nonay, 1993; Schaaper, 1993; Jin et al., 2001; Fortune et al., 2005). Thus, in the absence of proofreading, polymerases can slowly extend many mispaired primer termini, creating base-base mismatches or small bulges in duplex DNA.
Distortions in the DNA due to replication errors are substrates for post-replicative mismatch repair (MMR) [reviewed in (Kunkel and Erie, 2005; Iyer et al., 2006)]. MMR complexes bind mismatched DNA and nick the nascent strand, providing entry points for enzymes that remove and resynthesize the error-containing DNA strand (Kadyrov et al., 2006; Kadyrov et al., 2009). If left unrepaired, mismatches lead to permanent mutations in the next round of replication when the mutated strand serves as a template. Defects in proofreading or MMR increase mutation rates between 10- and 1000-fold in organisms as diverse as phage, bacteria, yeast, mice, and humans (Muzyczka et al., 1972; Simon et al., 1991; Fishel et al., 1993; Morrison et al., 1993; Parsons et al., 1993; Schaaper, 1993; Reitmair et al., 1997; Goldsby et al., 2002; Albertson et al., 2009). Simultaneous inactivation of both pathways increases mutation rates by more than 10,000-fold (Morrison et al., 1993; Schaaper, 1993). Thus, polymerase nucleotide selection, proofreading, and MMR act sequentially to achieve an accuracy of better than 10−9 mutations/base pair/cell division (Drake et al., 1998).
DISCOVERY OF ANTIMUTATORS
Given the low cellular mutation rate, the isolation of mutants with improved replication fidelity poses distinct challenges. Whereas mutators emerge when cells are subjected to adaptive pressure [reviewed in (Sturtevant, 1937; Drake et al., 1998; Giraud et al., 2001b; de Visser, 2002; Elena and Lenski, 2003; Denamur and Matic, 2006)], few selection schemes exist for antimutators. Furthermore, many processes contribute to the residual background mutation rate observed in wild-type cells so that an antimutator allele in any one pathway may not significantly improve overall fidelity (Drake, 1993). Cells contend with various forms of DNA damage through translesion DNA synthesis, base-excision repair, nucleotide excision repair, and recombination (Friedberg et al., 2006). The actual contributions of individual pathways to background mutation rate vary with cell type, genetic background, and environmental conditions. In some cases, one pathway may predominate. For instance, in yeast, DNA polymerase zeta (Pol ζ) generates between 50 and 75% of all background mutations (Quah et al., 1980; Roche et al., 1994; Datta et al., 2000), complicating the isolation of antimutators in other pathways. For the above reasons, most antimutators have been identified by laborious screening procedures or as suppressors of a mutator phenotype or other deleterious DNA replication-associated defects.
T4 Pol
Drake uncovered the very first antimutator DNA polymerase variants while screening a collection of bacteriophage T4 gene 43 mutations for mutator phenotypes (Drake and Allen, 1968; Drake et al., 1969). Gene 43 encodes the viral replicase, a B-family DNA polymerase with homology to eukaryotic replicative polymerases (Pol α, Pol δ, and Pol ε). Gene 43 mutant phage strains were monitored for reversion of rII alleles containing premature stop codons that prevent T4 from replicating in Escherichia coli that are lysogenic for phage lambda. The rII mutant growth defect provided an exquisitely sensitive assay for mutation frequency, allowing detection of a single mutant in 109 – 1010 phage. Two temperature-sensitive mutants exhibited lower rates of reversion than wild-type T4 (Drake and Allen, 1968; Drake et al., 1969), providing evidence that DNA polymerase fidelity could be improved by single amino-acid substitutions (Figure 2). This revelation was startling at the time: given the deleterious nature of most mutations, why hadn’t evolution favored these higher fidelity variants?
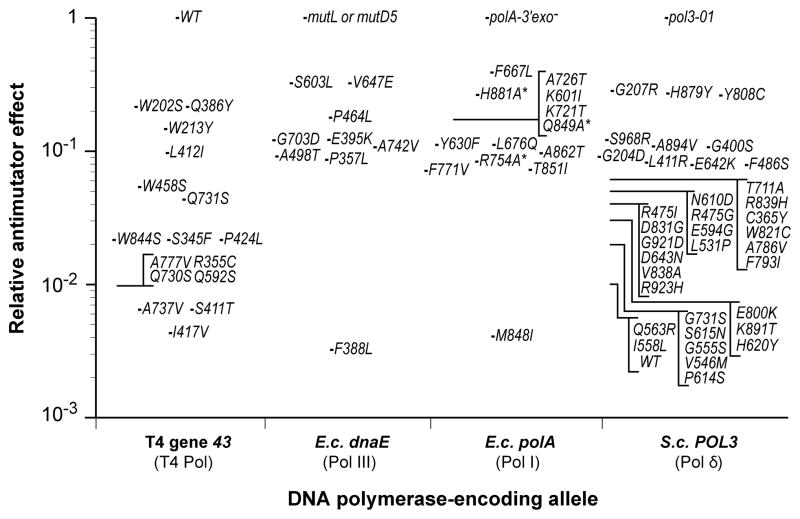
Figure 2.
Antimutator effects of DNA polymerase variants. The relative mutation frequency or rate conferred by T4 Pol, E. coli Pol III and Pol I, and yeast Pol δ antimutator variants are shown. Each variant is indicated by the single amino-acid substitution that confers antimutator activity. The T4 Pol antimutator effects are based on reversion frequency of rII UV199oc relative to wild-type T4 Pol (Reha-Krantz, 1988; Reha-Krantz and Nonay, 1993; Reha-Krantz and Nonay, 1994; Reha-Krantz and Wong, 1996). The value for the S411T single mutant is inferred from the reversion frequency of the S411T, L412M double mutant (Reha-Krantz and Nonay, 1993). Pol III antimutator effects are based on the average degree of mutL and/or mutD5 mutator suppression, as measured by the frequency of Rifampicin-resistance, Nalidixic acid-resistance, GalK2 (ochre) reversion and/or lacZ118 (ochre) reversion (Fijalkowska et al., 1993; Fijalkowska and Schaaper, 1993; Fijalkowska and Schaaper, 1995; Fijalkowska and Schaaper, 1996). The average background reversion frequency for WT cells is reported to be 3 × 10−4 relative to mutD5 (Schaaper, 1988). All Pol I antimutators were engineered into an E. coli Pol I variant lacking proofreading capability (D424A), with the exception of the Taq Pol I antimutator F667L (Suzuki et al., 2000). The relative antimutator activities of Pol I variants marked with an asterisk are based on in vitro gap-filling mutation rate determinations (Minnick et al., 1999). All other Pol I values are based on reversion frequencies of a premature nonsense codon in a plasmid-borne β-lactamase gene (Suzuki et al., 2000; Shinkai and Loeb, 2001; Shinkai et al., 2001; Loh et al., 2007). The effect of Pol δ antimutators are based on suppression of the proofreading-defective pol3-01 mutator allele, as measured by the rate of FOA-resistance mutations (Herr et al., 2011).
Early biochemical analyses in the Bessman and Nossal labs established a paradigm for how changes to the peptide sequence of a DNA polymerase could increase fidelity. T4 Pol contains a functional proofreading domain (Muzyczka et al., 1972). The first antimutator variants, later shown to carry alanine to valine substitutions at positions 737 or 777 in the thumb domain (Figure 3A,H), hydrolyzed more dNTPs per base-pair synthesized than wild-type polymerase (Muzyczka et al., 1972). Further analysis of the A737V variant revealed the polymerase to be less processive and efficient at strand-displacement synthesis (Gillin and Nossal, 1976a; Gillin and Nossal, 1976b). In contrast, mutator polymerase variants hydrolyzed less dNTPs per base-pair synthesized than wild type, strongly suggesting that replication fidelity was determined by the balance between proofreading and primer extension (Muzyczka et al., 1972). The high rate of dNTP hydrolysis by the antimutator polymerases indicates that proofreading frequently removes even correctly paired 3′ primer termini. As a sign of this hyper-editing, T4 antimutator phage replicate poorly in an optA1 E. coli strain, which has lower dNTP pools due to high levels of a cellular dGTPase (Gauss et al., 1983; Reha-Krantz and Nonay, 1994). The demonstration that increased proofreading efficiency comes with a pronounced cost in fitness provides one explanation as to why evolution has not favored the higher fidelity variants. Studies of the antimutator alleles using other mutation reporter sites revealed another answer. The antimutator polymerases lowered mutation rates for AT→GC transitions, but not for GC→AT transitions or transversions (Drake et al., 1969; Ripley, 1975). Indeed, the rate of transversions was slightly higher at some sites. Thus, gains in fidelity conferred by some antimutators may be specific to certain sequence contexts. Moreover, these same polymerase variants may cause decreased fidelity in other contexts (Drake, 1993). For this reason, the relative magnitude of antimutator activity based on reversion assays reported in Figure 2 should be interpreted with caution. Forward mutation rate assays based on gene inactivation, which have numerous potential mutation sites, give a more balanced assessment of overall replication fidelity.
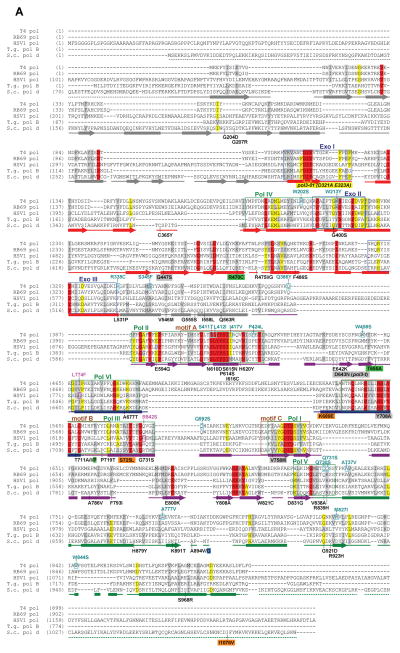
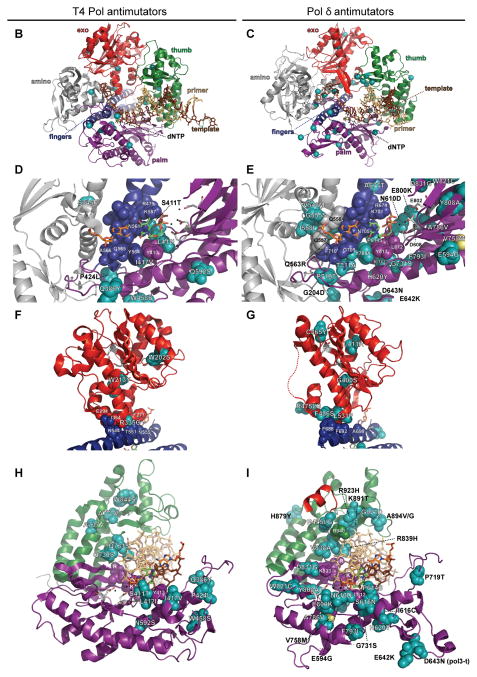
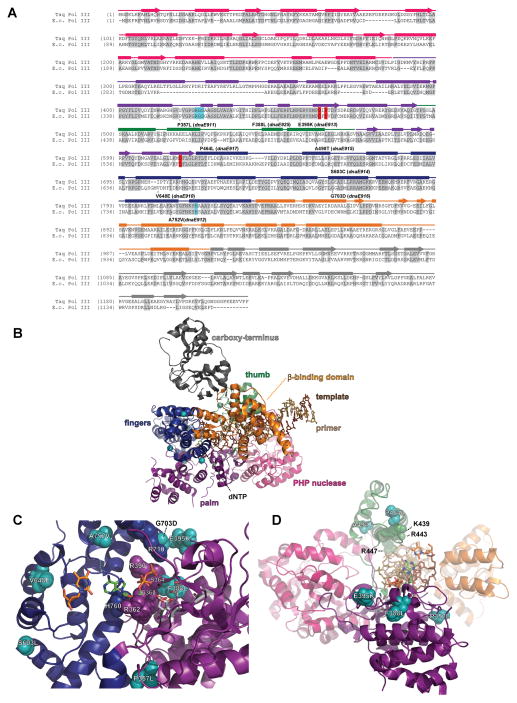
Figure 3.
B-family polymerase antimutators. (A) Aligned sequences of five B-family DNA polymerases are shown: bacteriophage T4 (T4 Pol), bacteriophage RB69 (RB69 pol), Herpes Simplex Virus 1 (HSV1 pol), Thermococcus gorgonarius (T.g. pol B), and Saccharomyces cerevisiae (S.c. pol d). Secondary structural elements of yeast Pol δ (Swan et al., 2009) are indicated below the alignment and color coded to depict their locations in the amino (gray), exo (red), palm (purple), fingers (blue) and thumb (green) domains of the protein: rectangles, α-helices; arrows, β-strands; solid lines, loops; dashed lines, structure not solved. Highlighted amino-acid residues in the alignment correspond to the following: red, absolutely conserved; yellow, identical in majority of sequences; gray, similar in majority of sequences. Conserved polymerase and exonuclease motifs are indicated by colored frames: blue, Exo motifs; green, Pol motifs (Wang et al., 1989); brown, motifs A, B, C (Delarue et al., 1990). Amino-acid substitutions of interest are placed above or below the alignment at the relevant position, highlighted according to the following scheme: aqua, T4 Pol antimutators (Reha-Krantz, 1995a; Reha-Krantz and Wong, 1996; Reha-Krantz, 2010); magenta, HSV1 Pol antimutators (Hall et al., 1984; Gibbs et al., 1988; Hwang et al., 2004; Tian et al., 2009); yellow, yeast pol3-01; no highlight, antimutators isolated as suppressors of the pol3-01 mutator phenotype; green, antimutators isolated as suppressors of the pol3-D407A mutator phenotype; orange, antimutators isolated as suppressors of the pol3-F406A mutator phenotype (three substitutions in the same mutant); blue, antimutator isolated as a suppressor of the pol3-D463A mutator phenotype; gray, previously reported antimutators in Pol δ (pol3-t (D643N), G447S, Y708A, and V758M) (Tran et al., 1999a; Hadjimarcou et al., 2001; Pavlov et al., 2004; Li et al., 2005). The antimutators affecting pol3-01, pol3-D407A, pol3-F406A, and pol3-D463A are from Herr et al. (Herr et al., 2011). (B-I) Locations of antimutator amino-acid substitutions in T4 Pol [B, D, F, and H; mapped onto RB69Pol (Franklin et al., 2001), Protein Data Bank accession code 1IG9] and Pol δ [C, E, G, and I; mapped onto Pol δ (Swan et al., 2009), PDB accession code 3IAY]. The proteins are shown as schematic diagrams of the α-carbon backbone with α-carbons or amino acids of interest depicted as space-filling spheres. Wild type amino-acid residues corresponding to antimutator mutations are shown as teal spheres and labeled to indicate the antimutator substitutions. Structural domains are color coded as in the alignment in panel A. The incoming dNTPs are denoted by green CPK sticks and the template nucleotides by orange CPK sticks. Active-site carboxylate side chains are gray CPK sticks coming out of the purple (palm) or red (exo) ribbons. Metal ions are small black spheres. Important non-mutated residues proximal to the antimutator substitutions are also shown as space-filling spheres and color coded according to domain. (B and C) Distribution of antimutator substitutions. The α-carbons of wild type residues corresponding to antimutators are shown as teal spheres. (D and E) Antimutator substitutions near the polymerase active site. The fingers and thumb domains have been removed for clarity. T4 Pol S345 and Pol δ residues V546, G555, I558, and Q563 from the amino domain are in three closely associated α-helices that bind the template and buttress the fingers domain. The antimutator substitution (V758M) that conferred PAA-resistance to a Pol δ-L612M strain is shown in yellow. (F and G) Antimutator substitutions in the exo domain. All T4 Pol antimutator variants occurred in the context of a functional exonuclease, unlike Pol δ antimutator polymerases, which carried D321A and D323A substitutions that inactivate the exonuclease. The dotted red line in (G) corresponds to a missing loop in the Pol δ structure (amino acids 491–496). (H and I) Antimutator substitutions in the DNA binding track. The primer (gold) and template (brown) are held by a series of interactions along the DNA minor groove. Minor-groove ‘sensing’ residues in the KKRYA motif of the palm domain (T4 Pol K702, K703, R704; Pol δ K813, K814 and R815) are shown as space-filling spheres labeled K and R. Unpaired 5′ nucleotides of the template along with the amino, exo, and fingers domains have been removed for clarity. A color version of this figure is available online.
After this early work, Linda Reha-Krantz isolated a large collection of new antimutator T4 Pol variants [reviewed in (Reha-Krantz, 1995b)]. The first variants were identified by growing T4 gene 43 amber mutants in bacterial strains expressing different suppressor tRNAs (Reha-Krantz, 1988). By varying the suppressor tRNA, the influence of different amino acids at the stop codon position could be assessed. Eight stop codon/suppressor tRNA combinations produced an antimutator phenotype (Figure 2). Those with the strongest effect (Q730S, Q731S, and W844S) clustered together in the thumb domain with A737V and A777V (Figure 3A,H), suggesting that they shared a common mechanism.
A strong antimutator substitution (I417V) in the palm domain of T4 Pol was discovered in a mutator phage encoding eight other amino-acid substitutions. Analysis of each substitution’s effect on polymerase fidelity revealed the presence of both mutator and antimutator changes in the same T4 Pol variant, suggesting that the I417V-encoding mutation may have arisen to mollify the mutator (Reha-Krantz, 1988). I417V affects motif A, which is highly conserved in DNA polymerases (Figure 3A) and helps to form the dNTP binding site (Figure 3D). In theory, the mutation could elevate replication fidelity by increasing nucleotide selectivity and/or reducing polymerase processivity, thereby allowing more time for proofreading. The I417V mutant phage grows poorly on the optA1 E. coli strain, and the purified polymerase displays reduced processivity (Reha-Krantz and Nonay, 1994). These observations are consistent with increased fidelity due to hyper-editing, although a concomitant effect on nucleotide selectivity has not been excluded.
Additional antimutagenic changes emerged with the development of the first genetic selection for antimutators. The selection utilized a T4 Pol variant allele encoding an L412M substitution, which was originally identified as a suppressor mutation of the optA1 sensitivity conferred by I417V (Reha-Krantz and Nonay, 1994; Stocki et al., 1995). As a single amino-acid substitution in T4 Pol, L412M elevates mutation rate moderately and confers sensitivity to the antiviral drug and pyrophosphate analogue, phosphonoacetic acid (PAA) (Reha-Krantz et al., 1993; Reha-Krantz and Nonay, 1994). A PAA-resistant Herpes Simplex Virus I (HSV I) mutant polymerase encoding an R842S amino-acid substitution (Figure 3A) was previously shown to be an antimutator (Hall et al., 1984; Gibbs et al., 1988). On the hunch that PAA-resistance was linked to antimutagenesis, Reha-Krantz selected for PAA-resistant variants of the L412M T4 mutant phage as well as a second PAA-sensitive mutant involving duplication of D836 (Reha-Krantz and Nonay, 1994; Reha-Krantz and Wong, 1996). They identified substitutions in motif A (S411T and L412I) and elsewhere in the polymerase (S335C and S345F) that bestowed antimutator activity even in the absence of the PAA-sensitizing mutation (Reha-Krantz and Nonay, 1994; Reha-Krantz and Wong, 1996). As expected, S335C also confers optA1 sensitivity. Selection for genetic suppressors revealed a P424L substitution that restored the ability of the S335C phage to propagate on the optA1 strain. Remarkably, the P424L single-mutant phage is an antimutator and unable to grow on optA1 E. coli (Li et al., 2010a). The S335C,P424L double-mutant phage retains substantial antimutator properties, although the mutation rate is modestly higher than either of the single mutants alone (Li et al., 2010a). The ability of the double mutant to reduce the antimutator and optA1 sensitivity phenotypes of the single mutants suggests that the amino-acid substitutions affect a common mechanism that influences fidelity. Genetic suppressor analysis of the optA1-sensitive phenotype of P424L mutant phage suggests that this mechanism may relate to closure of the fingers domain (Li et al., 2010a).
Together, these elegant genetic and biochemical experiments with T4 Pol serve as a reference for all other antimutator studies, and indicate that a wide variety of single amino-acid substitutions can increase replication fidelity (Figures 2, 3). Moreover, the recurrent phenotype of optA1-sensitivity and PAA-resistance argues that most if not all T4 antimutator polymerases increase replication fidelity by shifting the balance from polymerization to proofreading.
E. coli Pol III
Studies with E. coli Pol III in the 1990’s by Roel Schaaper and colleagues were the first to indicate that amino-acid substitutions in DNA polymerases can elevate replication fidelity in the absence of proofreading activity. Because Pol III performs the bulk of both leading- and lagging-strand DNA synthesis in E. coli (Johnson and O’Donnell, 2005), mutator variants are especially detrimental. The polymerase activity resides in the α subunit of Pol III, encoded by the dnaE gene, while the proofreading activity is found in the tightly-associated ε subunit, encoded by the adjacent gene on the chromosome, dnaQ. The first Pol III antimutator variant was found as a suppressor of the strong dnaQ mutator allele, mutD5 (Schaaper and Cornacchio, 1992). mutD5 exhibits a conditional mutator phenotype, producing moderate mutation rates in minimal media and high rates in rich conditions (Maruyama et al., 1983; Scheuermann et al., 1983), where polymerase errors partially saturate MMR (Schaaper, 1988; Schaaper, 1989; Schaaper and Radman, 1989). Mutation accumulation in mutD5 cells slows bacterial growth, and Schaaper isolated a spontaneous, faster growing clone of cells. Phage P1 transduction of mutD5 from this clone resulted in both large and small colonies, suggesting that the isolate contained a suppressor mutation linked to dnaQ (Schaaper and Cornacchio, 1992). The mutation (later shown to encode a V648E substitution) mapped to dnaE and suppressed not only the growth defect, but also the mutD5 mutator phenotype (Figure 2), as determined by stop codon reversion and ‘forward’ mutation frequency assays, which score mutations at multiple sites.
With the discovery that changes to the polymerase subunit of Pol III can offset proofreading defects, Schaaper and co-workers addressed whether Pol III antimutators suppress defects in MMR. In bacterial MMR, a MutS homodimeric protein recognizes the mismatched DNA and recruits a MutL homodimer, which in turn recruits the MutH endonuclease that nicks the nascent DNA and initiates repair (Iyer et al., 2006). Mutagenized DNA from the dnaE-dnaQ chromosomal region was transduced into a mutL mutator strain, and mutants with an attenuated mutator phenotype were identified by scoring reversion frequency with a papillation assay (Fijalkowska et al., 1993). Colonies with lower numbers of papillae also exhibited reduced mutation frequencies by forward mutation assays, confirming their antimutator character. The mutants carried mutations in dnaE that encoded single amino-acid substitutions in the polymerase subunit (Figures 2, 4) (Fijalkowska and Schaaper, 1993). The ability to isolate dnaE-encoded suppressors of mutL indicates that most mutations in this strain derive from Pol III errors that escape proofreading. Thus, the antimutators reduce the frequency of the most common Pol III-derived MMR substrates.
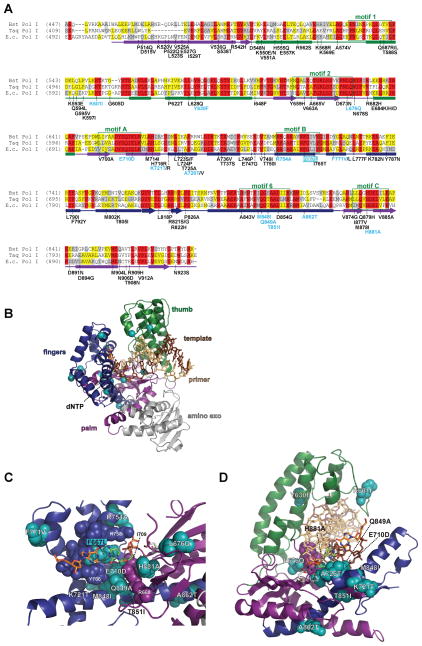
Figure 4.
Pol III antimutators. (A) Alignment of Pol III peptide sequences from Thermus aquaticus (Taq Pol III) and Escherichia coli (E.c. Pol III). Secondary structural elements based on the Taq Pol III structure (Wing et al., 2008) (PDB accession code 3E0D) are indicated below the alignment and color coded to depict their location (PHP, magenta; palm, purple; fingers, blue; thumb, green; β-binding, orange; carboxy-terminus, charcoal): rectangles, α-helices; arrows, β-strands; solid lines, loops. Amino acids shaded in gray are conserved between Taq and E. coli. Aspartates that coordinate metal ions for catalysis are shaded in red. Residues that stack with ribose of incoming dNTP are shaded in cyan. E. coli dnaE amino-acid changes and allele numbers are depicted below the alignment. (B) Distribution of antimutator substitutions. The α-carbons of antimutator positions are shown as teal spheres. (C) Antimutators near the Pol III active site. The RGS motif (362–364) and H760 cradle the incoming dNTP, while R390 and R710 interact with the triphosphate. (D) Antimutators in the Pol III Thumb. A498T and P464L affect the positioning of an α-helix that includes three basic residues that interact with the primer•template backbone (K439, R443, R447). A color version of this figure is available online.
The mechanism of the mutL Pol III antimutators could be interpreted within the paradigm established by the T4 studies in which antimutators lower mutation rates by increasing proofreading efficiency. In order to determine whether proofreading activity of the ε subunit was required for their antimutator phenotype, Schaaper and colleagues moved the new dnaE antimutators into the mutD5 strain (Fijalkowska and Schaaper, 1995). The new antimutators effectively suppressed the mutator phenotype of the proofreading mutant (Figure 2), suggesting that these antimutators did not require the ε subunit for their gains in replication fidelity. However, the underlying mutation in the mutD5 allele encodes a T to I change (DTET to DTEI, underlined) adjacent to two highly conserved acidic residues required for catalysis (in bold). Because these essential catalytic residues were still intact, it remained possible that the antimutators simply increased the efficacy of the residual proofreading activity of the mutant ε subunit. Complete elimination of proofreading activity by substitution of the essential aspartate and glutamate residues to alanine (ATAT) produced a very strong dominant mutator phenotype that was lethal when the mutant protein was the sole source of the ε subunit (Fijalkowska and Schaaper, 1996). The antimutator substitutions in the α subunit rescued this lethality, confirming decisively that the amino-acid alterations in Pol III lower mutation rates independently of the proofreading activity of the ε subunit (Fijalkowska and Schaaper, 1996).
Some Pol III antimutators may suppress polymerase errors by increasing nucleotide selectivity. Evidence of altered base selectivity comes from the observation that certain Pol III antimutators suppress the mutator phenotype of mutT, which encodes an 8-oxo-dGTPase. By ‘cleansing’ nucleotide pools, the mutT protein minimizes transversions caused by A 8-oxo-dGTP base pairs (Schaaper, 1996). Schaaper identified two new Pol III antimutator variants that suppressed the mutT mutator phenotype 10-fold, and these were also effective in suppressing the mutL mutator phenotype. One antimutator encoded by the dnaE940 allele (mutation unknown) was three times more effective at suppressing mutT than other Pol III antimutators that were equally effective in suppressing mutL (Schaaper, 1996). The disproportionate effect of this allele on mutT suggests that the corresponding polymerase contains changes in the active site that excludes A 8-oxo-dGTP base pairs. Whether improved base-selectivity accounts for all Pol III antimutator effects is uncertain. As recognized by Schaaper, the antimutators improve replication fidelity in the context of the cellular milieu and may promote polymerase dissociation from the DNA, allowing an alternate proofreading activity to edit the error (Fijalkowska and Schaaper, 1995).
Recent reports indicate that the PHP domain of the Thermus thermophilus Pol III α subunit exhibits a Zn2+-dependent 3′→5′ exonuclease activity capable of editing mispaired primer termini (Stano et al., 2006). Substitution of two aspartates in the presumptive PHP active site from Thermus aquaticus Pol III inactivated nuclease activity, facilitating co-crystallization of the enzyme with primer•template DNA (Wing et al., 2008). The resulting polymerase structure revealed the PHP active site is a full turn of the DNA helix away from the polymerase active site (Figure 4B), suggesting that dissociation of the duplex or significant unwinding of the primer strand would be required for PHP editing of the 3′ end (Wing et al., 2008). Is the E. coli Pol III PHP domain an active nuclease and could it contribute to antimutagenesis in Schaaper’s mutants? Arginine residues replace two histidines in the E. coli PHP active site (R10 and R203; Figure 4A) (Aravind and Koonin, 1998). Moreover, purified recombinant E. coli Pol III α subunit did not degrade a primer•template with a terminal mismatch (Mo and Schaaper, 1996). These observations question whether the E. coli PHP is indeed a nuclease. However, it remains possible that the E. coli PHP degrades single-stranded DNA or requires the context of the holoenzyme to be active. A key test for this or any other candidate proofreader is to determine whether substitution of the active site aspartates (see T. aquaticus, above) counteracts the effects of the antimutators.
E. coli Pol I
The T4 Pol and E. coli Pol III polymerases play essential roles in the replication of their respective genomes, thereby facilitating the use of genetic approaches to identify active antimutator variants. In contrast, E. coli Pol I is not essential for survival of wild type cells (Kornberg and Baker, 1992). Thus, Pol I antimutator variants were initially identified by site-directed mutagenesis of an exonuclease-deficient Pol I allele (pol3A-3′exo−) followed by screening for biochemical activity and alterations in mutation rate (Minnick et al., 1999). Using this approach, three amino-acid substitutions were found (R754A, Q849A, and H881A) that individually confer significant antimutator activity (Figure 2) (Minnick et al., 1999).
To facilitate the study of Pol I fidelity, Loeb and colleagues cleverly combined random mutagenesis of Pol I (Suzuki et al., 2000; Patel et al., 2001; Shinkai and Loeb, 2001; Loh et al., 2007) with a genetic selection strategy first used to identify functional Pol β variants (Sweasy and Loeb, 1993). Pol I participates in DNA repair as well as in the initiation of ColE1 plasmid replication (Kornberg and Baker, 1992). Functional derivatives of Pol I were identified by their ability to complement a temperature-sensitive Pol I allele (polA12) in a synthetically lethal interaction with recA718 in E. coli. The polA12 recA718 double-mutant strain exhibits persistent single-stranded gaps in genomic DNA at the non-permissive temperature, suggesting that the complementing mutant polymerases perform gap repair (Witkin and Roegner-Maniscalco, 1992). Once functional Pol I variants were identified, they were screened for altered fidelity in vitro by a gap-filling forward mutation assay or in vivo by scoring reversion of a nonsense codon in the β-lactamase gene inserted close to the origin of replication on a ColE1 plasmid.
The first screen, targeting motif B of the fingers domain (O-helix) of Thermus aquaticus (Taq) Pol I, which is naturally proofreading-deficient, identified an F667L substitution that lowered the frequency of transversions in vitro (Suzuki et al., 2000). The second mutagenesis screen, targeting motif A of proofreading-proficient E. coli Pol I, identified an E710D substitution that lowered β-lactamase reversion three-fold (Shinkai and Loeb, 2001; Shinkai et al., 2001). A related screen targeting motif A of Taq Pol I identified a mutant with multiple changes (L605R, L606M, V607K, A608S, L609I, S612R) that encoded a higher fidelity enzyme (Patel et al., 2001). Finally, Loeb and colleagues mutagenized the entire polymerase domain of exonuclease-deficient E. coli Pol I (Loh et al., 2007). A surprising 12% of mutants suppressed β-lactamase reversion at least 10-fold. Some antimutator polymerases sustained single amino-acid changes (Y630F, L676Q, K721T, F771V, M848I, T851I, and A862T). But most enzymes carried multiple substitutions, which complicates the interpretation. Analysis of one of these complex variants found that both amino-acid substitutions in the clone (K601I and A726T) individually confer an antimutator phenotype (Loh et al., 2007). Moreover, the substitutions acted in an additive fashion in the double-mutant enzyme, suggesting that they lower mutation rates by independent means. Thus, other complex Pol I variants may also carry multiple antimutator amino-acid substitutions.
The single amino-acid changes sufficient for Pol I antimutagenesis cluster with substitutions encoded in the complex mutants (Figure 5A). Mutations in several other regions were repeatedly found in the complex antimutator clones, highlighting candidate regions for further study. These include sequences upstream of motif 1 (514–574), upstream of motif 6 (818–826), and downstream of motif C (904–912) (Figure 5A). The wide distribution of antimutator substitutions in Pol I mirrors the diverse locations of antimutator substitutions in T4 Pol and Pol III. As with the Pol III antimutators, increases in replication fidelity occurs independently of the associated proofreading exonuclease, which is inactivated in these screens. Thus, Pol I antimutator amino-acid substitutions may either increase nucleotide selectivity or reduce extension of mispaired primer termini.
Figure 5.
Pol I antimutators. (A) Alignment of Pol I peptide sequences from Bacillus stearothermophilus (Bst Pol I), Thermus aquaticus (Taq Pol I), and Escherichia coli (E.c. Pol I). Secondary structural elements based on the Taq Pol I structure (Li et al., 1998) (PDB accession code 3KTQ) are indicated below the alignment and color coded to depict their locations in the amino (the proofreading exonuclease in E. coli Pol I; gray); palm (purple), thumb (green) and fingers (blue) domains of the protein: rectangles, α-helices; arrows, β-strands; solid lines, loops. Highlighted amino acid residues in the alignment correspond to the following: red, absolutely conserved; yellow, identical in majority of sequences; gray, similar in majority of sequences. Conserved Pol I motifs are indicated by green frames. Amino-acid substitutions of interest are placed below the alignment at the relevant position, color coded according to the following scheme: teal, single amino acid antimutator substitutions in E. coli Pol I (Minnick et al., 1999; Loh et al., 2007); black, substitutions found in complex mutant alleles from Loh et. al (Loh et al., 2007); white lettering with teal shaded box, Taq Pol I antimutator, (Suzuki et al., 2000). (B) Distribution of antimutator substitutions. The α-carbons of antimutator positions are shown as teal spheres. (C) Pol I active site antimutator substitutions. (D) Pol I antimutators that may influence DNA binding. Purple spheres next to L676Q correspond to adjacent amino acids from Motif 2 that bind the -3 base pair of the primer•template (PNL676QN). A color version of this figure is available online.
Kinetic analyses suggest that mutant Pol I enzymes with R754A and Q849A substitutions are less prone to extending mismatched primer termini than wild type Pol I (Minnick et al., 1999). Likewise, the complex motif A antimutator variant of Taq Pol I (L605R, L606M, V607K, A608S, L609I, S612R) also exhibited reduced mispair extension (Patel et al., 2001). In failing to extend mispairs efficiently, these mutant polymerases may lower apparent error rates by preventing misinsertions from being extended into duplex DNA; rare unextended mispairs are not detected in the gap-filling assay. A failure to extend mispairs could also account for reductions in mutation rates in the β-lactamase reversion assay in vivo. Pol III normally replaces Pol I following initiation of ColE1 plasmid replication (Kornberg and Baker, 1992). This usually occurs at a primasome assembly site (Kornberg and Baker, 1992), but can occur in its absence (Camps et al., 2003). The reversion site used in these studies is positioned 230 nucleotides downstream of the origin. Thus, any defect that promotes Pol I switching to Pol III, at or before the reversion site, would produce an antimutator phenotype.
Yeast Pol δ
In current models of eukaryotic DNA replication, Pol δ performs the bulk of lagging-strand DNA synthesis, while Pol ε synthesizes the leading strand [reviewed in (Kunkel and Burgers, 2008)]. The first report of a eukaryotic antimutator polymerase variant was in yeast and resulted from combining the Pol δ frameshift mutator allele pol3-t (Tran et al., 1995) with the proofreading-defective allele pol3-01 (Morrison et al., 1993). Rather than producing a compound mutator phenotype, the double mutant exhibited a three-fold reduction in mutation rate relative to pol3-01 (Tran et al., 1999a). Pavlov et al. demonstrated that another weak mutator allele (pol3-Y708A) suppressed pol3-01 frameshift mutations, although not base-substitution errors (Pavlov et al., 2004). Curiously, the analogous substitution (Y831A) in proofreading-defective Pol ε (encoded by pol2-4) suppressed both types of errors (Pavlov et al., 2004). The mutator phenotypes of pol3-t and pol3-Y708A partially depend on monubiquitination of PCNA at K164 and the activity of the TLS polymerase, Pol ζ (Northam et al., 2006), suggesting that the amino-acid substitutions in these pol3 mutants promote remodeling of the replication fork. The observation that pol3-t and pol3-Y708A suppress the pol3-01 mutator phenotype suggests that error-free pathways may also gain access to primer termini in the mutant strains. Thus, the contrasting effects of antimutator substitutions on proofreading-defective Pol δ and Polε may reflect intrinsic differences between the polymerases or the ability of alternate replication enzymes to correct or extend errors on the leading and lagging strands.
Additional antimutator alleles of Pol δ were identified by Reha-Krantz and colleagues, who extended their T4 studies into yeast. Introduction of a G447S substitution into a β-hairpin structure in the exonuclease domain of Pol δ produced a frameshift-suppression phenotype (Hadjimarcou et al., 2001). The analogous change in T4 Pol (G255S) had been isolated as a suppressor of the optA1-sensitivity conferred by an antimutator (Stocki et al., 1995). In T4 Pol, the substitution decreases fidelity by inhibiting hyper-partitioning of the primer strand to the exonuclease domain (Stocki et al., 1995). This suggests that yeast Pol δ G447S suppresses misalignment errors that cause frameshifts by minimizing primer DNA dissociation. In a separate study, Reha-Krantz demonstrated that Pol δ could be made PAA-sensitive by the introduction of an L612M substitution (analogous to T4 Pol L412M; see above) (Li et al., 2005). Selection for PAA-resistance in the pol3-L612M strain identified a V758M substitution that suppressed the L612M mutator phenotype in an MMR-deficient (msh2Δ) mutant background (Li et al., 2005). Intriguingly, substitution of the L612-equivalent in human Pol δ with lysine (L606K) increases polymerase fidelity in vitro in the presence or absence of proofreading activity (Schmitt et al., 2010). Taken together, these observations indicate that Pol δ and T4 Pol have common antimutagenic pathways. This conclusion is strongly supported by the large collection of antimutator changes we recently isolated in Pol δ that suppress the proofreading defect encoded by pol3-01 [(Herr et al., 2011); Figures 2 and 3A].
We exploited the observation that in yeast, as in E. coli, excessive mutation accumulation undermines colony formation. Evidence for this ‘replication error-induced extinction’ came from early studies by Morrison and Sugino who demonstrated that proofreading and MMR acted sequentially to correct DNA polymerase errors (Morrison et al., 1993; Morrison and Sugino, 1994; Tran et al., 1999b). Inactivation of the integral proofreading activity of Pol δ (e.g., pol3-01) increased mutation rates by nearly two orders of magnitude, as did inactivation of MMR (Simon et al., 1991; Morrison et al., 1993; Prolla et al., 1994). Diploids defective in both Pol δ proofreading and MMR (pol3-01/pol3-01 pms1/pms1) exhibited a 104-fold increase (Morrison and Sugino, 1993; Tran et al., 1999b). Double-mutant haploid spores derived from POL3/pol3-01 PMS1/pms1 diploids germinated, but formed microcolonies of around 100 cells arrested at all stages of the cell cycle (Morrison and Sugino, 1993; Tran et al., 1999b). A pure replication defect would have arrested cells immediately in S-phase or perhaps at the G2/M transition, but more than 40% of pol3-01 pms1 cells exhibited a G1-arrest phenotype, while cells arrested in S-phase comprised only 5% (Morrison and Sugino, 1993). The relative abundances of each morphology in the pol3-01 pms1 microcolonies correlate with the distribution of cell-cycle arrest phenotypes observed in a survey of null phenotypes of all essential yeast genes (Yu et al., 2006), suggesting that random mutations in essential genes drive extinction of the double-mutant mutator yeast.
To define the maximal mutation rate in haploids, we explored the synthetic relationship between Pol δ proofreading and MMR using a plasmid shuffling scheme (Boeke et al., 1984; Simon et al., 1991) that created a conditional mutator phenotype. The yeast strain for these studies carried a deletion of the chromosomal gene for MSH6 (msh6Δ), an essential MMR component for repair of most base-base mismatches (Marsischky et al., 1996). We shuffled in a collection of mutant pol3 plasmids carrying hypomorphic proofreading-defective alleles of POL3, as well as null alleles, such as pol3-01. Some pol3 msh6Δ combinations were viable and displayed soaring mutation rates (1000-fold increase). However, the strongest pol3 mutators were lethal in combination with msh6Δ, consistent with previous observations of synthetic lethality between pol3-01 and msh6Δ (Sokolsky and Alani, 2000). By measuring mutation rates of the viable combinations and calculating the theoretical mutation rates of the lethal combinations, we determined that the threshold for error-induced extinction is around 1 × 10−3 can1 mutants/cell division. Assuming CAN1 (1500 base pairs) represents an average gene, this mutation rate predicts a mutational target size of 1000 essential genes, which is very close to the number of essential genes in yeast determined by systematic gene inactivation (Winzeler et al., 1999). Thus, accumulation of mutations throughout the genome, in many different contexts, likely drives replication error-induced extinction in yeast.
While defining the upper limits of mutagenesis, we isolated numerous mutants that survived the replication error-induced extinction induced by combining msh6Δ with pol3-01 (D321A E323A), pol3-F406A, or pol3-D407A (Figure 3A). Mutants with increased growth capacity also arose in an msh6Δ strain expressing a high, but sub-lethal mutator phenotype induced by pol3-D463A. A large-scale screen for pol3-01 msh6Δ suppressors revealed that one third of these error-induced extinction (eex) mutants carried mutations that encode single amino-acid changes in Pol δ. Importantly, the rest mapped to the nuclear genome, implicating other genes in antimutagenesis. Some of these chromosomal mutations may alter checkpoint signaling genes (e.g., DUN1) responsible for a heightened pol3-01 mutator state (Datta et al., 2000). Others may influence the fidelity of the polymerase directly. The amino-acid substitutions in Pol δ conferred a range of antimutator activities, suppressing pol3-01 between 3- and 50-fold in forward mutation rate assays at CAN1 and URA3 (Figure 2). DUN1 deficiency reduces the pol3-01 mutator phenotype 5- to 10-fold (Datta et al., 2000). Thus, the strongest antimutators from our study likely influence the fidelity of the polymerase directly, in addition to any effects on checkpoint signaling.
Many antimutator substitutions affected polymerase features previously implicated in antimutagenesis in the T4 studies (Figure 3A). Five mutants encoded changes to motif A of the palm domain (N610D, P614S, S615N, I616C, and H620Y). Additional suppressors affected regions of the palm domain with less conservation (A786V, F793I, E800K, Y808A, and W821C). Three substitutions affected motif Pol V at the boundary between the palm and the thumb domains, including two (V838A and R839H) that corresponded precisely with the T4 Pol Q730S and Q731S antimutators. As observed in the T4 studies, numerous antimutator substitutions affected other parts of the thumb domain (H879Y, K891T, A894V/G, G921D, R923H and S968R). And surprisingly, even though the Pol δ exonuclease was inactivated in the pol3-01 msh6Δ strain, numerous suppressors altered the exonuclease domain (C365Y, G400S, R475I/G, F486S, and L531P), in a manner reminiscent of the T4 Pol antimutators W202S, W213Y, and R335C. Intriguingly, Pol δ L531P aligns with T4 Pol R335C at the boundary between the exonuclease domain and a sequence just before the palm domain that folds with the amino-terminal domain. Our screen revealed four additional substitutions in this linker sequence (V546M, G555S, I558L, and Q563R). These were among the strongest suppressors (Figure 2), suggesting that this region of the polymerase has an important role in replication fidelity.
MMR-proficient strains expressing the mutant alleles as the sole source of Pol δ grew normally, indicating that the mutant polymerases retain sufficient activity to fulfill the essential role of Pol δ in lagging-strand DNA synthesis. Because error-induced extinction is due to random mutations in multiple genes, the antimutators likely reduce Pol δ errors in many different sequence contexts. Moreover, like the Pol III and Pol I antimutators, the gains in replication fidelity must occur without the aid of the proofreading activity of the polymerase, which was inactivated in these screens.
STRUCTUAL IMPLICATIONS OF ANTIMUTATORS
The antimutagenic amino-acid substitutions described above are distributed throughout the structures of their respective DNA polymerases. But far from random, the underlying changes affect functionally important features that evoke distinct, yet potentially overlapping antimutator mechanisms. Antimutators affect polymerase active sites, exonuclease domains, and structural elements that bind double-stranded DNA.
Antimutators in the Active Site
The active sites of all DNA polymerases are found at the junction between the fingers and palm domains. In A- and B-family polymerases, the catalytic carboxylates reside in β-strands and loops from motifs A and C that contribute to the palm β-sheet surface (Figures 3A,5A). The ribose of the incoming dNTP stacks at the edge of the β-sheet surface on motif A residues proximal to the catalytic carboxylates (T4 Pol: L412, Y413; Pol δ: L612, Y613; E. coli Pol I: I709, E710; Figures 3D,E and 5C). These dNTP stacking residues are at one end of the motif A α-helix, which angles down from the β-sheet surface, wedged between two conserved α-helices. One interacting helix is from motif B of the fingers domain (Figures 3A and 5A) that rises above the plane of the β-sheet, defining the binding pocket for the dNTP base, together with the template nucleotide and the last primer•template base-pair. In a large domain movement, the fingers domain close over the incoming dNTP and the binding pocket from this motif B helix interrogates the incoming base. Basic residues from the fingers domain (T4 Pol R479, K557; Pol δ R674, K701; E. coli Pol I R754, K758) sandwich the triphosphate against metal ions coordinated by the catalytic carboxylates. This stabilizes fingers domain closure and helps to create the catalytic center for phosphoryl transfer. The second interacting helix immediately follows motif B in B-family polymerases (Figure 3A) and corresponds to motif 6 in the A-family in Pol I (Figure 5). Note that these two α-helices are structurally homologous, although classified as part of the fingers domain in the A-family polymerases and the palm domain in the B-family. Each helix runs at right angles to the motif A helix, supporting the dNTP sugar binding surface. Thus, three conserved, interacting α-helices create a dynamic dNTP binding site adjacent to the essential catalytic residues.
Alterations to the structure of the catalytic surface promote antimutagenesis in both A- and B-family DNA polymerases. Antimutators in Pol δ (N610D, A786V, and E800K) affect amino acids that influence formation of the β-sheet involved in metal ion coordination (Figure 3E). E800 is adjacent to E802, which interacts with Metal C (Swan et al., 2009). A double-mutant enzyme bearing E800A and E802A substitutions exhibited reduced catalytic efficiency, which led to the conclusion that E802 coordination of Metal C contributes to pyrophosphate formation (Swan et al., 2009). However, E800 plays a key structural role in knitting the β-sheet together, and the double-mutant enzyme may suffer defects to β-sheet stability as well as Metal C coordination. E800, supported by A786, interacts with N610 from the adjacent β-strand, which lies between the motif A helix and the Metal B-binding aspartate, D608 (Figure 3A,E,I) (Swan et al., 2009). Both E800K and N610D antimutator substitutions replace the H-bonding interaction that normally occurs between these residues with charge repulsion, disrupting an interaction also observed in the analogous positions in RB69 Pol (D684 with T413 via H2O) and Thermococcus gorgonarius Pol (E578 with R406) (Wang et al., 1997; Hopfner et al., 1999; Franklin et al., 2001). Alteration of the catalytic surface of the palm domain also induces an antimutagenic effect in Pol I. H881 (mutated to A) is adjacent to the Metal A-binding aspartate, D882 (Figure 5A,C) and stacks with the ribose of the primer terminus, helping to orient the 3′ OH for chemistry. Thus, disruptions to the architecture of the β-sheet active-site surface can be antimutagenic.
A related avenue of antimutagenesis involves modification of the dNTP binding site. Most striking perhaps are the numerous antimutator substitutions found in motif A in T4 Pol (S411T, L412I, I417V, P424L; Figure 3A,D,H), Pol δ (P614S, S615N, I616C, H620Y; Figure 3A,E,I), and Pol I (E710D; Figure 5). The antimutators T4 Pol L412I and Pol I E710D alter the surface of the binding pocket that contacts the sugar of the incoming nucleotide. Pol δ Y708A of motif B affects the same surface, contacting the minor-grove side of the base. The analogous mutant in RB69 Pol (Y567A) is a strong mutator, but this phenotype is suppressed by a second substitution in the dNTP binding site, S565G (Trzemecka et al., 2010; Xia et al., 2011). Structural and biochemical analyses show that G565 in the S565G,Y567A double mutant limits the flexibility of the template base position (Xia et al., 2011) and that, under certain conditions in vitro, the double-mutant polymerase dissociates more rapidly from duplexes with mispaired primer termini (Trzemecka et al., 2010). Thus, changes to the binding pocket may influence not only nucleotide selectivity, but also stability of the replication complex. Other antimutators influencing the dNTP binding pocket include Pol δ P614S, which positions N705 of motif B next to the base of the incoming dNTP, and Taq Pol I F667L, which aligns structurally with Pol δ N705 (compare Figures 3E and 5C). Some antimutators alter residues that support the minor groove binding surface or mediate inter-helical contacts that help create the functional architecture. For instance, Pol δ I616 of motif A, in addition to supporting amino acids that contact the dNTP sugar (L612, Y613), abuts the antimutator position G731 (mutated to S) from the helix immediately following motif B (Figure 3A,E). Similarly, Pol I substitutions in motif 6 (M848I, Q849A, and T851I; Figure 5A,C) affect residues within 4 A of E710 at the interface between motifs A and 6. Q849 also plays a role in shaping the dNTP binding pocket by interacting with the last primer•template base-pair (Kiefer et al., 1998). Two other Pol I antimutators (K721T and A726T; Figure 5) affect helix M, which is an A-family-specific structure in the fingers domain that supports the motif A helix from the opposite side of motif 6 (Loh et al., 2007). Thus, structural perturbations throughout the dNTP binding site promote antimutagenesis in both A- and B-family polymerases.
A cluster of T4 Pol and Pol δ antimutator substitutions affect amino acids in a linker sequence between the exonuclease and palm domains. This sequence, which folds with the amino domain, binds the template and buttresses the fingers domain (Figure 3A,D,E). Pol δ Q563R alters a residue that contacts the fingers in the closed conformation (Figure 3E), suggesting that antimutator substitutions in this region may destabilize closure of the fingers domain. The importance of fingers closure for fidelity is also indicated by the A677T antimutator, which lies at the interface between the two α-helicies of the fingers domain, one turn of the helix from the dNTP-interacting residue, R674 (Figure 3E). Other substitutions in the exonuclease-palm linker (Pol δ V546M and I558L) interact in the hydrophobic core of the fold (Figure 3E). Intriguingly, T4 Pol S345F occupies almost the same position in the structure as Pol δ V546M (compare Figures 3D and 3E). G555S in Pol δ adds a side chain to this core, immediately adjacent to two amino acids that interact with the template nucleotide: Q556 contacts the template phosphate, while Q557 helps define the shape of the binding pocket for the template sugar (Figure 3E). The location of these B-family antimutators is especially intriguing, because this region aligns structurally with the pre-insertion site for the template nucleotide observed in crystal structures of the A-family B. stearothermophilus Pol I (Johnson et al., 2003). Closure of the Pol I fingers flips the template nucleotide from this location into the active site. The existence of an analogous pre-insertion site remains to be demonstrated for B-family polymerases, but the phenotype of these Pol δ and T4 Pol antimutators suggests that this region influences polymerase fidelity.
Seven out of nine E. coli Pol III antimutator substitutions are found in the palm or fingers domains despite occurring in a polymerase with very different architecture (Figure 4). During Pol III synthesis the ribose of the incoming dNTP stacks on R362 (E. coli numbering) of the RGS motif in the palm and H760 of the fingers (Figure 4C) (Wing et al., 2008). The triphosphate interacts with G363, S364, R390, R710, and the metals bound by the catalytic aspartates, D403 and D405 (Figure 4C). The strongest dnaE antimutator alters F388 (mutated to L) (Figure 2), which brings the helices containing the RGS motif and R390 into close proximity through a hydrophobic interaction (Figure 4C). Two other dnaE antimutators (E395K and G703D) alter amino acids that pin the palm and the fingers domains together, positioning R710 to interact with the dNTP (Figure 4C). Thus, in all three polymerase families, modifications to the network of interactions that position the incoming dNTP serve to increase replication fidelity.
Antimutagenic changes to the active sites of DNA polymerases could increase fidelity in several ways. Variants may increase negative selectivity in the binding pocket against non-Watson-Crick base-pairs. However, such variants may be rare if the benefits of improved selectivity against one non-Watson-Crick base-pair are blunted by less stringent selection against a different incorrect base-pair. Amino-acid substitutions may also increase accuracy without modifying the dNTP binding pocket. Antimutators affecting the structure of the catalytic surface may promote delays in phosphoryl transfer, providing more time for incorrect nucleotides to dissociate. Other changes may enhance the ability of the polymerase to adopt a conformation that promotes release of incorrect dNTPs. Similarly, destabilization of fingers closure may allow incorrect nucleotides to dissociate more rapidly than normal.
In addition to any increases in dNTP selectivity, antimutator substitutions in the active site likely promote partitioning of the primer strand to the exonuclease domain. All T4 Pol antimutators exhibit a hyper-editing phenotype that exhausts dNTP pools in the optA1 strain (Reha-Krantz, 2010). This can be explained by frequent partitioning of correctly paired primer termini to the exonuclease domain and degradation. Primer partitioning in a polymerase poised for misincorporation would be especially efficient due to inherently slow catalysis, leading to dissociation of the incorrect dNTP. In enzymes with defective proofreading, the same mechanism could occur without costly degradation of the 3′ end, allowing error-free synthesis to resume upon reannealing back into the polymerase active site. In the presence of a terminal mispair, more complete dissociation of the duplex DNA from the polymerase may allow alternate editing enzymes to correct the error in trans (Perrino and Loeb, 1990; Morrison and Sugino, 1994; Fijalkowska and Schaaper, 1995; Pavlov et al., 2004; Albertson and Preston, 2006; Pavlov et al., 2006a).
The vast majority of antimutators affecting the polymerase active site were identified using genetic screens in replicating organisms. Detailed biochemical, structural and kinetic analyses of purified variant polymerases are needed to determine their mechanisms of increased fidelity.
B-family Antimutators That Change the Exonuclease Domain
Several T4 Pol and Pol δ antimutator substitutions involve amino acids with structural roles in the exonuclease domain. W202S and W213Y in T4 Pol and C365Y, G400S, and L411R in Pol δ change amino acids that contribute to the hydrophobic core (Figure 3F,G). Pol δ F486S and L531P alter a hydrophobic interaction between two α-helices of the exonuclease domain and F688 from the fingers domain (Figure 3G). The relevance of this interaction for antimutagenesis is reinforced by the T4 Pol antimutator R335C, which also lies at the interface between the fingers and exonuclease domains (Figure 3F). The positions of these antimutators, together with the Pol δ Q563R substitution described above, support the idea that noncovalent interactions between the fingers domain and other parts of the polymerase may stabilize the closed conformation and influence fidelity. It should be emphasized that the Pol δ antimutator substitutions occur in the context of a catalytically dead exonuclease domain. The substitutions are unlikely to restore editing activity; indeed, two of the Pol δ substitutions (G400S and F486S) cause weak mutator phenotypes when the exonuclease catalytic residues are restored (Herr et al., 2011), consistent with reduced proofreading efficiency. The structural defects to the core of the exonuclease domain may also alter closure of the fingers or positioning of the β-hairpin loop that interacts with the template strand (Hogg et al., 2007; Swan et al., 2009). Or during attempts to proofread an error, these structural changes may reduce the affinity of the exonuclease domain for the primer and promote polymerase dissociation.
Antimutator Substitutions Along the DNA Binding Track
Dissociation-based models best explain the antimutagenic properties of substitutions that change amino acids along the DNA binding track. Pol δ V838 (mutated to A) sits next to the primer backbone at the -3 base-pair1. R839 (mutated to H) interacts with the minor groove at base pairs -4 and -5 (Figure 3I)(Swan et al., 2009). These positions correspond precisely with the T4 Pol Q730S and Q731S antimutator substitutions (Figure 3A,H). Other B-family antimutator substitutions affect amino acids that support adjacent DNA-interacting residues. In Pol δ, D831 (mutated to G) forms a salt bridge with K833, positioning the carbon chain of the lysine against the deoxyribose of the -2 primer position (Figure 3I). R475 (mutated to I or G) from the exonuclease domain overlays R840, which interacts with the -3 phosphate1 of the primer strand. W821C and Y808A affect β-strands on either side of the KKRYA loop, which interrogates the minor groove of base-pairs -2 (via K814) and -5 (via R815) (Swan et al., 2009). Finally, E594 (mutated to G) helps to position K813 of the KKRYA motif to interact with the template phosphate backbone at the -4 position. The Pol I L676Q antimutator may also diminish DNA binding. L676 packs against residues from motif 2 (N675 and N678) that interact with the minor grove of the -3 base pair (Figure 5D). Motif 2 forms a β-hairpin turn that contours the template backbone (positions -2 through -5). The extensive interactions of DNA polymerases with the minor groove of the first several base pairs of DNA duplex has been hypothesized to facilitate proofreading of misinsertions even after they have been extended (Franklin et al., 2001; Johnson and Beese, 2004; Swan et al., 2009). Thus, by reducing overall binding affinity, these antimutator polymerases may be especially prone to releasing mispaired primer-templates.
In addition to mutations affecting minor-groove binding close to the active site, antimutator substitutions affect the flexible tip of the thumb domain, which clamps down on the DNA in the elongation mode and moves with the primer strand during partitioning to the exonuclease active site (Franklin et al., 2001). Two Pol δ antimutator substitutions in this subdomain affect residues that interact directly with the DNA (Figure 3I). R923 (mutated to H) interacts with the -4 phosphate of the primer stand, and K891 (mutated to T) extends out across the minor groove towards the template strand. Although not in position to interact directly with the DNA, H879Y alters an amino acid near the hinge of the thumb tip. The T4 Pol A737V, A777V, and W844S antimutators (Figure 3H) change positions that appear to be involved in the folding of the thumb tip or stabilization of the ‘clamped down’ conformation, which is important for elongation (Franklin et al., 2001).
Antimutator substitutions in Pol I and Pol III also alter residues in the thumb. Pol I K601 (mutated to I) resides in a loop of the thumb tip that folds back on itself and interacts with DNA (Figure 5D). K601 may stabilize the fold of this loop by forming a hydrogen bond to a carbonyl of the peptide backbone on the opposing strand of the loop. Pol I Y630F resembles the classic A737V T4 Pol antimutator. Located in the main body of the thumb, Y630 interacts with residues from motif 1 in the thumb tip, which may stabilize the closed conformation. The Pol III thumb domain (Figure 4B) is markedly different from the thumb domains of A- and B-family polymerases, yet performs a similar function in interacting with the primer•template duplex along the minor groove. The two antimutator substitutions in the Pol III thumb domain (P464L and A498T) affect amino acids may stabilize the position of an α-helix containing three basic residues (K439, R443, and R447) that contact the duplex DNA (Figure 4D).
Summary and Perspective
Although isolated from different organisms and different DNA polymerases, the antimutator substitutions described here exhibit commonalities that point to general mechanisms of DNA polymerase fidelity and antimutagenesis (Figure 6). Antimutator substitutions may improve nucleotide selectivity by making the dNTP binding pocket of the polymerase more restrictive, by slowing the rate of catalysis or destabilizing the catalytic complex so that incorrect dNTPs dissociate before misincorporation, or by enhancing polymerase dissociation from mispaired primer termini so that errors can be edited more efficiently either by an intrinsic exonuclease or by a cellular proofreading activity in trans.
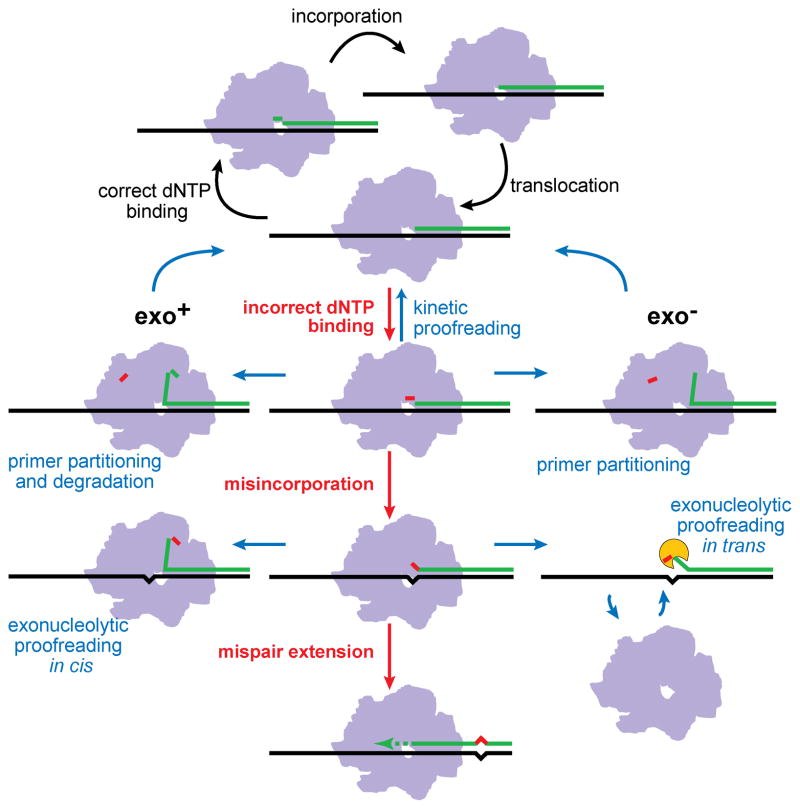
Figure 6.
Mechanisms of DNA polymerase fidelity and antimutagenesis. A reiterative cycle of correct dNTP binding, nucleotide incorporation, and translocation comprises accurate DNA replication (top, black arrows). Antimutator mechanisms (blue arrows) enhance existing processes that oppose incorrect dNTP binding and misincorporation (red arrows) in DNA polymerases with functional (Exo+) or defective intrinsic proofreading (Exo−). Antimutator substitutions may increase the effectiveness of kinetic proofreading, by reducing incorrect dNTP binding stability or by enhancing partitioning of correctly paired primer strands to the exonuclease active site in the presence of bound, incorrect dNTPs. In proofreading-proficient polymerases, partitioning to the exonuclease domain may lead to degradation of correct primer ends, as exemplified by T4 Pol optA1 sensitivity, as well as improved editing of misincorporations. In the absence of intrinsic proofreading, antimutators may enhance polymerase dissociation from the primer•template DNA, leading to editing by other cellular 3′→5′ exonucleases (proofreading in trans). A color version of this figure is available online.
The myriad of antimutagenic pathways revealed by these genetic studies is consistent with incremental optimization of DNA polymerase activity and fidelity during evolution. Coordination between polymerases and proofreading exonucleases represents perhaps the ultimate antimutagenic innovation, but not the only one. Archaea have evolved B-family DNA polymerases with a novel strategy to avoid DNA damage-induced mutations. These polymerases scan the incoming template DNA for uracil and hypoxanthine residues, which result from deamination of cytidine and adenine and can lead to transition mutations (Greagg et al., 1999). Uracil and hypoxanthine flip into a tight binding pocket in the amino domain and stall replication, likely leading to dissociation of the polymerase and initiation of DNA repair (Fogg et al., 2002; Connolly et al., 2003; Shuttleworth et al., 2004; Gill et al., 2007; Firbank et al., 2008; Killelea et al., 2010). Although this scanning functionality does not appear to extend to B-family polymerases from other Kingdoms (Wardle et al., 2008), it illustrates that antimutagenic changes in DNA polymerases may optimize their response to environmental damage in addition to influencing inherent error rates.
In this review we focused on antimutator variants of DNA polymerases. However, mutator phenotypes are also shaped by cellular responses to DNA replication errors. For example, in yeast the mutator phenotype conferred by loss of Pol δ proofreading (pol3-01 allele) depends in part on DUN1, a protein kinase gene that affects downstream events after S-phase checkpoint activation (Datta et al., 2000). This DUN1-dependency suggests that pausing at mispaired primer termini by proofreading-defective Pol δ activates the S-phase checkpoint through Mec1, which stabilizes the stalled replication fork (Friedel et al., 2009) and activates Dun1 through Rad53 (Chen et al., 2010). Once activated, Dun1 phosphorylates a number of downstream targets (Chen et al., 2010), including negative regulators of ribonucleotide reductase (Zhou and Elledge, 1992; Huang et al., 1998b; Zhao and Rothstein, 2002; Lee et al., 2008b; Wu and Huang, 2008). This signaling cascade ultimately increases dNTP pool concentrations in the cell, which helps DNA polymerases extend mispaired primer termini and copy past blocking lesions (Reichard, 1988; Goodman et al., 1993; Chabes et al., 2003; Mathews, 2006; Kumar et al., 2011). The pol3-01 mutator phenotype does not depend on error-prone synthesis by TLS polymerases Pol ζ or Pol η (Datta et al., 2000). Thus, loss of DUN1 may suppress the pol3-01 mutator phenotype by maintaining low dNTP levels, providing time for Pol δ to dissociate from mispaired primer termini and alternate repair pathways to correct 3′ errors. Defects in other S-phase checkpoint molecules may have a similar antimutator effect. Therefore, in addition to direct effects on polymerase fidelity, alleles in polymerase or checkpoint genes may suppress mutator phenotypes by limiting Dun1 activation.
The plasticity of DNA replication fidelity may be important during times of intense selection pressure, which select for mutators that more quickly adapt to changing environments (Sturtevant, 1937; Drake et al., 1998; Giraud et al., 2001b; de Visser, 2002; Elena and Lenski, 2003; Denamur and Matic, 2006). During tumorigenesis, for instance, cells acquire multiple genetic changes that release restrictions on proliferation (Hanahan and Weinberg, 2011). Mutator phenotypes were predicted more than 30 years ago to accelerate tumorigenesis (Loeb et al., 1974), and recent experiments suggest that many human tumors exhibit a mutator phenotype (Bielas et al., 2006). Consistent with this hypothesis, defects in MMR, Pol δ proofreading, or Pol ε proofreading accelerate discrete cancer phenotypes in mice (de Wind et al., 1995; Reitmair et al., 1996; Goldsby et al., 2001; Goldsby et al., 2002; Albertson et al., 2009). Defects in MMR and Pol δ fidelity are also associated with human tumors (Fishel et al., 1993; Parsons et al., 1993; Daee et al., 2010). Once the requisite mutations for tumorigenesis are obtained, continued expression of a strong mutator phenotype may be deleterious for cancer cells as they are in microbial populations. The discovery of robust pathways of antimutagenesis offers one mechanism by which mutator phenotypes can be suppressed to tolerable levels that allow cancer cells to maintain replicative fitness.
Acknowledgments
We thank our colleagues Larry Loeb, Scott Kennedy, and Mark Prindle for critical reading of the manuscript.
Footnotes
Here we refer to nucleotide positions relative to the direction of DNA synthesis, where zero (0) corresponds to the template nucleotide dNTP (T0•dNTP) and -1, -2, -3, etc. correspond to base pairs in the duplex DNA that are 1, 2, 3, etc. nucleotides ‘upstream’ of T0 dNTP. Phosphates in the 5′ position are designated by the same numbering (e.g., the phosphate between the -3 and -4 bases on the primer strand is designated the -3 phosphate.
DECLARATION OF INTEREST
The authors report no conflicts of interest. The authors alone are responsible for the content and writing of the paper. This work was supported by the National Institutes of Health (R01 ES09927, R01 CA098243, R01 CA111582, P20 CA103728, P30 AG13280, and P01 AG01751). Lindsey Williams was supported by a Public Health Service, National Research Service Award (T32 GM07270). Alan J. Herr, Ph.D., holds a Hitchings-Elion Fellowship from the Burroughs-Wellcome Fund (BWF 1000415.02).
References
- Albertson TM, Ogawa M, Bugni JM, Hays LE, Chen Y, Wang Y, Treuting PM, Heddle JA, Goldsby RE, Preston BD. DNA polymerase ε and δ proofreading suppress discrete mutator and cancer phenotypes in mice. Proc Natl Acad Sci USA. 2009;106:17101–17104. doi: 10.1073/pnas.0907147106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Albertson TM, Preston BD. DNA replication fidelity: proofreading in trans. Curr Biol. 2006;16:R209–211. doi: 10.1016/j.cub.2006.02.031. [DOI] [PubMed] [Google Scholar]
- Aravind L, Koonin EV. Phosphoesterase domains associated with DNA polymerases of diverse origins. Nucleic Acids Res. 1998;26:3746–3752. doi: 10.1093/nar/26.16.3746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bailey S, Wing RA, Steitz TA. The structure of T. aquaticus DNA polymerase III is distinct from eukaryotic replicative DNA polymerases. Cell. 2006;126:893–904. doi: 10.1016/j.cell.2006.07.027. [DOI] [PubMed] [Google Scholar]
- Barry ER, Bell SD. DNA replication in the archaea. Microbiol Mol Biol Rev. 2006;70:876–887. doi: 10.1128/MMBR.00029-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beard WA, Wilson SH. Structural Insights into the origins of DNA polymerase fidelity. Structure. 2003;11:489–496. doi: 10.1016/s0969-2126(03)00051-0. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Analyzing fidelity of DNA polymerases. Methods Enzymol. 1995;262:217–232. doi: 10.1016/0076-6879(95)62020-6. [DOI] [PubMed] [Google Scholar]
- Bebenek K, Kunkel TA. Streisinger revisited: DNA synthesis errors mediated by substrate misalignments. Cold Spring Harb Symp Quant Biol. 2000;65:81–91. doi: 10.1101/sqb.2000.65.81. [DOI] [PubMed] [Google Scholar]
- Beckman J, Kincaid K, Hocek M, Spratt T, Engels J, Cosstick R, Kuchta RD. Human DNA polymerase alpha uses a combination of positive and negative selectivity to polymerize purine dNTPs with high fidelity. Biochemistry. 2007;46:448–460. doi: 10.1021/bi061243s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beese LS, Derbyshire V, Steitz TA. Structure of DNA polymerase I Klenow fragment bound to duplex DNA. Science. 1993;260:352–355. doi: 10.1126/science.8469987. [DOI] [PubMed] [Google Scholar]
- Bielas JH, Loeb KR, Rubin BP, True LD, Loeb LA. Human cancers express a mutator phenotype. Proc Natl Acad Sci USA. 2006;103:18238–18242. doi: 10.1073/pnas.0607057103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boeke JD, LaCroute F, Fink GR. A positive selection for mutants lacking orotidine-5′-phosphate decarboxylase activity in yeast: 5-fluoro-orotic acid resistance. Mol Gen Genet. 1984;197:345–346. doi: 10.1007/BF00330984. [DOI] [PubMed] [Google Scholar]
- Camps M, Naukkarinen J, Johnson BP, Loeb LA. Targeted gene evolution in Escherichia coli using a highly error-prone DNA polymerase I. Proc Natl Acad Sci USA. 2003;100:9727–9732. doi: 10.1073/pnas.1333928100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavanaugh NA, Urban M, Beckman J, Spratt TE, Kuchta RD. Identifying the features of purine dNTPs that allow accurate and efficient DNA replication by herpes simplex virus I DNA polymerase. Biochemistry. 2009;48:3554–3564. doi: 10.1021/bi8022202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chabes A, Georgieva B, Domkin V, Zhao X, Rothstein R, Thelander L. Survival of DNA damage in yeast directly depends on increased dNTP levels allowed by relaxed feedback inhibition of ribonucleotide reductase. Cell. 2003;112:391–401. doi: 10.1016/s0092-8674(03)00075-8. [DOI] [PubMed] [Google Scholar]
- Chao L, Cox EC. Competition between high and low mutating strains of Escherichia coli. Evolution. 1983;37:125–134. doi: 10.1111/j.1558-5646.1983.tb05521.x. [DOI] [PubMed] [Google Scholar]
- Chen SH, Albuquerque CP, Liang J, Suhandynata RT, Zhou H. A proteome-wide analysis of kinase-substrate network in the DNA damage response. J Biol Chem. 2010;285:12803–12812. doi: 10.1074/jbc.M110.106989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiaramonte M, Moore CL, Kincaid K, Kuchta RD. Facile polymerization of dNTPs bearing unnatural base analogues by DNA polymerase α and Klenow fragment (DNA polymerase l) Biochemistry. 2003;42:10472–10481. doi: 10.1021/bi034763l. [DOI] [PubMed] [Google Scholar]
- Connolly BA, Fogg MJ, Shuttleworth G, Wilson BT. Uracil recognition by archaeal family B DNA polymerases. Biochem Soc Trans. 2003;31:699–702. doi: 10.1042/bst0310699. [DOI] [PubMed] [Google Scholar]
- Daee DL, Mertz TM, Shcherbakova PV. A cancer-associated DNA polymerase δ variant modeled in yeast causes a catastrophic increase in genomic instability. Proc Natl Acad Sci USA. 2010;107:157–162. doi: 10.1073/pnas.0907526106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datta A, Schmeits JL, Amin NS, Lau PJ, Myung K, Kolodner RD. Checkpoint-dependent activation of mutagenic repair in Saccharomyces cerevisiae pol3–01 mutants. Mol Cell. 2000;6:593–603. doi: 10.1016/s1097-2765(00)00058-7. [DOI] [PubMed] [Google Scholar]
- de Visser JA. The fate of microbial mutators. Microbiology. 2002;148:1247–1252. doi: 10.1099/00221287-148-5-1247. [DOI] [PubMed] [Google Scholar]
- de Wind N, Dekker M, Berns A, Radman M, te Riele H. Inactivation of the mouse Msh2 gene results in mismatch repair deficiency, methylation tolerance, hyperrecombination, and predisposition to cancer. Cell. 1995;82:321–330. doi: 10.1016/0092-8674(95)90319-4. [DOI] [PubMed] [Google Scholar]
- Delarue M, Poch O, Tordo N, Moras D, Argos P. An attempt to unify the structure of polymerases. Protein Eng. 1990;3:461–467. doi: 10.1093/protein/3.6.461. [DOI] [PubMed] [Google Scholar]
- Denamur E, Matic I. Evolution of mutation rates in bacteria. Mol Microbiol. 2006;60:820–827. doi: 10.1111/j.1365-2958.2006.05150.x. [DOI] [PubMed] [Google Scholar]
- Ding J, Das K, Hsiou Y, Sarafianos SG, Clark AD, Jacobo-Molina A, Tantillo C, Hughes SH, Arnold E. Structure and functional implications of the polymerase active site region in a complex of HIV-1 RT with a double-stranded DNA template-primer and an antibody fab fragment at 2.8 A resolution. J Mol Biol. 1998;284:1095–1111. doi: 10.1006/jmbi.1998.2208. [DOI] [PubMed] [Google Scholar]
- Doublie S, Tabor S, Long AM, Richardson CC, Ellenberger T. Crystal structure of a bacteriophage T7 DNA replication complex at 2.2 A resolution. Nature. 1998;391:251–258. doi: 10.1038/34593. [DOI] [PubMed] [Google Scholar]
- Drake JW. General antimutators are improbable. J Mol Biol. 1993;229:8–13. doi: 10.1006/jmbi.1993.1002. [DOI] [PubMed] [Google Scholar]
- Drake JW, Allen EF. Antimutagenic DNA polymerases of bacteriophage T4. Cold Spring Harb Symp Quant Biol. 1968;33:339–344. doi: 10.1101/sqb.1968.033.01.039. [DOI] [PubMed] [Google Scholar]
- Drake JW, Allen EF, Forsberg SA, Preparata RM, Greening EO. Genetic control of mutation rates in bacteriophage T4. Nature. 1969;221:1128–1132. [PubMed] [Google Scholar]
- Drake JW, Charlesworth B, Charlesworth D, Crow JF. Rates of spontaneous mutation. Genetics. 1998;148:1667–1686. doi: 10.1093/genetics/148.4.1667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Echols H, Goodman MF. Fidelity mechanisms in DNA replication. Annu Rev Biochem. 1991;60:477–511. doi: 10.1146/annurev.bi.60.070191.002401. [DOI] [PubMed] [Google Scholar]
- Elena SF, Lenski RE. Evolution experiments with microorganisms: the dynamics and genetic bases of adaptation. Nat Rev Genet. 2003;4:457–469. doi: 10.1038/nrg1088. [DOI] [PubMed] [Google Scholar]
- Eom SH, Wang J, Steitz TA. Structure of Taq polymerase with DNA at the polymerase active site. Nature. 1996;382:278–281. doi: 10.1038/382278a0. [DOI] [PubMed] [Google Scholar]
- Fijalkowska IJ, Dunn RL, Schaaper RM. Mutants of Escherichia coli with increased fidelity of DNA replication. Genetics. 1993;134:1023–1030. doi: 10.1093/genetics/134.4.1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Schaaper RM. Antimutator mutations in the α subunit of Escherichia coli DNA polymerase III: identification of the responsible mutations and alignment with other DNA polymerases. Genetics. 1993;134:1039–1044. doi: 10.1093/genetics/134.4.1039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Schaaper RM. Effects of Escherichia coli dnaE antimutator alleles in a proofreading-deficient mutD5 strain. J Bacteriol. 1995;177:5979–5986. doi: 10.1128/jb.177.20.5979-5986.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fijalkowska IJ, Schaaper RM. Mutants in the Exo I motif of Escherichia coli dnaQ: defective proofreading and inviability due to error catastrophe. Proc Natl Acad Sci USA. 1996;93:2856–2861. doi: 10.1073/pnas.93.7.2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Firbank SJ, Wardle J, Heslop P, Lewis RJ, Connolly BA. Uracil recognition in archaeal DNA polymerases captured by X-ray crystallography. J Mol Biol. 2008;381:529–539. doi: 10.1016/j.jmb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- Fishel R, Lescoe MK, Rao MR, Copeland NG, Jenkins NA, Garber J, Kane M, Kolodner R. The human mutator gene homolog MSH2 and its association with hereditary nonpolyposis colon cancer. Cell. 1993;75:1027–1038. doi: 10.1016/0092-8674(93)90546-3. [DOI] [PubMed] [Google Scholar]
- Fogg MJ, Pearl LH, Connolly BA. Structural basis for uracil recognition by archaeal family B DNA polymerases. Nat Struct Biol. 2002;9:922–927. doi: 10.1038/nsb867. [DOI] [PubMed] [Google Scholar]
- Fortune JM, Pavlov YI, Welch CM, Johansson E, Burgers PM, Kunkel TA. Saccharomyces cerevisiae DNA polymerase δ: high fidelity for base substitutions but lower fidelity for single- and multi-base deletions. J Biol Chem. 2005;280:29980–29987. doi: 10.1074/jbc.M505236200. [DOI] [PubMed] [Google Scholar]
- Franklin MC, Wang J, Steitz TA. Structure of the replicating complex of a pol α family DNA polymerase. Cell. 2001;105:657–667. doi: 10.1016/s0092-8674(01)00367-1. [DOI] [PubMed] [Google Scholar]
- Friedberg EC, Walker GC, Siede W, Wood RD, Schultz RA, Ellenberger T. DNA Repair and Mutagenesis. Washington, D.C.: ASM Press; 2006. [Google Scholar]
- Friedel AM, Pike BL, Gasser SM. ATR/Mec1: coordinating fork stability and repair. Curr Opin Cell Biol. 2009;21:237–244. doi: 10.1016/j.ceb.2009.01.017. [DOI] [PubMed] [Google Scholar]
- Gauss P, Doherty DH, Gold L. Bacterial and phage mutations that reveal helix-unwinding activities required for bacteriophage T4 DNA replication. Proc Natl Acad Sci USA. 1983;80:1669–1673. doi: 10.1073/pnas.80.6.1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbs JS, Chiou HC, Bastow KF, Cheng YC, Coen DM. Identification of amino acids in herpes simplex virus DNA polymerase involved in substrate and drug recognition. Proc Natl Acad Sci USA. 1988;85:6672–6676. doi: 10.1073/pnas.85.18.6672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gill S, O’Neill R, Lewis RJ, Connolly BA. Interaction of the family-B DNA polymerase from the archaeon Pyrococcus furiosus with deaminated bases. J Mol Biol. 2007;372:855–863. doi: 10.1016/j.jmb.2007.07.015. [DOI] [PubMed] [Google Scholar]
- Gillin FD, Nossal NG. Control of mutation frequency by bacteriophage T4 DNA polymerase. I. The CB120 antimutator DNA polymerase is defective in strand displacement. J Biol Chem. 1976a;251:5219–5224. [PubMed] [Google Scholar]
- Gillin FD, Nossal NG. Control of mutation frequency by bacteriophage T4 DNA polymerase. II. Accuracy of nucleotide selection by the L88 mutator, CB120 antimutator, and wild type phage T4 DNA polymerases. J Biol Chem. 1976b;251:5225–5232. [PubMed] [Google Scholar]
- Giraud A, Matic I, Tenaillon O, Clara A, Radman M, Fons M, Taddei F. Costs and benefits of high mutation rates: adaptive evolution of bacteria in the mouse gut. Science. 2001a;291:2606–2608. doi: 10.1126/science.1056421. [DOI] [PubMed] [Google Scholar]
- Giraud A, Radman M, Matic I, Taddei F. The rise and fall of mutator bacteria. Curr Opin Microbiol. 2001b;4:582–585. doi: 10.1016/s1369-5274(00)00254-x. [DOI] [PubMed] [Google Scholar]
- Goldsby RE, Hays LE, Chen X, Olmsted EA, Slayton WB, Spangrude GJ, Preston BD. High incidence of epithelial cancers in mice deficient for DNA polymerase δ proofreading. Proc Natl Acad Sci USA. 2002;99:15560–15565. doi: 10.1073/pnas.232340999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldsby RE, Lawrence NA, Hays LE, Olmsted EA, Chen X, Singh M, Preston BD. Defective DNA polymerase-δ proofreading causes cancer susceptibility in mice. Nat Med. 2001;7:638–639. doi: 10.1038/88963. [DOI] [PubMed] [Google Scholar]
- Goodman MF, Creighton S, Bloom LB, Petruska J. Biochemical basis of DNA replication fidelity. Crit Rev Biochem Mol Biol. 1993;28:83–126. doi: 10.3109/10409239309086792. [DOI] [PubMed] [Google Scholar]
- Greagg MA, Fogg MJ, Panayotou G, Evans SJ, Connolly BA, Pearl LH. A read-ahead function in archaeal DNA polymerases detects promutagenic template-strand uracil. Proc Natl Acad Sci USA. 1999;96:9045–9050. doi: 10.1073/pnas.96.16.9045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hadjimarcou MI, Kokoska RJ, Petes TD, Reha-Krantz LJ. Identification of a mutant DNA polymerase δ in Saccharomyces cerevisiae with an antimutator phenotype for frameshift mutations. Genetics. 2001;158:177–186. doi: 10.1093/genetics/158.1.177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall JD, Coen DM, Fisher BL, Weisslitz M, Randall S, Almy RE, Gelep PT, Schaffer PA. Generation of genetic diversity in herpes simplex virus: an antimutator phenotype maps to the DNA polymerase locus. Virology. 1984;132:26–37. doi: 10.1016/0042-6822(84)90088-6. [DOI] [PubMed] [Google Scholar]
- Hanahan D, Weinberg Robert A. Hallmarks of cancer: the next generation. Cell. 2011;144:646–674. doi: 10.1016/j.cell.2011.02.013. [DOI] [PubMed] [Google Scholar]
- Henneke G, Flament D, Hubscher U, Querellou J, Raffin JP. The hyperthermophilic euryarchaeota Pyrococcus abyssi likely requires the two DNA polymerases D and B for DNA replication. J Mol Biol. 2005;350:53–64. doi: 10.1016/j.jmb.2005.04.042. [DOI] [PubMed] [Google Scholar]
- Herr AJ, Ogawa M, Lawrence NA, Williams LN, Eggington JM, Singh M, Smith RA, Preston BD. Mutator suppression and escape from replication error-induced extinction in yeast. PLoS Genet. 2011 doi: 10.1371/journal.pgen.1002282. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hogg M, Aller P, Konigsberg W, Wallace SS, Doublie S. Structural and biochemical investigation of the role in proofreading of a β hairpin loop found in the exonuclease domain of a replicative DNA polymerase of the B family. J Biol Chem. 2007;282:1432–1444. doi: 10.1074/jbc.M605675200. [DOI] [PubMed] [Google Scholar]
- Hopfner KP, Eichinger A, Engh RA, Laue F, Ankenbauer W, Huber R, Angerer B. Crystal structure of a thermostable type B DNA polymerase from Thermococcus gorgonarius. Proc Natl Acad Sci USA. 1999;96:3600–3605. doi: 10.1073/pnas.96.7.3600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang H, Chopra R, Verdine GL, Harrison SC. Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science. 1998a;282:1669–1675. doi: 10.1126/science.282.5394.1669. [DOI] [PubMed] [Google Scholar]
- Huang M, Zhou Z, Elledge SJ. The DNA replication and damage checkpoint pathways induce transcription by inhibition of the Crt1 repressor. Cell. 1998b;94:595–605. doi: 10.1016/s0092-8674(00)81601-3. [DOI] [PubMed] [Google Scholar]
- Hwang YT, Zuccola HJ, Lu Q, Hwang CB. A point mutation within conserved region VI of herpes simplex virus type 1 DNA polymerase confers altered drug sensitivity and enhances replication fidelity. J Virol. 2004;78:650–657. doi: 10.1128/JVI.78.2.650-657.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iyer RR, Pluciennik A, Burdett V, Modrich PL. DNA mismatch repair: functions and mechanisms. Chem Rev. 2006;106:302–323. doi: 10.1021/cr0404794. [DOI] [PubMed] [Google Scholar]
- Jin YH, Obert R, Burgers PM, Kunkel TA, Resnick MA, Gordenin DA. The 3′→5′ exonuclease of DNA polymerase δ can substitute for the 5′ flap endonuclease Rad27/Fen1 in processing Okazaki fragments and preventing genome instability. Proc Natl Acad Sci USA. 2001;98:5122–5127. doi: 10.1073/pnas.091095198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson A, O’Donnell M. Cellular DNA replicases: components and dynamics at the replication fork. Annu Rev Biochem. 2005;74:283–315. doi: 10.1146/annurev.biochem.73.011303.073859. [DOI] [PubMed] [Google Scholar]
- Johnson KA. The kinetic and chemical mechanism of high-fidelity DNA polymerases. Biochim Biophys Acta. 2010;1804:1041–1048. doi: 10.1016/j.bbapap.2010.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson SJ, Beese LS. Structures of mismatch replication errors observed in a DNA polymerase. Cell. 2004;116:803–816. doi: 10.1016/s0092-8674(04)00252-1. [DOI] [PubMed] [Google Scholar]
- Johnson SJ, Taylor JS, Beese LS. Processive DNA synthesis observed in a polymerase crystal suggests a mechanism for the prevention of frameshift mutations. Proc Natl Acad Sci USA. 2003;100:3895–3900. doi: 10.1073/pnas.0630532100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joyce CM, Benkovic SJ. DNA polymerase fidelity: kinetics, structure, and checkpoints. Biochemistry. 2004;43:14317–14324. doi: 10.1021/bi048422z. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Dzantiev L, Constantin N, Modrich P. Endonucleolytic function of MutLα in human mismatch repair. Cell. 2006;126:297–308. doi: 10.1016/j.cell.2006.05.039. [DOI] [PubMed] [Google Scholar]
- Kadyrov FA, Genschel J, Fang Y, Penland E, Edelmann W, Modrich P. A possible mechanism for exonuclease 1-independent eukaryotic mismatch repair. Proc Natl Acad Sci USA. 2009;106:8495–8500. doi: 10.1073/pnas.0903654106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiefer JR, Mao C, Braman JC, Beese LS. Visualizing DNA replication in a catalytically active Bacillus DNA polymerase crystal. Nature. 1998;391:304–307. doi: 10.1038/34693. [DOI] [PubMed] [Google Scholar]
- Killelea T, Ghosh S, Tan SS, Heslop P, Firbank SJ, Kool ET, Connolly BA. Probing the interaction of archaeal DNA polymerases with deaminated bases using X-ray crystallography and non-hydrogen bonding isosteric base analogues. Biochemistry. 2010;49:5772–5781. doi: 10.1021/bi100421r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kincaid K, Beckman J, Zivkovic A, Halcomb RL, Engels JW, Kuchta RD. Exploration of factors driving incorporation of unnatural dNTPs into DNA by Klenow fragment (DNA polymerase I) and DNA polymerase α. Nucleic Acids Res. 2005;33:2620–2628. doi: 10.1093/nar/gki563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kool ET. Active site tightness and substrate fit in DNA replication. Annu Rev Biochem. 2002;71:191–219. doi: 10.1146/annurev.biochem.71.110601.135453. [DOI] [PubMed] [Google Scholar]
- Kornberg A, Baker TA. DNA Replication. New York: W.H. Freeman and Company; 1992. [Google Scholar]
- Kumar D, Abdulovic AL, Viberg J, Nilsson AK, Kunkel TA, Chabes A. Mechanisms of mutagenesis in vivo due to imbalanced dNTP pools. Nucleic Acids Res. 2011;39:1360–1371. doi: 10.1093/nar/gkq829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA. Evolving views of DNA replication (in)fidelity. Cold Spring Harb Symp Quant Biol. 2009;74:91–101. doi: 10.1101/sqb.2009.74.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Burgers PM. Dividing the workload at a eukaryotic replication fork. Trends Cell Biol. 2008;18:521–527. doi: 10.1016/j.tcb.2008.08.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel TA, Erie DA. DNA mismatch repair. Annu Rev Biochem. 2005;74:681–710. doi: 10.1146/annurev.biochem.74.082803.133243. [DOI] [PubMed] [Google Scholar]
- Leconte AM, Hwang GT, Matsuda S, Capek P, Hari Y, Romesberg FE. Discovery, characterization, and optimization of an unnatural base pair for expansion of the genetic alphabet. J Am Chem Soc. 2008;130:2336–2343. doi: 10.1021/ja078223d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee HR, Helquist SA, Kool ET, Johnson KA. Importance of hydrogen bonding for efficiency and specificity of the human mitochondrial DNA polymerase. J Biol Chem. 2008a;283:14402–14410. doi: 10.1074/jbc.M705007200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee YD, Wang J, Stubbe J, Elledge SJ. Dif1 is a DNA-damage-regulated facilitator of nuclear import for ribonucleotide reductase. Mol Cell. 2008b;32:70–80. doi: 10.1016/j.molcel.2008.08.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li L, Murphy KM, Kanevets U, Reha-Krantz LJ. Sensitivity to phosphonoacetic acid: a new phenotype to probe DNA polymerase δ in Saccharomyces cerevisiae. Genetics. 2005;170:569–580. doi: 10.1534/genetics.104.040295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li V, Hogg M, Reha-Krantz LJ. Identification of a new motif in family B DNA polymerases by mutational Analyses of the bacteriophage T4 DNA polymerase. J Mol Biol. 2010a;400:295–308. doi: 10.1016/j.jmb.2010.05.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Y, Korolev S, Waksman G. Crystal structures of open and closed forms of binary and ternary complexes of the large fragment of Thermus aquaticus DNA polymerase I: structural basis for nucleotide incorporation. EMBO J. 1998;17:7514–7525. doi: 10.1093/emboj/17.24.7514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Santangelo TJ, Čuboňová L, Reeve JN, Kelman Z. Affinity purification of an archaeal DNA replication protein network. mBio. 2010b;1:e00221–00210. doi: 10.1128/mBio.00221-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ling H, Boudsocq F, Woodgate R, Yang W. Crystal structure of a Y-family DNA polymerase in action: a mechanism for error-prone and lesion-bypass replication. Cell. 2001;107:91–102. doi: 10.1016/s0092-8674(01)00515-3. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Bielas JH, Beckman RA. Cancers exhibit a mutator phenotype: clinical implications. Cancer Res. 2008;68:3551–3557. doi: 10.1158/0008-5472.CAN-07-5835. [DOI] [PubMed] [Google Scholar]
- Loeb LA, Springgate CF, Battula N. Errors in DNA replication as a basis of malignant changes. Cancer Res. 1974;34:2311–2321. [PubMed] [Google Scholar]
- Loh E, Choe J, Loeb LA. Highly tolerated amino acid substitutions increase the fidelity of Escherichia coli DNA polymerase I. J Biol Chem. 2007;282:12201–12209. doi: 10.1074/jbc.M611294200. [DOI] [PubMed] [Google Scholar]
- Mao EF, Lane L, Lee J, Miller JH. Proliferation of mutators in a cell population. J Bacteriol. 1997;179:417–422. doi: 10.1128/jb.179.2.417-422.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsischky GT, Filosi N, Kane MF, Kolodner R. Redundancy of Saccharomyces cerevisiae MSH3 and MSH6 in MSH2-dependent mismatch repair. Genes Dev. 1996;10:407–420. doi: 10.1101/gad.10.4.407. [DOI] [PubMed] [Google Scholar]
- Maruyama M, Horiuchi T, Maki H, Sekiguchi M. A dominant (mutD5) and a recessive (dnaQ49) mutator of Escherichia coli. J Mol Biol. 1983;167:757–771. doi: 10.1016/s0022-2836(83)80109-0. [DOI] [PubMed] [Google Scholar]
- Mathews CK. DNA precursor metabolism and genomic stability. FASEB J. 2006;20:1300–1314. doi: 10.1096/fj.06-5730rev. [DOI] [PubMed] [Google Scholar]
- Matsuda S, Henry AA, Romesberg FE. Optimization of unnatural base pair packing for polymerase recognition. J Am Chem Soc. 2006;128:6369–6375. doi: 10.1021/ja057575m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCulloch SD, Kunkel TA. The fidelity of DNA synthesis by eukaryotic replicative and translesion synthesis polymerases. Cell Res. 2008;18:148–161. doi: 10.1038/cr.2008.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mildvan AS. Mechanism of enzyme action. Annu Rev Biochem. 1974;43:357–399. doi: 10.1146/annurev.bi.43.070174.002041. [DOI] [PubMed] [Google Scholar]
- Minnick DT, Bebenek K, Osheroff WP, Turner RM, Jr, Astatke M, Liu L, Kunkel TA, Joyce CM. Side chains that influence fidelity at the polymerase active site of Escherichia coli DNA polymerase I (Klenow fragment) J Biol Chem. 1999;274:3067–3075. doi: 10.1074/jbc.274.5.3067. [DOI] [PubMed] [Google Scholar]
- Mo J-Y, Schaaper RM. Fidelity and error specificity of the α catalytic subunit of Escherichia coli DNA polymerase III. J Biol Chem. 1996;271:18947–18953. doi: 10.1074/jbc.271.31.18947. [DOI] [PubMed] [Google Scholar]
- Morales JC, Kool ET. Efficient replication between non-hydrogen-bonded nucleoside shape analogs. Nat Struct Biol. 1998;5:950–954. doi: 10.1038/2925. [DOI] [PubMed] [Google Scholar]
- Morales JC, Kool ET. Varied molecular interactions at the active sites of several DNA polymerases: nonpolar nucleoside isosteres as probes. J Am Chem Soc. 2000;122:1001–1007. doi: 10.1021/ja993464+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moran S, Ren RX, Kool ET. A thymidine triphosphate shape analog lacking Watson-Crick pairing ability is replicated with high sequence selectivity. Proc Natl Acad Sci USA. 1997;94:10506–10511. doi: 10.1073/pnas.94.20.10506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Johnson AL, Johnston LH, Sugino A. Pathway correcting DNA replication errors in Saccharomyces cerevisiae. EMBO J. 1993;12:1467–1473. doi: 10.1002/j.1460-2075.1993.tb05790.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Morrison A, Sugino A. DNA polymerase II, the epsilon polymerase of Saccharomyces cerevisiae. Prog Nucleic Acid Res Mol Biol. 1993;46:93–120. doi: 10.1016/s0079-6603(08)61019-3. [DOI] [PubMed] [Google Scholar]
- Morrison A, Sugino A. The 3′→5′ exonucleases of both DNA polymerases δ and ε participate in correcting errors of DNA replication in Saccharomyces cerevisiae. Mol Gen Genet. 1994;242:289–296. doi: 10.1007/BF00280418. [DOI] [PubMed] [Google Scholar]
- Muzyczka N, Poland RL, Bessman MJ. Studies on the biochemical basis of spontaneous mutation. I. A comparison of the deoxyribonucleic acid polymerases of mutator, antimutator, and wild type strains of bacteriophage T4. J Biol Chem. 1972;247:7116–7122. [PubMed] [Google Scholar]
- Nilsson AI, Kugelberg E, Berg OG, Andersson DI. Experimental adaptation of Salmonella typhimurium to mice. Genetics. 2004;168:1119–1130. doi: 10.1534/genetics.104.030304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Northam MR, Garg P, Baitin DM, Burgers PM, Shcherbakova PV. A novel function of DNA polymerase ζ regulated by PCNA. EMBO J. 2006;25:4316–4325. doi: 10.1038/sj.emboj.7601320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parsons R, Li GM, Longley MJ, Fang WH, Papadopoulos N, Jen J, de la Chapelle A, Kinzler KW, Vogelstein B, Modrich P. Hypermutability and mismatch repair deficiency in RER+ tumor cells. Cell. 1993;75:1227–1236. doi: 10.1016/0092-8674(93)90331-j. [DOI] [PubMed] [Google Scholar]
- Patel PH, Kawate H, Adman E, Ashbach M, Loeb LA. A single highly mutable catalytic site amino acid is critical for DNA polymerase fidelity. J Biol Chem. 2001;276:5044–5051. doi: 10.1074/jbc.M008701200. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Frahm C, McElhinny SA, Niimi A, Suzuki M, Kunkel TA. Evidence that errors made by DNA polymerase α are corrected by DNA polymerase δ. Curr Biol. 2006a;16:202–207. doi: 10.1016/j.cub.2005.12.002. [DOI] [PubMed] [Google Scholar]
- Pavlov YI, Maki S, Maki H, Kunkel TA. Evidence for interplay among yeast replicative DNA polymerases alpha, delta and epsilon from studies of exonuclease and polymerase active site mutations. BMC Biol. 2004;2:11. doi: 10.1186/1741-7007-2-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pavlov YI, Shcherbakova PV, Rogozin IB. Roles of DNA polymerases in replication, repair, and recombination in eukaryotes. Int Rev Cytol. 2006b;255:41–132. doi: 10.1016/S0074-7696(06)55002-8. [DOI] [PubMed] [Google Scholar]
- Perrino FW, Loeb LA. Hydrolysis of 3′-terminal mispairs in vitro by the 3′ → 5′ exonuclease of DNA polymerase δ permits subsequent extension by DNA polymerase α. Biochemistry. 1990;29:5226–5231. doi: 10.1021/bi00474a002. [DOI] [PubMed] [Google Scholar]
- Petruska J, Goodman MF. Enthalpy-entropy compensation in DNA melting thermodynamics. J Biol Chem. 1995;270:746–750. doi: 10.1074/jbc.270.2.746. [DOI] [PubMed] [Google Scholar]
- Potapova O, Chan C, DeLucia AM, Helquist SA, Kool ET, Grindley ND, Joyce CM. DNA polymerase catalysis in the absence of Watson-Crick hydrogen bonds: analysis by single-turnover kinetics. Biochemistry. 2006;45:890–898. doi: 10.1021/bi051792i. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Preston BD, Albertson TM, Herr AJ. DNA replication fidelity and cancer. Semin Cancer Biol. 2010;20:281–293. doi: 10.1016/j.semcancer.2010.10.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prolla TA, Christie DM, Liskay RM. Dual requirement in yeast DNA mismatch repair for MLH1 and PMS1, two homologs of the bacterial mutL gene. Mol Cell Biol. 1994;14:407–415. doi: 10.1128/mcb.14.1.407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quah SK, von Borstel RC, Hastings PJ. The origin of spontaneous mutation in Saccharomyces cerevisiae. Genetics. 1980;96:819–839. doi: 10.1093/genetics/96.4.819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ. Amino acid changes coded by bacteriophage T4 DNA polymerase mutator mutants. Relating structure to function. J Mol Biol. 1988;202:711–724. doi: 10.1016/0022-2836(88)90552-9. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz LJ. Learning about DNA polymerase function by studying antimutator DNA polymerases. Trends Biochem Sci. 1995a;20:136–140. doi: 10.1016/s0968-0004(00)88987-2. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz LJ. Use of genetic analyses to probe structure, function, and dynamics of bacteriophage T4 DNA polymerase. Methods Enzymol. 1995b;262:323–331. doi: 10.1016/0076-6879(95)62027-3. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz LJ. DNA polymerase proofreading: Multiple roles maintain genome stability. Biochim Biophys Acta. 2010;1804:1049–1063. doi: 10.1016/j.bbapap.2009.06.012. [DOI] [PubMed] [Google Scholar]
- Reha-Krantz LJ, Nonay RL. Genetic and biochemical studies of bacteriophage T4 DNA polymerase 3′→5′-exonuclease activity. J Biol Chem. 1993;268:27100–27108. [PubMed] [Google Scholar]
- Reha-Krantz LJ, Nonay RL. Motif A of bacteriophage T4 DNA polymerase: role in primer extension and DNA replication fidelity. Isolation of new antimutator and mutator DNA polymerases. J Biol Chem. 1994;269:5635–5643. [PubMed] [Google Scholar]
- Reha-Krantz LJ, Nonay RL, Stocki S. Bacteriophage T4 DNA polymerase mutations that confer sensitivity to the PPi analog phosphonoacetic acid. J Virol. 1993;67:60–66. doi: 10.1128/jvi.67.1.60-66.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reha-Krantz LJ, Wong C. Selection of bacteriophage T4 antimutator DNA polymerases: a link between proofreading and sensitivity to phosphonoacetic acid. Mutat Res. 1996;350:9–16. doi: 10.1016/0027-5107(95)00085-2. [DOI] [PubMed] [Google Scholar]
- Reichard P. Interactions between deoxyribonucleotide and DNA synthesis. Annu Rev Biochem. 1988;57:349–374. doi: 10.1146/annurev.bi.57.070188.002025. [DOI] [PubMed] [Google Scholar]
- Reitmair AH, Cai JC, Bjerknes M, Redston M, Cheng H, Pind MT, Hay K, Mitri A, Bapat BV, Mak TW, Gallinger S. MSH2 deficiency contributes to accelerated APC-mediated intestinal tumorigenesis. Cancer Res. 1996;56:2922–2926. [PubMed] [Google Scholar]
- Reitmair AH, Risley R, Bristow RG, Wilson T, Ganesh A, Jang A, Peacock J, Benchimol S, Hill RP, Mak TW, Fishel R, Meuth M. Mutator phenotype in Msh2-deficient murine embryonic fibroblasts. Cancer Res. 1997;57:3765–3771. [PubMed] [Google Scholar]
- Ripley LS. Transversion mutagenesis in bacteriophage T4. Mol Gen Genet. 1975;141:23–40. doi: 10.1007/BF00332376. [DOI] [PubMed] [Google Scholar]
- Roche H, Gietz RD, Kunz BA. Specificity of the yeast rev3Δ antimutator and REV3 dependency of the mutator resulting from a defect (rad1Δ) in nucleotide excision repair. Genetics. 1994;137:637–646. doi: 10.1093/genetics/137.3.637. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rothwell PJ, Mitaksov V, Waksman G. Motions of the fingers subdomain of klentaq1 are fast and not rate limiting: implications for the molecular basis of fidelity in DNA polymerases. Mol Cell. 2005;19:345–355. doi: 10.1016/j.molcel.2005.06.032. [DOI] [PubMed] [Google Scholar]
- Saenger W. Principles of Nucleic Acid Structure. New York: Springer-Verlag; 1984. [Google Scholar]
- Sarafianos SG, Marchand B, Das K, Himmel DM, Parniak MA, Hughes SH, Arnold E. Structure and function of HIV-1 reverse transcriptase: molecular mechanisms of polymerization and inhibition. J Mol Biol. 2009;385:693–713. doi: 10.1016/j.jmb.2008.10.071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sawaya MR, Prasad R, Wilson SH, Kraut J, Pelletier H. Crystal structures of human DNA polymerase β complexed with gapped and nicked DNA: evidence for an induced fit mechanism. Biochemistry. 1997;36:11205–11215. doi: 10.1021/bi9703812. [DOI] [PubMed] [Google Scholar]
- Schaaper RM. Mechanisms of mutagenesis in the Escherichia coli mutator mutD5: role of DNA mismatch repair. Proc Natl Acad Sci USA. 1988;85:8126–8130. doi: 10.1073/pnas.85.21.8126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM. Escherichia coli mutator mutD5 is defective in the mutHLS pathway of DNA mismatch repair. Genetics. 1989;121:205–212. doi: 10.1093/genetics/121.2.205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM. Base selection, proofreading, and mismatch repair during DNA replication in Escherichia coli. J Biol Chem. 1993;268:23762–23765. [PubMed] [Google Scholar]
- Schaaper RM. Suppressors of Escherichia coli mutT: antimutators for DNA replication errors. Mutat Res. 1996;350:17–23. doi: 10.1016/0027-5107(95)00086-0. [DOI] [PubMed] [Google Scholar]
- Schaaper RM, Cornacchio R. An Escherichia coli dnaE mutation with suppressor activity toward mutator mutD5. J Bacteriol. 1992;174:1974–1982. doi: 10.1128/jb.174.6.1974-1982.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaaper RM, Radman M. The extreme mutator effect of Escherichia coli mutD5 results from saturation of mismatch repair by excessive DNA replication errors. EMBO J. 1989;8:3511–3516. doi: 10.1002/j.1460-2075.1989.tb08516.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scheuermann R, Tam S, Burgers PM, Lu C, Echols H. Identification of the ε-subunit of Escherichia coli DNA polymerase III holoenzyme as the dnaQ gene product: a fidelity subunit for DNA replication. Proc Natl Acad Sci USA. 1983;80:7085–7089. doi: 10.1073/pnas.80.23.7085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitt MW, Venkatesan RN, Pillaire MJ, Hoffmann JS, Sidorova JM, Loeb LA. Active site mutations in mammalian DNA polymerase δ alter accuracy and replication fork progression. J Biol Chem. 2010;285:32264–32272. doi: 10.1074/jbc.M110.147017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinkai A, Loeb LA. In vivo mutagenesis by Escherichia coli DNA polymerase I. Ile709 in motif A functions in base selection. J Biol Chem. 2001;276:46759–46764. doi: 10.1074/jbc.M104780200. [DOI] [PubMed] [Google Scholar]
- Shinkai A, Patel PH, Loeb LA. The conserved active site motif A of Escherichia coli DNA polymerase I is highly mutable. J Biol Chem. 2001;276:18836–18842. doi: 10.1074/jbc.M011472200. [DOI] [PubMed] [Google Scholar]
- Shuttleworth G, Fogg MJ, Kurpiewski MR, Jen-Jacobson L, Connolly BA. Recognition of the pro-mutagenic base uracil by family B DNA polymerases from archaea. J Mol Biol. 2004;337:621–634. doi: 10.1016/j.jmb.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Simon M, Giot L, Faye G. The 3′ to 5′ exonuclease activity located in the DNA polymerase δ subunit of Saccharomyces cerevisiae is required for accurate replication. EMBO J. 1991;10:2165–2170. doi: 10.1002/j.1460-2075.1991.tb07751.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Johnson T, Shaver A. The evolution of mutation rates: separating causes from consequences. Bioessays. 2000;22:1057–1066. doi: 10.1002/1521-1878(200012)22:12<1057::AID-BIES3>3.0.CO;2-W. [DOI] [PubMed] [Google Scholar]
- Sniegowski PD, Gerrish PJ, Lenski RE. Evolution of high mutation rates in experimental populations of E. coli. Nature. 1997;387:703–705. doi: 10.1038/42701. [DOI] [PubMed] [Google Scholar]
- Sokolsky T, Alani E. EXO1 and MSH6 are high-copy suppressors of conditional mutations in the MSH2 mismatch repair gene of Saccharomyces cerevisiae. Genetics. 2000;155:589–599. doi: 10.1093/genetics/155.2.589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stano NM, Chen J, McHenry CS. A coproofreading Zn(2+)-dependent exonuclease within a bacterial replicase. Nat Struct Mol Biol. 2006;13:458–459. doi: 10.1038/nsmb1078. [DOI] [PubMed] [Google Scholar]
- Steitz TA, Smerdon SJ, Jager J, Joyce CM. A unified polymerase mechanism for nonhomologous DNA and RNA polymerases. Science. 1994;266:2022–2025. doi: 10.1126/science.7528445. [DOI] [PubMed] [Google Scholar]
- Stocki SA, Nonay RL, Reha-Krantz LJ. Dynamics of bacteriophage T4 DNA polymerase function: identification of amino acid residues that affect switching between polymerase and 3′ → 5′ exonuclease activities. J Mol Biol. 1995;254:15–28. doi: 10.1006/jmbi.1995.0595. [DOI] [PubMed] [Google Scholar]
- Sturtevant AH. Essays on evolution. I. On the effects of selection on mutation rate. Q Rev Biol. 1937;12:464–467. [Google Scholar]
- Suzuki M, Yoshida S, Adman ET, Blank A, Loeb LA. Thermus aquaticus DNA polymerase I mutants with altered fidelity. Interacting mutations in the O-helix. J Biol Chem. 2000;275:32728–32735. doi: 10.1074/jbc.M000097200. [DOI] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. Structural basis of high-fidelity DNA synthesis by yeast DNA polymerase δ. Nat Struct Mol Biol. 2009;16:979–986. doi: 10.1038/nsmb.1663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sweasy JB, Loeb LA. Detection and characterization of mammalian DNA polymeraseβ mutants by functional complementation in Escherichia coli. Proc Natl Acad Sci USA. 1993;90:4626–4630. doi: 10.1073/pnas.90.10.4626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tian W, Hwang YT, Lu Q, Hwang CBC. Finger domain mutation affects enzyme activity, DNA replication efficiency, and fidelity of an exonuclease-deficient DNA polymerase of herpes simplex virus type 1. J Virol. 2009;83:7194–7201. doi: 10.1128/JVI.00632-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Degtyareva NP, Gordenin DA, Resnick MA. Genetic factors affecting the impact of DNA polymerase δ proofreading activity on mutation avoidance in yeast. Genetics. 1999a;152:47–59. doi: 10.1093/genetics/152.1.47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Degtyareva NP, Koloteva NN, Sugino A, Masumoto H, Gordenin DA, Resnick MA. Replication slippage between distant short repeats in Saccharomyces cerevisiae depends on the direction of replication and the RAD50 and RAD52 genes. Mol Cell Biol. 1995;15:5607–5617. doi: 10.1128/mcb.15.10.5607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tran HT, Gordenin DA, Resnick MA. The 3′→5′ exonucleases of DNA polymerases δ and ε and the 5′→3′ exonuclease Exo1 have major roles in postreplication mutation avoidance in Saccharomyces cerevisiae. Mol Cell Biol. 1999b;19:2000–2007. doi: 10.1128/mcb.19.3.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trobner W, Piechocki R. Selection against hypermutability in Escherichia coli during long term evolution. Mol Gen Genet. 1984;198:177–178. doi: 10.1007/BF00328720. [DOI] [PubMed] [Google Scholar]
- Trostler M, Delier A, Beckman J, Urban M, Patro JN, Spratt TE, Beese LS, Kuchta RD. Discrimination between right and wrong purine dNTPs by DNA polymerase I from Bacillus stearothermophilus. Biochemistry. 2009;48:4633–4641. doi: 10.1021/bi900104n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trzemecka A, Jacewicz A, Carver GT, Drake JW, Bebenek A. Reversal of a mutator activity by a nearby fidelity-neutral substitution in the RB69 DNA polymerase binding pocket. J Mol Biol. 2010;404:778–793. doi: 10.1016/j.jmb.2010.09.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai Y-C, Johnson KA. A new paradigm for DNA polymerase specificity. Biochemistry. 2006;45:9675–9687. doi: 10.1021/bi060993z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang J, Sattar AK, Wang CC, Karam JD, Konigsberg WH, Steitz TA. Crystal structure of a pol α family replication DNA polymerase from bacteriophage RB69. Cell. 1997;89:1087–1099. doi: 10.1016/s0092-8674(00)80296-2. [DOI] [PubMed] [Google Scholar]
- Wang TS-F, Wong SW, Korn D. Human DNA polymerase α: predicted functional domains and relationships with viral DNA polymerases. FASEB J. 1989;3:14–21. doi: 10.1096/fasebj.3.1.2642867. [DOI] [PubMed] [Google Scholar]
- Wardle J, Burgers PMJ, Cann IKO, Darley K, Heslop P, Johansson E, Lin L-J, McGlynn P, Sanvoisin J, Stith CM, Connolly BA. Uracil recognition by replicative DNA polymerases is limited to the archaea, not occurring with bacteria and eukarya. Nucleic Acids Res. 2008;36:705–711. doi: 10.1093/nar/gkm1023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Genetical implications of the structure of deoxyribonucleic acid. Nature. 1953a;171:964–967. doi: 10.1038/171964b0. [DOI] [PubMed] [Google Scholar]
- Watson JD, Crick FH. Molecular structure of nucleic acids; a structure for deoxyribose nucleic acid. Nature. 1953b;171:737–738. doi: 10.1038/171737a0. [DOI] [PubMed] [Google Scholar]
- Wing RA, Bailey S, Steitz TA. Insights into the replisome from the structure of a ternary complex of the DNA polymerase III α-subunit. J Mol Biol. 2008;382:859–869. doi: 10.1016/j.jmb.2008.07.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winzeler EA, Shoemaker DD, Astromoff A, Liang H, Anderson K, Andre B, Bangham R, Benito R, Boeke JD, Bussey H, Chu AM, Connelly C, Davis K, Dietrich F, Dow SW, El Bakkoury M, Foury F, Friend SH, Gentalen E, Giaever G, Hegemann JH, Jones T, Laub M, Liao H, Liebundguth N, Lockhart DJ, Lucau-Danila A, Lussier M, M’Rabet N, Menard P, Mittmann M, Pai C, Rebischung C, Revuelta JL, Riles L, Roberts CJ, Ross-MacDonald P, Scherens B, Snyder M, Sookhai-Mahadeo S, Storms RK, Veronneau S, Voet M, Volckaert G, Ward TR, Wysocki R, Yen GS, Yu K, Zimmermann K, Philippsen P, Johnston M, Davis RW. Functional characterization of the S. cerevisiae genome by gene deletion and parallel analysis. Science. 1999;285:901–906. doi: 10.1126/science.285.5429.901. [DOI] [PubMed] [Google Scholar]
- Witkin EM, Roegner-Maniscalco V. Overproduction of DnaE protein (α subunit of DNA polymerase III) restores viability in a conditionally inviable Escherichia coli strain deficient in DNA polymerase I. J Bacteriol. 1992;174:4166–4168. doi: 10.1128/jb.174.12.4166-4168.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Huang M. Dif1 controls subcellular localization of ribonucleotide reductase by mediating nuclear import of the R2 subunit. Mol Cell Biol. 2008;28:7156–7167. doi: 10.1128/MCB.01388-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia S, Wang M, Lee HR, Sinha A, Blaha G, Christian T, Wang J, Konigsberg W. Variation in mutation rates caused by RB69pol fidelity mutants can be rationalized on the basis of their kinetic behavior and crystal structures. J Mol Biol. 2011;406:558–570. doi: 10.1016/j.jmb.2010.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu L, Pena Castillo L, Mnaimneh S, Hughes TR, Brown GW. A survey of essential gene function in the yeast cell division cycle. Mol Biol Cell. 2006;17:4736–4747. doi: 10.1091/mbc.E06-04-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao X, Rothstein R. The Dun1 checkpoint kinase phosphorylates and regulates the ribonucleotide reductase inhibitor Sml1. Proc Natl Acad Sci USA. 2002;99:3746–3751. doi: 10.1073/pnas.062502299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou Z, Elledge SJ. Isolation of crt mutants constitutive for transcription of the DNA damage inducible gene RNR3 in Saccharomyces cerevisiae. Genetics. 1992;131:851–866. doi: 10.1093/genetics/131.4.851. [DOI] [PMC free article] [PubMed] [Google Scholar]