Abstract
Mechanisms by which aniline exposure elicits splenotoxicity, especially a tumorigenic response, are not well-understood. Earlier, we have shown that aniline exposure leads to oxidative DNA damage and up-regulation of OGG1 and NEIL1/2 DNA glycosylases in rat spleen. However, the contribution of endonuclease III homolog 1 (NTH1) and apurinic/apyrimidinic endonuclease 1 (APE1) in the repair of aniline-induced oxidative DNA damage in the spleen is not known. This study was, therefore, focused on examining whether NTH1 and APE1 contribute to the repair of oxidative DNA lesions in the spleen, in an experimental condition preceding tumorigenesis. To achieve this, male SD rats were subchronically exposed to aniline (0.5 mmol/kg/day via drinking water for 30 days), while controls received drinking water only. By quantitating the cleavage products, the activities of NTH1 and APE1 were assayed using substrates containing thymine glycol (Tg) and tetrahydrofuran, respectively. Aniline treatment led to significant increases in NTH1- and APE1-mediated BER activity in the nuclear extracts of spleen of aniline-treated rats compared to the controls. NTH1 and APE1 mRNA expression in the spleen showed 2.9- and 3.2-fold increases, respectively, in aniline-treated rats compared to controls. Likewise, Western blot analysis showed that protein expression of NTH1 and APE1 in the nuclear extracts of spleen from aniline-treated rats was 1.9- and 2.7-fold higher than controls, respectively. Immunohistochemistry indicated that aniline treatment also led to stronger immunoreactivity for both NTH1 and APE1 in the spleens, confined to the red pulp areas. These results, thus, show that aniline exposure is associated with induction of NTH1 and APE1 in the spleen. The increased repair activity of NTH1 and APE1 could be an important mechanism for the removal of oxidative DNA lesions. These findings thus identify a novel mechanism through which NTH1 and APE1 may regulate the repair of oxidative DNA damage in aniline-induced splenic toxicity.
Keywords: Oxidative DNA damage, NTH1, APE1, DNA base excision repair, Aniline, Spleen
Introduction
Aniline, a toxic aromatic amine, is an extensively used industrial chemical. Exposure to aniline leads to toxic damages in the spleen, which is characterized by splenomegaly, increased erythropoietic activity, hyperpigmentation, hyperplasia, fibrosis, and a variety of primary sarcomas of the spleen after chronic exposure in rats (Goodman et al., 1984; Weinberger et al., 1985; Bus and Popp, 1987; Khan et al., 1993, 1997, 1999, 2006; Pauluhn, 2004). However, the molecular mechanisms by which aniline exerts its toxicological and tumorigenic effects in the spleen are not well understood. Earlier studies from our laboratory have shown that aniline exposure results in iron overload and oxidative stress in the spleen (Khan et al., 1997, 1999, 2003a, 2003b). More importantly, our studies have shown increased oxidative DNA damage in the spleen of rats following aniline exposure (Wu et al., 2005; Ma et al., 2008, 2011), which could potentially lead to mutagenic and/or carcinogenic responses.
Reactive oxygen species (ROS) and reactive nitrogen species (RNS) can directly oxidize and damage macromolecules such as proteins, lipids and DNA (Marcon et al., 2009). ROS is believed to play an important role in the pathogenesis of several diseases, such as cancer, rheumatoid arthritis, cardiovascular disease, and aging (Ames et al., 1993; Gotz et al., 1994; Hazra et al., 2007; Saha et al., 2010). In order to repair oxidative DNA damage, organisms possess multiple glycosylases which recognize damaged bases and initiate base excision repair (BER) (Liu et al., 2010). The BER pathway is the major pathway by which oxidative DNA lesions are removed from the genome, thus, representing a critical step in the maintenance of genomic stability. BER is a stepwise process initiated by specialized DNA glycosylases. Accordingly, this pathway is critical for preventing diseases resulting from oxidative DNA damage. In mammalian cells, there are several different DNA glycosylases with overlapping substrate specificities that remove oxidative DNA base lesions. These glycosylases include 8-oxoguanine glycosylase 1 (OGG1), endonuclease III homologue 1 (NTH1), and the Nei-like DNA glycosylases (NEIL1/2) (Rolseth et al., 2008; Mori et al., 2009; Ma et al., 2011). Excision of DNA damaged bases by OGG1 and NTH1 glycosylases is followed by a common pathway involving an AP-endonuclease (APE) to generate 3’ OH terminus at the damage site. APE, the apurinic/apyrimidinic endonuclease 1 (APE1), can play a central role in BER by removing 3’ blocking groups produced after base excision by β-elimination (Zhao et al., 2008, Marcon et al., 2009). APE1 is not only an essential enzyme involved in BER pathway, but also involved in reduction-oxidation regulation (Yuk et al., 2009), and acts as a major redox-signaling factor that has a wide variety of important cellular functions including transcription factor regulation, oxidative signaling and cell cycle control (Zhang et al., 2009). These multiple functions of mammalian APE1, both in DNA repair and gene regulation, warrant extensive analysis of its own regulation and dissection of the mechanisms of action (Bhakat et al., 2009).
Thymine glycol (Tg) is one of the major toxic oxidative DNA lesions generated by ROS (Takao et al., 2002); it blocks both DNA replication and transcription, causing cell death (Trzeciak et al., 2004). Tg is repaired in mammalian cells by NTH1. NTH1 has both DNA glycosylase activity and apurinic/apyrimidinic (AP) lyase activity, both of which function in the initiation of BER (Goto et al., 2009). NTH1 can cleave the Tg damaged DNA via β–elimination much more efficiently than OGG1 (Izumi et al., 2003). However, the role of NTH1 in aniline-induced oxidative DNA damage is not known.
We have previously shown in the spleen of rats exposed to aniline that increased oxidative DNA damage was associated with an up-regulation of OGG1, a specific DNA glycosylase involved in the removal of 8-hydroxy-2’-deoxyguanosine (8-OHdG) adducts (Ma et al., 2008). Furthermore, we have also shown that NEILs, which are distinct from OGG1 in structural features and reaction mechanisms, and which act on different substrates (Dou et al., 2003; Englander and Ma, 2006, Ma et al., 2011), are also involved in the repair of aniline-induced DNA damage in the spleen of aniline-treated rats. Although the role of NTH1 and APE1 in DNA repair and transcriptional regulation has been widely investigated, the roles of NTH1 and APE1 in the regulation of aniline-induced splenotoxic response have not been evaluated. Furthermore, the molecular mechanisms by which NTH1 and APE1 may contribute to aniline-induced tumorigenesis remain largely unknown. Therefore, the current study is focused on examining the contribution of NTH1 and APE1 on repair of oxidative DNA damage in the spleen following aniline insult.
Materials and methods
Animal and treatment
Sprague-Dawley rats (Male, ~ 200 g), obtained from Harlan (Indianapolis, IN), were housed in wire-bottom cages over absorbent paper with free access to tap water and Purina rat chow. The animals were acclimatized in a controlled-environment animal room (temperature, 22 °C; relative humidity, 50%; photoperiod, 12-h light/dark cycle) for 7 days prior to treatment. The experiments were performed in accordance with the guidelines of the National Institutes of Health and were approved by the Institutional Animal Care and Use Committee of University of Texas Medical Branch. The animals were divided into two groups of six rats each. One group of animals was given 0.5 mmol/kg/day of aniline (~97%; Sigma-Aldrich, Milwaukee, WI) via drinking water (pH of the solution adjusted to ~6.8) (Khan et al., 1993, 1999; Ma et al., 2008, 2011), whereas the other group received drinking water only and served as controls. The choice of aniline dose was based on our earlier studies that showed significant increases in lipid peroxidation, protein oxidation and oxidative DNA damage (oxidative stress) in the spleen (Khan et al., 1999; Ma et al., 2008, 2011). After 30 days, the rats were euthanized under nembutal (sodium pentobarbital) anesthesia and the spleens were removed immediately, blotted, weighed and stored at −80 °C until further analysis. A portion of the spleen was snap-frozen in liquid nitrogen and stored at −80 °C for RNA isolation. Also, portions of the spleen from control and aniline-treated rats were fixed in 10% neutral buffered formalin for immunohistological processing.
Oligonucleotide 5′ end-labeling for NTH1 and APE1 BER assay
The 30- and 21-mer oligonucleotides containing thymine glycols (Tg) and tetrahydrofuran (THF) at positions 13 and 9 from the 5’-end (sequences shown below) were obtained from Sigma (St. Louis, MO) and Trevigen (Gaithersburg, MD), respectively. Oligonucleotides (5.5 pmol) were end-labeled using 5 µCi of [γ-32P]ATP (3000 Ci/mmol, Perkin-Elmer Life & Analytical Sciences, Boston, MA) and T4 polynucleotide kinase (New England BioLabs, Ipswich, MA) and then passed through a G-25 spin column (GE Healthcare, Piscataway, NJ) for purifying the radiolabeled oligonucleotides, and annealed with 1.5-2 fold complementary oligonucleotide by gradual cooling to room temperature. NTH1 oligonucleotide substrate sequence:
5’– GAT CCT CTA GAG TgCG ACC TGC AGG CAT GCA – 3’
3’– CTA GGA GAT CTC AGC TGG ACG TCC GTA CGT – 5’
APE1 oligonucleotide substrate sequence:
5’– CCT GCC CTGTHF GCA GCT GTG GC – 3’
3’– GGA CGG GAC A CGT CGA CAC CG – 5’
Preparation of nuclear extracts (NEs) for NTH1 and APE1 BER activities and Western blotting
The nuclear protein extracts (NEs) were prepared according to the method of Ma et al. (2008, 2011) with minor modifications. Spleen tissues (from control and aniline-treated rats) were cut into smaller pieces, homogenized briefly with a loose glass pestle in cold hypotonic buffer [10 mM HEPES-KOH, 10 mM KCl, 100 µM EDTA, 100 µM EGTA, 1 mM DTT, 0.5 mM PMSF, 2 µg/ml pepstatin, and a complete protease inhibitor cocktail (Roche, Germany)], and incubated on ice for 25 min. Tissues were then homogenized with a tight pestle and centrifuged at 800 g for 4 min to obtain nuclear pellets. Pellets were gently washed two times with homogenizing buffer. Nuclear proteins were extracted in a high salt buffer (20 mM HEPES-KOH, 400 mM NaCl, 1 mM EDTA, 1 mM EGTA, 1 mM DTT, 1 mM PMSF, 2 µg/ml pepstatin, and protease inhibitor cocktail) by incubating for 45 min on ice with reverse mixing at intervals of 10 min. The NEs were cleared by centrifugation (16,000 g, 10 min), mixed with glycerol (15%, final concentration) and stored at −80 °C until further analysis.
Cleavage assay
Glycosylase and endonuclease activities were measured in vitro in the NEs with synthetic end-labeled double-stranded oligonucleotide substrates containing Tg (targeted by NTH1) and THF (targeted by APE1), respectively. Glycosylase assay for NTH1 was done as described earlier (Ma et al., 2008, 2011) with slight modifications. Briefly, the reactions were done in 20 µl with 40 µg NEs, 1 nM end-labeled substrate, and reaction buffer (25 mM Hepes-KOH, pH 8.0, 2 M EDTA, 1 M dithiothreitol, 150 mM KCl, 0.5 µg/µl BSA, 1.5% glycerol). Incubation was done at 37 °C for indicated times and was terminated with 5X alkaline loading buffer (0.5 M NaOH, 97% formamide, 10 mM EDTA, pH 8, 0.025% bromophenol blue, 0.025% xylene cyanol) and heated at 90 °C for 4 min. For positive control, recombinant NTH1 purchased from Trevigen was used. The endonuclease assays of APE1 were done as described earlier (Englander and Ma, 2006) with minor modifications. Briefly, the reactions were assembled with 1 nM end-labeled substrate, 20 ng NEs and the reaction buffer containing MgCl2 (100 mM Tris–HCl pH 7.6, 1 mM DTT, 0.5 mM MgCl2, 0.2 mM EDTA, 0.1 mg/ml BSA) and terminated with 5X loading buffer (97% formamide, 10 mM EDTA pH 8, 0.025% bromophenol blue, 0.025% xylene cyanol) without heating. For positive control, recombinant hAPE1 obtained from Trevigen was used. Reaction mixtures were resolved on 15% polyacrylamide-7 M urea gels in Tris-borate buffer (89 mM Tris-HCl, 89 mM H3BO3, 2 mM EDTA, pH ~8.3) at 16 mA for 150 min, and products were visualized by autoradiography and quantified on Phosphorimager (GE Healthcare, Piscataway, NJ). Phosphorimager values, representing percent of substrate cleavage within the linear range of each reaction, were converted into cleavage-product amounts. Values from 6 individual rats and 3 cleavage assays per extract were averaged and plotted as means ± SD.
RNA isolation, cDNA synthesis and real-time PCR for NTH1 and APE1
Total RNA isolation from spleen tissues, cDNA synthesis and the real-time PCR were performed as described in our earlier studies (Ma et al., 2008, 2011) with minor modifications.
RNA isolation
Total RNA was isolated from spleen tissues using RiboPure kit (Ambion, Austin, TX) as per the manufacturer's instructions. To eliminate contaminating genomic DNA, RNA preparation was treated with RNase free DNase I (DNA-free kit, Ambion). The total RNA concentration was determined by measuring the absorbance at 260 nm. RNA integrity was verified electrophoretically by ethidium bromide staining and by measuring A260/A280 ratio.
Real-time PCR
Real-time PCR was performed essentially as described earlier (Wang et al., 2005; Ma et al., 2008, 2011). Briefly, first-strand cDNA was prepared from isolated RNA by using SuperScript III First-Strand Synthesis Kit (Invitrogen, Carisbad, CA) as described earlier (Wang et al., 2005). Quantitative real-time PCR employing a two-step cycling protocol (denaturation and annealing/extension) was carried out using Mastercycler Realplex as per manufacturer's instructions (Eppendorf North America, Westburry, NY). The sequences of the forward and the reverse primers of NTH1 and APE1 for real-time PCR were 5’-GGCCTATGAGGTTGCTAACG- 3’ and 5’- GTGCGTCCTTCTTGCTTCTC- 3’ for NTH1 and 5’-TCAGAAAACGTCAGCCAGTG-3’ and 5’-CGGGAGTTTGTTCTCTGAGC- 3’ for APE1, respectively. For each cDNA sample, parallel reactions were performed in triplicate for the detection of NTH1, APE1 and 18 S. The reaction samples in a final volume of 25 µl contained 2 µl of cDNA templates, 2 µl primer pair, 12.5 µl iQ SYBR Green Supermix and 8.5 µl water. Amplification conditions were identical for all reactions: 95 °C for 2 min for template denaturation and hot start prior to PCR cycling. A typical cycling protocol consisted of three stages: 15 s at 95 °C for denaturation, 30 s at 63 °C for annealing, 30 s at 72 °C for extension, and an additional 6 s hold for fluorescent signal acquisition. To avoid the non-specific signal from primer-dimers, the fluorescence signal was detected 2 °C below the melting temperature (Tm) of individual amplicon and above the Tm of the primer-dimers (Rajeevan et al., 2001 and Simpson et al., 2000). A total of 40 cycles were performed for the studies. After the final cycle of the PCR, a melt curve analysis (Ririe et al., 1997 and Wei et al., 2003) was performed for each sample. The reactions were cooled to 60 °C and then heated up to 95 °C at the ramp rate of 0.2 °C/s to denature double-stranded PCR products. The fluorescent signal recorded during DNA melting was plotted against temperature to generate the melt curve for each reaction. The Tm of individual amplicon was identified from the melt curve analysis using the Mastercycler Realplex software (Eppendorf). The specificity and purity of an amplicon for a particular target gene was confirmed by electrophoresis on 2% agarose gel stained with ethidium bromide, which showed a single prominent band of expected size on gel.
Quantitation of PCR was done using the comparative CT method as described in User Bulletin No. 2 of Applied Biosystems (Foster City, CA), and reported as fold difference relative to the calibrator cDNA (QuantumRNA Universal 18 S Standards, Ambion). The fold change in cDNA (target gene) of NTH1 and APE1 relative to the 18 S endogenous control was determined by: Fold change = 2−ΔΔCT, where ΔΔCT = (CT Aniline – CT 18S) - (CT Control – CT 18S).
Western blotting for NTH1 and APE1 in splenic NEs
Protein extracts (60 µg) were denatured by heating at 90 °C for 5 min and subjected to 12% sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE, GenScript, Piscataway, NJ). The separated proteins were transferred onto polyvinylidene difluoride (PVDF) microporous membrane (Millipore Corporation, Billerica, MA) using a transfer buffer (25 mM Tris-HCl, 190 mM glycine, pH 8.4, 10% methanol). The membrane was then blocked with 10% non-fat dry milk and incubated overnight at 4 °C with anti-NTH1 antibodies (1:200 dilution, Santa Cruz, CA) or anti-APE1 (1:200 dilution; Novus, Littleton, CO). The membrane was washed three times with PBS-Tween 20 buffer. After incubation with the secondary antibody [anti-goat IgG-horseradish peroxidase (HRP) for NTH1 and anti-rabbit IgG-HRP for APE1], the membranes were washed and developed using a chemiluminescence detection kit (ECL, GE Healthcare). As a control, after stripping membrane with stripping buffer (Boston BioProducts, Worcester, MA), it was re-probed with anti-actin antibody (Sigma) and developed as above. NTH1 and APE1 bands were quantitated by densitometry and normalized using the actin signal to correct for differences in loading of proteins from control and experimental groups. For densitometric analysis, the protein bands on the blot were measured by Eagle Eye II software (Stratagene, Inc., La Jolla, CA).
Immunohistochemical localization of NTH1 and APE1
Immunohistochemical localization of NTH1 and APE1 was done using the method described earlier (Ma et al., 2008, 2011). Briefly, paraffin sections were cut and deparaffinized at 55 °C for 1 h and treated with xylene and various concentrations of ethanol, then rehydrated with water. The slides were incubated with sodium citrate buffer at 95 °C for 20 min for antigen retrieval and subsequently incubated with reagents for blocking the non-specific binding sites, which included quenching of endogenous peroxidase activity with 0.3% H2O2 in water for 15 min, 10% normal serum (Sigma) for 30 min, and avidin and biotin block solutions (Vector Laboratories, Burlingame, CA) for 15 min each. The sections were then incubated with primary antibody (NTH1 antibody, 1:100, Novus; APE1 antibody, 1:100, ProteinTech Group, Chicago, IL) overnight at 4 °C. Immunoreactivity was detected by the ABC method (Vectastain Elite ABC Kit, Vector Laboratories, Burlingame, CA) with color development using the substrate 3,3′-diaminobenzidine (DAB). Mayer's Hematoxylin was then added as a counterstain for 1 min. The negative controls were immunostained as above, but with goat serum instead of primary antibodies. Histological evaluation of staining was done at 400x under an OLYMPUS BX51 Microscope (Leads Instruments, Inc., Irving, TX).
Statistical analysis
Data are expressed as means ± SD of six animals in each group. Comparison between the groups was made by p value determination using Student's two-tailed t-test (GraphPad InStat 3 software, La Jolla, CA). A p value of < 0.05 was considered to be statistically significant.
Results
NTH1- and APE1-mediated BER in response to aniline exposure
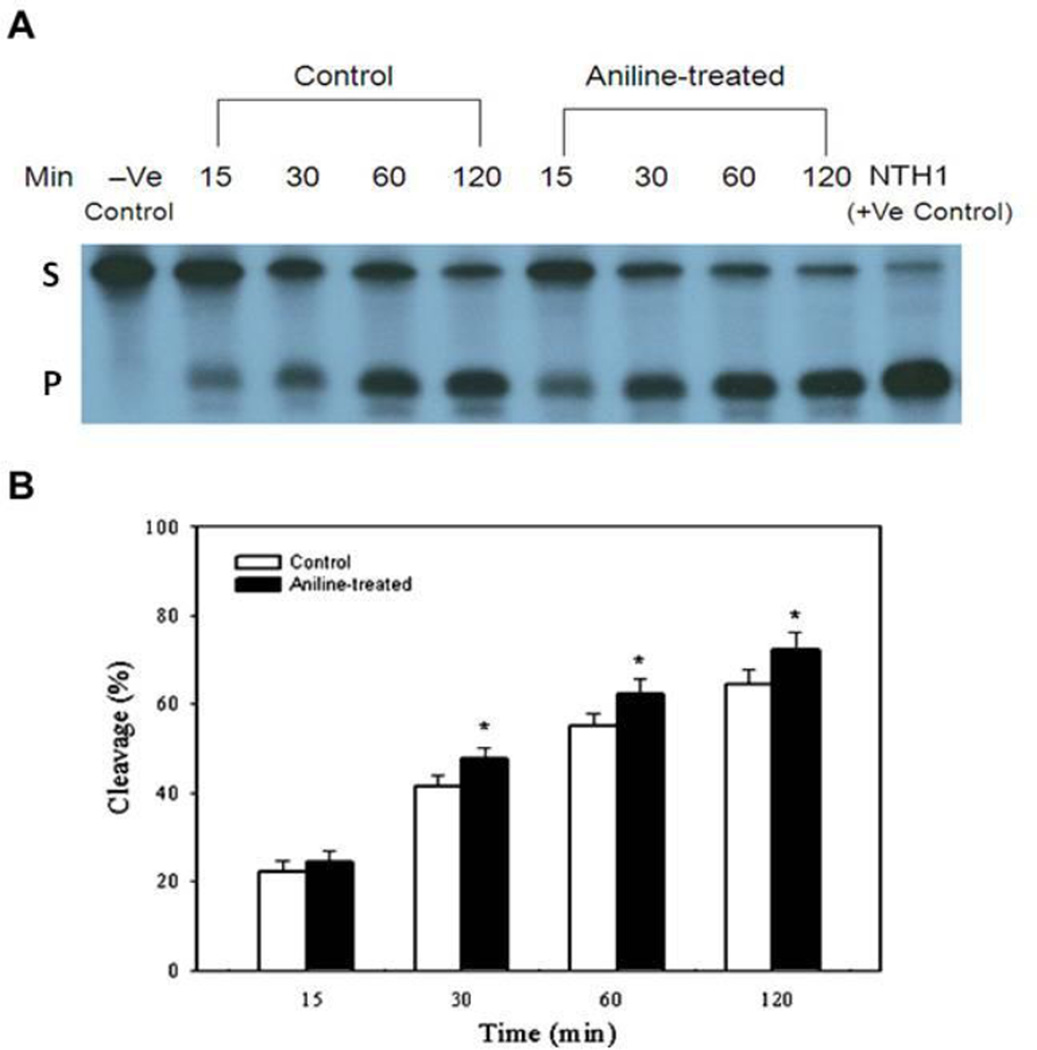
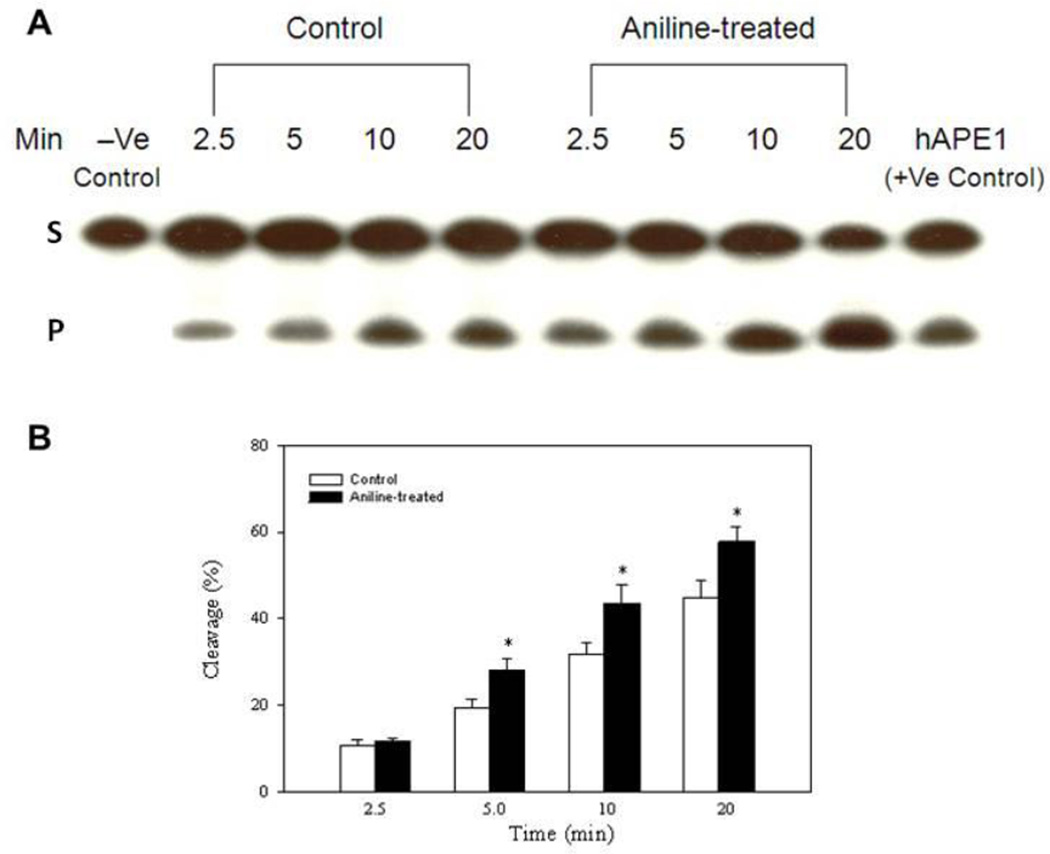
The capacity for excision of Tg and abasic sites (AP) with end-labeled oligonucleotide substrates (targeted by NTH1 and APE1, respectively) was evaluated in the NEs from control and aniline-treated rats. The cleavage activities were calculated from the cleavage yields generated in the linear range of each reaction. NTH1- and APE1-mediated BER activities of NEs were determined by quantifying radioactivity generated over time in cleavage products of end-labeled oligonucleotides (Fig. 1A, 2A). The average excision activity for NTH1 was 0.64 fmol/µg protein/h for control animals, whereas for aniline-treated animals it was 0.74 fmol/µg protein/h (Fig. 1B, Table 1). Average excision activity for APE1 was 2.03 fmol/µg protein/h for control animals, whereas for aniline-treated animals it was 2.63 fmol/µg protein/h ( Fig. 2B, Table 2). Also the patterns of cleavage over time are shown in Fig. 1B and 2B for NTH1 and APE1, respectively, with the values for each time point. Our data, thus, show significant increases of 1.2- and 1.3-fold in NTH1 and APE1 BER activities in the splenic NEs of aniline-treated rats, respectively, suggesting increased capacity to excise the Tg:G and AP(THF):A structured substrates, which are preferentially targeted by NTH1 and APE1 (Englander and Ma, 2006, Yndestad et al., 2009).
Fig. 1.
BER activity in the splenic NEs of control and aniline-treated rats. The assay was conducted with end-labeled oligonucleotides (Tg:G) which are targeted by NTH1 (see methods for details). (A) Autoradiogram of incision products (P) generated over time by cleavage of end-labeled double-stranded oligonucleotides carrying the Tg lesion adducts (S). Negative control (without NEs) and positive control (recombinant NTH1) are included in the external lanes. (B) Values from Phosphorimager quantitation of products, generated with NEs from 5 rats per group and 3 cleavage assays per extract, were averaged and plotted as means ± SD. * Indicates that the cleavage is significantly different from controls for each respective time point (p < 0.05).
Fig. 2.
BER activity in the splenic NEs of control and aniline-treated rats. The assay was conducted with end-labeled oligonucleotides (THF:A) which are targeted by APE1 (see methods for details). (A) Autoradiogram of incision products (P) generated over time by cleavage of end-labeled double-stranded oligonucleotides carrying the THF lesion adducts (S). Negative control (without NEs) and positive control (recombinant hAPE1) are included in the external lanes. (B) Values from Phosphorimager quantitation of products, generated with NEs from 5 rats per group and 3 cleavage assays per extract, were averaged and plotted as means ± SD. * Indicates that the cleavage is significantly different from controls for each respective time point (p < 0.05).
Table 1.
NTH1 activity in the splenic nuclear proteins of control (C) and aniline-treated (A) rats
| Time point (min) | NTH1 Activity* (C) | NTH1 Activity* (A) |
|---|---|---|
| 15 | 0.89 ± 0.10 | 0.98 ± 0.09 |
| 30 | 0.82 ± 0.05 | 0.97 ± 0.05Δ |
| 60 | 0.55 ± 0.03 | 0.64 ± 0.04Δ |
| 120 | 0.32 ± 0.02 | 0.36 ± 0.02Δ |
Values are mean ± S.D and are expressed as fmol/µg protein/h;
Values are significantly different from controls (p<0.05).
Table 2.
APE1 activity in the splenic nuclear proteins of control (C) and aniline-treated (A) rats
| Time point (min) | APE1 Activity* (C) | APE1 Activity* (A) |
|---|---|---|
| 2.5 | 2.57 ± 0.13 | 2.82 ± 0.10 |
| 5.0 | 2.31 ± 0.26 | 3.36 ± 0.33Δ |
| 10 | 1.96 ± 0.16 | 2.62 ± 0.24Δ |
| 20 | 1.34 ± 0.12 | 1.73 ± 0.10Δ |
Values are mean ± S.D and are expressed as fmol/µg protein/h.
Values are significantly different from controls (p<0.05).
Effect of aniline exposure on mRNA expression of NTH1 and APE1
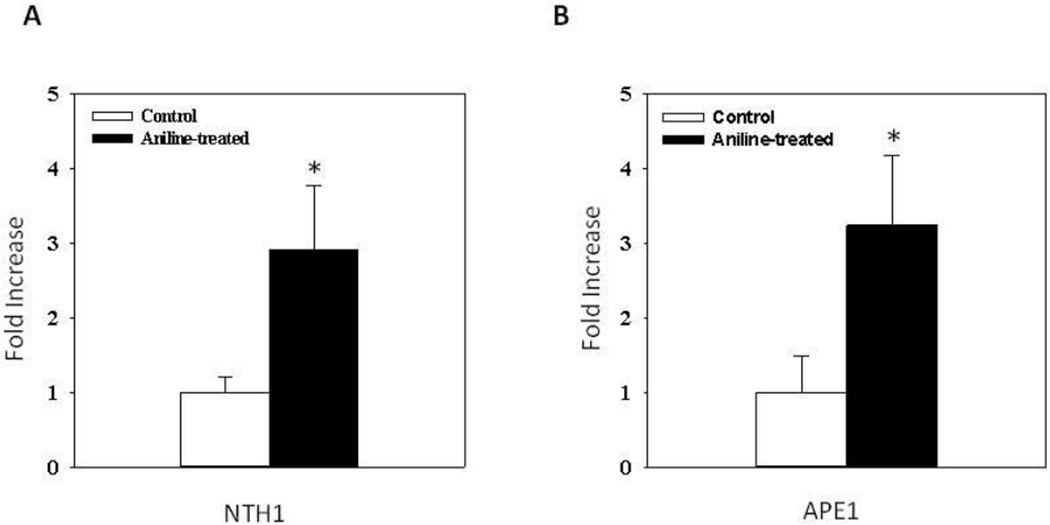
For determining changes in expression of NTH1 and APE1 in the spleen following aniline exposure, NTH1 and APE1 mRNA levels were analyzed by real-time PCR. As shown in Fig. 3, aniline exposure resulted in 2.9- and 3.2-fold increases in NTH1 (A) and APE1 (B) mRNA expression, respectively, compared to controls (p<0.05).
Fig. 3.
Real-time PCR analysis of NTH1 (A) and APE1 (B) gene expression in the spleens of control and aniline-treated rats. Total RNA was extracted from spleen tissues, real-time PCR was performed, and the fold change in mRNA expression was determined. Values are means ± SD. *p < 0.05.
NTH1 and APE1 protein expression in the splenic NEs following aniline exposure
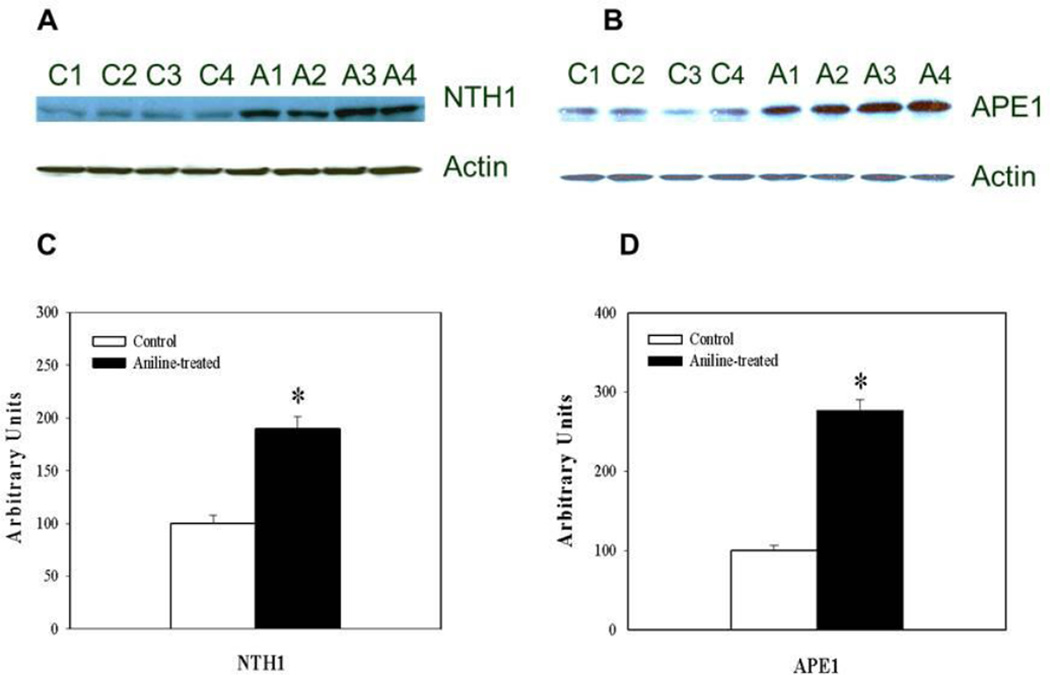
To determine whether increases in the gene expression were also associated with increases in corresponding protein levels, NTH1 and APE1 protein expression in the spleens was determined by Western blotting. As evident from Fig. 4, NTH1 and APE1 protein expression in the NEs of aniline-treated rats were 1.9- and 2.7-fold higher, respectively, than the controls (p<0.05).
Fig. 4.
Western blot detection of NTH1 (A) and APE1 (B) in the NEs from control and aniline-treated rats. Lanes C1-C4: controls; lanes A1-A4: aniline-treated. Densitometric analyses of NTH1 and APE1 bands from control and aniline-treated rats (C, D). The densitometric analysis of the protein bands was done using Eagle Eye II software. Data from aniline-treated spleen samples were statistically compared to untreated controls, which were arbitrarily set at 100. Values are means ± SD of four determinations. *p < 0.05.
Immunohistochemical assessment of NTH1 and APE1 in the spleen of rats exposed to aniline
To localize NTH1 and APE1 expression in the spleen of experimental and control animals, immunohistochemical studies were conducted. While control spleens showed sparse immunostaining for NTH1 and APE1, significantly increased number of cells immunostained for both NTH1 and APE1 in the spleens of aniline-treated rats were observed (Figs. 5A and 5B: NTH1; 5C and 5D: APE1). The immunoreactivity for NTH1 and APE1 appeared predominantly in the red pulp areas of the spleen. APE1 showed much stronger immunoactivity compared to NTH1 following aniline insult with regard to both number of cells stained and overall staining intensity.
Fig. 5.
Immunohistochemistry of NTH1 and APE1 in the spleens of control rats (A and C) vs. aniline-treated rats (B and D). Control spleen showed sparse immunoreactivity for NTH1 and APE1, whereas spleen from aniline-treated rats showed increased immunoreactive cells and increased intensity of staining for NTH1 and APE1, confined to the red pulp areas. Also, APE1 showed much stronger immunoactivity compared to NTH1 following aniline insult with regard to both number of cells stained and overall staining intensity.
Discussion
ROS-induced DNA damage can result in a wide variety of DNA lesions, such as oxidized bases, abasic sites, and single strand breaks. This can lead to mutations, cell death, cancer and agerelated pathophysiology (Izumi et al., 2003; Nowsheen et al., 2009). The repair of oxidized DNA damage due to ROS insult is mainly dependent on the activities of BER enzymes, including OGG1, NTH1, NEILs, APE1 and polynucleotide kinase (PNK), which can effectively remove such lesions. For example, 8-OHdG is repaired by OGG1, 5-hydroxyuracil (5-OHU) is removed preferentially by NEILs, Tg and other pyrimidine derives are removed by NTH1, creating AP sites that are then repaired by APE (Saha et al., 2010; Redrejo-Rodriguez et al., 2011; Ma et al., 2008, 2011). The generation of Tg in DNA occurs through oxidative attack on the pyrimidine base. Tg is a potentially fatal DNA lesion produced by chemical oxidants and other environmental toxicants (Takao et al., 2002). Removal of these lesions result in abasic sites that are one of the most frequent endogenous lesions in cellular DNA and their repair is critical for genomic stability and cellular survival (Alseth et al., 2004). NTH1 possesses not only DNA glycosylase activity but also AP lyase activity (Izumi et al., 2003), and the involvement of NTH1 indicates the repair of damage occurring in duplex DNA, possibly during transcription and/or replication. In mammalian cells, APE1 primarily removes the 3’ phosphors, α, β- unsaturated aldehyde generated by the β-lyase activity of OGG1 and NTH1 (Hazra et al., 2007). Furthermore, APE1 is also involved in reduction-oxidation regulation (Yuk et al., 2009), and has been shown to be a pluripotent factor and may be a key effector of the relationships between mammalian BER, oxidative signaling, transcription regulation, and cell cycle control (Evans et al., 2000; Kelly and Parsons 2001).
Contribution of oxidative mechanisms, including oxidative DNA damage, is well established in the splenic toxicity of aniline (Khan et al., 1999, 2006; Wu et al., 2005; Ma et al., 2008, 2011). An imbalance in DNA damage and repair activites could be critical in the initiation of a mutagenic and/or carcinogenic response in the spleen, and thus, could provide a mechanistic explanation for aniline-induced sarcomas of the spleen (Ma et al., 2008). Our previous studies showed that aniline-induced oxidative stress in spleen is associated with increases in 8-OHdG levels, and induction of 8-OHdG-specific lyase (OGG1) activity (Ma et al., 2008). Based on these results, it has become clear that aniline-induced oxidative DNA damage mainly involves BER pathway (Ma et al., 2008, 2011). We also found that aniline exposure leads to induction of NEIL1/2-associated BER activity and NEIL1/2 gene up-regulation in the spleen (Ma et al., 2011). The increased BER activities of NEILs could indicate an adaptive response against ROS-induced DNA damage resulting from aniline exposure, and could represent another important mechanism for the removal of oxidative DNA lesions, especially in transcribed DNA following aniline insult (Ma et al., 2011).
In the current study, we provide evidence of the involvement of other BER enzymes, i.e., NTH1 and APE1. Our data showed that NTH1- and APE1-mediated repair activities were 1.2- and 1.3- fold increased, respectively, in the spleens of aniline-treated rats compared to the controls (Fig. 1 and 2, Tables 1 and 2), clearly indicating their involvement in the repair of aniline-induced DNA damage. Our findings on NTH1 and APE1 are consistent with our earlier findings on OGG1 and NEILs which are involved in the repair of aniline-induced DNA damage in the spleen. APE1 also has redox activity and can enhance DNA-binding activity of several transcription factors, including NF-κB, AP-1, through reduction of their critical cysteine residues (Ando et al., 2008). Earlier, we have shown aniline-induced activation of both NF-κB (Wang et al., 2005) and AP-1 (Khan et al., 2006, Wang et al., 2008) in the spleen. Results of the current study on APE1 would imply that APE1 plays an important role in enhancing DNA-binding activity and transcriptional activities of NF-κB and AP-1 (Ando et al., 2008). Our findings may also suggest that APE1 biological activities are exerted by two functionally distinct domains, the N-terminus containing the nuclear localization signal sequence is principally devoted to the redox transcriptional co-activition activity, and the C-terminus exerts the enzymatic activity on the abasic sites of DNA. Aniline exposure in this study led to increased NTH1- and APE1- associated BER activity in the nuclear extract of spleens, clearly suggesting that NTH1 and APE1 have roles in the repair of aniline-induced oxidative DNA lesions in the spleen. Interestingly, NTH1 has also an AP lyase activity (Izumi et al., 2003). Our findings indicate that aniline toxicity is mediated by ROS and are well supported by other studies that demonstrate activation of NTH1 and APE1 in response to ROS (Alseth et al., 2004; Tell et al., 2005; Hazra et al., 2007). AP sites, if left unrepaired, could often give rise to point mutation and sometimes more complex mutations leading possibly to sarcomas/carcinogenesis.
The increases in NTH1 and APE1 BER activities were coupled with increases in NTH1 and APE1 gene and protein expression, suggesting that aniline exposure affected the transcriptional and translational regulation of NTH1 and APE1 in the spleen. NTH1 and APE1 mRNA expression in the spleen of aniline-treated rats showed 2.9- and 3.2-fold increases, respectively. Interestingly, their protein expression also showed a similar pattern (1.9- and 2.7-fold increases in NTH1 and APE1 expression, respectively) in the NEs of spleen from aniline-treated rats. Thus, it is evident from our data that aniline can exert its effects at both the transcriptional and translational level. Our data also show that APE1 was affected more than NTH1 by aniline exposure. This is not surprising as AP sites (target of APE1) are generated by both OGG1 and NTH1. The increases in BER activities mediated by NTH1 and APE1 are also supported by our immunohistochemistry data which showed relatively stronger immunoreactivity for NTH1 and APE1 in the spleens of aniline-treated rats. It is interesting to note that in comparison to NTH1, APE1 showed greater mRNA and protein expression, higher BER activity, and stronger immunoreactivity, suggesting greater production of AP sites that are substrates for APE1 following aniline exposure.
Aniline exposure could induce a variety of oxidized DNA lesions including foramidopyrimidines as well as oxidation products of cytosine in addition to 8-OHdG. There is still a limited understanding of the mechanism by which BER involving NTH1 and APE1 could provide protection against spleen-induced sarcomas/carcinogenesis of aniline. The splenotoxic effects and tumorigenic responses resulting from exposure to aniline deserves in-depth studies to provide mechanistic explanations.
In conclusion, to our knowledge, this is the first study to report induction of NTH1- and APE1- associated BER activity and NTH1 and APE1 gene up-regulation in the spleen following aniline exposure. NTH1 and APE1, like OGG1 and NEILs, are also associated with the repair of DNA damage in the spleen of rats after aniline exposure. Spleens from aniline-treated rats had greater NTH1 and APE1 BER activity, mRNA and protein levels, and strong NTH1 and APE1 immunoreactivity, especially in the red pulp areas. Results of this study as well as our earlier findings on the induction of OGG1- and NEIL-associated BER activity (Ma et al., 2008, 2011) suggest that aniline induces a number of lesions (repaired by OGG1, NTH1 and other glycosylases), thus creating the AP sites repaired by APE1. The increased BER activity of NTH1 and APE1 and previously reported OGG1 and NEILs ((Ma et al., 2008, 2011) may represent an adaptive response against ROS-induced DNA damage resulting from aniline exposure, and could be an important mechanism for the removal of oxidative DNA lesions. These data could lead to a better understanding of the role of NTH1 and APE1 in regulation of aniline-induced splenic toxicity in rats, and a better understanding of the relationship between cleaved oxidative DNA damage and DNA repair in relation to pathogenesis of sarcomas of the spleen after aniline exposure. The involvement of NTH1 and APE1 may also support repair of damage occurring in duplex DNA possibly during transcription and/or replication.
Highlights.
Aniline exposure led to greater NTH1- and APE1-mediated BER activity in rat spleen.
Aniline exposure was also associated with greater mRNA and protein levels of NTH1 and APE1 in the spleen.
The increased BER activity of NTH1 and APE1 represents an adaptive response against ROS-induced DNA damage resulting from aniline exposure.
Increased repair activity of NTH1 and APE1 is an important mechanism for the removal of oxidative DNA lesions.
Acknowledgments
This publication was made possible by grant ES06476 from National Institute of Environmental Health Sciences (NIEHS), National Institutes of Health (NIH). Its contents are solely the responsibility of the authors and do not necessarily represent the official views of NIEHS, NIH.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Conflict of interest statement
The authors declare that there is no conflict of interest.
References
- Alseth I, Korvald H, Osman F, Seeberg E, Bjørås M. A general role of the DNA glycosylase Nth1 in the abasic sites cleavage step of base excision repair in Schizosaccharomyces pombe. Nucleic Acids Res. 2004;32:5119–5125. doi: 10.1093/nar/gkh851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ames BN, Shigenaga MK, Hagen TM. Oxidants, antioxidants, and the degenerative diseases of aging. Proc. Natl. Acad. Sci. USA. 1993;90:7915–7922. doi: 10.1073/pnas.90.17.7915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ando K, Hirao S, Kabe Y, Ogura Y, Sato I, Yamaguchi Y, Wada T, Handa H. A new APE1/Ref-1-dependent pathway leading to reduction of NF-kappaB and AP-1, and activation of their DNA-binding activity. Nucleic Acids Res. 2008;36:4327–4336. doi: 10.1093/nar/gkn416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bhakat KK, Mantha AK, Mitra S. Transcriptional regulatory functions of mammalian AP-endonuclease (APE1/Ref-1), an essential multifunctional protein. Antioxid. Redox Signal. 2009;11:621–638. doi: 10.1089/ars.2008.2198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bus JS, Popp JA. Perspectives on the mechanism of action of the splenic toxicity of aniline and structurally-related compounds. Food Chem. Toxicol. 1987;25:619–626. doi: 10.1016/0278-6915(87)90024-x. [DOI] [PubMed] [Google Scholar]
- Dou H, Mitra S, Hazra TK. Repair of oxidized bases in DNA bubble structures by human DNA glycosylases NEIL1 and NEIL2. J. Biol. Chem. 2003;278:49679–49684. doi: 10.1074/jbc.M308658200. [DOI] [PubMed] [Google Scholar]
- Englander EW, Ma H. Differential modulation of base excision repair activities during brain ontogeny: Implications for repair of transcribed DNA. Mech. Ageing Dev. 2006;127:64–69. doi: 10.1016/j.mad.2005.09.008. [DOI] [PubMed] [Google Scholar]
- Evans AR, Limp-Foster M, Kelley MR. Going APE over ref-1. Mutat. Res. 2000;461:83–108. doi: 10.1016/s0921-8777(00)00046-x. [DOI] [PubMed] [Google Scholar]
- Goodman DG, Ward JM, Reichardt WD. Splenic fibrosis and sarcomas in F344 rats fed diets containing aniline hydrochloride, p-chloroaniline, azobenzene, o-toluidine hydrochloride, 4,4'-sulfonyldianiline, or D & C red No. 9. J. Natl. Cancer Inst. 1984;73:265–273. [PubMed] [Google Scholar]
- Goto M, Shinmura K, Igarashi H, Kobayashi M, Konno H, Yamada H, Iwaizumi M, Kageyama S, Tsuneyoshi T, Tsugane S, Sugimura H. Altered expression of the human base excision repair gene NTH1 in gastric cancer. Carcinogenesis. 2009;30:1345–1352. doi: 10.1093/carcin/bgp108. [DOI] [PubMed] [Google Scholar]
- Götz ME, Künig G, Riederer P, Youdim MB. Oxidative stress: free radical production in neural degeneration. Pharmacol. Ther. 1994;63:37–122. doi: 10.1016/0163-7258(94)90055-8. [DOI] [PubMed] [Google Scholar]
- Hazra TK, Das A, Das S, Choudhury S, Kow YW, Roy R. Oxidative DNA damage repair in mammalian cells: a new perspective. DNA Repair. 2007;6:470–480. doi: 10.1016/j.dnarep.2006.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Izumi T, Wiederhold LR, Roy G, Roy R, Jaiswal A, Bhakat KK, Mitra S, Hazra TK. Mammalian DNA base excision repair proteins: their interactions and role in repair of oxidative DNA damage. Toxicology. 2003;193:43–65. doi: 10.1016/s0300-483x(03)00289-0. [DOI] [PubMed] [Google Scholar]
- Kelley MR, Parsons SH. Redox regulation of the DNA repair function of the human AP endonuclease Ape1/ref-1. Antioxid. Redox Signal. 2001;3:671–683. doi: 10.1089/15230860152543014. [DOI] [PubMed] [Google Scholar]
- Khan MF, Gu Y, Alock NW, Boor PJ, Ansari GAS. Oxidative stress in splenotoxicity of aniline. Fundam. Appl. Toxicol. 1997;35:22–30. doi: 10.1006/faat.1996.2259. [DOI] [PubMed] [Google Scholar]
- Khan MF, Kannan S, Wang J. Activation of transcription factor AP-1 and mitogen-activated protein kinases in aniline-induced splenic toxicity. Toxicol. Appl. Pharmacol. 2006;210:86–93. doi: 10.1016/j.taap.2005.08.006. [DOI] [PubMed] [Google Scholar]
- Khan MF, Kaphalia BS, Boor PJ, Ansari GAS. Subchronic toxicity of aniline hydrochloride in rats. Arch. Environ. Contant. Toxicol. 1993;24:368–374. doi: 10.1007/BF01128736. [DOI] [PubMed] [Google Scholar]
- Khan MF, Wu X, Ansari GA, Boor PJ. Malondialdehyde-protein adducts in the spleens of aniline-treated rats: immunochemical detection and localization. J. Toxicol. Environ. Health, Part A. 2003b;66:93–102. doi: 10.1080/15287390306464. [DOI] [PubMed] [Google Scholar]
- Khan MF, Wu X, Boor PJ, Ansari GAS. Oxidative modification of proteins and lipids in aniline-induced splenic toxicity. Toxicol. Sci. 1999;48:134–140. doi: 10.1093/toxsci/48.1.134. [DOI] [PubMed] [Google Scholar]
- Khan MF, Wu X, Kaphalia BS, Boor PJ, Ansari GA. Nitrotyrosine formation in splenic toxicity of aniline. Toxicology. 2003a;194:95–102. doi: 10.1016/j.tox.2003.08.008. [DOI] [PubMed] [Google Scholar]
- Liu M, Bandaru V, Bond JP, Jaruga P, Zhao X, Christov PP, Burrows CJ, Rizzo CJ, Dizdaroglu M, Wallace SS. The mouse ortholog of NEIL3 is a functional DNA glycosylase in vitro and in vivo. Proc. Natl. Acad. Sci. USA. 2010;107:4925–4930. doi: 10.1073/pnas.0908307107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wang J, Abdel-Rahman S, Boor PJ, Khan MF. Oxidative DNA damage and its repair in rat spleen following subchronic exposure to aniline. Toxicol. Appl. Pharmacol. 2008;233:247–253. doi: 10.1016/j.taap.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma H, Wang J, Abdel-Rahman SZ, Hazra TK, Boor PJ, Khan MF. Induction of NEIL1 and NEIL2 DNA glycosylases in aniline-induced splenic toxicity. Toxicol. Appl. Pharmacol. 2011;251:1–7. doi: 10.1016/j.taap.2010.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcon G, Tell G, Perrone L, Garbelli R, Quadrifoglio F, Tagliavini F, Giaccone G. APE1/Ref-1 in Alzheimer's disease: an immunohistochemical study. Neurosci. Lett. 2009;466:124–127. doi: 10.1016/j.neulet.2009.09.039. [DOI] [PubMed] [Google Scholar]
- Marenstein DR, Chan MK, Altamirano A, Basu AK, Boorstein RJ, Cunningham RP, Teebor GW. Substrate specificity of human endonuclease III (hNTH1). Effect of human APE1 on hNTH1 activity. J Biol. Chem. 2003;278:9005–9012. doi: 10.1074/jbc.M212168200. [DOI] [PubMed] [Google Scholar]
- Mori H, Ouchida R, Hijikata A, Kitamura H, Ohara O, Li Y, Gao X, Yasui A, Lloyd RS, Wang JY. Deficiency of the oxidative damage-specific DNA glycosylase NEIL1 leads to reduced germinal center B cell expansion. DNA Repair. 2009;8:1328–1332. doi: 10.1016/j.dnarep.2009.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowsheen S, Wukovich RL, Aziz K, Kalogerinis PT, Richardson CC, Panayiotidis MI, Bonner WM, Sedelnikova OA, Georgakilas AG. Accumulation of oxidatively induced clustered DNA lesions in human tumor tissues. Mutat. Res. 2009;674:131–136. doi: 10.1016/j.mrgentox.2008.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Redrejo-Rodríguez M, Saint-Pierre C, Couve S, Mazouzi A, Ishchenko AA, Gasparutto D, Saparbaev M. New insights in the removal of the hydantoins, oxidation product of pyrimidines, via the base excision and nucleotide incision repair pathways. PLoS One. 2011;6:e21039. doi: 10.1371/journal.pone.0021039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolseth V, Rundén-Pran E, Luna L, McMurray C, Bjørås M, Ottersen OP. Widespread distribution of DNA glycosylases removing oxidative DNA lesions in human and rodent brains. DNA Repair. 2008;7:1578–1588. doi: 10.1016/j.dnarep.2008.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saha T, Rih JK, Roy R, Ballal R, Rosen EM. Transcriptional regulation of the base excision repair pathway by BRCA1. J Biol. Chem. 2010;285:19092–19105. doi: 10.1074/jbc.M110.104430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takao M, Kanno S, Shiromoto T, Hasegawa R, Ide H, Ikeda S, Sarker AH, Seki S, Xing JZ, Le XC, Weinfeld M, Kobayashi K, Miyazaki J, Muijtjens M, Hoeijmakers JH, van der Horst G, Yasui A. Novel nuclear and mitochondrial glycosylases revealed by disruption of the mouse Nth1 gene encoding an endonuclease III homolog for repair of thymine glycols. EMBO J. 2002;21:3486–3493. doi: 10.1093/emboj/cdf350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tell G, Damante G, Caldwell D, Kelley MR. The intracellular localization of APE1/Ref-1: more than a passive phenomenon? Antioxid. Redox Signal. 2005;7:367–384. doi: 10.1089/ars.2005.7.367. [DOI] [PubMed] [Google Scholar]
- Trzeciak AR, Nyaga SG, Jaruga P, Lohani A, Dizdaroglu M, Evans MK. Cellular repair of oxidatively induced DNA base lesions is defective in prostate cancer cell lines, PC-3 and DU-145. Carcinogenesis. 2004;25:1359–1370. doi: 10.1093/carcin/bgh144. [DOI] [PubMed] [Google Scholar]
- Wang J, Kannan S, Li H, Khan MF. Cytokine gene expression and activation of NF-kappa B in aniline-induced splenic toxicity. Toxicol. Appl. Pharmacol. 2005;203:36–44. doi: 10.1016/j.taap.2004.07.012. [DOI] [PubMed] [Google Scholar]
- Wang J, Wang G, Ansari GA, Khan MF. Activation of oxidative stress-responsive signaling pathways in early splenotoxic response of aniline. Toxicol. Appl. Pharmacol. 2008;230:227–234. doi: 10.1016/j.taap.2008.02.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weinberger MA, Albert RH, Montgomery SB. Splenotoxicity associated with splenic sarcomas in rats fed high doses of DRC Red No. 9 or aniline hydrochloride. J. Natl. Cancer Inst. 1985;75:681–690. [PubMed] [Google Scholar]
- Wu X, Kannan S, Ramanujam VM, Khan MF. Iron release and oxidative DNA damage in splenic toxicity of aniline. J. Toxicol. Environ. Health, Part A. 2005;68:657–666. doi: 10.1080/15287390590921757. [DOI] [PubMed] [Google Scholar]
- Yndestad A, Neurauter CG, Oie E, Forstrøm RJ, Vinge LE, Eide L, Luna L, Aukrust P, Bjørås M. Up-regulation of myocardial DNA base excision repair activities in experimental heart failure. Mut. Res. 2009;666:32–38. doi: 10.1016/j.mrfmmm.2009.03.008. [DOI] [PubMed] [Google Scholar]
- Yuk JM, Yang CS, Shin DM, Kim KK, Lee SK, Song YJ, Lee HM, Cho CH, Jeon BH, Jo EK. A dual regulatory role of apurinic/apyrimidinic endonuclease 1/redox factor-1 in HMGB1-induced inflammatory responses. Antioxid. Redox Signal. 2009;11:575–588. doi: 10.1089/ars.2008.2196. [DOI] [PubMed] [Google Scholar]
- Zhang Y, Wang J, Xiang D, Wang D, Xin X. Alterations in the expression of the apurinic/apyrimidinic endonuclease-1/redox factor-1 (APE1/Ref-1) in human ovarian cancer and identification of the therapeutic potential of APE1/Ref-1 inhibitor. Int. J. Oncol. 2009;35:1069–1079. [PubMed] [Google Scholar]
- Zhao J, Gao F, Zhang Y, Wei K, Liu Y, Deng X. Bcl2 inhibits a basic site repair by down-regulating APE1 endonuclease activity. J Biol. Chem. 2008;283:9925–9932. doi: 10.1074/jbc.M708345200. [DOI] [PMC free article] [PubMed] [Google Scholar]