Abstract
In utero priming to malaria antigens renders cord blood mononuclear cells (CBMC) more susceptible to productive HIV infection in vitro in the absence of exogenous stimulation. This provides a unique model to better understand mechanisms affecting lymphocyte susceptibility to HIV infection in vivo. Effector memory CD3+CD4+ T cells (TEM) were the exclusive initial targets of HIV with rapid spread to central memory cells. HIV susceptibility correlated with increased expression of CD25 and HLA-DR on TEM. Virus entered all samples equally, however gag/pol RNA was only detected in HIV susceptible samples, suggesting regulation of proviral gene transcription. Targeted analysis of human genes in memory T cells showed greater expression of IFNG, NFATc1, IRF1, FOS, and PPIA and decreased expression YY1 and TFCP2 in HIV susceptible samples. Thus fetal priming to exogenous antigens enhances specific proviral gene transcription pathways in effector memory cells that may increase risk of vertical transmission of HIV.
Keywords: Human immunodeficiency virus, HIV replication in cord blood memory T cells, effector memory T cells, susceptibility to HIV
Introduction
Mother-to-child transmission (MTCT) of the virus remains an important global public health concern despite advances in targeted antiretroviral therapy among HIV infected pregnant women and newborns (UNAIDS, 2010). Maternal coinfections with pathogens such as malaria, tuberculosis and helminthes may influence the risk of transmitting the virus to offspring and contribute to high rates of MTCT in many parts of the world (Ayisi et al., 2004; Brahmbhatt et al., 2003; Gallagher et al., 2005; Gupta et al., 2011; Inion et al., 2003; Steiner et al., 2010; van Eijk et al., 2003). These coinfections may affect the risk of MTCT of HIV by several mechanisms including increased viral loads in pregnancy, formation of genital lesions and/or by activation of fetal lymphocytes. We have previously shown that cord blood mononuclear cells (CBMC) collected from newborns having evidence of in utero exposure and immune activation to malaria demonstrated increased in vitro susceptibility to HIVBaL infection (Steiner et al., 2010). In this in vitro HIV infection model, HIVBaL infected 50% of malaria sensitized CBMC without exogenous activation.
HIV infection in neonates and infants differs from adults. Children infected early in life often exhibit higher levels of viremia, more rapid progression to AIDS, and increased frequency of opportunitistic infections (Mawhinney and Pagano, 1994; Resino et al., 2000; Tovo et al., 1992). The mechanisms underlying this differential disease course remain unclear. A contributing factor may be a higher total number of circulating T lymphocytes, predominantly naïve CD3+CD4+ cells, undergoing homeostatic proliferation to fill different lymphocyte compartments (Clement, 1992). Several studies have demonstrated increased susceptibility to HIV in CBMC compared to adult peripheral blood mononuclear cells (PBMC) (Ahmad et al., 2011; Sundaravaradan et al., 2006). Both naïve (CD45RA+) and memory (CD45RO+) CD3+CD4+ cells isolated from CBMC showed increased capacity to sustain viral gene transcription and replication compared to adult PBMC (Ahmad et al., 2011). This increased susceptibility correlated with differential mRNA expression of a variety of host genes associated with the HIV viral lifecycle including upregulation of several STAT family signal transducers, transcription factors such as NF-κB, E2F, and HAT, and several matrix metalloproteinases while the repressive transcription factor YY1 was downregulated (Sundaravaradan et al., 2010). A significant limitation to these in vitro studies is the necessity of mitogenic activation of CBMC prior to virus exposure to allow HIV infection. This stimulation likely alters target cell phenotype, gene expression profiles, and HIV susceptibility of CBMC.
Our finding that in utero activation of fetal lymphocytes to malaria blood stage antigens renders CBMC susceptible to HIV infection in vitro in the absence of mitogenic stimulation enables us to characterize the targets and mechanisms that may affect lymphocyte susceptibility to HIV infection in vivo. We demonstrate that CD3+CD4+ cells are the sole targets of HIV in our system and show that only CD45RO+ memory cells are productively infected. Further, by utilizing the protease inhibitor saquinavir, we demonstrate that CD45RO+CD27− effector memory cells are the primary initial targets of HIVBaL with rapid spread to other memory cells including those of the CD45RO+CD27+ central memory phenotype. We show that differences in activation marker expression and gene expression profiles within the memory cell compartment correlate with susceptibility to HIVBaL infection and provide evidence that viral entry is not the limiting step in establishing productive HIV infection in CBMC.
Results
HIVBaL enters only CD3+CD4+ T cells
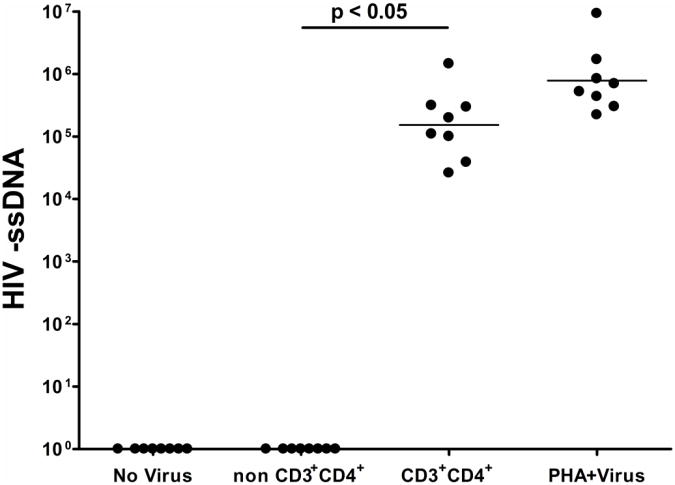
We have previously demonstrated that fifty percent of CBMC samples from infants demonstrating evidence of prenatal immune sensitization to malaria antigens were capable of sustaining viral replication detected by reverse transcriptase activity assay at day fifteen following virus exposure in the absence of any exogenous mitogenic stimulation (Steiner et al., 2010). These samples capable of sustaining viral replication have been termed “HIV susceptible” while those in which HIV replication was not detected have been termed “HIV not susceptible.” Further, we have previously shown that CD3+CD4+ T cells from HIV susceptible samples represent the primary reservoir of replicating virus at day four post exposure to HIVBaL as determined by flow cytometric detection of p24 (Steiner et al., 2010). In this model it is possible that HIVBaL could infect non-CD3+CD4+ T cells but not lead to sufficient viral replication to be detectable by flow cytometric methods. To determine whether HIVBaL invades CD3+CD4+ T cells only or also other cell types, frozen aliquots of CBMC from HIV susceptible samples were thawed and exposed to HIVBaL at the same multiplicity of infection (MOI) as previously described (Steiner et al., 2010). The CD3+CD4+ and non- CD3+CD4+ cellular subsets were subsequently isolated using magnetic bead separation (Miltenyi) twenty-four hours after virus exposure. Virus entered and initiated reverse transcription only in CD3+CD4+ T cells based on the detection of HIV minus strand strong stop DNA (HIV - ssDNA) in this cellular compartment and its absence in the non- CD3+CD4+ compartment in all samples tested (Figure 1). HIVBaL entered unstimulated CD3+CD4+ T cells similarly compared to total CBMC that had been cultured in the presence of PHA for three days prior to virus exposure, providing further evidence that virus preferentially enters CD3+CD4+ T cells in this in vitro model system.
Figure 1. HIVBaL minus strand strong-stop DNA is detectable in CD3+CD4+ cells and absent in non CD3+CD4+ cells twenty-four hours post virus exposure.
HIV –ssDNA normalized to β-globin. Specific conditions indicated in the figure: i) CBMC cultured without virus, ii) Non CD3+CD4+ and iii) CD3+CD4+ conditions represent cell fractions following magnetic bead separation twenty-four hours after whole CBMC were exposed to virus, iv) PHA+Virus condition represents whole CBMC cultured in the presence of PHA for three days prior to virus exposure. n= 8 HIV susceptible samples. Lines represent median for each condition.
CD3+CD4+CD45RO+CD27- effector memory cells are the initial target of infection
In order to further specify the initial cellular targets of HIVBaL infection in our system, flow cytometric phenotyping of p24 expressing cells was performed four days after infection. Cell surface markers CD45RO and CD27 were used to divide the CD3+CD4+ T cell population into naïve (CD45RO−CD27+), central memory (CD45RO+CD27+, TCM) and effector memory (CD45RO+CD27−, TEM) subsets, as previously described (Sallusto et al., 1999; Ferrando-Martinez et al., 2010; Yonkers et al., 2011). Though the naïve subset consistently represented greater than 92% of CD3+CD4+ T cells, HIV p24 was not detectable in this compartment (Table 1). In contrast, p24 was detectable in both the central memory (median of 4.7% of CD45RO+CD27+ cells, Table 1) and effector memory (median of 10.6% of CD45RO+CD27- cells). Therefore, both central and effector memory CD3+CD4+ cells are capable of sustaining HIVBaL replication at day four in our model system, but naïve CD4+ T cells are not (Table 1).
Table I.
Frequency of CD3+CD4+ T lymphocytes expressing p24 at 96 hours post HIVBaL exposure and in the presence or absence of protease inhibitor saquinavir.
| HIV susceptible samples | p24 expression in CD45RO−CD27+ (naïve) | p24 expression in CD45RO+CD27+ (TCMa) | p24 expression in CD45RO+CD27− (TEMb) | ||
|---|---|---|---|---|---|
| − saquinavir | + saquinavir | − saquinavir | + saquinavir | ||
| 1 | 0 | 3.2 | 0 | 13.6 | 6.7 |
| 2 | 0 | 4.7 | 0 | 8.6 | 5.3 |
| 3 | 0 | 2.5 | 0 | 11.9 | 4.2 |
| 4 | 0 | 4.8 | 0 | 10.6 | 6.8 |
| 5 | 0 | 5.1 | 0 | 9.0 | 3.4 |
central memory
effector memory
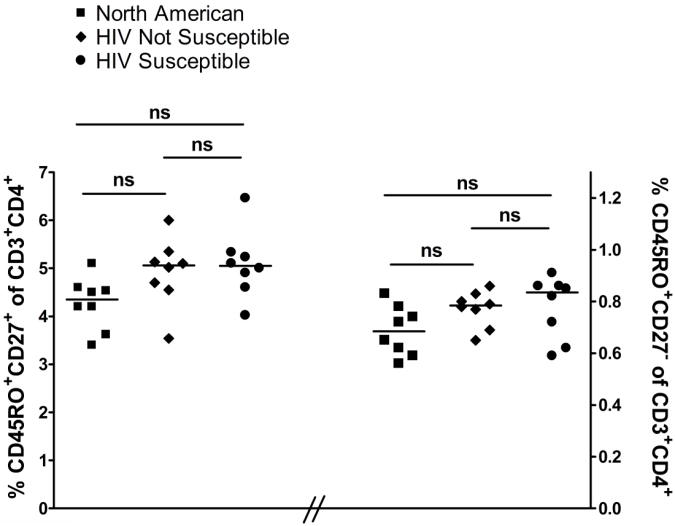
In order to determine which memory CD3+CD4+ T cell subset was the initial target of infection, experiments were performed in which the protease inhibitor saquinavir was present throughout the four days of cell culture. Saquinavir does not interfere with first round viral infection events detectable by p24 expression, but prevents subsequent spreading and second round infection. In the presence of saquinavir, p24 expression in the CD45RO+CD27+ central memory compartment was never detected while the frequency of p24 expressing CD45RO+CD27− effector memory cells was half of that detected in the absence of saquinavir (Table 1). There was no difference in the frequency of TCM and TEM among CBMC of HIV susceptible, HIV not susceptible, and North American subjects (Figure 2), excluding the possibility that differences in frequency of these memory cell subsets could account for differences in susceptibility to HIVBaL infection. Thus, TEM appear to be the primary initial targets of HIVBaL infection with subsequent spread to both TCM and additional TEM.
Figure 2. Frequency of central memory or effector memory CD4+ cell subsets does not significantly vary between HIV susceptible, HIV not susceptible, and North American samples.
Frequency of CD45RO+CD27+ (central memory) and CD45RO+CD27− (effector memory) cells were determined after gating on viable CD3+CD4+ lymphocytes. Lines represent median values. N=8 for each group.
Expression of activation markers on memory cells correlates with HIV susceptibility
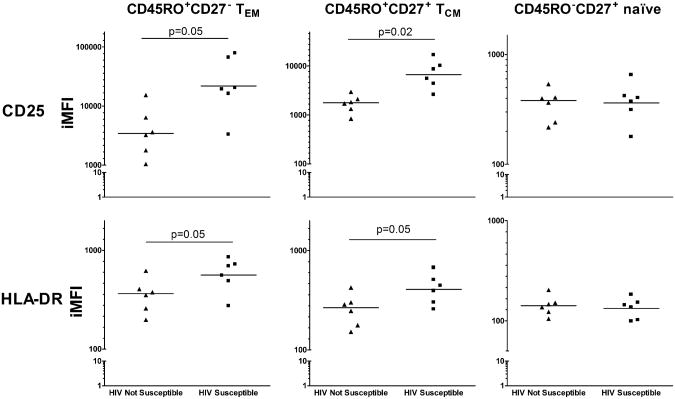
We next evaluated whether TCM and TEM from HIV susceptible samples showed increased activation as measured by CD25 and HLA-DR expression compared to TCM and TEM HIV not susceptible samples. Frequency and expression of CD25 were increased on both CD45RO+CD27− TEM (IFMI= 21,843 versus 3,600, p=0.05) and CD45RO+CD27+ TCM (IMFI=6,715 versus 1,676, p= 0.02) from HIV susceptible compared to HIV not susceptible samples respectively (Figure 3). Similarly, HIV susceptible samples demonstrated higher frequency and expression of HLA-DR for both TEM (IMFI=563 versus 346 for TEM, p= 0.05) and TCM (IMFI=408 versus 250 for TCM, p= 0.05). CCR5 expression did not differ between HIV susceptible and not susceptible samples for either TEM (IMFI=1686 versus 1583, p=.14) or TCM (IMFI= 875 versus 824, p=0.62). Similarly, expression of CD4 did not differ between HIV susceptible and not susceptible samples for either CD3+CD4+ TEM (geoMFI= 1834 versus 1766, p=0.63) or CD3+CD4+ TCM (1820 versus 1859, p=0.78). CD25 and HLA-DR expression were similar for CD45RO−CD27+ naïve cells from HIV susceptible versus HIV not susceptible samples.
Figure 3. Expression of activation markers on TCM and TEM cells correlates with HIV susceptibility.
Integrated mean fluorescent indices (iMFI) for CD25 (top) and HLA-DR (bottom) gated on viable CD3+CD4+ lymphocytes and appropriate CD45RO and CD27 expression. N=6 for each group. Lines indicate geometric mean of iMFI. Statistics calculated by t test.
HIV entry does not account for differences in HIV susceptibility
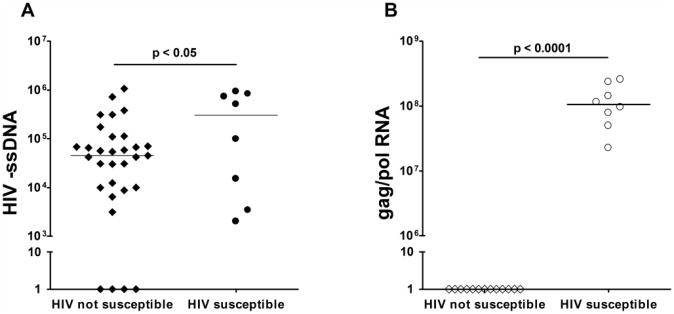
We sought to determine whether differences in susceptibility to HIV infection were reflected in the ability of HIV to enter cells or a later viral life cycle event. HIV susceptible or HIV not susceptible CBMC were thawed and exposed to virus as described above. DNA and RNA were isolated from total CBMC twenty-four hours post infection and both qPCR for minus strand strong-stop DNA (-ssDNA) and RTq-PCR for gag/pol gene expression was performed. Although HIV –ssDNA was detected in all HIV susceptible samples, HIV –ssDNA was also amplified in 27 of 31 (87%) HIV not susceptible samples with the range of detectable expression levels similar to that of the HIV susceptible group (3100-1.07×106 vs. 2000-9.28×105, respectively, Figure 4). In contrast, gag/pol expression was detected in all HIV susceptible samples analyzed, but in none of the HIV not susceptible samples. These results suggest that while HIV is capable of entering CBMC from Kenyan samples to a similar degree in vitro, only in samples that are susceptible to productive infection does the viral life cycle progress to proviral gene transcription.
Figure 4. Detection of minus strand strong-stop DNA is similar between HIV susceptible and HIV not susceptible CBMC twenty-four hours post virus exposure, but gag/pol RNA is only detected in HIV susceptible CBMC.
A) HIV –ssDNA normalized to β-globin. Sample sizes: HIV not susceptible, n=31; HIV susceptible, n=8. B) gag/pol RNA normalized to R18. Sample sizes: HIV not susceptible, n=13; HIV susceptible, n=8. Lines indicate median.
A distinct set of genes relevant to proviral gene transcription are differentially expressed in memory T cells and correlate with HIV susceptibility in vitro
To further address possible differences between HIV susceptible and not susceptible memory T cells, gene expression levels of a subset of host genes relevant to HIV infection were assessed using the HIV Infection and Host Response PCR array (SA Biosciences/Qiagen). This array analyzes the expression of eighty-four host genes selected for their described role in HIV viral entry, reverse transcription, integration, proviral gene transcription, and virion assembly as well as host immune responses to infection. This targeted array system was chosen because we wished to specifically determine expression profiles in the CD4+CD45RO+ memory subset and limited cell numbers precluded whole genome analysis.
Of the eighty-four genes analyzed, eight demonstrated greater than two-fold differences in expression between at least two of the comparison groups (Figure 5) while seventy-six genes showed no difference between groups (Supplemental table). Notably, gene expression of proinflammatory cytokines TNF and IFNγ were elevated in HIV susceptible samples along with transcription factors and regulators NFATc1, IRF1, FOS and the cellular cofactor PPIA compared to HIV not susceptible and North American CBMC (Fig. 5 A&C). In contrast, the expression of YY1 and TFCP2, two transcription factors that have been associated with repression of HIV viral replication, were 9-fold lower in HIV susceptible memory T cells compared to both HIV not susceptible and North American memory T cells. There was little difference in gene expression between HIV not susceptible and North American CBMC (Fig. 5B). These results show HIV susceptible memory T cells express greater levels of specific genes that facilitate HIV infection and replication, but most notably have impaired expression of suppressor genes for the viral life cycle.
Figure 5. Differential expression of host genes in total memory CD3+CD4+ cells from HIV susceptible, HIV not susceptible and North American CBMC.
A: HIV susceptible vs North American. B: HIV not susceptible vs North American. C: HIV susceptible vs HIV not susceptible. N=4 for all groups. * p=0.07-0.14, ** p=0.05.
Discussion
Our previous study established that CBMC from fifty percent of malaria sensitized Kenyan infants were capable of being productively infected with HIV in vitro without prior exogenous stimulation to provide a relevant model that may better recapitulate host susceptibility factors to HIV in vivo (Steiner et al., 2010). Here we extend these observations to demonstrate that CD3+CD4+ cells are the only cellular compartment in which HIVBaL enters and initiates reverse transcription. Further, while both central (TCM) and effector (TEM) memory T cells are capable of sustaining detectable viral replication following virus exposure, in this in vitro model of CBMC infection, naïve CD4+ T cells are not. By utilizing a protease inhibitor to pharmacologically prevent second-round infection, we were able to demonstrate that the initial cellular target of HIV in our model system is the TEM with rapid spread to TCM. Both TEM and TCM from HIV susceptible CBMC demonstrated evidence of increased cellular activation as demonstrated by increased frequency and expression of surface CD25 and HLA-DR compared to HIV not susceptible samples. Analysis of gene expression profiles for eighty-four host genes relevant to HIV infection within the CD4+CD45RO+ total memory subset revealed enhanced gene expression of cytokines TNF and IFNγ, transcription factors and regulators NFATc1, IRF1, and FOS, as well as PPIA while expression of YY1 and TFCP2 were markedly decreased in HIV susceptible samples. Interestingly, no difference was observed in HIV –ssDNA expression between HIV susceptible and HIV not susceptible CBMC, suggesting that cellular activation is not necessary for HIV entry. In contrast, detection of HIV gag/pol RNA was exclusively detected in HIV susceptible CBMC. Together, these findings support a model in which HIV binding and invasion of CBMC is not the primary difference between HIV susceptible and not susceptible CBMC. Rather, differential cellular activation and expression of host genes that influence HIV gene transcription enhances viral replication and spread within memory CD4+CD45RO+ T cells, initially in TEM and subsequently in TCM.
Other studies have examined mechanisms of susceptibility of CBMC to HIV in infection in vitro. These studies have only compared CBMC with adult peripheral blood mononuclear cells (PBMC) to susceptibility to HIV in vitro (Ahmad et al., 2011; Sundaravaradan et al., 2006; Sundaravaradan et al., 2010). They have also required pre-activation to allow HIV infection in vitro. Interestingly, these studies demonstrated enhanced HIV replication in monocytes/macrophages and total T lymphocytes, as well as more specifically in both CD4+CD45RA+ naïve T cell and CD4+CD45RO+ memory T cell subsets from CBMC compared to PBMC (Ahmad et al., 2011; Sundaravaradan et al., 2006). This is in contrast to our observations in which HIV –ssDNA is only detected in CD3+CD4+ T cells twenty-four hours after virus exposure and further only CD4+CD45RO+ memory cells exhibit productive viral replication as determined by flow cytometic detection of p24. The required prestimulation may have artificially enhanced viral replication in cells that may not have been susceptible in vivo, thereby explaining the more restricted profile of HIV susceptible cells in our model system. These studies also used primary HIV isolates, R5 and X4 HIV strains to infect. In support of our observations, previous studies have demonstrated both predominant HIV infection of CD4+CD45RO+ T cells in infants as well as preferential replication of HIV in the memory T cell subset (Schnittman et al., 1990; Sleasman et al., 1996; Spina et al., 1997).
We demonstrate that TEM are the primary initial target of infection in our model system. This observation is consistent with several other studies that have shown initial HIV infection of TEM in diverse settings such as cervico-vaginal tissue, dendritic cell mediated transmission, and a humanized mouse model system (Groot et al., 2006; Nie et al., 2009; Saba et al., 2010). Preferential depletion of the TEM compartment is a characteristic feature of early infection in both human HIV and SIV model systems, particularly in mucosal sites such as the gut associated lymphoid tissue (Brenchley, 2004; Douek et al., 2003; Mehandru, 2004). The preference of HIV infection for TEM compared to TCM may be related to the lower activation threshold of TEM and their presence in a variety of peripheral tissues compared to restriction of TCM in lymph nodes (Sallusto, et al 1999). T cell activation is determined by the amount and duration of antigen exposure as well as the cytokine milieu. In cord blood antigen concentrations are likely to be low thus preferentially activating and infecting TEM. Importantly, while CD4+CD45RO−CD27+ naïve cells are not an important source of replicating virus in our model system, this does not preclude infection of naïve T cells as HIV infection progresses and spreads. Indeed, HIV proviral transcripts may be detected in naïve as well as TCM and TEM cells from patients with established HIV infection (Ostrowski et al., 1999).
It has been hypothesized that effector memory CD4+ T cells are more easily infected due to increased cell surface expression of CCR5, the primary co-receptor mediating infection of R5 type HIV (Bleul et al., 1997; Groot et al., 2006). Interestingly, we did not observe any difference in surface CCR5 or CD4 expression on memory T cells from HIV susceptible compared to not susceptible samples. We did, however, observe increased expression of the cell surface activation markers CD25 and HLA-DR on both TEM and TCM from HIV susceptible samples. Further, total memory cells from HIV susceptible samples had increased mRNA expression of genes associated with cellular activation. These included TNF, which enhances HIV viral replication through downstream effects of NF-κB, and IFNγ, which has been shown to possess both inhibitory and activating effects on HIV replication (Kedzierska and Crowe, 2001; Kedzierska et al., 2003; Osborn et al., 1989). Transcription regulators FOS, NFATc1, and IRF1 were also upregulated in memory T cells from HIV susceptible samples. FOS, a component of the transcription factor AP-1, and NFATc1 have been described to enhance HIV proviral gene transcription directly by binding to sites in both the HIV long terminal repeat (LTR) region and intragenic regions as well as by synergizing with NF-κB pathways (Colin et al., 2011; Kinoshita et al., 1997; Kinoshita et al., 1998; Roebuck et al., 1993; Roebuck et al., 1996; Romanchikova et al., 2003). IRF1 binds directly to target sites within the HIV sequence and forms a functional complex with NF-κB at the LTR kappa sites thereby enhancing the viral gene transcription and viral replication (Battistini et al., 2002; Marsili et al., 2003; Sgarbanti et al., 2008). Interestingly, the expression of transcriptional repressors YY1 and TFCP2 were both dramatically decreased in CD4+CD45RO+ memory CBMC from HIV susceptible samples compared to HIV not susceptible and North American samples. TFCP2 has been demonstrated to recruit YY1 to the HIV LTR where they cooperatively repress proviral gene transcription by the recruitment of histone deacetylase to the viral promoter region (Coull et al., 2000; He and Margolis, 2002). Recently decreased YY1 expression has been observed and correlated with increased HIV susceptibility in CBMC compared to adult PBMC, further supporting a potentially important role of these repressive transcription factors in modulating viral replication in CBMC (Sundaravaradan et al., 2010).
An important limitation to the current study is the exclusive use of a lab-adapted R5 HIV isolate, BaL, for infection experiments because of limited availability of CBMC from offspring of malaria exposed women. It is possible that primary isolates or X4 or X4/R5 HIV strains may have different primary targets of infection. Our rationale for using BaL are prior observations that R5 strains often represent the dominant viral population detected during the early stages of clinical HIV-1 infection whereas X4 strains are typically detected in later stages of infection (De Jong, et al, 1992).
In conclusion, this study demonstrates that CD45RO+CD27− effector memory cells are initial targets of HIVBaL in this in vitro model system with rapid spread to other memory cells including those of the CD45RO+CD27+ central memory phenotype. HIV susceptibility was further correlated with increased immune activation as measured by both cell surface activation marker expression and differing gene expression profiles within the CD4+CD45RO+ compartment. While this study does not specifically address the cause of this observed increase in immune activation in HIV susceptible samples, we have previously demonstrated that CBMC from infants primed in utero to malaria antigens were more susceptible to in vitro HIV infection (Steiner et al., 2010). This model system is unique in that we observe differences between CBMC samples in susceptibility to HIV at low MOI and in the absence of prestimulation, which may allow further studies to reveal novel pathways mediating susceptibility to HIV infection.
Materials and Methods
Study population
Characteristics of study population and CBMC sample collection and handling were performed as described previously (Steiner et al., 2010). Briefly, samples were obtained from newborns born at the Msambweni District Hospital, Coast Province, Kenya an area endemic for malaria, schistosomiasis, lymphatic filariasis and intestinal helminthes (Malhotra et al., 1997; Malhotra et al., 2009). All women were tested for HIV with the offspring of HIV positive women excluded from the current study. Additional CBMC were prepared from healthy North American newborns delivered at University Hospitals of Cleveland (Cleveland, OH) following the same procedures. Ethical approval was obtained from the Institutional Review Boards of University Hospitals of Cleveland and the Kenya Medical Research Institute in Nairobi and informed consent for the study was obtained from study subjects.
In vitro HIV infection
HIV-1 Bal strain was initially obtained from NIH AIDS reagent repository and a large amount virus propagated in CD4-transduced and CCR5 expressing human glioma cell line U87/CD4-CCR5 (kindly provided by Dr. E. J. Arts, Case Western Reserve University) and cryopreserved. An aliquot from a secondary passage from the Arts lab was again amplified in U87/CD4-CCR5 and a 10 ml culture supernatant were removed every three days, and frozen in separate aliquots. RT assay was performed to determine which aliquots demonstrated the highest level of viable virus. These aliquots were pooled. TCID50 was determined for this pool and small aliquots used for all subsequent experiments.
Ficoll-Hypaque density gradient separation was used to isolate CBMC which were promptly cyropreserved. Cryopreserved CBMC were thawed (all samples showed >85% viable cell recovery with preservation of immune function as indicated by similar recall responses to malaria antigens between fresh and cryopreserved aliquots from the same cord blood sample) and rested overnight in RPMI-1640 + L-glutamine supplemented with penicillin/streptomycin (100 units/100mg/ml), HEPES (10 mM), and 10% pooled human AB serum (cRPMI+10%). From this point forward, culture media consisted of cRPMI+10% supplemented with 1.0 ng/ml recombinant human IL-2 (rhIL-2, BD Biosciences). Infection with the R5 tropic virus HIVBaL (kindly provided by Dr. Eric Arts, Case/UHC Center for AIDS Research) was performed at an experimentally determined optimal multiplicity of infection (MOI) of 0.001 for four hours at 37°C after which cells were centrifuged, washed with fresh media and resuspended according to appropriate experimental culture conditions (Steiner et al., 2010). The protease inhibitor saquinavir (5 ng/ml) was added to cultures following resuspension as appropriate.
Isolation of CD3+CD4+ cell subset
Twenty-four hours after virus exposure, the CD3+CD4+ cell subset was separated from the total cultured CBMC using the CD4+ T Cell Isolation Kit II (Miltenyi Biotech) following manufacturer's instructions. Both the flow through (CD3+CD4+) and column-bound (non CD3+CD4+) cells were collected. Flow cytometric analysis consistently demonstrated greater than 95% purity of the CD3+CD4+ fraction (data not shown).
Nucleic Acid Extraction
Twenty-four hours after virus exposure, cells were centrifuged and culture supernatant removed. Cells were harvested using TRI Reagent (Molecular Research Center, Inc., Cincinnati, OH) to allow efficient extraction of both RNA and DNA performed according to the manufacturer's recommended protocol. RNA and DNA concentrations were determined spectrophotometrically with dilution in Phosphate Buffer (Molecular Research Center, Inc., Cincinnati, OH).
Minus strand strong stop DNA and gag/pol RNA detection by RTQ-PCR
Quantification of HIVBaL minus-strand strong stop DNA (-ssDNA) was performed using real time quantitative PCR methods. Primers and probes were as follows: for -ssDNA [probe (6FAM-AACAGACGGG CACACACTACTTGAAGCA-TAMRA) and primers (5′-GCCTCAATAAA- GCTTGCCTGA-3′, and 5′-CTGAGGGATCTCTAGTTACCAGAGTCA-3′)], and for β-globin DNA [probe (6FAM-CACCTTTGCTACACTGAGTGACCTGCACTG-TAMRA), and primers (5′-GTTATGCTCACGGATGACCTCAA-3′ and 5′-GGTCCACGTGCAGCTTGTT-3′)]. Quantities of DNA was determined by serial dilution of cloned target cDNA in each assay and normalized to the copy number of β-globin. Quantification of HIV gag/pol RNA expression was performed using real time quantitative PCR methods. Primers were as follows: for HIV gag/pol [(5′-CATGTTTTCAGCATTATCAGAAGGA-3′) and (5′-CCACTGTGTTTAGCATGGTGTTTAA-3′)] and for the R18 RNA polymerase ribozyme [(5′-CGCCGCTAGAGGTGAAATTC-3′) and (5′-CATTCTTGGCAAATGCTTTCG-3′). SYBERGreen (Applied Biosystems) was used for detection of PCR product. Results were normalized to the copy number of the R18 . The real-time PCR quantification was performed using Applied Biosystems 7300 Real Time PCR System.
Flow Analysis
One million cells were washed and stained with fluorochrome labeled antibodies for 30 minutes in the dark at 4°C. For analysis of HIV infection the following antibodies were used: CD3-APC-eFluor780, CD4-PE, CD45RO-APC, CD27-PE-Cy7 (ebiosciences), and p24-FITC (Beckman Coulter, gift of Dr. Helene Bernstein). For analysis of activation markers on isolated total CD3+CD4+ memory cells, the following antibodies were used: CD4-PE, CD45RO-APC, CD27-FITC, CD25- APC-eFluor780, HLA-DR-PE-Cy7(ebiosciences); or CD4-Alexa700, CD45RO-APC, CD27-FITC, CCR5-PE (ebiosciences); Samples were processed (50,000 events) on a BD LSRII flow cytometer and data analyzed with FlowJo software (Tree Star, Inc). Gating was determined by fluorescence minus one (FMO) staining and compensation calculated using Comp Beads (BD Biosciences) and compensation platform in FlowJo. Frequency was determined as percentage of cells with fluorescence greater than FMO gate while geometric mean fluorescence intensity (geoMFI) was used to evaluate expression levels. Integrated mean fluorescent indices (iMFI) were calculated as the product of percent positive cells and the geoMFI of the positive cells. Statistical comparisons of iMFI were performed using Student's t test.
Host gene expression determination by PCR array in total memory CD3+CD4+ cells
The total memory CD3+CD4+CD45RO+ subset was isolated from three samples each of Kenyan HIV susceptible, Kenyan HIV not susceptible, and North American samples for which greater than 120×106 cryopreserved CBMC were available using the Memory CD4+ T cell Isolation Kit (Miltenyi Biotech), following manufacturer's instructions. Flow cyotmetric analysis of isolated cells consistently demonstrated >93% purity as determined by CD4 and CD45RO staining (data not shown). RNA was isolated from the isolated CD4+CD45RO+ subset using Qiagen RNeasy mini prep and purity and quantity was determined. HIV Infection and Host Response PCR Array (SA Biosciences/Qiagen) was used according to manufacturer's instructions. Relative gene expression was calculated by ΔΔCt method. The real-time PCR quantification was performed using Applied Biosystems 7300 Real Time PCR System.
Gene expression levels that show a >2-fold difference between experimental groups were considered as potentially biologically relevant. Significant levels were assessed using ANOVA with correction for multiple comparisons (Statistical package provided by SA Biosciences Micrarray (Qiagen Inc.).
Supplementary Material
Highlight.
In utero priming increases susceptibility to HIV without exogenous stimulation
Effector memory T cells are primary initial target of infection in this system
Susceptibility to HIV correlates with increased activation of memory cells
HIV gene expression but not virus entry correlated with susceptibility
Host gene expression differed between susceptible and not susceptible samples
Acknowledgments
The authors wish to thank medical superintendent Dr. Dawood Mwaura, and nurses Victoria Saidi, Hashora Mwanguku, Zaituni Mwakileo, Fatuma Ngare, Ruth Notina, and Florence Wambua, for assisting with recruitment of women to the study, collection of cord blood samples, and care of the women and their newborns. We are grateful for the help of Elton Mzungu, Kefar Wambua, Charles NgaNga, and Alex Osore who performed many of the immunologic assays and parasitological examinations, and Grace Methenge and Christine Lucas for data entry and management. This work was supported in part by NIH grants AI064687 and MH080601. Technical assistance was provided by the Case/UHC Center for Aids Research (NIH grant number: AI36219). Drs. Zahra Toossi and Alan Levine provided helpful editing and critical review of this manuscript. Finally we appreciate the willingness of the women residing in the Msambweni location to participate in the study.
Role of the funding source: Funding for this work was provided in part by NIH grants AI064687, MH080601 and Merit Review Award from Veterans Affairs Research Service. Kevin Steiner was supported in part by the Case MSTP (NIH grant T32 GM07250) and the Case Geographic Medicine and Infectious Diseases Training Grant (NIH grant T32 A107024). Funding providers did not contribute in any way to the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
Disclosure Statement: Potential conflicts of interest: none reported for any author.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Ahmad N, Mehta R, Harris DT. HIV-1 replication and gene expression occur at higher levels in neonatal blood naive and memory T-lymphocytes compared with adult blood cells. Virology. 2011;413(1):39–46. doi: 10.1016/j.virol.2011.01.033. [DOI] [PubMed] [Google Scholar]
- Ayisi JG, van Eijk AM, Newman RD, ter Kuile FO, Shi YP, Yang C, Kolczak MS, Otieno JA, Misore AO, Kager PA, Lal RB, Steketee RW, Nahlen BL. Maternal malaria and perinatal HIV transmission, western Kenya. Emerg Infect Dis. 2004;10(4):643–652. doi: 10.3201/eid1004.030303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Battistini A, Marsili G, Sgarbanti M, Ensoli B, Hiscott J. IRF regulation of HIV-1 long terminal repeat activity. Journal of Interferon & Cytokine Research: The Official Journal of the International Society for Interferon and Cytokine Research. 2002;22(1):27–37. doi: 10.1089/107999002753452638. [DOI] [PubMed] [Google Scholar]
- Bleul CC, Wu L, Hoxie JA, Springer TA, Mackay CR. The HIV coreceptors CXCR4 and CCR5 are differentially expressed and regulated on human T lymphocytes. Proc Natl Acad Sci U S A. 1997;94(5):1925–1930. doi: 10.1073/pnas.94.5.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brahmbhatt H, Kigozi G, Wabwire-Mangen F, Serwadda D, Sewankambo N, Lutalo T, Wawer MJ, Abramowsky C, Sullivan D, Gray R. The effects of placental malaria on mother-to-child HIV transmission in Rakai, Uganda. AIDS. 2003;17(17):2539–2541. doi: 10.1097/00002030-200311210-00020. [DOI] [PubMed] [Google Scholar]
- Brenchley JM. CD4+ T Cell Depletion during all Stages of HIV Disease Occurs Predominantly in the Gastrointestinal Tract. Journal of Experimental Medicine. 2004;200(6):749–759. doi: 10.1084/jem.20040874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clement LT. Isoforms of the CD45 common leukocyte antigen family: markers for human T-cell differentiation. Journal of Clinical Immunology. 1992;12(1):1–10. doi: 10.1007/BF00918266. [DOI] [PubMed] [Google Scholar]
- Colin L, Vandenhoudt N, de Walque S, Van Driessche B, Bergamaschi A, Martinelli V, Cherrier T, Vanhulle C, Guiguen A, David A, Burny A, Herbein G, Pancino G, Rohr O, Van Lint C. The AP-1 Binding Sites Located in the pol Gene Intragenic Regulatory Region of HIV-1 Are Important for Viral Replication. In: Sommer P, editor. PLoS ONE. 4. Vol. 6. 2011. p. e19084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coull JJ, Romerio F, Sun JM, Volker JL, Galvin KM, Davie JR, Shi Y, Hansen U, Margolis DM. The human factors YY1 and LSF repress the human immunodeficiency virus type 1 long terminal repeat via recruitment of histone deacetylase 1. Journal of Virology. 2000;74(15):6790–6799. doi: 10.1128/jvi.74.15.6790-6799.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Jong JJ, De Ronde A, Keulen W, Tersmette M, Goudsmit J. Minimal requirements for the human immunodeficiency virus type 1 V3 domain to support the syncytium-inducing phenotype: analysis by single amino acid substitution. Journal of Virology. 1992;66:6777–6780. doi: 10.1128/jvi.66.11.6777-6780.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Douek DC, Picker LJ, Koup RA. T cell dynamics in HIV-1 infection. Annual Review of Immunology. 2003;21:265–304. doi: 10.1146/annurev.immunol.21.120601.141053. [DOI] [PubMed] [Google Scholar]
- van Eijk AM, Ayisi JG, ter Kuile FO, Misore AO, Otieno JA, Rosen DH, Kager PA, Steketee RW, Nahlen BL. HIV increases the risk of malaria in women of all gravidities in Kisumu, Kenya. AIDS. 2003;17(4):595–603. doi: 10.1097/00002030-200303070-00015. [DOI] [PubMed] [Google Scholar]
- Ferrando-Martinez S, Ruiz-Mateos E, Leal M. CD27 and CCR7 expression on naive T cells, are both necessary? Immunology Letters. 2010;127(2):157–158. doi: 10.1016/j.imlet.2009.10.001. [DOI] [PubMed] [Google Scholar]
- Gallagher M, Malhotra I, Mungai PL, Wamachi AN, Kioko JM, Ouma JH, Muchiri E, King CL. The effects of maternal helminth and malaria infections on mother-to-child HIV transmission. AIDS. 2005;19(16):1849–1855. doi: 10.1097/01.aids.0000189846.90946.5d. [DOI] [PubMed] [Google Scholar]
- Groot F, van Capel TMM, Schuitemaker J, Berkhout B, de Jong EC. Differential susceptibility of naïve, central memory and effector memory T cells to dendritic cell-mediated HIV-1 transmission. Retrovirology. 2006;3:52. doi: 10.1186/1742-4690-3-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta A, Bhosale R, Kinikar A, Gupte N, Bharadwaj R, Kagal A, Joshi S, Khandekar M, Karmarkar A, Kulkarni V, Sastry J, Mave V, Suryavanshi N, Thakar M, Kulkarni S, Tripathy S, Sambarey P, Patil S, Paranjape R, Bollinger RC, Jamkar A for the Six Week Extended-Dose Nevirapine (SWEN) India Study Team. Maternal Tuberculosis: A Risk Factor for Mother-to-Child Transmission of Human Immunodeficiency Virus. Journal of Infectious Diseases. 2011;203(3):358–362. doi: 10.1093/jinfdis/jiq064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He G, Margolis DM. Counterregulation of chromatin deacetylation and histone deacetylase occupancy at the integrated promoter of human immunodeficiency virus type 1 (HIV-1) by the HIV-1 repressor YY1 and HIV-1 activator Tat. Molecular and Cellular Biology. 2002;22(9):2965–2973. doi: 10.1128/MCB.22.9.2965-2973.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inion I, Mwanyumba F, Gaillard P, Chohan V, Verhofstede C, Claeys P, Mandaliya K, Van Marck E, Temmerman M. Placental malaria and perinatal transmission of human immunodeficiency virus type 1. J Infect Dis. 2003;188(11):1675–1678. doi: 10.1086/379737. [DOI] [PubMed] [Google Scholar]
- Kedzierska K, Crowe S, Turville S, Cunningham A. The influence of cytokines, chemokines and their receptors on HIV-1 replication in monocytes and macrophages. Rev Med Virol. 2003;13:39–56. doi: 10.1002/rmv.369. [DOI] [PubMed] [Google Scholar]
- Kedzierska K, Crowe SM. Cytokines and HIV-1: interactions and clinical implications. Antiviral Chemistry & Chemotherapy. 2001;12(3):133–150. doi: 10.1177/095632020101200301. [DOI] [PubMed] [Google Scholar]
- Kinoshita S, Chen BK, Kaneshima H, Nolan GP. Host control of HIV-1 parasitism in T cells by the nuclear factor of activated T cells. Cell. 1998;95(5):595–604. doi: 10.1016/s0092-8674(00)81630-x. [DOI] [PubMed] [Google Scholar]
- Kinoshita S, Su L, Amano M, Timmerman LA, Kaneshima H, Nolan GP. The T cell activation factor NF-ATc positively regulates HIV-1 replication and gene expression in T cells. Immunity. 1997;6(3):235–244. doi: 10.1016/s1074-7613(00)80326-x. [DOI] [PubMed] [Google Scholar]
- Malhotra I, Dent A, Mungai P, Wamachi A, Ouma JH, Narum DL, Muchiri E, Tisch DJ, King CL. Can Prenatal Malaria Exposure Produce an Immune Tolerant Phenotype?: A Prospective Birth Cohort Study in Kenya. PLoS Med. 2009;6(7):e1000116. doi: 10.1371/journal.pmed.1000116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Malhotra I, Ouma J, Wamachi A, Kioko J, Mungai P, Omollo A, Elson L, Koech D, Kazura JW, King CL. In utero exposure to helminth and mycobacterial antigens generates cytokine responses similar to that observed in adults. J Clin Invest. 1997;99(7):1759–1766. doi: 10.1172/JCI119340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marsili G, Borsetti A, Sgarbanti M, Remoli AL, Ridolfi B, Stellacci E, Ensoli B, Battistini A. On the role of interferon regulatory factors in HIV-1 replication. Annals of the New York Academy of Sciences. 2003;1010:29–42. doi: 10.1196/annals.1299.005. [DOI] [PubMed] [Google Scholar]
- Mawhinney S, Pagano M. Distribution of the latency period for perinatally acquired AIDS. Statistics in Medicine. 1994;13(19-20):2031–2042. doi: 10.1002/sim.4780131913. [DOI] [PubMed] [Google Scholar]
- Mehandru S. Primary HIV-1 Infection Is Associated with Preferential Depletion of CD4+ T Lymphocytes from Effector Sites in the Gastrointestinal Tract. Journal of Experimental Medicine. 2004;200(6):761–770. doi: 10.1084/jem.20041196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nie C, Sato K, Misawa N, Kitayama H, Fujino H, Hiramatsu H, Heike T, Nakahata T, Tanaka Y, Ito M, Koyanagi Y. Selective infection of CD4+ effector memory T lymphocytes leads to preferential depletion of memory T lymphocytes in R5 HIV-1-infected humanized NOD/SCID/IL-2Rgammanull mice. Virology. 2009;394(1):64–72. doi: 10.1016/j.virol.2009.08.011. [DOI] [PubMed] [Google Scholar]
- Osborn L, Kunkel S, Nabel GJ. Tumor necrosis factor alpha and interleukin 1 stimulate the human immunodeficiency virus enhancer by activation of the nuclear factor kappa B. Proc Natl Acad Sci U S A. 1989;86(7):2336–2340. doi: 10.1073/pnas.86.7.2336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ostrowski MA, Chun TW, Justement SJ, Motola I, Spinelli MA, Adelsberger J, Ehler LA, Mizell SB, Hallahan CW, Fauci AS. Both memory and CD45RA+/CD62L+ naive CD4(+) T cells are infected in human immunodeficiency virus type 1-infected individuals. Journal of Virology. 1999;73(8):6430–6435. doi: 10.1128/jvi.73.8.6430-6435.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Resino S, Gurbindo D, Cano JM, Sanchez-Ramón S, Muõz-Fernández MA. Predictive markers of clinical outcome in vertically HIV-1-infected infants. A prospective longitudinal study. Pediatric Research. 2000;47(4 Pt 1):509–515. doi: 10.1203/00006450-200004000-00016. [DOI] [PubMed] [Google Scholar]
- Roebuck KA, Brenner DA, Kagnoff MF. Identification of c-fos-responsive elements downstream of TAR in the long terminal repeat of human immunodeficiency virus type-1. The Journal of Clinical Investigation. 1993;92(3):1336–1348. doi: 10.1172/JCI116707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roebuck KA, Gu DS, Kagnoff MF. Activating protein-1 cooperates with phorbol ester activation signals to increase HIV-1 expression. AIDS (London, England) 1996;10(8):819–826. doi: 10.1097/00002030-199607000-00004. [DOI] [PubMed] [Google Scholar]
- Romanchikova N, Ivanova V, Scheller C, Jankevics E, Jassoy C, Serfling E. NFAT transcription factors control HIV-1 expression through a binding site downstream of TAR region. Immunobiology. 2003;208(4):361–365. doi: 10.1078/0171-2985-00283. [DOI] [PubMed] [Google Scholar]
- Saba E, Grivel JC, Vanpouille C, Brichacek B, Fitzgerald W, Margolis L, Lisco A. HIV-1 sexual transmission: early events of HIV-1 infection of human cervico-vaginal tissue in an optimized ex vivo model. Mucosal Immunology. 2010;3(3):280–290. doi: 10.1038/mi.2010.2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sallusto F, Lenig D, Forster R, Lipp M, Lanzavecchia A. Two subsets of memory T lymphocytes with distinct homing potentials and effector functions. Nature. 1999;401(6754):708–712. doi: 10.1038/44385. [DOI] [PubMed] [Google Scholar]
- Schnittman SM, Lane HC, Greenhouse J, Justement JS, Baseler M, Fauci AS. Preferential infection of CD4+ memory T cells by human immunodeficiency virus type 1: evidence for a role in the selective T-cell functional defects observed in infected individuals. Proceedings of the National Academy of Sciences of the United States of America. 1990;87(16):6058–6062. doi: 10.1073/pnas.87.16.6058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sgarbanti M, Remoli AL, Marsili G, Ridolfi B, Borsetti A, Perrotti E, Orsatti R, Ilari R, Sernicola L, Stellacci E, Ensoli B, Battistini A. IRF-1 is required for full NF-kappaB transcriptional activity at the human immunodeficiency virus type 1 long terminal repeat enhancer. Journal of Virology. 2008;82(7):3632–3641. doi: 10.1128/JVI.00599-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sleasman JW, Aleixo LF, Morton A, Skoda-Smith S, Goodenow MM. CD4+ memory T cells are the predominant population of HIV-1-infected lymphocytes in neonates and children. AIDS (London, England) 1996;10(13):1477–1484. doi: 10.1097/00002030-199611000-00004. [DOI] [PubMed] [Google Scholar]
- Spina CA, Prince HE, Richman DD. Preferential replication of HIV-1 in the CD45RO memory cell subset of primary CD4 lymphocytes in vitro. J Clin Invest. 1997;99(7):1774–1785. doi: 10.1172/JCI119342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Steiner K, Myrie L, Malhotra I, Mungai P, Muchiri E, Dent A, King CL. Fetal Immune Activation to Malaria Antigens Enhances Susceptibility to In Vitro HIV Infection in Cord Blood Mononuclear Cells. Journal of Infectious Diseases. 2010;202(6):899–907. doi: 10.1086/655783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundaravaradan V, Mehta R, Harris DT, Zack JA, Ahmad N. Differential expression and interaction of host factors augment HIV-1 gene expression in neonatal mononuclear cells. Virology. 2010;400(1):32–43. doi: 10.1016/j.virol.2010.01.018. [DOI] [PubMed] [Google Scholar]
- Sundaravaradan V, Saxena SK, Ramakrishnan R, Yedavalli VR, Harris DT, Ahmad N. Differential HIV-1 replication in neonatal and adult blood mononuclear cells is influenced at the level of HIV-1 gene expression. Proc Natl Acad Sci U S A. 2006;103(31):11701–11706. doi: 10.1073/pnas.0602185103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tovo PA, de Martino M, Gabiano C, Cappello N, D'Elia R, Loy A, Plebani A, Zuccotti GV, Dallacasa P, Ferraris G. Prognostic factors and survival in children with perinatal HIV-1 infection. The Italian Register for HIV Infections in Children. Lancet. 1992;339(8804):1249–1253. doi: 10.1016/0140-6736(92)91592-v. [DOI] [PubMed] [Google Scholar]
- UNAIDS. UNAIDS Report on the global AIDS epidemic 2010. Geneva: UNAIDS; 2010. [Google Scholar]
- Yonkers NL, Sieg S, Rodriguez B, Anthony DD. Reduced Naive CD4 T Cell Numbers and Impaired Induction of CD27 in Response to T Cell Receptor Stimulation Reflect a State of Immune Activation in Chronic Hepatitis C Virus Infection. Journal of Infectious Diseases. 2011;203(5):635–645. doi: 10.1093/infdis/jiq101. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.