Abstract
Previous studies have identified two zebrafish mutants, cloche and groom of cloche, which lack the majority of the endothelial lineage at early developmental stages. However, at later stages, these avascular mutant embryos generate rudimentary vessels, indicating that they retain the ability to generate endothelial cells despite this initial lack of endothelial progenitors. To further investigate molecular mechanisms that allow the emergence of the endothelial lineage in these avascular mutant embryos, we analyzed the gene expression profile using microarray analysis on isolated endothelial cells. We find that the expression of the genes characteristic of the mesodermal lineages are substantially elevated in the kdrl+ cells isolated from avascular mutant embryos. Subsequent validation and analyses of the microarray data identifies Sox11b, a zebrafish ortholog of SRY-related HMG box 11 (SOX11), which have not previously implicated in vascular development. We further define the function sox11b during vascular development, and find that Sox11b function is essential for developmental angiogenesis in zebrafish embryos, specifically regulating sprouting angiogenesis. Taken together, our analyses illustrate a complex regulation of endothelial specification and differentiation during vertebrate development.
Keywords: cloche, groom of cloche, sox11b, vascular development, zebrafish
INTRODUCTION
Endothelial cells are a major component of the vascular system, which is essential for the development, growth, and survival of an individual. Failures in regulating the development of endothelial lineage contribute to a wide variety of pathological conditions, including cancer, psoriasis, arthritis, congenital or inherited diseases, as well as heart and brain ischemia, neurodegeneration, and osteoporosis (Carmeliet, 2005). During development, the endothelial lineage arises from mesodermal tissues. It has been reported that diverse mesodermal tissues including lateral plate mesoderm (Pardanaud et al., 1996), blood islands within the yolk sac (Ferkowicz and Yoder, 2005; Risau and Flamme, 1995), allantois (Caprioli et al., 2001), somitic mesoderm (Wilting et al., 1995), as well as placenta (Demir et al., 2007; Yamaguchi et al., 1993), can produce endothelial cells during development. Moreover, the entire mesoderm excluding notochord and prechordal mesoderm can serve as sources for endothelial cells (Noden, 1989), suggesting that angiogenic potential might be one of the intrinsic properties of the developing mesoderm. Subsequently, endothelial cells further differentiate as arterial, venous or lymphatic endothelial cells, each of which possesses unique molecular and cellular characteristics.
Specification and differentiation of the endothelial lineage are regulated by arrays of signaling pathways and transcription factors. Previous research has identified key signaling pathways that modulate the differentiation of the endothelial lineages from the nascent mesodermal progenitors, including vascular endothelial growth factor (VEGF) (Keck et al., 1989; Leung et al., 1989), fibroblast growth factor (FGF) (Abraham et al., 1986; Gospodarowicz et al., 1983), Wnt (Ishikawa et al., 2001), and Bone Morphogenetic Protein (BMP) (Yamashita et al., 1997), as well as essential transcription factors such as ETS transcription factor family member, Etv2/ER71 and FLI1 (Wernert et al., 1992), and T-cell acute lymphocytic leukemia protein 1 (TAL1) (Visvader et al., 1998). Given the developmental heterogeneity, it is not surprising that cellular responses to these factors vary among subsets of endothelial cells. For instance, we have recently reported that BMP2 signaling selectively activates venous endothelial cells without influencing arterial endothelial cells (Wiley and Jin, 2011). Similarly, Wnt signaling regulates the formation of the endothelial lineage within the tailbud mesoderm, without obvious effects on the lateral plate mesoderm (Martin and Kimelman, 2012). Therefore, identification of additional factors that regulate specification and differentiation of the endothelial lineage will help us to further delineate the heterogeneity of endothelial cells.
To identify additional factors involved in vascular development, we have previously performed a large scale forward genetic screen using Tg(kdrl:eGFP)s843 transgenic zebrafish, which labels all endothelial cells with eGFP (Jin et al., 2005). From the screen, we have identified a novel mutant, groom of cloche (grc), which lacks the majority of the endothelial lineage at early stage (Jin et al., 2007), which is reminiscent of previously isolated mutant, cloche (clo) that lack both endothelial and hematopoietic lineages (Stainier et al., 1995). Despite the lack of endothelial cells at early stages, these avascular mutant embryos can generate endothelial cells at later stages (Jin et al., 2007), suggesting that distinct molecular mechanisms may be used to modulate the emergence of the endothelial lineage in these embryos.
In this report, we performed microarray analysis using the endothelial cells isolated from late stage avascular mutant embryos and compared the expression profile of transcription factors with endothelial cells isolated from wild-type embryos. We find that the expression level of 43 transcription factors is significantly up-regulated in endothelial cells isolated from avascular mutant embryos. The majority of transcription factors we identified in our microarray have not been implicated in vascular development. We further analyze the function of one of these transcription factors, SRY-related HMG Box 11b (Sox11b), in endothelial differentiation and subsequent vascular patterning. We find that Sox11b is expressed in endothelial cells during development, and is essential for sprouting angiogenesis in zebrafish. Our results demonstrate that developmental ontogeny of the endothelial lineage is far more complex than previously thought.
MATERIALS AND METHODS
Zebrafish husbandry and heatshock treatment
Zebrafish (Danio Rerio) embryos were raised as previously described (Westerfield, 1989). The following transgenic and mutant fish lines were utilized: Tg(kdrl:eGFP)s843 (Jin et al., 2005), cloche (clo)s5 (Stainier et al., 1995), Casanova (cas)s4 (Alexander et al., 1998), groom of cloche (grc)s635 (Jin et al., 2007), Tg(hsp70l:bmp2b)fr13 (Chocron et al., 2007), Tg(hsp70l: noggin3)fr14 (Chocron et al., 2007). Heatshock treatment was administered by incubating 24hpf embryos at 42°C for 30 min.
Florescent activated cell sorting (FACS) and RNA isolation
The 18.5hpf Tg(kdrl:eGFP)s843 embryos were dissociated in HBSS with 5% FBS and subsequently incubated with 100 μg/ml Liberase solution (Roche) for 15 min at 37°C. Embryos were then triturated and the resulting suspension was pushed through a 40 μM cell culture filter (BD Biosciences) and the reaction was stopped using 5 mM EDTA, pH 8.0 in HBSS minus Ca2+ and Mg2+. Gates for flow cytometry were selected based on the Phycoerythrin versus FITC plot. Double sorts indicated an enrichment to > 95% GFP+ cells. RNA was extracted from isolated cells using Trizol (Invitrogen) and the accompanying protocol. Multiple rounds of flow cytometry were performed and RNA for each biological replicate was pooled. From the heatshock treated embryos, endothelial cells were harvested at 32 hpf.
Microarray analyses and quantitative RT-PCR
The WT ovation Pico Kit was used to amplify the RNA samples to satisfactory RNA integrity score (RIN) score (Schroeder et al., 2006). Otherwise, gene expression profiling was performed as previously described (Lobenhofer et al., 2008) using an Agilent Zebrafish array version 2. Using the Statistical Analysis of Microarrays (SAM), the raw data for wild-type, grc, clo and cas was analyzed. We disregarded genes whose expression was down-regulated in cas, which would represent genes expressed in pharyngeal endoderm. Genes highly significantly up-regulated (q = 0, fold change > 2) in both grc and clo mutants were further analyzed.
For qRT-PCR, RNA was extracted using the QIagen RNeasy mini kit and accompanying protocol opting to add 300 ng of carrier RNA to each sample. The iScript cDNA synthesis kit (Bio-Rad) was used to transcribe entire RNA extracts, immediately after RNA extraction. cDNA samples were then diluted to a volume of 300 μl. Using 2X Power syber mastermix, 640 nM of each primer, and 8 μl of cDNA in a 25 μl reaction, amplification of transcript amplicon was monitored on a Bio-Rad cfx96 system. Gene expression was normalized to either 18S rRNA or B-actin housekeeping genes. Melting curve analysis was performed on all reactions. Ct versus cDNA concentration plots were also used to determine that there was a linear ratio of amplification of housekeeping genes to gene-of-interest at a particular cDNA concentration. Data was analyzed using the 2−ΔΔCT method (Livak and Schmittgen, 2001). At least three biological replicates of three technical replicates were performed for each conditon. Primers for qRT-PCR were generated using quantprime (REF#890). Primers used were: 18s rRNA (5′-CACTTGTCCCTCTAAGAAGTTGCA-3′ and 5′-GG TTGATTCCGATAACGAACGA-3′), sox11b (5′-CGAGTTCCC GGACTATTGCA-3′ and 5′ TCTCCCGCGATCATCTCACT-3′), zfhx4 (5′-CTCCTTTGTGTGGGAAGCAT-3′ and 5′-CCCTG AATGTGGAACAGCAT-3′), and klf5l (5′-AACCCGCAGTGAG AATCGCAAC-3′ and 5′-ATCCATCTCCATCCGTGTCTGAGC-3′).
In situ probe synthesis
Probes were synthesized using the SP6/T7 DIG-UTP labeling kit (Roche) from linearized template. RNA was quantified, monitored by agarose gel electrophoresis for a singular product, diluted in in situ hybridization solution to 100 ng/μl and stored at −20°C.
Morpholino knockdown of sox11b
Previously reported sox11b morpholino (5′-CATGTTCAAACACACTTTTCCCTCT-3′), which blocks peptide synthesis, and control morpholino (5′-CCTCTTACCTCAGTTACAATTTATA-3′) were used (Veldman et al., 2007). All embryos were injected with 4.6 nL of injection mix containing 5 μM HEPES, pH 7.6 and 0.05% Phenol red as a tracer.
RESULTS AND DISCUSSION
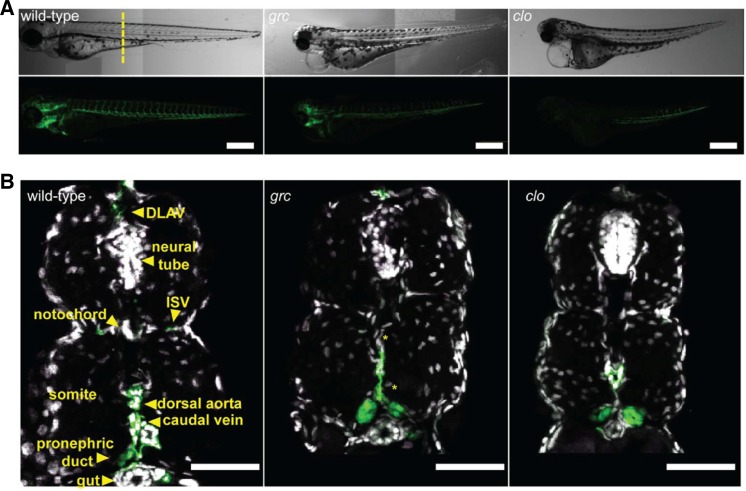
As previously reported, both clo and grc homozygous mutant embryos lack endothelial cells at 18 hpf (Jin et al., 2007; Stainier et al., 1995) (data not shown). However, at 72 hpf, kdrl+ cells were present in these avascular mutant embryos (Figs. 1A–1C). Interestingly, counterstaining with DAPI in this experiment also showed that the midline region where are exclusively populated by kdrl+ cells in wild-type embryos also contains a substantial number of kdrl− cells in avascular mutant embryos (Fig. 1A, yellow asterisks), alluding that vascular progenitors in these embryos may fail to undergo proper differentiation.
Fig. 1.
Avascular mutant embryos generate endothelial cells at later stages. (A) Gross morphology of 72 hpf wild-type (left), groom of cloche (grc) (middle), and cloche (clo) embryos in Tg(kdrl:eGFP) background. Both bright-field (top rows) and epifluorescent (bottom rows) images are shown. Scale bar = 250 μm. (B) Transverse section of 72 hpf embryos taken from the area marked by dashed line in (A). GFP+ endothelial cells are shown in green and nuclei stained with DAPI are shown in white. Scale bar = 50 μm.
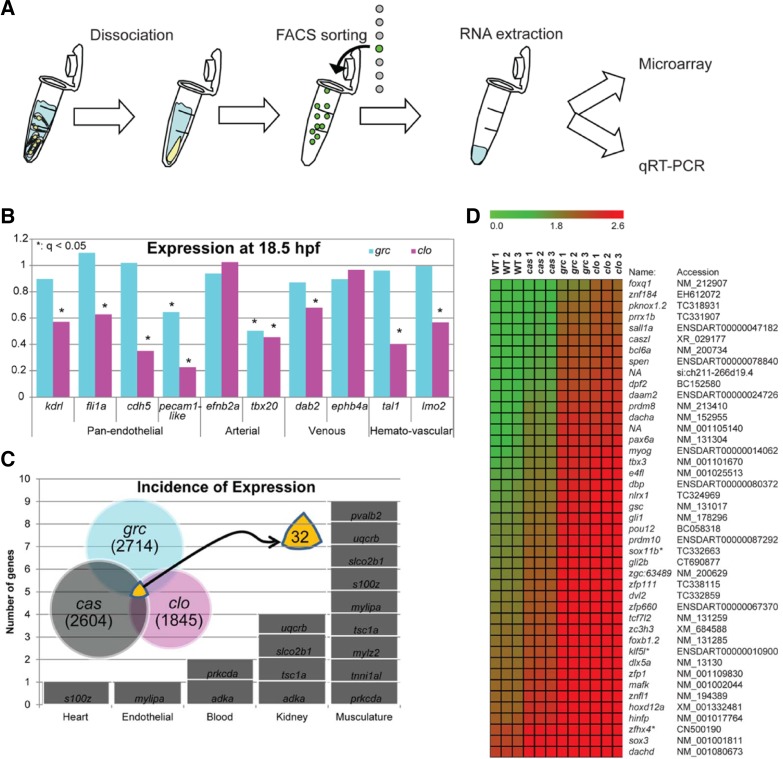
To better understand molecular mechanisms underlying the recovery of endothelial lineage, we analyzed the transcriptional profile of kdrl+ cells in wild-type and avascular mutant embryos by microarray analyses (Fig. 2A). Since kdrl, a zebrafish ortholog of vascular endothelial growth factor receptor 2 (VEGFR2) (Bussmann et al., 2008), is also expressed in pharyngeal endoderm (Jin et al., 2005), it is possible that a significant portion of kdrl+ cells isolated in avascular mutant embryos may represent non-endothelial lineage. Therefore, we used homozygous cas embryos wherein the entire presumptive endoderm fails to specify with little apparent effect on the vasculature (Jin et al., 2005). Genes down-regulated in kdrl+ cells isolated from homozygous cas mutant embryos were discarded prior to further analyses and validation.
Fig. 2.
Expression profile of kdrl+ cells isolated from avascular mutant embryos. (A) Schematic diagram for molecular profiling. (B) Expression profile of known lineage specific markers in microarray (*q < 0.05). (C) Characteristics of genes which are up-regulated in endothelial cells in all three avascular mutant embryos. Total of 32 genes were shown to be up-regulated. (D) Expression profiles of putative transcription factors of which function have not previously implicated in the endothelial lineage. These genes were up in grc and clo (q = 0), but not down-regulated in cas.
We found that the expression level of endothelial-enriched genes were largely unaltered in kdrl+ cells of homozygous grc mutant embryos. In contrast, the majority of these genes were down-regulated in the same population from homozygous clo mutant embryos (Fig. 2A), suggesting that a locus affected by grc mutation may be only required in a subset of endothelial cells. A small subset of endothelial-enriched genes was markedly down-regulated in both homozygous grc and clo mutant embryos. For instance, we found that an arterial specific marker, tbx20 (Ahn et al., 2000), as well as a putative zebrafish ortholog of mammalian Platelet Endothelial Cell Adhesion Molecule (PECAM), ENSDART00000084729, was significantly down-regulated in kdrl+ cells isolated from both homozygous grc and clo mutant embryos (Fig. 2A). Interestingly, we found that genes up-regulated (q = 0) in all three mutants were found to be characteristic of other mesodermal, non-endothelial lineages such as somite, blood, or kidney (Fig. 2B). For instance, we found protein kinase c delta a (prkcda), which are expressed in blood and somitic lineages (Patten et al., 2007), and adenosine kinase a (adka), which are expressed in blood and pronephric lineages (ZFIN direct data submission by Thisse et al., 2001), was up-regulated in kdrl+ cells from avascular mutant embryos. Taken together, our microarray data suggest that kdrl+ cells found in avascular mutant embryos may retain more mesodermal characteristics than those from wild-type embryos.
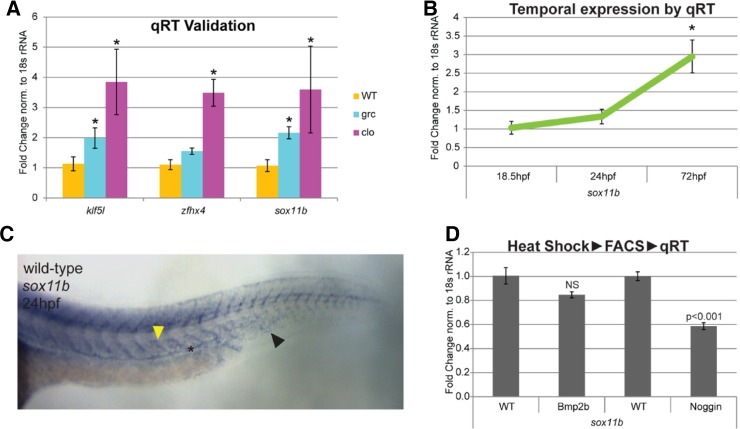
To better understand molecular characteristics of the kdrl+ cells in avascular mutant embryos, we analyzed the expression level of transcription factors in our microarray data (Fig. 2C). We found that total of 43 transcription factors were up-regulated in kdrl+ cells from avascular mutant embryos (Fig. 2D) at q = 0. Among these transcription factors, we further analyze the function of sox11b, a zebrafish ortholog of SRY-related HMG Box 11 (SOX11) (Veldman et al., 2007), which is a member of SOXC subgroup (Bowles et al., 2000). Previously, it has been shown that Sox11b is essential for mediating retinal development and neuronal regeneration in zebrafish (Veldman et al., 2007). However, its role in endothelial cells and vascular development has not been investigated. Up-regulation of sox11b in kdrl+ cells in avascular mutant embryos was confirmed by quantitative RT-PCR (Fig. 3A).
Fig. 3.
Sox11b expression is elevated in kdrl+ cells isolated from avascular mutant embryos. (A) Quantitative RT-PCR analyses confirmed the up-regulated expression of sox11b in endothelial cells of avascular mutant embryos. Two additional transcription factor, klf5l and zfhx4, were used as positive controls. (B) Temporal expression change of sox11b expression in endothelial cells. (C) In situ hybridization of sox11b at 24hpf wild-type embryo. In addition to neural tube and somite, axial vessels express sox11b. Anterior somite, axial vessel, and caudal vein are indicated by the yellow arrow, asterisk, and black arrow, respectively. (D) Effects of BMP signaling on sox11b expression. A decreased activity of BMP signaling significantly reduces the expression of sox11b in endothelial cells.
During development, sox11b is highly expressed in multiple tissues including neurons, somites, and retina as previously proposed. In addition, approximately at 24hpf, sox11b expression was detectable in developing posterior axial vessels in wild-type embryos (Fig. 3B). To analyze temporal changes in sox11b expression within endothelial cells, kdrl+ cells were isolated from wild-type embryos and quantitative RT-PCR was performed. We found that sox11b expression can be detected as early as 18 hpf, and the level of expression gradually increases until 72 hpf within endothelial cells, consistent with our in situ hybridization result (Fig. 3C). Interestingly, the expression of sox11b appears to be induced by bone morphogenetic protein (Bmp) signaling, as over-expression of Noggin3, an endogenous antagonist of Bmp signaling, led to a substantially decrease on the level of sox11b transcript level (Fig. 3D). Considering that Bmp signaling functions as a context-dependent pro-angiogenic cue (Kim et al., 2012; Wiley and Jin, 2011), it is possible that Sox11b may function as one of the effectors in this process.
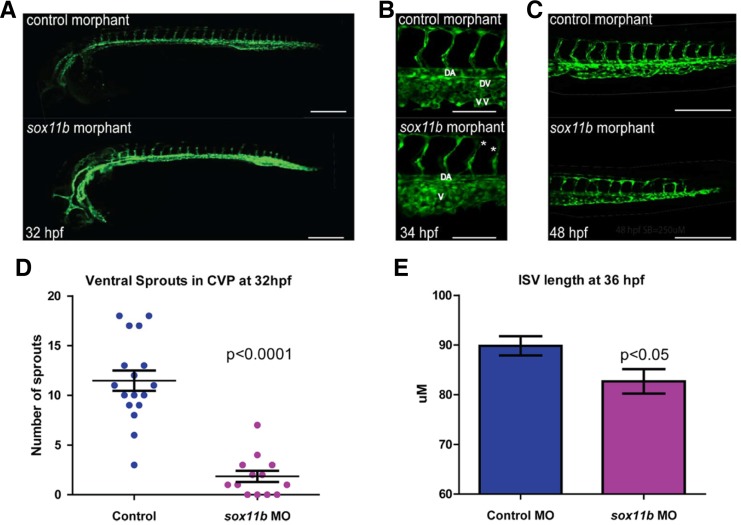
To better assess the function of Sox11b during vascular development, we attenuated the activity of Sox11b by injecting morpholino (MO) anti-sense oligonucleotide as previously reported (Nasevicius and Ekker, 2000). Embryos injected with sox11b MO displayed discernible defects in vascular development, compared to control MO injected embryo (Fig. 4A). At 32 hpf, the length of intersegmental vessels, which sprout from the dorsal aorta at this stage (Isogai et al., 2001), was substantially reduced in sox11b MO injected embryos (Fig. 4B). While control MO injected embryos had an average length of 89.82 ± 1.92 μm (N = 139 ISVs), sox11b MO injected embryos intersegmental vessels were significantly shorter, 82.67 ± 2.46 μm (N = 156 ISVs, N = 8 embryos; Figs. 4E and 4F), indicating that the function of Sox11b is essential for the morphogenesis of sprouting intersegmental vessels during development. Since a mammalian ortholog of Sox11b, SOX11 is known to promote transcription of key cell cycle regulators including Cyclin-dependent kinase CDKN2B and Histones (Wang et al., 2010), as well as arrays of Actin binding proteins which modulate cell motility (Wang et al., 2010), it is possible down-regulation of Sox11b by MO injection led to a decreased endothelial proliferation and/or migration.
Fig. 4.
Sox11b regulates sprouting angiogenesis during development. (A) Epifluorescent micrographs of control (top) or sox11b (bottom) morpholino (MO) injected embryos. Trunk regions posterior to the end of yolk extension are shown. Scale bar = 250 μm. (B) Intersegmental vessel and caudal vein defect in sox11b MO injected embryos at 34 hpf. Scale bar = 100 μm. (C) Truncation of ISVs and plexus defects at 48hpf. Scale bar = 250 μm. (D) Decreased venous sprouting angiogenesis in the caudal vein plexus (CVP) of sox11b MO injected embryos at 32hpf is quantified. (E) The effect of sox11b MO on the length of intersegmental vessels is quantified (N = 139, N = 10 embryos).
Since intersegmental vessels at 24 hpf are arterial in nature (Isogai et al., 2001), we investigated whether Sox11b preferentially influences migration of arterial endothelial cells, by analyzing the effects of Sox11b knock-down on sprouting angiogenesis of caudal vein plexus (CVP). Previously, we reported that the CVP undergoes morphogenetic changes starting at 30 hpf by forming extensive ventral sprouts (Wiley and Jin, 2011). In sox11b MO injected embryos, the number of angiogenic sprouts was drastically reduced compared to control MO injected embryos at 32 hpf (1.85 ± 0.56 in sox11b MO injected embryos and 11.4 ± 1.0 in control MO injected embryos; Figs. 4G and 4H). Morphologically, the CVP in sox11b MO injected embryos failed to undergo proper morphogenesis to generate the dorsal vein and the ventral vein as in wild-type embryos (Fig. 4G), reflecting the attenuated sprouting angiogenesis in these embryos.
Our results indicate that kdrl+ cells in avascular mutant embryos express a unique transcriptional profile that allow them to circumvent the initial failure of endothelial specification, which led to the formation of rudimentary vascular structure in these embryos. We found that a number of transcription factors were selectively up-regulated in the kdrl+ cells of avascular mutant embryos, indicating that these transcription factors may guide an alternative mechanism to generate the endothelial lineage. We analyzed the function of one of the transcription factors isolated from our microarray, Sox11b, and found that Sox11b plays an important role in early morphogenesis of the vasculature. Taken together, our data provides a compelling evidence of developmental heterogeneity of the endothelial lineage.
Acknowledgments
The authors would like to thank members of Jin lab for helpful discussion. This work has been supported by Developmental Biology Training Grant to C.E.S. and American Heart Association Pre-doctoral Fellowship to M.J.W., and grants from the NIH (HL090960) and the American Heart Association Scientist Development Award to S.-W.J.
REFERENCES
- Abraham J.A., Mergia A., Whang J.L., Tumolo A., Friedman J., Hjerrild K.A., Gospodarowicz D., Fiddes J.C. Nucleotide sequence of a bovine clone encoding the angiogenic protein, basic fibroblast growth factor. Science. 1986;233:545–548. doi: 10.1126/science.2425435. [DOI] [PubMed] [Google Scholar]
- Ahn D.G., Ruvinsky I., Oates A.C., Silver L.M., Ho R.K. tbx20, a new vertebrate T-box gene expressed in the cranial motor neurons and developing cardiovascular structures in zebrafish. Mech. Dev. 2000;95:253–258. doi: 10.1016/s0925-4773(00)00346-4. [DOI] [PubMed] [Google Scholar]
- Alexander J., Stainier D.Y., Yelon D. Screening mosaic F1 females for mutations affecting zebrafish heart induction and patterning. Dev. Genet. 1998;22:288–299. doi: 10.1002/(SICI)1520-6408(1998)22:3<288::AID-DVG10>3.0.CO;2-2. [DOI] [PubMed] [Google Scholar]
- Bowles J., Schepers G., Koopman P. Phylogeny of the SOX family of developmental transcription factors based on sequence and structural indicators. Dev. Biol. 2000;227:239–255. doi: 10.1006/dbio.2000.9883. [DOI] [PubMed] [Google Scholar]
- Bussmann J., Lawson N., Zon L., Schulte-Merker S. Zebrafish VEGF receptors: a guideline to nomenclature. PLoS Genet. 2008;4:e1000064. doi: 10.1371/journal.pgen.1000064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli A., Minko K., Drevon C., Eichmann A., Dieterlen-Lievre F., Jaffredo T. Hemangioblast commitment in the avian allantois: cellular and molecular aspects. Dev. Biol. 2001;238:64–78. doi: 10.1006/dbio.2001.0362. [DOI] [PubMed] [Google Scholar]
- Carmeliet P. Angiogenesis in life, disease and medicine. Nature. 2005;438:932–936. doi: 10.1038/nature04478. [DOI] [PubMed] [Google Scholar]
- Chocron S., Verhoeven M.C., Rentzsch F., Hammerschmidt M., Bakkers J. Zebrafish Bmp4 regulates left-right asymmetry at two distinct developmental time points. Dev. Biol. 2007;305:577–588. doi: 10.1016/j.ydbio.2007.03.001. [DOI] [PubMed] [Google Scholar]
- Demir R., Seval Y., Huppertz B. Vasculogenesis and angiogenesis in the early human placenta. Acta Histochem. 2007;109:257–265. doi: 10.1016/j.acthis.2007.02.008. [DOI] [PubMed] [Google Scholar]
- Ferkowicz M.J., Yoder M.C. Blood island formation: longstanding observations and modern interpretations. Exp. Hematol. 2005;33:1041–1047. doi: 10.1016/j.exphem.2005.06.006. [DOI] [PubMed] [Google Scholar]
- Gospodarowicz D., Cheng J., Lirette M. Bovine brain and pituitary fibroblast growth factors: comparison of their abilities to support the proliferation of human and bovine vascular endothelial cells. J. Cell Biol. 1983;97:1677–1685. doi: 10.1083/jcb.97.6.1677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishikawa T., Tamai Y., Zorn A.M., Yoshida H., Seldin M.F., Nishikawa S., Taketo M.M. Mouse Wnt receptor gene Fzd5 is essential for yolk sac and placental angiogenesis. Development. 2001;128:25–33. doi: 10.1242/dev.128.1.25. [DOI] [PubMed] [Google Scholar]
- Isogai S., Horiguchi M., Weinstein B.M. The vascular anatomy of the developing zebrafish: an atlas of embryonic and early larval development. Dev. Biol. 2001;230:278–301. doi: 10.1006/dbio.2000.9995. [DOI] [PubMed] [Google Scholar]
- Jin S.W., Beis D., Mitchell T., Chen J.N., Stainier D.Y. Cellular and molecular analyses of vascular tube and lumen formation in zebrafish. Development. 2005;132:5199–5209. doi: 10.1242/dev.02087. [DOI] [PubMed] [Google Scholar]
- Jin S.W., Herzog W., Santoro M.M., Mitchell T.S., Frantsve J., Jungblut B., Beis D., Scott I.C., D’Amico L.A., Ober E.A., et al. A transgene-assisted genetic screen identifies essential regulators of vascular development in vertebrate embryos. Dev. Biol. 2007;307:29–42. doi: 10.1016/j.ydbio.2007.03.526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keck P.J., Hauser S.D., Krivi G., Sanzo K., Warren T., Feder J., Connolly D.T. Vascular permeability factor, an endothelial cell mitogen related to PDGF. Science. 1989;246:1309–1312. doi: 10.1126/science.2479987. [DOI] [PubMed] [Google Scholar]
- Kim J.D., Kang H., Larrivee B., Lee M.Y., Mettlen M., Schmid S.L., Roman B.L., Qyang Y., Eichmann A., Jin S.W. Context-dependent proangiogenic function of bone morphogenetic protein signaling is mediated by disabled homolog 2. Dev. Cell. 2012;23:441–448. doi: 10.1016/j.devcel.2012.07.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leung D.W., Cachianes G., Kuang W.J., Goeddel D.V., Ferrara N. Vascular endothelial growth factor is a secreted angiogenic mitogen. Science. 1989;246:1306–1309. doi: 10.1126/science.2479986. [DOI] [PubMed] [Google Scholar]
- Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-delta delta C(T)) method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- Lobenhofer E.K., Auman J.T., Blackshear P.E., Boorman G.A., Bushel P.R., Cunningham M.L., Fostel J.M., Gerrish K., Heinloth A.N., Irwin R.D., et al. Gene expression response in target organ and whole blood varies as a function of target organ injury phenotype. Genome Biol. 2008;9:R100. doi: 10.1186/gb-2008-9-6-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin B.L., Kimelman D. Canonical Wnt signaling dynamically controls multiple stem cell fate decisions during vertebrate body formation. Dev. Cell. 2012;22:223–232. doi: 10.1016/j.devcel.2011.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nasevicius A., Ekker S.C. Effective targeted gene ‘knockdown’ in zebrafish. Nat. Genet. 2000;26:216–220. doi: 10.1038/79951. [DOI] [PubMed] [Google Scholar]
- Noden D.M. Embryonic origins and assembly of blood vessels. Am. Rev. Respir. Dis. 1989;140:1097–1103. doi: 10.1164/ajrccm/140.4.1097. [DOI] [PubMed] [Google Scholar]
- Pardanaud L., Luton D., Prigent M., Bourcheix L.M., Catala M., Dieterlen-Lievre F. Two distinct endothelial lineages in ontogeny, one of them related to hemopoiesis. Development. 1996;122:1363–1371. doi: 10.1242/dev.122.5.1363. [DOI] [PubMed] [Google Scholar]
- Patten S.A., Sihra R.K., Dhami K.S., Coutts C.A., Ali D.W. Differential expression of PKC isoforms in developing zebrafish. Int. J. Dev. Neurosci. 2007;25:155–164. doi: 10.1016/j.ijdevneu.2007.02.003. [DOI] [PubMed] [Google Scholar]
- Risau W., Flamme I. Vasculogenesis. Annu. Rev. Cell Dev. Biol. 1995;11:73–91. doi: 10.1146/annurev.cb.11.110195.000445. [DOI] [PubMed] [Google Scholar]
- Schroeder A., Mueller O., Stocker S., Salowsky R., Leiber M., Gassmann M., Lightfoot S., Menzel W., Granzow M., Ragg T. The RIN: an RNA integrity number for assigning integrity values to RNA measurements. BMC Mol. Biol. 2006;7:3. doi: 10.1186/1471-2199-7-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier D.Y., Weinstein B.M., Detrich H.W., 3rd, Zon L.I., Fishman M.C. Cloche, an early acting zebrafish gene, is required by both the endothelial and hematopoietic lineages. Development. 1995;121:3141–3150. doi: 10.1242/dev.121.10.3141. [DOI] [PubMed] [Google Scholar]
- Veldman M.B., Bemben M.A., Thompson R.C., Goldman D. Gene expression analysis of zebrafish retinal ganglion cells during optic nerve regeneration identifies KLF6a and KLF7a as important regulators of axon regeneration. Dev. Biol. 2007;312:596–612. doi: 10.1016/j.ydbio.2007.09.019. [DOI] [PubMed] [Google Scholar]
- Visvader J.E., Fujiwara Y., Orkin S.H. Unsuspected role for the T-cell leukemia protein SCL/tal-1 in vascular development. Genes Dev. 1998;12:473–479. doi: 10.1101/gad.12.4.473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Bjorklund S., Wasik A.M., Grandien A., Andersson P., Kimby E., Dahlman-Wright K., Zhao C., Christensson B., Sander B. Gene expression profiling and chromatin immunoprecipitation identify DBN1, SETMAR and HIG2 as direct targets of SOX11 in mantle cell lymphoma. PLoS One. 2010;5:e14085. doi: 10.1371/journal.pone.0014085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wernert N., Raes M.B., Lassalle P., Dehouck M.P., Gosselin B., Vandenbunder B., Stehelin D. c-ets1 proto-oncogene is a transcription factor expressed in endothelial cells during tumor vascularization and other forms of angiogenesis in humans. Am. J. Pathol. 1992;140:119–127. [PMC free article] [PubMed] [Google Scholar]
- Westerfield M. The Zebrafish Book: A Guide for the Laboratory Use of Zebrafish (Brachydanio rerio) Eugene, USA: University of Oregon Press; 1989. [Google Scholar]
- Wiley D.M., Jin S.W. Bone morphogenetic protein functions as a context-dependent angiogenic cue in vertebrates. Semin. Cell Dev. Biol. 2011;22:1012–1018. doi: 10.1016/j.semcdb.2011.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilting J., Brand-Saberi B., Huang R., Zhi Q., Kontges G., Ordahl C.P., Christ B. Angiogenic potential of the avian somite. Dev. Dyn. 1995;202:165–171. doi: 10.1002/aja.1002020208. [DOI] [PubMed] [Google Scholar]
- Yamaguchi T.P., Dumont D.J., Conlon R.A., Breitman M.L., Rossant J. flk-1, an flt-related receptor tyrosine kinase is an early marker for endothelial cell precursors. Development. 1993;118:489–498. doi: 10.1242/dev.118.2.489. [DOI] [PubMed] [Google Scholar]
- Yamashita H., Shimizu A., Kato M., Nishitoh H., Ichijo H., Hanyu A., Morita I., Kimura M., Makishima F., Miyazono K. Growth/differentiation factor-5 induces angiogenesis in vivo. Exp. Cell Res. 1997;235:218–226. doi: 10.1006/excr.1997.3664. [DOI] [PubMed] [Google Scholar]