Abstract
Ciliates are an ancient and diverse group of microbial eukaryotes that have emerged as powerful models for RNA-mediated epigenetic inheritance. They possess extensive sets of both tiny and long noncoding RNAs that, together with a suite of proteins that includes transposases, orchestrate a broad cascade of genome rearrangements during somatic nuclear development. This Review emphasizes three important themes: the remarkable role of RNA in shaping genome structure, recent discoveries that unify many deeply diverged ciliate genetic systems, and a surprising evolutionary “sign change” in the role of small RNAs between major species groups.
Introduction
The molecular biology of ciliated protists can be disorienting. One reason is that ciliates show an iconoclastic disregard for textbook models of genome architecture. Two defining features of ciliates are the presence of hair-like superstructures known as cilia, anchored in the cell cortex, and nuclear dimorphism: each cell contains two kinds of nuclei, each with a different genome. At various times in the life cycle, a single cell can contain dozens or, in some species, hundreds of actual nuclei, as genomes are restructured, degraded, fragmented, rebuilt, and amplified to high copy number (Prescott, 1994). In addition to novel cell and genome architecture, thousands of genes in some lineages are scrambled into pieces that are cut and precisely rejoined to create functional coding sequences. Furthermore, this process is epigenetically regulated by RNA, introducing a new perspective on DNA’s role as the primary source of heritable information and variation (Mochizuki et al., 2002; Nowacki et al., 2008; Yao et al., 2003). Some organisms jettison up to 98% of their genomes on the pathway toward restoring functional genes, and the rearranged chromosomes of some species are gene sized, containing telomeres but lacking centromeres (Prescott, 1994; Swart et al., 2013). Even the genetic code has been rewired more often in ciliates than in any other lineage, proving that the code is a far cry from a frozen accident of evolutionary history (Lozupone et al., 2001). Furthermore, Euplotes demonstrates frequent programmed ribosomal frameshifting (reviewed in Klobutcher and Farabaugh, 2002). In fundamental ways, these organisms challenge our model-centered view of eukaryotes and leave us wondering whether we, as members of the more recently diverged evolutionary lineage, might actually be the odd ones out on the genetic playground.
The morphological diversity of ciliates is vast, with 4,500 known species and possibly an order of magnitude more still undescribed (Finlay, 1998; Foissner et al., 2008). Correspondingly, the level of genetic diversity within ciliates dwarfs that among plants, animals, and fungi (Prescott, 1994). Together with diatoms, ciliates comprise a major portion of the world’s plankton and thus play important ecological roles (Caron et al., 2012). Though most are single celled, ciliates can be huge, with some species forming branched colonies and cells of the long, trumpet-shaped Stentor reaching more than 1 mm (Finlay, 1998). Ciliate niches can be fresh or salt water, photosynthetic or heterotrophic, free swimming or benthic, and psychrophillic or other extreme environments, and some species have even adapted to the anaerobic environment of cockroach (Ricard et al., 2008) and frog intestines (Wichterman, 1937). Although a few species are parasitic (Coyne et al., 2011), most feed on algae, bacteria or other ciliates, and some even harbor algal or bacterial symbionts (Finlay and Esteban, 2001; Fokin, 2012).
Nuclear dimorphism, a unifying feature of ciliates, provides a mechanism to segregate two genetic functions within the same cell: a micronucleus (MIC) provides the germline, constituting the only DNA passed from parent to progeny during sexual reproduction, whereas the macronucleus (MAC) performs the somatic functions of transcription and translation for all vegetative growth and a sexual division, which is also the only means by which populations increase in number (Prescott, 1994). Ciliates thus accomplish a division of labor of somatic and germline functions despite being unicellular. Sexual reproduction between compatible mating types initiates genome rearrangement after exchange of haploid micronuclei (Figure 1). The parental MAC is discarded at this point and is gradually replaced by a rearranged copy of the fertilized (zygotic) MIC. Most DNA in the vegetative MIC is not only transcriptionally inactive but is also interrupted by nongenic sequences (internal eliminated sequences, or IESs), which must be removedto create functional chromosomes in the MAC. The retained sequences are known as macronuclear-destined sequences, or MDSs. Some spirotrichous ciliates, such as Oxytricha and Stylonychia (Figure 2), also possess thousands of scrambled genes in their micronuclei, with MDS segments present in an encrypted order or inversely oriented in the micronucleus. This necessitates the precise reordering and splicing together of hundreds of thousands of gene segments to restore coding regions. These organisms eliminate in total well over 90% of the DNA in the MIC during the process of genome rearrangement. Germline transposons (but not all transposase genes; Swart et al., 2013) are also removed during production of a functional MAC, as well as satellite repeats and other MIC-limited noncoding DNA (Prescott, 1994). Genome rearrangements in the oligohymenophorean ciliates Tetrahymena and Paramecium (Figure 2) are less severe but still require deletion of roughly 6,000 or 45,000 IESs, respectively (Arnaiz et al., 2012; Fass et al., 2011), with ~25% of the genome eliminated in Paramecium (Arnaiz et al., 2012) and at least 10%–20% (Yao and Gorovsky, 1974) but as much as 33% eliminated in Tetrahymena (Tetrahymena Comparative Sequencing Project, Broad Institute of Harvard and MIT, http://www.broadinstitute.org/annotation/genome/Tetrahymena/GenomeStats.html) (Coyne et al., 2012).
Figure 1. Simplified Ciliate Life Cycles.

(A and H) Reproductive, vegetative growth occurs a sexually by cell division, including mitosis of the germline micronucleus (MIC, indicated by a circle) and amitosis of the somatic macronucleus (MAC, indicated by an oval).
(B) Starvation induces conjugation between compatible mating types, initiating the nonreproductive sexual cycle.
(C) Meiosis of the MIC produces haploid gametic nuclei.
(D and E) (D) Exchange of haploid micronuclei and fertilization produces two new, diploid, zygotic nuclei (E).
(F) Mitosis of zygotic nuclei produces two identical micronuclei, and one nucleus begins to differentiate into a new MAC.
(G and H) (G) Degradation of the old MAC occurs during differentiation of the new MAC. Mature cells (H) enter again into vegetative growth (Nowacki et al., 2009).
Figure 2. Cladogram of Ciliate Genera Discussed in the Text, along with Other Representative Eukaryotic Genera.
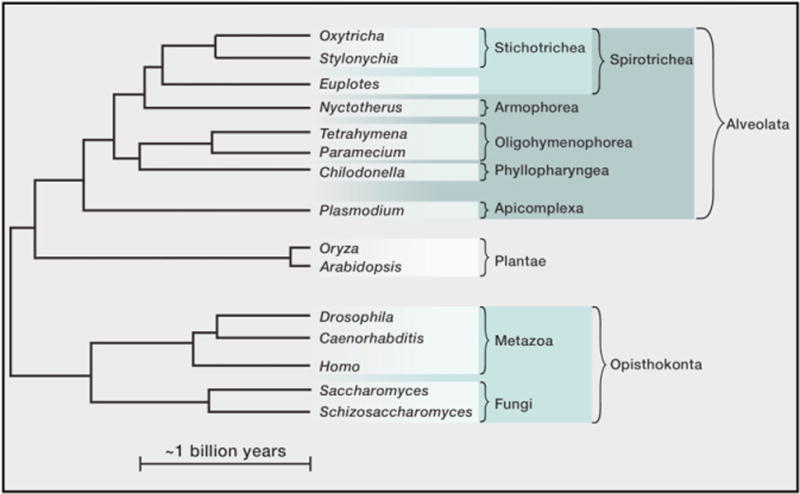
The branching order, branch lengths, and approximate scale bar, for reference, are based on Parfrey et al. (2011), with the addition of Euplotes based on Chang et al. (2005).
Programmed Pruning of the Genome: Ancient Origins from Mobile DNA
An emerging theme in ciliate genome rearrangements is that transposases play central roles in programmed DNA deletion, as well as transposon excision from the micronucleus. Tetrahymena and Paramecium both require a domesticated, single-copy macronuclear PiggyBac family transposase gene for IES removal (Baudry et al., 2009; Cheng et al., 2010). Oxytricha, on the other hand, recruits the services of thousands of micronuclear Tc1/mariner family transposases (Doak et al., 1994; Herrick et al., 1985). RNA interference (RNAi) experiments inhibiting expression of TBE transposase strongly inhibit not only the transposons’ own clearance but also the process of genome rearrangement, leading to accumulation of high-molecular-weight DNA, as well as transposons (Nowacki et al., 2009). The Tec transposons of another spirotrich, Euplotes, are highly abundant and carry the same target sequence duplication (TA) as Euplotes IESs (Jacobs and Klobutcher, 1996; Jacobs et al., 2003; Jahn et al., 1993), suggesting a common mechanism of excision. These observations led Klobutcher and Herrick (1997) to propose that IESs originated as transposons that went through a period of active replication and dispersal through the genome (a bloom phase), but most have since become inactive and degenerated (or faded), retaining only those sequence elements necessary for excision (Klobutcher and Herrick, 1997).
In some species, the sequence similarities between IESs and transposons extend beyond TA repeats. Short IESs in Euplotes crassus have ~8 bp inverted repeats, including the TA duplication. Their consensus sequence, 5′-TATrGCRN-3′, is notably similar to the inverted repeats at the ends of Tec elements (5′-TATAGAGG-3′), and these are also echoed in the Paramecium IES inverted repeat 5′-TAYAGYNR-3′. Together, these sequences also bear similarity to the ends of Tc1/mariner transposons (5′-TACAGTKS-3′; K = G or T, S = C or G, R = G or A) (Jaraczewski and Jahn, 1993; Klobutcher and Herrick, 1995, 1997).
The use of TA repeats to demarcate at least some precisely excised IESs appears universal among the well-studied ciliates (Figures 3A and 3B). Stichotrich species in general, such as Oxytricha, have short regions of microhomology, called pointers, at all sites of programmed IES elimination and MDS recombination. Although pointer length can vary from 2 to 20 bp or more, there is a bias for TA among all 2 bp pointers in Oxytricha (X. Chen and L.F.L., unpublished data). Not only does Paramecium possess TA repeats at all IES termini, but a genome-wide study in Tetrahymena (Fass et al., 2011) uncovered a limited number of small (< 500 bp) precisely excised IESs flanked by TTAA repeats (Fass et al., 2011), whereas most larger, imprecisely excised IESs are flanked by 1–8 bp direct repeats (Yao et al., 2002) reminiscent of the variable pointers in stichotrichs. Furthermore, TTAA is identical to the consensus sequence recognized by PiggyBac transposases in Paramecium and Tetrahymena that have been co-opted for IES excision (Baudry et al., 2009; Cheng et al., 2010; Fraser et al., 1996).
Figure 3. Structural Elements of IES Regions and Transposon-Derived Genes or Systems.
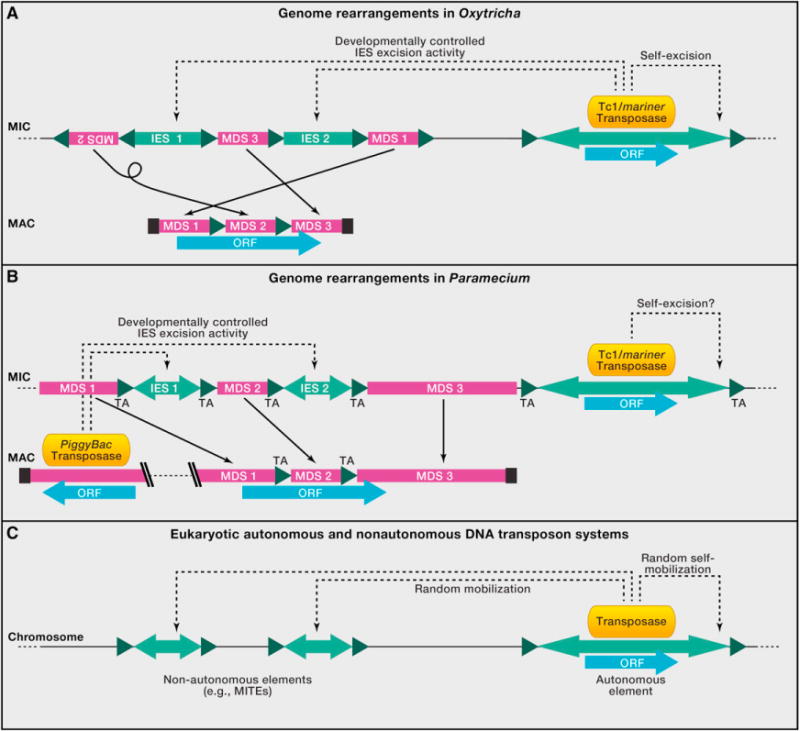
Magenta boxes, macronuclear-destined sequence (MDS); light green, internal eliminated sequences (IES) or transposable elements. Inverted light green arrowheads indicate inverted repeats at ends of deleted sequences. Dark green triangles indicate short direct repeats (pointer sequences). Blue bars indicate open reading frames (ORFs). Solid thin arrows show correspondence between DNA sequences in the MIC and MAC, and black boxes indicate telomeres on MAC chromosomes.
(A) Shown on the left is a schematic scrambled gene in Oxytricha or Stylonychia. In Oxytricha, thousands of MIC-encoded Tc1/mariner transposases (orange) likely participate in removal of IESs (indicated by dotted arrows) as well as themselves (Nowacki et al., 2009).
(B) In oligohymenophorean ciliates, represented by Paramecium, IES excision requires a domesticated PiggyBac transposase encoded in the MAC. Paramecium has only TA dinucleotides as pointers and also flanking Tc1/mariner transposons in the MIC (Arnaiz et al., 2012). Euplotes crassus, a spirotrich, similarly has nonscrambled IESs with TA dinucleotides flanking both IESs and Tec transposons (Jacobs and Klobutcher, 1996). Tetrahymena differs from Paramecium in having mostly imprecise excision of longer intergenic IESs, with some precisely excised IESs flanked by TTAA repeats.
(C) Schematic illustration of eukaryotic nonautonomous and autonomous DNA transposons. The autonomous elements (right) encode transposases that can mobilize truncated and simplified nonautonomous elements (left) throughout the genome. The structure of these elements mirrors the structure of ciliate IESs (nonautonomous elements) and their controlling transposons (autonomous elements), most likely reflecting their evolutionary origins and development (see Klobutcher and Herrick, 1997).
Together, these observations highlight programmed elimination of short IESs flanked by TA (or, in Tetrahymena, TTAA) repeats as a common theme in ciliate genome rearrangements (Figures 3A and 3B). This feature may be ancestral to ciliates, and the expanded variants observed in Tetrahymena and Oxytricha could represent later lineage-specific innovations. Because the preferred integration site of Tc1/mariner class transposons is TA (Plasterk et al., 1999), it is likely that some IESs originated as this type of transposon (Klobutcher and Herrick, 1997). In support of this model, most characterized species contain transposable elements of the Tc1/mariner family in their micronuclear genomes (Arnaiz et al., 2012; Chalker and Yao, 2011; Doak et al., 1994; Jahn et al., 1993; Le Mouël et al., 2003) (Figures 3A and 3B). Though Tetrahymena’s germline transposons are currently incompletely described, the MIC genome assembly (www.broadinstitute.org) contains several BLAST matches to mariner elements (E value cutoff of 10−12), and Eisen et al. (2006) describe several micronuclear-limited Tc1/mariner elements. At some point in the common ancestor of Tetrahymena and Paramecium (class Oligohymenophorea), a PiggyBac-type transposase was captured in the macronucleus and pressed into service for IES excision, usurping a role perhaps previously performed by germline Tc1/mariner transposases, which retained their excision function as they expanded in the germlines of spirotrichs. Over time, most transposons themselves degenerated because they were inactive, retaining only the sequences necessary for efficient excision, leading to the modern transposon-IES systems observed in ciliates.
The transposable element field has long described two types of transposable elements: those that are autonomous, encoding the machinery sufficient for their transposition and those that are nonautonomous and have lost the ability to move themselves but can be moved and replicated by the machinery encoded in the autonomous elements (Casacuberta and Santiago, 2003; Wessler, 2006; Yang et al., 2009) (Figure 3C). One class of nonautonomous elements, known as miniature inverted repeat transposable elements, or MITEs, are several hundred bp in length but have much higher activity and copy number than their cognate full-length transposable elements (Casacuberta and Santiago, 2003; Wessler 2006; Yang et al., 2009). Ciliate transposons and IESs can be viewed as analogs of the autonomous/nonautonomous systems in other eukaryotes (Figure 3). However, ciliate IES are less constrained than MITE elements because they only need to excise themselves from the genome, and they appear in some cases to have become so simplified that sequence similarity to the original transposons has been lost (Figure 3) (Klobutcher and Herrick, 1997). Of course, whether IESs originally invaded as autonomous, full-length elements or as shorter, MITE-like elements remains unknown, but it is possible that there may be some IES that are still actively mobile. Indeed, there are some transposons that interrupt functional genes in Oxytricha’s MIC and are effectively IESs.
This model suggests that nuclear dimorphism and the complex genome rearrangements in ciliates may have emerged as a solution to the problem of transposon invasion. By removing transposons and then reconstructing the genome at every round of sexual division, the organism and its lineage can neatly bypass a transposon’s potentially catastrophic effects, recovering stability at the cost of a more complex genetic system. Furthermore, it is striking that ciliates coevolved with and are now dependent on the products of the parent transposon—namely an efficient transposase—to facilitate the elaborate process of genome remodeling itself and to maintain genome integrity over time.
Discovery of Cytoplasmic, RNA-Mediated Maternal Inheritance in Paramecium
Historically, Paramecium has provided key insights into epigenetics and the roles of noncoding RNA in genome rearrangements. In 1984, a survey of X-ray-induced mutants uncovered strain d48, which lacks a gene for the A surface antigen in its macronucleus (Epstein and Forney, 1984). Macronuclei of wild-type cells contain the gene, and the micronuclei of both wild-type and mutant strains are identical (Epstein and Forney, 1984). This genetic difference is stable and maternally inherited—the presence or absence of the gene in the macronucleus confers the presence or absence of the gene in the macronucleus of the next sexual generation through the process of development (Epstein and Forney, 1984). Moreover, this effect is general—mating type is controlled in a similar fashion (Nanney, 1953), and there are similar maternal effects at other genes (Duharcourt et al., 1995; Scott et al., 1994). Transformation of the d48 macronucleus with segments of the A surface antigen is sufficient to restore the gene to the macronucleus of subsequent generations (Koizumi and Kobayashi, 1989), and retention of some IESs can be induced in a similar manner (Duharcourt et al., 1998). Because the developing, new macronucleus never physically interacts with the old macronucleus, these studies established the existence of diffusible, cytoplasmic trans-nuclear factors that regulate genome rearrangement and mediate epigenetic inheritance.
However, these previous results in Paramecium were challenged by data that appear to demonstrate the opposite effect. In some cases, transformation of the macronucleus induces deletion of the homologous sequences from the macronuclear genome of subsequent generations (Meyer, 1992; Meyer et al., 1997). The observation that deletions correlate with the production of small RNAs from the transgenes, whereas retention does not, offered some clarity (Garnier et al., 2004). Convincing support for this model came from the demonstration that deletions could be programmed by simply feeding Paramecium with E. coli expressing double-stranded RNA that gets processed into small RNAs in the ciliate (Garnier et al., 2004), highlighting the importance of noncoding RNA in the DNA elimination pathway. Furthermore, direct injection of small RNAs is also sufficient to induce elimination of the corresponding DNA sequence, apparently via two possible mechanisms: one inducing degradation of the long, noncoding RNAs that sequester scan RNAs (scnRNAs, described in detail in the next section) in the parental macronucleus and the second by directly targeting elimination of homologous sequences in the developing macronucleus (Lepère et al., 2008).
Tetrahymena and the Origin of an RNAi-Based Model for Genome Rearrangement in Oligohymenophorean Ciliates
Studies of Tetrahymena have contributed many landmark discoveries, ranging from ribozymes (Kruger et al., 1982) (Nobel prize in Chemistry, 1989), telomeres, and telomerase (Blackburn and Gall, 1978; Greider and Blackburn, 1985) (Nobel prize in Physiology or Medicine, 2009) to the purification of dynein (Gibbons and Rowe, 1965) and the discovery of the first histone-modifying enzyme (Brownell et al., 1996).
While investigations of maternal inheritance were underway in Paramecium, Mochizuki and colleagues discovered a vital role for small RNA-binding proteins (and their small RNA cargo) in genome rearrangement in Tetrahymena (Mochizuki et al., 2002). Their seminal work contributed to the small RNA revolution, revealing the first class of Piwi-associated small RNAs. Knockout of Twi1p, a Piwi family member and small RNA-binding protein, blocks genome rearrangement at an early stage (Mochizuki et al., 2002). Since then, many details of the relevant small RNA pathway, known as the scan RNA or scnRNA pathway, have come into focus in studies conducted in both ciliate models (Figure 4A) (reviewed in Duharcourt et al., 2009; Kataoka and Mochizuki, 2011; Mochizuki, 2010).
Figure 4. Two Models of RNA-Mediated Genome Rearrangements in Ciliates.
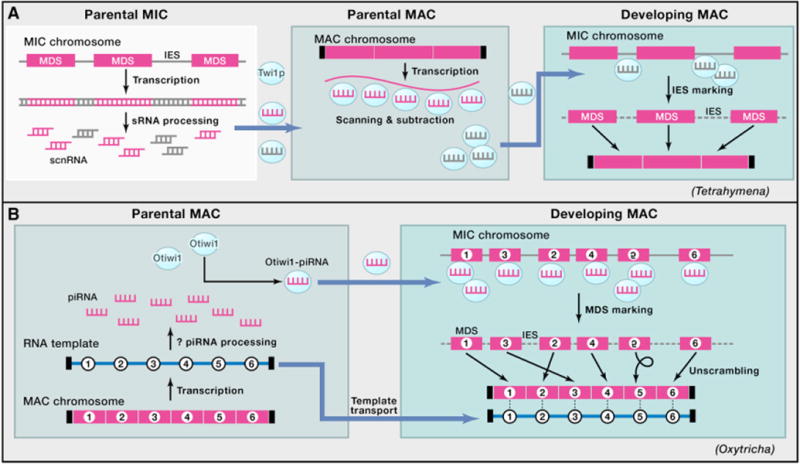
(A) In Tetrahymena, bidirectional transcription of the MIC genome produces double-stranded RNA, which is processed into scnRNA duplexes. The Piwi protein Twi1p loads scnRNAs in the cytoplasm and transports them into the parental MAC to scan the somatic transcriptome. This process enriches for scnRNAs that do not pair with homologous sequences from the parental MAC. The MIC-limited scnRNAs are then transported into the developing MAC, where they recognize and mark IES regionsonMIC chromosomes. IES excision and telomere addition produce mature MAC chromosomes. In both panels, magenta rectangles denote MDSs, and combs indicate sRNA.
(B) In Oxytricha, transcription of either strand of gene-sized chromosomes in the parental MAC produces telomere-containing template RNAs (blue numbered line) during conjugation. Either these template RNAs or other long noncoding RNAs are processed into 27 nt piRNAs. These form a complex with the Piwi protein Otiwi1 that transports them into the newly developing MAC, where the piRNAs recognize and mark the MDS portions of the MIC chromosome that are retained during genome rearrangement. The maternal template RNAs are also transported to the developing MAC, where they guide correct MDS ordering of numbered segments 1–6 (including inversion of segment 5) and DNA repair at recombination junctions to produce mature somatic chromosomes that are capped with short telomeres (small vertical black bars).
In the scnRNA model, the entire micronuclear genome apparently is transcribed in both sense and antisense directions, producing double-stranded RNAs that are substrates for a Dicer-like protein (Carmell and Hannon, 2004; Chalker and Yao, 2001; Lepère et al., 2008). Recent work, however, has demonstrated that the biogenesis of scnRNAs is biased toward IESs at the level of transcription (Schoeberl et al., 2012). Methylation by a Hen1 homolog stabilizes the resulting small RNAs (scnRNAs) (Kurth and Mochizuki, 2009), which are then transported to the parental macronucleus where a “scanning” process (hence the name) selectively degrades small RNAs that hybridize to the parental macronuclear genome (possibly via noncoding RNAs that derive from that genome [Aronica et al., 2008]) (Figure 4A). This scanning step depletes MDS-targeted small RNAs, enriching for those that target IESs (Schoeberl et al., 2012). The discovery that microinjection of double-stranded RNA targeting coding regions induces their deletion in Tetrahymena—effectively reprogramming the coding regions as IES sequence (Yao et al., 2003)—supports this model. Given the presence of functional RNAi machinery in Tetrahymena (Lee and Collins, 2006), the microinjected double-stranded RNA could be processed into novel scnRNAs that are capable of directing the deletion of cognate DNA sequences.
Accumulating data indicate that scnRNAs guide the deposition of chromatin marks, specifically on histones, for DNA elimination. The central player in this process is the histone methyltransferase enhancer of zeste homolog Ezl1, which is required for methylation of histone H3 at lysine 27 (Liu et al., 2007). Methylation of both H3K9 and H3K27 is essential for proper DNA elimination and production of viable progeny (Chung and Yao, 2012; Liu et al., 2004, 2007). Two chromodomain proteins, Pdd1p (Madireddi et al., 1996) and Pdd3p (Nikiforov et al., 2000), are part of the machinery that recognizes these repressive chromatin marks (Liu et al., 2007; Taverna et al., 2002). In turn, these proteins recruit other factors, including Lia1p (Rexer and Chalker, 2007) and other novel genes (Yao et al., 2007), as well as Pdd2p (Smothers et al., 1997). Importantly, simply tethering Pdd1p to a genetic sequence is sufficient to drive DNA elimination (Taverna et al., 2002), suggesting that the primary role of scnRNAs—and the histone modifications that they direct—could be to target Pdd1p to eliminated sequences.
Subsequent DNA processing events may be similar in both Tetrahymena and Paramecium, where domesticated PiggyBac-related transposases localize to developing macronuclei; functional inactivation of these genes inhibits DNA cleavage in both organisms, strongly supporting the mechanistic role of these enzymes as molecular scissors (Baudry et al., 2009; Cheng et al., 2010). Proteins of the nonhomologous end-joining (NHEJ) pathway, Ku80 (Tetrahymena) (Lin et al., 2012), XRCC4 (Paramecium), and Ligase IV (Paramecium) (Kapusta et al., 2011) are vital for repair of cut DNA ends, at least in the oligohymenophorean ciliates.
Oxytricha and Stylonychia: Complicated DNA Rearrangements
Compared to oligohymenophorean ciliates, genome remodeling in stichotrichs is much more extensive. The somatic genome not only eliminates more than 90% of the germline but also severely fragments macronuclear chromosomes down to gene-size molecules, called nanochromosomes, that average just 3 kb (Swart et al., 2013). Furthermore, all surveyed stichotrich species have scrambled genes (Chang et al., 2005), with DNA segments often out of order in the germline nucleus, compared to their order in the functional versions in the macronucleus. These unique features make Oxytricha and Stylonychia important models to study noncoding RNAs that regulate genome remodeling, as well as the so-called “junk DNA” that occupies most of the dispensable portion of the germline.
Macronuclear development in Oxytricha and Stylonychia requires complex genome rearrangements to sort and reorder the tens of thousands of precursor DNA segments, with some genes assembled from ~50 or more pieces (Chang et al., 2005; Prescott, 1994). How cells achieve and maintain the high precision necessary for this unscrambling process has been an active area of study. The pointer repeats, short sequences of microhomology present at the junctions between every pair of rejoined segments, are proposed to participate in the recombination between segments, and this feature suggests further similarity to the NHEJ pathway. However, the short length of pointers (typically 2–20 bp) makes them insufficient to act as guides for accurate assembly of the whole genome. Moreover, the system also manages to tolerate a surprising level of errors of imprecise excision at other regions of microhomology (cryptic pointers) near splice junctions early in rearrangement (Möllenbeck et al., 2008). This led to a proposed need for proofreading during genome rearrangement and the development of a template-guided genome rearrangement model (Prescott et al., 2003; Angeleska et al., 2007) with later experimental support (Nowacki et al., 2008).
The RNA-mediated genome rearrangement pathway, discovered in Oxytricha, suggests that a maternal cache of long, noncoding RNAs—essentially an RNA copy of the somatic genome—transfers to the developing new macronucleus and instructs genome-wide unscrambling (Figure 4B). The proposed long, telomere-containing transcripts are specifically observed during conjugation, and RNAi against these molecules leads to abnormally rearranged chromosomes in the progeny. Furthermore, injection of artificial RNA templates can reprogram rearrangement of the corresponding gene (Nowacki et al., 2008), which provided the strongest support for the template-guided model. Remarkably, the reprogramming effect is stable across multiple sexual generations, underscoring the power of non-coding RNA to shape the genome and to mediate transgenerational epigenetic inheritance. Although the exact mechanism of RNA-guided DNA rearrangement needs more investigation, one clue comes from an unexpected finding that point substitutions close to DNA recombination junctions occasionally transfer from the RNA templates to the rearranged DNA, implicating RNA-directed DNA synthesis during rearrangement (Nowacki et al., 2008). This finding also provides a route for certain acquired somatic substitutions to transfer to the next generation, bypassing the traditional mode of DNA inheritance via the germline. For example, somatic mutations that accumulate during either vegetative growth or RNA template transcription can transfer to the somatic genome during genome rearrangements. Such phenomena may contribute to elevated substitution rates in proteins encoded in the macronucleus (Zufall et al., 2006) and also supply additional epigenetic variation that natural selection can amplify if it leads to faster growth or an increased rate or efficiency of conjugation.
An additional role for the maternal RNA templates is the regulation of chromosome copy number in the new macronucleus after sexual conjugation. In spirotrichs, gene-sized nanochromosomes are present in thousands of copies per cell, which might help accommodate growth of these large eukaryotic cells. In 2010, two studies in Oxytricha and Stylonychia (Heyse et al., 2010; Nowacki et al., 2010) found that injection of wild-type RNA templates during conjugation could increase DNA copy number of the corresponding genes in the progeny, whereas RNAi against a specific template decreased the DNA copy number. This RNA regulation of chromosome copy number effectively regulates gene dosage because there is almost no genetic linkage in the MAC. The effect is also heritable for multiple sexual generations, suggesting stable epigenetic inheritance via maternally expressed RNA and illustrating the multitasking roles of these long, noncoding RNAs.
A new twist on the model for RNA-mediated genome rearrangement comes with the discovery of PIWI-interacting RNAs (piRNAs) in Oxytricha. In contrast to scnRNAs in Tetrahymena and Paramecium, Oxytricha expresses a class of 27 nt short (or small) RNAs (sRNAs) that map only to the macronuclear genome (instead of the germline) (Fang et al., 2012; Zahler et al., 2012). Deep sequencing of these sRNAs suggests that they originate from the whole somatic genome rather than from specific loci, which is different from metazoans. Otiwi1, a Piwi protein that associates with these 27 nt piRNAs, relocalizes from the old to the new macronucleus, suggesting crosstalk between the two nuclei (Fang et al., 2012) (Figure 4B). Notably, injections of 27 nt synthetic RNAs that correspond to normally deleted regions lead to their retention during genome rearrangement, suggesting a protective role for these somatically derived piRNAs (Fang et al., 2012). This is also in striking contrast with the results of small RNA injection in Paramecium, where sRNAs can target corresponding DNA regions for deletion (Lepère et al., 2008). Moreover, the sRNA-induced DNA retention in Oxytricha is inherited across sexual generations, highlighting another stable, epigenetic effect of RNA on genomic DNA processing across multiple generations.
As mentioned in the section on transposon origins, another striking difference between Oxytricha and the oligohymenophoreans is that Oxytricha recruits the services of thousands of germline transposases instead of a single copy domesticated transposase in the MAC. Although the enzymatic mechanism that the transposases catalyze is unclear at present, one natural hypothesis is that they help introduce DNA cleavage, and this would be congruent with the step of RNA-directed DNA synthesis in the RNA template model discussed above. Furthermore, because piRNAs traditionally suppress transposons in other systems, we propose that the relationship that evolved in Oxytricha could be a new route through which piRNAs may antagonize transposon activity, with the piRNAs preventing transposase cleavage in the macronuclear-destined regions that they recognize in the zygotic micronucleus. Such an interaction would also prevent transposons from integrating into the somatic genome, thereby keeping the MAC transposon free. Transmission of heritable information via RNA, rather than directly through DNA, may more generally help exclude DNA transposons from the new somatic genome (Goldman and Landweber, 2012).
Searching for the Origin of Scrambled Genes: Euplotes, Chilodonella, and Nyctotherus
Euplotes sp., members of the class Spirotrichea, have been noted for the prevalence of +1 translational frameshifting (Klobutcher, 2005; Klobutcher and Farabaugh, 2002). Although Euplotes species have similar genome architectures to Oxytricha, to date they lack any evidence of scrambled genes (Prescott, 1994), although genome-wide micronuclear surveys are needed to be confident of this assertion. Similarly to Oxytricha and Stylonychia, the Euplotes crassus macronuclear genome is highly fragmented, containing ~10,000–20,000 gene-sized nanochromosomes (Vinogradov et al., 2012), each amplified to ~1,000 copies. The genetic content of the E. crassus macronucleus is ~40-fold reduced from the sequence complexity of its micronucleus (Baird et al., 1989), like that of Stylonychia. Similar to Paramecium, IESs in Euplotes crassus are short, typically 30–500 bp precisely excised regions flanked by direct TA repeats that initiate an 8 bp motif (see section “Programmed Pruning of the Genome: Ancient Origins from Mobile DNA” for details). It has been speculated that the presence of common TA-containing repeats at both IES and Tec element termini indicate a common mechanism of excision (Klobutcher and Herrick, 1995, 1997) potentially mediated by the Tec transposase itself. One could functionally test this by knocking down the Euplotes Tec transposase genes and measuring IES excision efficiencies, given that RNAi has been demonstrated to work in Euplotes (Paschka et al., 2003).
Taken together, a presumably ancient mechanism that precisely excises TA-flanked IESs appears to be shared by both oligohymenophorean and spirotrichous ciliates and facilitated by lineage-specific transposases. Further divergences in some lineages probably coevolved with the shift toward imprecise, intergenic IESs in Tetrahymena, and, independently, with relaxation of the TA requirement for pointers in Oxytricha’s lineage. This broadening of the sequences that can serve as pointers would have permitted an increase in the complexity of manipulations that the organism’s genetic system can support, creating the opportunity for the emergence of scrambled genes. With that in mind, we propose that investigations of the micronucleus of Euplotes octocarinatus would be valuable, as it is the only known Euplotes species whose pointers include longer, locally unique strings, as well as TA dinucleotides (Tan et al., 1999, 2001; Wang et al., 2005), suggesting that this species may also have the capacity to support complex genome rewiring.
Chilodonella, a member of a third ciliate class, Phyllopharyngea, also produces gene-sized macronuclear chromosomes and appears to be more closely related to oligohymophoreans than stichotrichs (Figure 2). Most notably, its MIC genome contains scrambled genes, like stichotrichs, including some with inversions (Katz and Kovner, 2010). This observation suggests that the origin of scrambled genes could have predated the split of more than one ciliate class and that the capacity was likely present in at least the common ancestor of Phyllopharyngea and Spirotrichea, making it all the more likely that one or more euplotid species (a basal spirotrich) might also have scrambled genes in its germline.
Another promising organism in the hunt for scrambled genes is Nyctotherus (Figure 2), an anaerobic ciliate genus that inhabits the hindgut of cockroaches (sp. ovalis) (Ricard et al., 2008) or frogs (sp. cordiformis) (Wichterman, 1937). These organisms belong to a fourth of 11 ciliate classes, Armophorea (Figure 2), that appears more closely related to the Spirotrichea than Phyl-lopharyngea, and N. ovalis is keenly noted for its replacement of mitochondria by hydrogenosomes (Boxma et al., 2005). Whereas little is known about the germline genome in N. ovalis, the macronuclear genome has been well surveyed and comprises highly fragmented nanochromosomes (Ricard et al., 2008), like Oxytricha, Stylonychia, Euplotes, and Chilodonella. It is possible that a fragmented somatic genome architecture may lead to relaxed constraints on other genomic features, permitting the acquisition of scrambled germline genes. The evolution of genome fragmentation itself appears to be polyphyletic (Riley and Katz, 2001), highlighting the plasticity of ciliate genome architectures.
Conclusions: Epigenetics and Noncoding RNA in Genome Rearrangements
Ciliate model systems offer surprising twists on eukaryotic biology that are often exaggerated phenomena present in many other systems. Viewed differently, many aspects of metazoans may be simplifications of a more universal biology elaborated in the ciliates, albeit altered and refined over two billion years of divergence (Parfrey et al., 2011) (Figure 2). For example, the autonomous/nonautonomous transposon conceptual framework neatly presages the origin and key features of IESs in ciliates (Klobutcher and Herrick, 1997) (Figure 3). As in other eukaryotes, piRNAs are involved in transposon control, but ciliates also take the extreme measure of deleting transposons entirely from their somatic genomes—the ultimate form of genetic silencing. Elements of ciliate biology that initially seem specialized, such as the template model for RNA-guided genome rearrangements, could even underlie some important but rare events in human biology related to cancer (Li et al., 2008; Rowley and Blumenthal, 2008), a situation itself that frequently involves thousands of genome rearrangements (Stephens et al., 2011). Though part of the normal developmental program in ciliates, the massive scale of such genome rearrangements could unleash genome instability in metazoa, highlighting the importance of understanding the mechanisms by which ciliates regulate their rearranging genomes and scrutinize them for accuracy. Lamprey (Smith et al., 2009, 2012) and Ascaris (Wang et al., 2012) provide just two examples in metazoa of carefully programmed somatic genome rearrangements that might offer some parallels to DNA rearrangements in ciliates.
A recurring theme of ciliate biology is the role of RNA as the driver in nucleic acid metabolism (Goldman and Landweber, 2012). Examples include myriad roles of long, noncoding RNAs (Chalker et al., 2005; Chalker and Yao, 2001; Heyse et al., 2010; Lepère et al., 2008; Nowacki et al., 2008, 2010) and small RNAs in the form of scanRNAs (Kataoka and Mochizuki, 2011; Lepère et al., 2009; Schoeberl et al., 2012) and piRNAs (Fang et al., 2012; Zahler et al., 2012) and their interaction. Long non-coding RNAs may serve as both molecular sponges in the macronucleus (Chalker and Yao, 2001; Lepère et al., 2008) and also as docking sites in the zygotic macronucleus for scanRNAs, making genome rearrangements dependent on RNA-RNA interactions at every step (Aronica et al., 2008; Lepère et al., 2008; Nowacki et al., 2011).
Furthermore, ciliates deploy a suite of epigenetic pathways, including RNA-regulated histone modification (reviewed in Chalker, 2008) and DNA methylation (reviewed in Gutierrez et al., 2000) to modulate genome structure. For example, Stylonychia has de novo cytosine methylation of transposable elements (Juranek et al., 2003), and recently, we reported a functional association of extensive cytosine methylation and hydroxymethylation with deletion of repetitive micronuclear elements, the old macronuclear genome, and potential errors of the DNA rearrangement pathway in Oxytricha (Bracht et al., 2012).
In sum, the functional roles of these epigenetic pathways reinforce the persistence of traits that are inherited from the soma but not directly encoded in the germline. The reduced role of the micronucleus is thus to provide the raw DNA material for somatically controlled rearrangement and expression. RNA-mediated transfer of somatic point substitutions (Nowacki et al., 2008) even provides a possible Lamarckian-type mechanism for the inheritance of acquired, nongenetic substitutions and may contribute to the observed acceleration of amino acid substitutions in ciliates (Zufall et al., 2006). The ciliate macronucleus therefore comprises a stably inherited epigenome, shaped by the action of RNA molecules over successive generations and any fitness advantages of the most successful epivariants. David Prescott, whose discoveries sowed the field of ciliate molecular biology, was fond of quoting Hamlet, “There are more things in heaven and earth, Horatio, than are dreamt of in your philosophy” (Hamlet Act 1, Scene 5) in reference to the surprises that ciliates have brought to molecular biology, but this was even before the roles of transposons and noncoding RNA were brought into the picture. We anticipate that these remarkable protists will continue to lead the way in unveiling fundamental biological phenomena, showcasing their epigenomes and programmed pathways for genome instability as extraordinary models of inheritance.
Acknowledgments
The authors acknowledge support from NSF grants 0923810 and 0900544 and NIH grants GM59708 (to L.F.L.) and 1F32GM099462 (to J.R.B.). W.F. was supported by DOD predoctoral fellowship W81XWH-10-1-0122, and A.D.G. is a NASA postdoctoral fellow. The content is solely the responsibility of the authors and does not necessarily represent the official views of any funding agency.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Angeleska A, Jonoska N, Saito M, Landweber LF. RNA-guided DNA assembly. J Theor Biol. 2007;248:706–720. doi: 10.1016/j.jtbi.2007.06.007. [DOI] [PubMed] [Google Scholar]
- Arnaiz O, Mathy N, Baudry C, Malinsky S, Aury JM, Wilkes CD, Garnier O, Labadie K, Lauderdale BE, Le Mouël A, et al. The Paramecium germline genome provides a niche for intragenic parasitic DNA: evolutionary dynamics of internal eliminated sequences. PLoS Genet. 2012;8:e1002984. doi: 10.1371/journal.pgen.1002984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronica L, Bednenko J, Noto T, DeSouza LV, Siu KW, Loidl J, Pearlman RE, Gorovsky MA, Mochizuki K. Study of an RNA helicase implicates small RNA-noncoding RNA interactions in programmed DNA elimination in Tetrahymena. Genes Dev. 2008;22:2228–2241. doi: 10.1101/gad.481908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baird SE, Fino GM, Tausta SL, Klobutcher LA. Micronuclear genome organization in Euplotes crassus: a transposonlike element is removed during macronuclear development. Mol Cell Biol. 1989;9:3793–3807. doi: 10.1128/mcb.9.9.3793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baudry C, Malinsky S, Restituito M, Kapusta A, Rosa S, Meyer E, Bétermier M. PiggyMac, a domesticated piggyBac transposase involved in programmed genome rearrangements in the ciliate Paramecium tetraurelia. Genes Dev. 2009;23:2478–2483. doi: 10.1101/gad.547309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blackburn EH, Gall JG. A tandemly repeated sequence at the termini of the extrachromosomal ribosomal RNA genes in Tetrahymena. J Mol Biol. 1978;120:33–53. doi: 10.1016/0022-2836(78)90294-2. [DOI] [PubMed] [Google Scholar]
- Boxma B, de Graaf RM, van der Staay GWM, van Alen TA, Ricard G, Gabaldón T, van Hoek AHAM, Moon-van der Staay SY, Koopman WJH, van Hellemond JJ, et al. An anaerobic mitochondrion that produces hydrogen. Nature. 2005;434:74–79. doi: 10.1038/nature03343. [DOI] [PubMed] [Google Scholar]
- Bracht JR, Perlman DH, Landweber LF. Cytosine methylation and hydroxymethylation mark DNA for elimination in Oxytricha trifallax. Genome Biol. 2012;13:R99. doi: 10.1186/gb-2012-13-10-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownell JE, Zhou JX, Ranalli T, Kobayashi R, Edmondson DG, Roth SY, Allis CD. Tetrahymena histone acetyltransferase A: a homolog to yeast Gcn5p linking histone acetylation to gene activation. Cell. 1996;84:843–851. doi: 10.1016/s0092-8674(00)81063-6. [DOI] [PubMed] [Google Scholar]
- Carmell MA, Hannon GJ. RNase III enzymes and the initiation of gene silencing. Nat Struct Mol Biol. 2004;11:214–218. doi: 10.1038/nsmb729. [DOI] [PubMed] [Google Scholar]
- Caron DA, Countway PD, Jones AC, Kim DY, Schnetzer A. Marine protistan diversity. Annu Rev Mar Sci. 2012;4:467–493. doi: 10.1146/annurev-marine-120709-142802. [DOI] [PubMed] [Google Scholar]
- Casacuberta JM, Santiago N. Plant LTR-retrotransposons and MITEs: control of transposition and impact on the evolution of plant genes and genomes. Gene. 2003;311:1–11. doi: 10.1016/s0378-1119(03)00557-2. [DOI] [PubMed] [Google Scholar]
- Chalker DL. Dynamic nuclear reorganization during genome remodeling of Tetrahymena. Biochim Biophys Acta. 2008;1783:2130–2136. doi: 10.1016/j.bbamcr.2008.07.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. Nongenic, bidirectional transcription precedes and may promote developmental DNA deletion in Tetrahymena thermophila. Genes Dev. 2001;15:1287–1298. doi: 10.1101/gad.884601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chalker DL, Yao MC. DNA elimination in ciliates: transposon domestication and genome surveillance. Annu Rev Genet. 2011;45:227–246. doi: 10.1146/annurev-genet-110410-132432. [DOI] [PubMed] [Google Scholar]
- Chalker DL, Fuller P, Yao MC. Communication between parental and developing genomes during Tetrahymena nuclear differentiation is likely mediated by homologous RNAs. Genetics. 2005;169:149–160. doi: 10.1534/genetics.104.032300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang WJ, Bryson PD, Liang H, Shin MK, Landweber LF. The evolutionary origin of a complex scrambled gene. Proc Natl Acad Sci USA. 2005;102:15149–15154. doi: 10.1073/pnas.0507682102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng CY, Vogt A, Mochizuki K, Yao MC. A domesticated piggyBac transposase plays key roles in heterochromatin dynamics and DNA cleavage during programmed DNA deletion in Tetrahymena thermophila. Mol Biol Cell. 2010;21:1753–1762. doi: 10.1091/mbc.E09-12-1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chung PH, Yao MC. Tetrahymena thermophila JMJD3 homolog regulates H3K27 methylation and nuclear differentiation. Eukaryot Cell. 2012;11:601–614. doi: 10.1128/EC.05290-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Hannick L, Shanmugam D, Hostetler JB, Brami D, Joardar VS, Johnson J, Radune D, Singh I, Badger JH, et al. Comparative genomics of the pathogenic ciliate Ichthyophthirius multifiliis, its free-living relatives and a host species provide insights into adoption of a parasitic lifestyle and prospects for disease control. Genome Biol. 2011;12:R100. doi: 10.1186/gb-2011-12-10-r100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coyne RS, Stover NA, Miao W. Whole genome studies of Tetrahymena. Methods Cell Biol. 2012;109:53–81. doi: 10.1016/B978-0-12-385967-9.00004-9. [DOI] [PubMed] [Google Scholar]
- Doak TG, Doerder FP, Jahn CL, Herrick G. A proposed superfamily of transposase genes: transposon-like elements in ciliated protozoa and a common “D35E” motif. Proc Natl Acad Sci USA. 1994;91:942–946. doi: 10.1073/pnas.91.3.942. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Butler A, Meyer E. Epigenetic self-regulation of developmental excision of an internal eliminated sequence on Paramecium tetraurelia. Genes Dev. 1995;9:2065–2077. doi: 10.1101/gad.9.16.2065. [DOI] [PubMed] [Google Scholar]
- Duharcourt S, Keller AM, Meyer E. Homology-dependent maternal inhibition of developmental excision of internal eliminated sequences in Paramecium tetraurelia. Mol Cell Biol. 1998;18:7075–7085. doi: 10.1128/mcb.18.12.7075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duharcourt S, Lepère G, Meyer E. Developmental genome rearrangements in ciliates: a natural genomic subtraction mediated by non-coding transcripts. Trends Genet. 2009;25:344–350. doi: 10.1016/j.tig.2009.05.007. [DOI] [PubMed] [Google Scholar]
- Eisen JA, Coyne RS, Wu M, Wu D, Thiagarajan M, Wortman JR, Badger JH, Ren Q, Amedeo P, Jones KM, et al. Macronuclear genome sequence of the ciliate Tetrahymena thermophila, a model eukaryote. PLoS Biol. 2006;4:e286. doi: 10.1371/journal.pbio.0040286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein LM, Forney JD. Mendelian and non-mendelian mutations affecting surface antigen expression in Paramecium tetraurelia. Mol Cell Biol. 1984;4:1583–1590. doi: 10.1128/mcb.4.8.1583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fang W, Wang X, Bracht J, Nowacki M, Landweber L. Piwi-interacting RNAs protect DNA against loss during Oxytricha genome rearrangement. Cell. 2012;151:1243–1255. doi: 10.1016/j.cell.2012.10.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fass JN, Joshi NA, Couvillion MT, Bowen J, Gorovsky MA, Hamilton EP, Orias E, Hong K, Coyne RS, Eisen JA, et al. Genome-scale analysis of programmed DNA elimination sites in Tetrahymena thermophila. G3 (Bethesda) 2011;1:515–522. doi: 10.1534/g3.111.000927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay BJ. The global diversity of protozoa and other small species. Int J Parasitol. 1998;28:29–48. doi: 10.1016/s0020-7519(97)00167-7. [DOI] [PubMed] [Google Scholar]
- Finlay BJ, Esteban GF. Exploring Leeuwenhoek’s legacy: the abundance and diversity of protozoa. Int Microbiol. 2001;4:125–133. doi: 10.1007/s10123-001-0027-y. [DOI] [PubMed] [Google Scholar]
- Foissner W, Chao A, Katz LA. Diversity and geographic distribution of ciliates (Protista: Ciliophora) Biodivers Conserv. 2008;17:345–363. [Google Scholar]
- Fokin SI. Frequency and biodiversity of symbionts in representatives of the main classes of Ciliophora. Eur J Protistol. 2012;48:138–148. doi: 10.1016/j.ejop.2011.12.001. [DOI] [PubMed] [Google Scholar]
- Fraser MJ, Ciszczon T, Elick T, Bauser C. Precise excision of TTAA-specific lepidopteran transposons piggyBac (IFP2) and tagalong (TFP3) from the baculovirus genome in cell lines from two species of Lepidoptera. Insect Mol Biol. 1996;5:141–151. doi: 10.1111/j.1365-2583.1996.tb00048.x. [DOI] [PubMed] [Google Scholar]
- Garnier O, Serrano V, Duharcourt S, Meyer E. RNA-mediated programming of developmental genome rearrangements in Paramecium tetraurelia. Mol Cell Biol. 2004;24:7370–7379. doi: 10.1128/MCB.24.17.7370-7379.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gibbons IR, Rowe AJ. Dynein:A protein with adenosine triphosphatase activity from Cilia. Science. 1965;149:424–426. doi: 10.1126/science.149.3682.424. [DOI] [PubMed] [Google Scholar]
- Goldman AD, Landweber LF. Oxytricha as a modern analog of ancient genome evolution. Trends Genet. 2012;28:382–388. doi: 10.1016/j.tig.2012.03.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greider CW, Blackburn EH. Identification of a specific telomere terminal transferase activity in Tetrahymena extracts. Cell. 1985;43:405–413. doi: 10.1016/0092-8674(85)90170-9. [DOI] [PubMed] [Google Scholar]
- Gutierrez JC, Callejas S, Borniquel S, Martin-Gonzalez A. DNA methylation in ciliates: implications in differentiation processes. Int Microbiol. 2000;3:139–146. [PubMed] [Google Scholar]
- Herrick G, Cartinhour S, Dawson D, Ang D, Sheets R, Lee A, Williams K. Mobile elements bounded by C4A4 telomeric repeats in Oxytricha fallax. Cell. 1985;43:759–768. doi: 10.1016/0092-8674(85)90249-1. [DOI] [PubMed] [Google Scholar]
- Heyse G, Jönsson F, Chang WJ, Lipps HJ. RNA-dependent control of gene amplification. Proc Natl Acad Sci USA. 2010;107:22134–22139. doi: 10.1073/pnas.1009284107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacobs ME, Klobutcher LA. The long and the short of developmental DNA deletion in Euplotes crassus. J Eukaryot Microbiol. 1996;43:442–452. doi: 10.1111/j.1550-7408.1996.tb04503.x. [DOI] [PubMed] [Google Scholar]
- Jacobs ME, Sánchez-Blanco A, Katz LA, Klobutcher LA. Tec3, a new developmentally eliminated DNA element in Euplotes crassus. Eukaryot Cell. 2003;2:103–114. doi: 10.1128/EC.2.1.103-114.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahn CL, Doktor SZ, Frels JS, Jaraczewski JW, Krikau MF. Structures of the Euplotes crassus Tec1 and Tec2 elements: identification of putative transposase coding regions. Gene. 1993;133:71–78. doi: 10.1016/0378-1119(93)90226-s. [DOI] [PubMed] [Google Scholar]
- Jaraczewski JW, Jahn CL. Elimination of Tec elements involves a novel excision process. Genes Dev. 1993;7:95–105. doi: 10.1101/gad.7.1.95. [DOI] [PubMed] [Google Scholar]
- Juranek S, Wieden HJ, Lipps HJ. De novo cytosine methylation in the differentiating macronucleus of the stichotrichous ciliate Stylonychia lemnae. Nucleic Acids Res. 2003;31:1387–1391. doi: 10.1093/nar/gkg233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapusta A, Matsuda A, Marmignon A, Ku M, Silve A, Meyer E, Forney JD, Malinsky S, Bétermier M. Highly precise and developmentally programmed genome assembly in Paramecium requires ligase IV-dependent end joining. PLoS Genet. 2011;7:e1002049. doi: 10.1371/journal.pgen.1002049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kataoka K, Mochizuki K. Programmed DNA elimination in Tetrahymena: a small RNA-mediated genome surveillance mechanism. Adv Exp Med Biol. 2011;722:156–173. doi: 10.1007/978-1-4614-0332-6_10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz LA, Kovner AM. Alternative processing of scrambled genes generates protein diversity in the ciliate Chilodonella uncinata. J Exp Zoolog B Mol Dev Evol. 2010;314:480–488. doi: 10.1002/jez.b.21354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA. Sequencing of random Euplotes crassus macronuclear genes supports a high frequency of +1 translational frameshifting. Eukaryot Cell. 2005;4:2098–2105. doi: 10.1128/EC.4.12.2098-2105.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G. Consensus inverted terminal repeat sequence of Paramecium IESs: resemblance to termini of Tc1-related and Euplotes Tec transposons. Nucleic Acids Res. 1995;23:2006–2013. doi: 10.1093/nar/23.11.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klobutcher LA, Herrick G. Developmental genome reorganization in ciliated protozoa: the transposon link. Prog Nucleic Acid Res Mol Biol. 1997;56:1–62. doi: 10.1016/s0079-6603(08)61001-6. [DOI] [PubMed] [Google Scholar]
- Klobutcher LA, Farabaugh PJ. Shifty ciliates: frequent programmed translational frameshifting in euplotids. Cell. 2002;111:763–766. doi: 10.1016/s0092-8674(02)01138-8. [DOI] [PubMed] [Google Scholar]
- Koizumi S, Kobayashi S. Microinjection of plasmid DNA encoding the A surface antigen of Paramecium tetraurelia restores the ability to regenerate a wild-type macronucleus. Mol Cell Biol. 1989;9:4398–4401. doi: 10.1128/mcb.9.10.4398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger K, Grabowski PJ, Zaug AJ, Sands J, Gottschling DE, Cech TR. Self-splicing RNA: autoexcision and autocyclization of the ribosomal RNA intervening sequence of Tetrahymena. Cell. 1982;31:147–157. doi: 10.1016/0092-8674(82)90414-7. [DOI] [PubMed] [Google Scholar]
- Kurth HM, Mochizuki K. 2′-O-methylation stabilizes Piwi-associated small RNAs and ensures DNA elimination in Tetrahymena. RNA. 2009;15:675–685. doi: 10.1261/rna.1455509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Le Mouël A, Butler A, Caron F, Meyer E. Developmentally regulated chromosome fragmentation linked to imprecise elimination of repeated sequences in paramecia. Eukaryot Cell. 2003;2:1076–1090. doi: 10.1128/EC.2.5.1076-1090.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee SR, Collins K. Two classes of endogenous small RNAs in Tetrahymena thermophila. Genes Dev. 2006;20:28–33. doi: 10.1101/gad.1377006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère G, Bétermier M, Meyer E, Duharcourt S. Maternal non-coding transcripts antagonize the targeting of DNA elimination by scanRNAs in Paramecium tetraurelia. Genes Dev. 2008;22:1501–1512. doi: 10.1101/gad.473008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lepère G, Nowacki M, Serrano V, Gout JF, Guglielmi G, Duharcourt S, Meyer E. Silencing-associated and meiosis-specific small RNA pathways in Paramecium tetraurelia. Nucleic Acids Res. 2009;37:903–915. doi: 10.1093/nar/gkn1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li H, Wang JL, Mor G, Sklar J. A neoplastic gene fusion mimics trans-splicing of RNAs in normal human cells. Science. 2008;321:1357–1361. doi: 10.1126/science.1156725. [DOI] [PubMed] [Google Scholar]
- Lin IT, Chao JL, Yao MC. An essential role for the DNA breakage-repair protein Ku80 in programmed DNA rearrangements in Tetrahymena thermophila. Mol Biol Cell. 2012;23:2213–2225. doi: 10.1091/mbc.E11-11-0952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu YF, Mochizuki K, Gorovsky MA. Histone H3 lysine 9 methylation is required for DNA elimination in developing macronuclei in Tetrahymena. Proc Natl Acad Sci USA. 2004;101:1679–1684. doi: 10.1073/pnas.0305421101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Taverna SD, Muratore TL, Shabanowitz J, Hunt DF, Allis CD. RNAi-dependent H3K27 methylation is required for heterochromatin formation and DNA elimination in Tetrahymena. Genes Dev. 2007;21:1530–1545. doi: 10.1101/gad.1544207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lozupone CA, Knight RD, Landweber LF. The molecular basis of nuclear genetic code change in ciliates. Curr Biol. 2001;11:65–74. doi: 10.1016/s0960-9822(01)00028-8. [DOI] [PubMed] [Google Scholar]
- Madireddi MT, Coyne RS, Smothers JF, Mickey KM, Yao MC, Allis CD. Pdd1p, a novel chromodomain-containing protein, links heterochromatin assembly and DNA elimination in Tetrahymena. Cell. 1996;87:75–84. doi: 10.1016/s0092-8674(00)81324-0. [DOI] [PubMed] [Google Scholar]
- Meyer E. Induction of specific macronuclear developmental mutations by microinjection of a cloned telomeric gene in Paramecium primaurelia. Genes Dev. 1992;6:211–222. doi: 10.1101/gad.6.2.211. [DOI] [PubMed] [Google Scholar]
- Meyer E, Butler A, Dubrana K, Duharcourt S, Caron F. Sequence-specific epigenetic effects of the maternal somatic genome on developmental rearrangements of the zygotic genome in Paramecium primaurelia. Mol Cell Biol. 1997;17:3589–3599. doi: 10.1128/mcb.17.7.3589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K. DNA rearrangements directed by non-coding RNAs in ciliates. Wiley Interdiscip. Rev RNA. 2010;1:376–387. doi: 10.1002/wrna.34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mochizuki K, Fine NA, Fujisawa T, Gorovsky MA. Analysis of a piwi-related gene implicates small RNAs in genome rearrangement in Tetrahymena. Cell. 2002;110:689–699. doi: 10.1016/s0092-8674(02)00909-1. [DOI] [PubMed] [Google Scholar]
- Möllenbeck M, Zhou Y, Cavalcanti ARO, Jönsson F, Higgins BP, Chang WJ, Juranek S, Doak TG, Rozenberg G, Lipps HJ, Landweber LF. The pathway to detangle a scrambled gene. PLoS ONE. 2008;3:e2330. doi: 10.1371/journal.pone.0002330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nanney DL. Mating Type Determination in Paramecium aurelia, a Model of Nucleo-Cytoplasmic Interaction. Proc Natl Acad Sci USA. 1953;39:113–119. doi: 10.1073/pnas.39.2.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nikiforov MA, Gorovsky MA, Allis CD. A novel chromodomain protein, pdd3p, associates with internal eliminated sequences during macronuclear development in Tetrahymena thermophila. Mol Cell Biol. 2000;20:4128–4134. doi: 10.1128/mcb.20.11.4128-4134.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Vijayan V, Zhou Y, Schotanus K, Doak TG, Landweber LF. RNA-mediated epigenetic programming of a genome-rearrangement pathway. Nature. 2008;451:153–158. doi: 10.1038/nature06452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Higgins BP, Maquilan GM, Swart EC, Doak TG, Landweber LF. A functional role for transposases in a large eukaryotic genome. Science. 2009;324:935–938. doi: 10.1126/science.1170023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Haye JE, Fang W, Vijayan V, Landweber LF. RNA-mediated epigenetic regulation of DNA copy number. Proc Natl Acad Sci USA. 2010;107:22140–22144. doi: 10.1073/pnas.1012236107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nowacki M, Shetty K, Landweber LF. RNA-mediated epigenetic programming of genome rearrangements. Annu Rev Genomics Hum Genet. 2011;12:367–389. doi: 10.1146/annurev-genom-082410-101420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parfrey LW, Lahr DJ, Knoll AH, Katz LA. Estimating the timing of early eukaryotic diversification with multigene molecular clocks. Proc Natl Acad Sci USA. 2011;108:13624–13629. doi: 10.1073/pnas.1110633108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paschka AG, Jonsson F, Maier V, Mollenbeck M, Paeschke K, Postberg J, Rupprecht S, Lipps HJ. The use of RNAi to analyze gene function in spirotrichous ciliates. Eur J Protistol. 2003;39:449–454. [Google Scholar]
- Plasterk RHA, Izsvák Z, Ivics Z. Resident aliens: the Tc1/mariner superfamily of transposable elements. Trends Genet. 1999;15:326–332. doi: 10.1016/s0168-9525(99)01777-1. [DOI] [PubMed] [Google Scholar]
- Prescott DM. The DNA of ciliated protozoa. Microbiol Rev. 1994;58:233–267. doi: 10.1128/mr.58.2.233-267.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prescott DM, Ehrenfeucht A, Rozenberg G. Template-guided recombination for IES elimination and unscrambling of genes in stichotrichous ciliates. J Theor Biol. 2003;222:323–330. doi: 10.1016/s0022-5193(03)00037-7. [DOI] [PubMed] [Google Scholar]
- Rexer CH, Chalker DL. Lia1p, a novel protein required during nuclear differentiation for genome-wide DNA rearrangements in Tetrahymena thermophila. Eukaryot Cell. 2007;6:1320–1329. doi: 10.1128/EC.00157-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ricard G, de Graaf RM, Dutilh BE, Duarte I, van Alen TA, van Hoek AHAM, Boxma B, van der Staay GWM, van der Staay SYM, Chang WJ, et al. Macronuclear genome structure of the ciliate Nyctotherus ovalis: Single-gene chromosomes and tiny introns. BMC Genomics. 2008;9:587. doi: 10.1186/1471-2164-9-587. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riley JL, Katz LA. Widespread distribution of extensive chromosomal fragmentation in ciliates. Mol Biol Evol. 2001;18:1372–1377. doi: 10.1093/oxfordjournals.molbev.a003921. [DOI] [PubMed] [Google Scholar]
- Rowley JD, Blumenthal T. The cart before the horse. Science. 2008;321:1302–1304. doi: 10.1126/science.1163791. [DOI] [PubMed] [Google Scholar]
- Schoeberl UE, Kurth HM, Noto T, Mochizuki K. Biased transcription and selective degradation of small RNAs shape the pattern of DNA elimination in Tetrahymena. Genes Dev. 2012;26:1729–1742. doi: 10.1101/gad.196493.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott JM, Mikami K, Leeck CL, Forney JD. Non-Mendelian inheritance of macronuclear mutations is gene specific in Paramecium tetraurelia. Mol Cell Biol. 1994;14:2479–2484. doi: 10.1128/mcb.14.4.2479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Antonacci F, Eichler EE, Amemiya CT. Programmed loss of millions of base pairs from a vertebrate genome. Proc Natl Acad Sci USA. 2009;106:11212–11217. doi: 10.1073/pnas.0902358106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith JJ, Baker C, Eichler EE, Amemiya CT. Genetic consequences of programmed genome rearrangement. Curr Biol. 2012;22:1524–1529. doi: 10.1016/j.cub.2012.06.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smothers JF, Mizzen CA, Tubbert MM, Cook RG, Allis CD. Pdd1p associates with germline-restricted chromatin and a second novel anlagen-enriched protein in developmentally programmed DNA elimination structures. Development. 1997;124:4537–4545. doi: 10.1242/dev.124.22.4537. [DOI] [PubMed] [Google Scholar]
- Stephens PJ, Greenman CD, Fu BY, Yang FT, Bignell GR, Mudie LJ, Pleasance ED, Lau KW, Beare D, Stebbings LA, et al. Massive genomic rearrangement acquired in a single catastrophic event during cancer development. Cell. 2011;144:27–40. doi: 10.1016/j.cell.2010.11.055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swart E, Bracht JR, Magrini V, Minx P, Chen X, Zhou Y, Khurana JS, Goldman AD, Nowacki M, Schotanus K, et al. The Oxytricha trifallax macronuclear genome: a complex eukaryotic genome with 16,000 tiny chromosomes. PLoS Biol. 2013;11:e1001473. doi: 10.1371/journal.pbio.1001473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan M, Heckmann K, Brünen-Nieweler C. The micronuclear gene encoding a putative aminoacyl-tRNA synthetase cofactor of the ciliate Euplotes octocarinatus is interrupted by two sequences that are removed during macronuclear development. Gene. 1999;233:131–140. doi: 10.1016/s0378-1119(99)00146-8. [DOI] [PubMed] [Google Scholar]
- Tan M, Heckmann K, Brünen-Nieweler C. Analysis of micronu-clear, macronuclear and cDNA sequences encoding the regulatory subunit of cAMP-dependent protein kinase of Euplotes octocarinatus: evidence for a ribosomal frameshift. J Eukaryot Microbiol. 2001;48:80–87. doi: 10.1111/j.1550-7408.2001.tb00418.x. [DOI] [PubMed] [Google Scholar]
- Taverna SD, Coyne RS, Allis CD. Methylation of histone h3 at lysine 9 targets programmed DNA elimination in Tetrahymena. Cell. 2002;110:701–711. doi: 10.1016/s0092-8674(02)00941-8. [DOI] [PubMed] [Google Scholar]
- Vinogradov DV, Tsoi OV, Zaika AV, Lobanov AV, Turanov AA, Gladishev VN, Gel’fand MS. Draft macronucleus genome of Euplotes crassus ciliate. Mol Biol. 2012;46:328–333. [PubMed] [Google Scholar]
- Wang W, Zhi H, Chai BF, Liang AH. Cloning and sequence analysis of the micronuclear and macronuclear gene encoding Rab protein of Euplotes octocarinatus. Biosci Biotechnol Biochem. 2005;69:649–652. doi: 10.1271/bbb.69.649. [DOI] [PubMed] [Google Scholar]
- Wang J, Mitreva M, Berriman M, Thorne A, Magrini V, Koutsovoulos G, Kumar S, Blaxter ML, Davis RE. Silencing of germline-ex pressed genes by DNA elimination in somatic cells. Dev Cell. 2012;23:1072–1080. doi: 10.1016/j.devcel.2012.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wessler SR. Transposable elements and the evolution of eukaryotic genomes. Proc Natl Acad Sci USA. 2006;103:17600–17601. doi: 10.1073/pnas.0607612103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wichterman R. Division and conjugation in Nyctotherus cordiformis (Ehr) Stein (Protozoa, Ciliata) with special reference to the nuclear phenomena. J Morphol. 1937;60:563–611. [Google Scholar]
- Yang G, Nagel DH, Feschotte C, Hancock CN, Wessler SR. Tuned for transposition: molecular determinants underlying the hyperactivity of a Stowaway MITE. Science. 2009;325:1391–1394. doi: 10.1126/science.1175688. [DOI] [PubMed] [Google Scholar]
- Yao MC, Gorovsky MA. Comparison of the sequences of macro- and micronuclear DNA of Tetrahymena pyriformis. Chromosoma. 1974;48:1–18. doi: 10.1007/BF00284863. [DOI] [PubMed] [Google Scholar]
- Yao MC, Duharcourt S, Chalker D. Genome-wide rearrangements of DNA in ciliates. In: Craig RCN, Gellert M, Lambowitz A, editors. Mobile DNA II. Washington, DC: ASM Press; 2002. pp. 730–758. [Google Scholar]
- Yao MC, Fuller P, Xi X. Programmed DNA deletion as an RNA-guided system of genome defense. Science. 2003;300:1581–1584. doi: 10.1126/science.1084737. [DOI] [PubMed] [Google Scholar]
- Yao MC, Yao CH, Halasz LM, Fuller P, Rexer CH, Wang SH, Jain R, Coyne RS, Chalker DL. Identification of novel chromatin-associated proteins involved in programmed genome rearrangements in Tetrahymena. J Cell Sci. 2007;120:1978–1989. doi: 10.1242/jcs.006502. [DOI] [PubMed] [Google Scholar]
- Zahler AM, Neeb ZT, Lin A, Katzman S. Mating of the stichotrichous ciliate Oxytricha trifallax induces production of a class of 27 nt small RNAs derived from the parental macronucleus. PLoS ONE. 2012;7:e42371. doi: 10.1371/journal.pone.0042371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zufall RA, McGrath CL, Muse SV, Katz LA. Genome architecture drives protein evolution in ciliates. Mol Biol Evol. 2006;23:1681–1687. doi: 10.1093/molbev/msl032. [DOI] [PubMed] [Google Scholar]


