Abstract
Friedreich's ataxia is the most common inherited autosomal recessive ataxia and is characterized by progressive degeneration of the peripheral and central nervous systems and cardiomyopathy. This disease is caused by the silencing of the FXN gene and reduced levels of the encoded protein, frataxin. Frataxin is a mitochondrial protein that functions primarily in iron-sulfur cluster synthesis. This small protein with an α/β sandwich fold undergoes complex processing and imports into the mitochondria, generating isoforms with distinct N-terminal lengths which may underlie different functionalities, also in respect to oligomerization. Missense mutations in the FXN coding region, which compromise protein folding, stability, and function, are found in 4% of FRDA heterozygous patients and are useful to understand how loss of functional frataxin impacts on FRDA physiopathology. In cells, frataxin deficiency leads to pleiotropic phenotypes, including deregulation of iron homeostasis and increased oxidative stress. Increasing amount of data suggest that oxidative stress contributes to neurodegeneration in Friedreich's ataxia.
1. Friedreich's Ataxia: Origin, Clinical Features, and Neurodegeneration
The main feature of Friedreich's ataxia (FRDA) disease is progressive and unremitting ataxia [1–3]. The first symptoms appear at puberty but onset of the disease can occur from infancy to after 60 years [4]. Major neurologic signs include gait and limb ataxia, tendon areflexia, dysarthria, sensory loss, and pyramidal signs [5]. Cardiomyopathy is a frequent symptom and is associated with a severe prognosis, particularly in young patients [6]. Some patients can also develop skeletal deformation, ocular abnormalities, hearing loss, and diabetes [5]. The neuropathology of FRDA involves degeneration of the dorsal root ganglia, peripheral nerves, the spinal cord, and the dentate nucleus in the cerebellum [7]. Patient nerves show axonal neuropathy with loss of large myelinated fibers and an increase in the number of small unmyelinated fibers [8, 9].
FRDA is caused by a GAA trinucleotide repeat expansion in the first intron of the FXN gene [10]. The majority of patients are homozygous for the trinucleotide expansion but in 4% of patients, one allele presents point mutations in the coding region. Expanded alleles lead to the inhibition of FXN expression resulting in decreased levels of the encoded protein, frataxin [10, 11]. The transcriptional repression of the FXN gene induced by the GAA expansion is due to arrest of RNA polymerase II progression and to heterochromatin-mediated gene silencing [12–16]. Frataxin is a mitochondrial protein involved in cellular iron use and maintenance of the redox status [4]. Although the function of frataxin has been a matter of debate since its discovery, it is now generally accepted that its primary function is in iron-sulfur cluster biosynthesis [17–20].
Frataxin is expressed in all cells of eukaryotic organisms. However, the levels of FXN mRNA and frataxin show tissue specificity that partially correlates with the sites of disease. In humans, the highest levels of expression are found in the heart and spinal cord and lower levels are observed in the cerebellum, liver, skeletal muscle, and pancreas [10]. The differential sensitivity of the tissues to frataxin deficiency is not clear but may depend on the cellular metabolism and/or on the somatic instability of expanded GAA triplet repeats [4, 21]. For dorsal root ganglia, one of the first tissues affected in FRDA patients, it was shown that somatic instability starts during embryonic development and continues throughout life, resulting in progressive, age-dependent accumulation of larger GAA triplet repeat expansions [21, 22]. Nevertheless, understanding why only certain tissues are sensitive to frataxin depletion will contribute to a better understanding of the pathophysiology of the disease. Oxidative stress has been suggested to be one of the major inducers of neurodegeneration, but the underlying mechanisms are not fully understood [4, 23, 24]. Antioxidant therapy has been tested since the discovery of the gene with different molecules with limited success in stopping the progression of the disease; however, it is a potential target to treat the disease, and new molecules are being tested.
2. The Frataxin Protein: Structure and Function
Human frataxin (FXN) is a small protein, which is involved in the mitochondrial biogenesis of iron-sulfur clusters (ISCs). These inorganic structures are essential redox cofactors found in several respiratory and metabolic enzymes within mitochondria. Although frataxin function remains to be fully elucidated at the molecular level, a wealth of biochemical data and the determination of three-dimensional structures from several homologues of FXN, particularly yeast frataxin (Yfh1), have contributed to substantial progress on understanding frataxin function [25–29].
The frataxin fold is characterized by a planar α-β sandwich motif, comprising two terminal α-helices, alongside with five antiparallel β-strands that make up two planes which are intersected by additional β-strands, thus composing the structure (Figure 1(a)). Several frataxins have been shown to undergo oligomerization reactions, assembling into trimers, hexamers, and larger order 24- and 48-mer assemblies, although there is no consensus in the community regarding the possibility of these reactions taking place in vivo or on their functional relevance (Figure 1(b)). Frataxin function involves participating in larger molecular assemblies with the several components from the mitochondrial ISC assembly machinery, and the molecular details of these interactions are only now starting to be elucidated with the disclosure of possible interactions taking place in such quaternary complexes [30–32]. Nevertheless, other still controversial roles have been proposed for frataxin, including delivery of iron to ferrochelatase for heme synthesis, recovery of the oxidatively damaged [3Fe4S] cluster in aconitase, and iron storage (reviewed in [17]).
Figure 1.
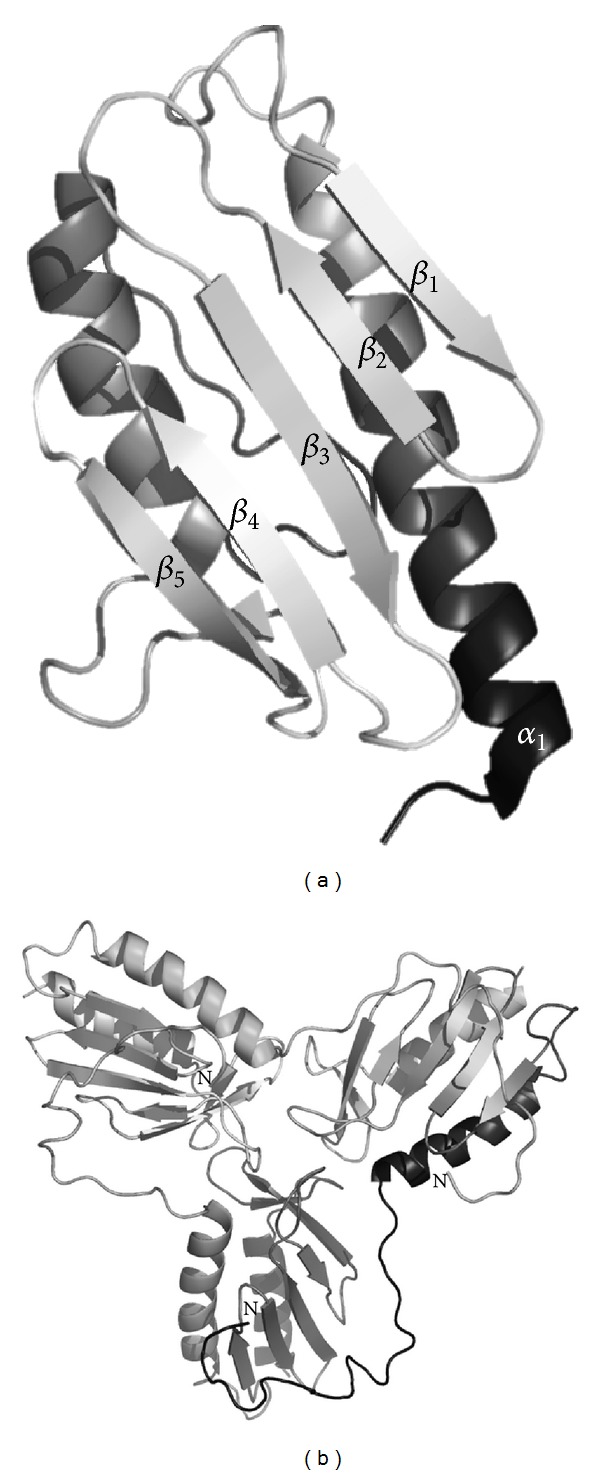
Structures of the frataxin monomer and trimer, denoting the typical α/β fold in which the α-helices pack against the β-sheet strands. (a) Structure of the human frataxin monomer (PDB: 3S4M). (b) Structure of the yeast frataxin trimer (PDB: 3OEQ). Note that in the trimer structure, unwinding of the N-terminal α-helix affords an interaction with a β-sheet from the nearby subunit.
One important region in the frataxin fold is the so-called acidic region, which comprises a set of acidic residues located within the first α-helix and the edge of the first β-strand (α1 and β1 in Figure 1(a)), that are involved in low affinity iron-binding [33]. Nevertheless, iron-binding is essential for the interaction between Yfh1 frataxin and the Isu scaffold protein from the ISC machinery [34]. Studies on Yfh1 focusing on a set of functional mutants in the acidic region have shown that charge-to-neutral alterations in the ridge do not abolish iron-binding, but rather decrease binding affinity [35]. Indeed, frataxin iron-binding capacity is quite robust, as even upon changing five of the most conserved residues from the putative iron-binding region, at least two iron atoms per monomer can be bound. This study has also elicited an interesting trade-off between frataxin function and stability of the fold. Although these negative charges have a functional role, at the same time, they significantly impair Yfh1 stability as their replacement results in a dramatic increase of protein stability while reducing conformational plasticity. The acidic ridge has thus evolved to favor function over stability, therefore highlighting its importance on frataxin structure. On the other hand, mutations of conserved residues of the portion of the acid ridge found in the β-sheet, affected interaction with the scaffold protein Isu, but did not affect overall protein stability [35].
Frataxin processing and cleavage upon import into the mitochondria results, in some cases, in the production of diverse isoforms with distinct structural properties, especially in what concerns oligomerization propensity [36–38]. Human frataxin is a 210-amino acid protein which upon expression in the cytosol is imported into the mitochondria matrix, where it undergoes a complex processing resulting in isoforms with different lengths at the N-terminus [36–38]. Identified isoforms include shorter frataxin versions (processed at positions 42- and 56-) and longer ones (processed at positions 78- and 81-). In humans, the most abundant products seem to be the monomeric form FXN(81-201) which predominates in fibroblasts, alongside with the oligomerization prone longer FXN(42-120) isoform, which is abundant in the heart, cerebellum, and dividing fibroblasts [39]. In fact, distinct N-terminal processing of human frataxin has been suggested to result in proteins with distinct functionalities in what concerns interactions with the ISC machinery proteins and iron-binding properties [17, 39]. In yeast, one major form of processed frataxin is detected in the mitochondria (corresponding to Yfh1 52-174), which seems to be present mostly in the form of trimers, unless protein overexpression takes place which then generates larger oligomers [40]. Stress and increased iron uptake seem also to induce the formation of higher order Yfh1 oligomers [41].
One aspect which remains to be fully investigated is whether frataxin may be itself a target for oxidative modifications in the context of FRDA in which very low frataxin levels are present with iron accumulation in an environment prone to oxidative reactions. To establish a proof of principle, the susceptibility of frataxin to amino acid carbonylation and nitration was investigated [42]. The results on FXN(91-210) showed that residues in the β-sheet surface (Tyr143, Tyr174, Tyr205, and Trp155) were preferential targets, and that modifications did not alter substantially iron binding or protein stability. Interestingly, the strictly conserved Trp155, which is mutated in early onset patients, is a hot spot for modifications leading to the speculation that this could be an effective mechanism to modulate frataxin interactions and thus its function [42].
3. FRDA-Related Frataxin Mutations Result in a Loss of Function Misfolding Disease
A small fraction of FRDA patients (around 4%) are compound heterozygotes for the intronic GAA trinucleotide repeat expansion, carrying in the other allele a missense mutation in the coding region of the FXN gene (Figure 2). These mutations are useful to understand how loss of functional frataxin impacts on FRDA physiopathology. Some of these mutations can be grouped according to FRDA symptoms severity: for example, whereas the FXN-p.Ile154Phe and FXN-p.Trp155Arg mutations lead to severe FRDA, the mutations FXN-p.Gly130Val and the FXN-p.Asp122Tyr account for milder clinical symptoms, although the latter has a very low prevalence [43]. Overall, the link between a point mutation in frataxin and the disease physiopathology remains unclear. Whereas first approaches shed light onto possible molecular mechanisms of disease by analyzing effects of mutations on protein folding and stability [44, 45], more recently mutations have also been studied in the context of cell models [46] and distinct frataxin isoforms with different N-terminal processing [47], and for some mutants, crystal structure information has been obtained [48].
Figure 2.
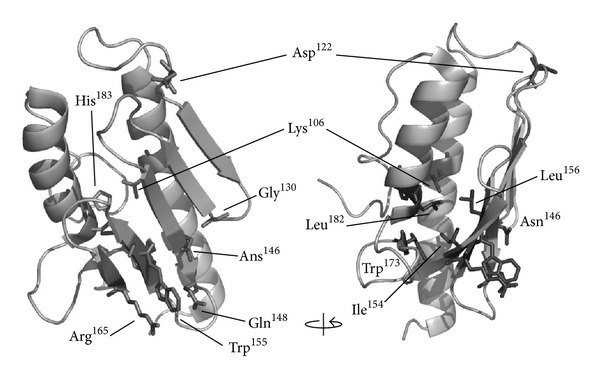
Mapping of amino acids mutated in FRDA compound heterozygous patients. FRDA mutations indicated on the human frataxin structure (PDB: 3S4M): Lys106Ser, Asp122Tyr, Gly130Val, Asn146Lys, Gln148Arg, Ile154Phe, Trp155Arg, Leu156Pro, Arg165Cys, Trp173Gly, Leu182Phe, Leu182His, and His183Arg. The arrow denotes a 90° rotation.
The initial studies on FRDA frataxin mutants focused on the aggressive phenotype FXN-p.Ile154Phe and FXN-p.Trp155Arg variants, studied in the background of the shorter processed FXN(91-210) protein [44]. The former is among the most common clinical mutation, and affects an isoleucine residue at the protein core, and thus is expected to directly affect the structure and stability of the protein fold. The latter affects a conserved tryptophan residue at the protein surface in the exposed region of a β-sheet. The study has shown that these FRDA frataxin variants retained the native fold under physiological conditions but were thermodynamically destabilized in respect to normal frataxin had a higher tendency towards proteolytic degradation, and iron-binding was only partly impaired, nevertheless with concomitant protein aggregation [44]. The implication of these findings was that the severe mutations did not abolish the expression of the FRDA frataxin variants, suggesting that these proteins are formed under physiological conditions. Interestingly, a FRDA murine fibroblast cell model based on the FXN-p.Ile154Phe has corroborated this observation in vivo, as transgenic expression of this pathological mutation partly rescued endogenous frataxin deficiency [46]. Further, these studies also evidenced decreased activity of iron-sulfur proteins and accumulation of iron [46], in agreement with the poorer iron-binding ability which had been observed in vitro [44].
The evidence that the pathogenic mechanism in FRDA mutant frataxin results from frataxin misfolding and instability suggests that, although quite different from other neurodegenerative diseases involving toxic aggregation and metal ions [49], FRDA in compound heterozygous patients can be classified as a protein misfolding disease [50]. In order to further explore this possibility, a broader study encompassing also frataxin mutations yielding the milder FRDA forms FXN-p.Asp122Tyr and FXN-p.Gly130Val was undertaken [45]. The conclusion from those studies showed that FRDA frataxins exhibit distinct degrees of conformational defects that impair folding and function. For example, protein degradation propensity does not correlate necessarily with disease severity, as the two milder mutations were found for example to undergo proteolysis at higher rates than the severe ones, which rather seem to expose more particular part of the protein. An exploration of folding defects caused by mutations resulting in a change in the soluble : insoluble ratio upon recombinant expression showed that the severe FXN-p.Trp155Arg and FXN-p.Ile154Phe variants are mostly expressed as insoluble peptides, indicating that these mutations affect early folding [45]. Interestingly these results were corroborated by an investigation of the effects of these mutations in the context of the isoforms FXN(42-210) and FXN(81-210) in cell-free and different cellular models [47]. In addition, this study has shown that the FXN-p.Trp155Arg mutation destabilizes FXN(42-210) to a greater extent, as the mutation in this form makes the protein more susceptible to in vivo degradation than in the FXN(81-210) form. Likewise, the FXN-p.Ile154Phe variant in FXN(42-210) has a strongly compromised solubility; however, in FXN(81-210) yields a folded polypeptide when expressed in different cells types [47], in agreement with the results obtained in vitro for the purified recombinant mutant variant [45].
4. Oxidative Stress in Frataxin-Deficient Cells and Neurodegeneration
Oxidative stress is a central feature of FRDA disease and still a privileged target for therapy [4, 23, 24]. An increasing amount of data from different organisms support the hypothesis that frataxin-deficiency causes a deregulation in the antioxidant defenses, which result in oxidative stress and pathology [4, 24, 51, 52]. Increased levels of prooxidant molecules such as H2O2 and superoxide have been detected in yeast, Drosophila and FRDA patient cells [40, 53–58]. In addition, frataxin deficiency increases the cellular sensitivity to a wide variety of prooxidants in yeast [58, 59], Caenorhabditis elegans [60], Drosophila [56, 61, 62], mouse [63], and patient FRDA cells [64–68].
The eukaryotic cellular response to oxidative stress involves the induction of detoxifying enzymes such as superoxide dismutases (SODs), an increase in glutathione and NADPH synthesis, a decrease in the reduced to oxidized glutathione ratio and glutathionylation of target proteins [69]. SODs convert superoxide into H2O2 and are upregulated upon oxidant insult. However, in fibroblasts from FRDA patients, unlike those from healthy controls, SODs are not upregulated in response to low doses of H2O2, oligomycin, and iron [57, 65, 66]. In yeast, the anaerobiosis to aerobiosis transition is an inducer of oxidative stress in Δyfh1 cells as a result of transcriptional repression of several genes encoding critical antioxidant enzymes (SODs, catalases, glutaredoxins, and thioredoxins) and of decrease of total glutathione levels [58, 70]. Glutathione is a major antioxidant molecule in eukaryotic cells. Furthermore, reversible protein S-glutathionylation is a post translational modification that provides protection of protein cysteines from irreversible oxidation and also functions in the transduction of redox signals [71]. Several studies show that frataxin deficiency leads to the impairment of glutathione homeostasis [52, 53, 72–75]. A significant increase in the glutathione pool bound to proteins was observed in blood samples, fibroblasts, and lymphoblasts from FRDA patients and yeast Δyfh1 cells [72, 74, 75]. In addition, Pastore et al. found that in patient fibroblasts actin was glutathionylated, which caused disassembly of the filaments [74]. The analysis of autopsy samples from the spinal cord of FRDA patients also showed abnormal microfilament polymerization [76].
The discovery of actin glutathionylation and the demonstration that the Nrf2-dependent Phase II antioxidant pathway is defective in patient fibroblasts provided the first mechanism for the reduction of antioxidant defenses in human frataxin-deficient cells [57]. Nuclear factor erythroid 2-related factor 2 (Nrf2) is a key transcription factor that responds to oxidants by inducing expression of antioxidant enzymes to restore redox homeostasis in the cell. The Nrf2 activity is regulated by the actin-associated Keap1 (Kelch-like ECH-associated protein 1), which is an adaptor protein for the Cul3-dependent E3 ubiquitin ligase complex. Under normal conditions, Keap1 sequesters Nrf2 in the cytoplasm and promotes its rapid degradation via ubiquitination [77]. Under oxidative stress conditions, the cysteines of the Keap1 protein became oxidized, and the activity of the Cul3-Keap1 ubiquitin E3 ligase complex is reduced. As a consequence, Nrf2 is stabilized and translocated to the nucleus where it binds to DNA sequences of the cis-acting antioxidant responsive element (ARE), activating the expression of genes encoding antioxidant enzymes like SODs, catalase, glutathione S-transferase, and NADH quinone oxidoreductase [77]. In fibroblasts from FRDA patients, Keap1 is not associated with actin, and Nrf2 is distributed diffusely in the cell [57]. In addition, in FRDA fibroblasts treated with oligomycin or tert-butylhydroquinone, Nrf2 fails to translocate to the nucleus and the antioxidant Phase II genes are not induced [57]. This phenotype can be reversed by 24 h treatment with the catalase mimetic Euk134; the actin stress fibers were reorganized, and Nrf2-signalling was restored, highlighting the role of H2O2 in FRDA pathophysiology. Recently, it was shown that the PIP5K1B gene that is located upstream of the FXN gene is also silenced in FRDA patient lymphocytes and fibroblasts [48]. The pip5k1β protein is a key regulatory factor of actin cytoskeleton dynamics, and its down-regulation in fibroblasts causes actin network destabilization [48]. Therefore, it is very likely that in FRDA patient cells, PIP5K1B gene silencing contributes to actin disassembly, but a direct evidence of actin glutathionylation in cells deficient for pip5k1β protein is missing.
Microarray analysis of dorsal root ganglia from YG8R frataxin-deficient mice and controls revealed significant differences in genes belonging to the thiol antioxidant, the myelination, and the axon transport functional categories [52]. In addition, the authors found not only a decreased expression of antioxidant genes that are regulated by Nrf2, but also Nrf2 (at mRNA and protein levels) in YG8R dorsal root ganglia compared to controls. However, their observations using different cell lines (HeLa, fibroblast, and ND7/23 dorsal root ganglion neuron cell lines and patient lymphoblasts) did not support deficient translocation of Nrf2 to the nucleus upon oxidant insult; Nrf2 is being mainly localized in the nucleus in frataxin-deficient cells. Consistent with these data, D'Oria et al. found no increase in Nrf2 expression in frataxin-silenced NSC34 cells (derived from the fusion of neuroblastoma cells and spinal cord motor neurons) compared to controls upon treatment with oxidized glutathione neither an increase in the Nrf2 nuclear fraction [78]. Neurons and fibroblasts are completely different cells types, and it is possible that in neurons antioxidant defense regulation depends primarily on Nrf2 levels while in fibroblasts it is the actin-driven translocation to the nucleus the major regulator.
Chronic inflammation and activity of glial cells are important factors that lead to neurodegeneration in other diseases, such as Alzheimer's disease and amyotrophic lateral sclerosis [79]. In FRDA, degeneration affects different types of neurons in the peripheral and central nervous systems, and it is likely that other cell types are also affected by frataxin deficiency. Several reports suggest that Schwann cells (SCs) could be specifically affected and that peripheral neuron loss would be a secondary event [9, 80]. Schwann cells are glial cells specialized in the myelination of axons in the peripheral nervous system. Signaling between neurons and SC plays an essential role in SC proliferation, survival, migration, and myelination [81]. On the opposite, SC also contributes to the preservation of axon integrity [82]. Analysis of the dorsal roots in patient autopsy samples shows differences in the myelination of thin fibers and reduced number of SC [9]. In addition, patient sural nerve autopsies also suggest participation of SC in neurodegeneration since although the large myelinated fibers are significantly reduced the total number of axons per unit area is similar to that of controls [83]. In agreement with these observations, in vitro studies using human SC lines showed frataxin knock-down by siRNA blocks cell cycle progression at G2M, and this is followed by an inflammatory response and an increase in cell death [80]. Treatment of these cells with antiinflammatory and antiapoptotic drugs rescued the death phenotype. Altogether, these results suggest that defects in SC could be at the origin of hypomyelination or demyelination and axon degeneration in FRDA patients.
5. Mitochondrial and Nuclear DNA Damage
Oxidative DNA damage is a natural consequence of aerobic metabolism that is exacerbated when cells are in oxidative stress conditions. ROS induce more than 20 lesions in the DNA, such as oxidized bases, apurinic/apyrimidinic (AP) sites, base deamination products, oxidized sugar fragments, and DNA strand breaks [84]. Damaged DNA bases, if not repaired, may have miscoding properties leading to mutation upon replication or by blocking progression of the replication fork [85, 86]. The most studied DNA lesion is the 8-oxo-7,8-dihydro-2′-deoxyguanosine (8-oxodG), a product of oxidation of guanine. Conflicting results have been obtained for the levels of urinary 8-oxodG between FRDA patients and controls, since a difference has not always been found [87, 88]. However, mitochondrial and nuclear DNA damage have been reported in human, mouse, and yeast frataxin-deficient cells [51, 89–92].
Mitochondrial DNA (mtDNA) loss was one of the first phenotypes reported for the Δyfh1 yeast cells [93–95] and for FRDA patient cells [96]. Karthikeyan et al. showed that strong frataxin depletion in yeast cells resulted in complete loss of mtDNA after 15 generations and that reduced frataxin levels resulted in only partial loss of mtDNA after 22 generations [91]. These results indicate that mtDNA loss is dependent on the frataxin levels in the cell. Recently, it was shown that in Δyfh1 yeast cells, mtDNA loss is oxygen exposition-dependent, with complete retention in anaerobic conditions [51]. Mitochondrial DNA lesions and significant loss in the heart, cerebellum, and dorsal root ganglia have been observed in FRDA patients [92, 96–98].
Decades ago, two reports showed evidence of nuclear DNA damage in skin fibroblasts and blood lymphocytes of FRDA patients in response to ionizing radiation and mutagens, respectively [99, 100]. These studies suggested that in these patients, an increased susceptibility to DNA damage and/or defective DNA repair pathways were present. In agreement with this hypothesis, the transcriptome profiling of total blood from 28 children with FRDA revealed the molecular signature of cell response to DNA damage [92]. Using quantitative PCR, the same authors showed increased number of mitochondrial and nuclear DNA lesions in blood cells from FRDA patients compared to controls. A link between frataxin expression, DNA-repair, and tumor initiation was observed in murine liver [89]. In addition, this work showed that overexpression of human frataxin in hamster fibroblasts was associated with a decrease in the nuclear mutation frequency.
Detailed studies on nuclear DNA damage and repair pathways have been performed in yeast. Analysis of diploid frataxin-deficient yeast cells showed evidence of chromosomal instability with higher levels of illegitimate mating, higher rate of spontaneous mutation, and increased sensitivity to the DNA-alkylating methyl methanesulfonate and to the replication inhibitor hydroxyurea than controls [90]. Furthermore, deletion of the glutathione peroxidase encoding gene GPX1 in frataxin-deficient cells resulted in a marked increase in the nuclear mutation rate. These results led the authors to suggest that the increased spontaneous nuclear damage in Δyfh1 cells was caused by H2O2 generated in the mitochondria [90]. Consistent with this hypothesis, the analysis of the exposure of anaerobically grown Δyfh1 cells to oxygen showed that frataxin-deficiency in yeast cells leads to increased nuclear DNA damage [51]. The effect of oxygen was very rapid, 15 min after Δyfh1 cell exposition to oxygen, antioxidant levels, were decreased and DNA strand breaks were visible. At 30 min, cell cycle was arrested at G1/S, and mutation frequency was increased in Δyfh1 cells. Two hours later, the cells adapted to oxygen since the cell cycle was reinitiated but were in a chronic oxidative stress state with a high spontaneous mutation rate. The nuclear DNA lesions detected in Δyfh1 cells were primarily caused by oxidized bases and single-strand breaks, and the Apn1 AP-endonuclease of the base excision repair pathway was essential for the repair these DNA lesions [51]. Altogether, these observations suggest that DNA damage and repair could be important features in FRDA disease progression.
6. Antioxidant Therapy
Different strategies for the discovery of effective treatments for FRDA disease are currently being developed or tested in clinical trials (see http://www.curefa.org/pipeline.html). Several groups have done significant advances in FXN gene replacement or frataxin replacement therapies [101, 102]. Other strategies target the expression of the GAA-expanded gene (e.g., the HDAC inhibitor RG2833 is in Phase I clinical trial) or the stabilization of the frataxin protein (e.g., erythropoietin is in Phase II clinical trial). In addition, molecules targeting physiological functions that are defective in patient cells, such as increase in mitochondrial functions and iron-sulfur cluster synthesis or decrease in oxidative stress and iron toxicity, are being developed or in clinical trials. The conclusion of a recent review of the results of all randomized controlled clinical trials with minimal duration of 12 months was that none of the pharmacological drugs tested, including idebenone, had a significant beneficial effect on the neurological symptoms in FRDA patients [103]. Nevertheless, several ongoing clinical trials are testing promising antioxidant molecules. The α-tocopheryl quinone EPI-A0001 is a potent antioxidant molecule that has been tested in a double-blind, randomized, placebocontrolled trial of two doses in 31 adults with FRDA for a short period (four weeks) [104]. Glucose tolerance was tested, and no statistical difference was observed in the Disposition Index, which is a measure of diabetic tendency. However, after four weeks of treatment a dose-dependent improvement in the Friedreich Ataxia Rating Scale score was observed indicating neurologic function improvement in the patients. The EPI-743 is a new drug based on vitamin E that modifies disease progression in patients suffering from inherited mitochondrial respiratory chain disorders [105]. A Phase II clinical trial is currently recruiting FRDA patients. OX1 (indole-3-propionic acid) is a naturally occurring drug compound that prevents oxidative stress by a combination of hydroxyl radical scavenging activity and metal chelation. This molecule is now in preclinical studies for toxicological safety.
7. Conclusion
In the last decade, major achievements have been obtained concerning the primary function of frataxin, and it is almost completely accepted that it is in iron-sulfur cluster synthesis. Also, the mechanisms that regulate FXN expression with expanded GAA repeats are well known. However, despite a strong international effort there is no proven therapy for FRDA that stops disease progression. One of the reasons could be that our knowledge of the physiology of frataxin-deficient cells, and in particular the neurons affected by the disease, is still reduced. A large number of studies show that frataxin-deficient cells present numerous pleiotropic secondary phenotypes, which arise from iron-sulfur clusters deficit, such as mitochondrial dysfunction, perturbed iron homeostasis, DNA damage and mutagenesis, and oxidative stress. However, other areas of research that are relevant for FRDA pathology and fundamental for the survival and functioning of neurons are still poorly studied; examples are mitochondria transport and dynamics, calcium homeostasis, nitric oxide signaling, and inflammation. A clearer picture of the effects of frataxin deficiency in neurons and other cell types that might participate in the degeneration process in FRDA disease is necessary.
The major problem until recently was the lack of good in vitro cellular and murine models. The mouse model that best recapitulates the disease features is the humanized mouse model (YAC transgenic mice containing the human FXN gene with 190 GAA repeats) [63]. However, in these mice the first phenotypes appear after 6 to 12 months of age, and there is a possibility of GAA trinucleotide repeat instability. New technologies that can allow the development of new and relevant models for FRDA disease research from stem cells have been developed. It is now possible to obtain individual types of neurons by patterning and differentiation of embryonic stem cells and induced pluripotent cells (iPS) or also by direct reprogramming of fibroblast or other cell types. A protocol for differentiation of peripheral nerve system neurons from human embryonic stem cells has been published [106]. The first reports of differentiation of neurons and cardiomyocytes from iPS cells from FRDA patients are encouraging, although the GAA trinucleotide repeats are unstable [107, 108]. Liu et al. differentiated sensory neurons and cardiomyocytes from iPS cells obtained from FRDA patients [107], and Hick et al. described mitochondrial deficits in the differentiated cells [109]. In the next years, the development of these new cell models will greatly contribute to our knowledge of the neurodegeneration process in FRDA disease.
Conflict of Interests
The authors declare that they have no conflict of interests.
Acknowledgments
Research on frataxin at the Gomes Laboratory was financed by the National Ataxia Foundation (USA) and by the ITQB Strategic Grant PEst-OE/EQB/LA0004/2011 (to ITQB-Laboratório Associado, Oeiras).
References
- 1.Friedreich N. Über degenerative Atrophie der spinalen Hinterstränge. Virchows Archiv fur Pathologische Anatomie und Physiologie und für klinische Medizin. 1863;27:1–26. [Google Scholar]
- 2.Friedreich N. Über degenerative Atrophie der spinalen Hinterstränge. Virchows Archiv fur Pathologische Anatomie und Physiologie und für klinische Medizin. 1863;26:391–419. [Google Scholar]
- 3.Friedreich N. Über degenerative Atrophie der spinalen Hinterstränge. Virchows Archiv fur Pathologische Anatomie und Physiologie und für klinische Medizin. 1863;26:433–459. [Google Scholar]
- 4.Santos R, Lefevre S, Sliwa D, Seguin A, Camadro J, Lesuisse E. Friedreich ataxia: molecular mechanisms, redox considerations, and therapeutic opportunities. Antioxidants and Redox Signaling. 2010;13(5):651–690. doi: 10.1089/ars.2009.3015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Harding AE. Friedreich’s ataxia: a clinical and genetic study of 90 families with an analysis of early diagnostic criteria and intrafamilial clustering of clinical features. Brain. 1981;104(3):589–620. doi: 10.1093/brain/104.3.589. [DOI] [PubMed] [Google Scholar]
- 6.Dürr A, Cossee M, Agid Y, et al. Clinical and genetic abnormalities in patients with Friedreich’s ataxia. The New England Journal of Medicine. 1996;335(16):1169–1175. doi: 10.1056/NEJM199610173351601. [DOI] [PubMed] [Google Scholar]
- 7.Pandolfo M. Friedreich ataxia: the clinical picture. Journal of Neurology. 2009;256(supplement 1):3–8. doi: 10.1007/s00415-009-1002-3. [DOI] [PubMed] [Google Scholar]
- 8.Hughes JT, Brownell B, Hewer RL. The peripheral sensory pathway in Friedreich’s ataxia: an examination by light and electron microscopy of the posterior nerve roots, posterior root ganglia, and peripheral sensory nerves in cases of Friedreich’s ataxia. Brain. 1968;91(4):803–818. doi: 10.1093/brain/91.4.803. [DOI] [PubMed] [Google Scholar]
- 9.Koeppen AH, Morral JA, Davis AN, et al. The dorsal root ganglion in Friedreich’s ataxia. Acta Neuropathologica. 2009;118(6):763–776. doi: 10.1007/s00401-009-0589-x. [DOI] [PubMed] [Google Scholar]
- 10.Campuzano V, Montermini L, Moltò MD, et al. Friedreich’s ataxia: autosomal recessive disease caused by an intronic GAA triplet repeat expansion. Science. 1996;271(5254):1423–1427. doi: 10.1126/science.271.5254.1423. [DOI] [PubMed] [Google Scholar]
- 11.Gellera C, Castellotti B, Mariotti C, et al. Frataxin gene point mutations in Italian Friedreich ataxia patients. Neurogenetics. 2007;8(4):289–299. doi: 10.1007/s10048-007-0101-5. [DOI] [PubMed] [Google Scholar]
- 12.Bidichandani SI, Ashizawa T, Patel PI. The GAA triplet-repeat expansion in Friedreich ataxia interferes with transcription and may be associated with an unusual DNA structure. American Journal of Human Genetics. 1998;62(1):111–121. doi: 10.1086/301680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Grabczyk E, Mancuso M, Sammarco MC. A persistent RNA·DNA hybrid formed by transcription of the Friedreich ataxia triplet repeat in live bacteria, and by T7 RNAP in vitro . Nucleic Acids Research. 2007;35(16):5351–5359. doi: 10.1093/nar/gkm589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Al-Mahdawi S, Pinto RM, Ismail O, et al. The Friedreich ataxia GAA repeat expansion mutation induces comparable epigenetic changes in human and transgenic mouse brain and heart tissues. Human Molecular Genetics. 2008;17(5):735–746. doi: 10.1093/hmg/ddm346. [DOI] [PubMed] [Google Scholar]
- 15.Herman D, Jenssen K, Burnett R, Soragni E, Perlman SL, Gottesfeld JM. Histone deacetylase inhibitors reverse gene silencing in Friedreich’s ataxia. Nature Chemical Biology. 2006;2(10):551–558. doi: 10.1038/nchembio815. [DOI] [PubMed] [Google Scholar]
- 16.Chan PK, Torres R, Yandim C, et al. Heterochromatinization induced by GAA-repeat hyperexpansion in Friedreich's ataxia can be reduced upon HDAC inhibition by vitamin B3. Human Molecular Genetics. 2013 doi: 10.1093/hmg/ddt115. [DOI] [PubMed] [Google Scholar]
- 17.Vaubel RA, Isaya G. Iron-sulfur cluster synthesis, iron homeostasis and oxidative stress in Friedreich ataxia. Molecular and Cellular Neuroscience. 55:50–61. doi: 10.1016/j.mcn.2012.08.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stemmler TL, Lesuisse E, Pain D, Dancis A. Frataxin and mitochondrial FeS cluster biogenesis. The Journal of Biological Chemistry. 2010;285(35):26737–26743. doi: 10.1074/jbc.R110.118679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rouault TA. Biogenesis of iron-sulfur clusters in mammalian cells: new insights and relevance to human disease. DMM Disease Models and Mechanisms. 2012;5(2):155–164. doi: 10.1242/dmm.009019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lill R, Hoffmann B, Molik S, et al. The role of mitochondria in cellular iron-sulfur protein biogenesis and iron metabolism. Biochimica et Biophysica Acta. 2012;1823(9):1491–1508. doi: 10.1016/j.bbamcr.2012.05.009. [DOI] [PubMed] [Google Scholar]
- 21.De Biase I, Rasmussen A, Endres D, et al. Progressive GAA expansions in dorsal root ganglia of Friedreich’s ataxia patients. Annals of Neurology. 2007;61(1):55–60. doi: 10.1002/ana.21052. [DOI] [PubMed] [Google Scholar]
- 22.De Biase I, Rasmussen A, Monticelli A, et al. Somatic instability of the expanded GAA triplet-repeat sequence in Friedreich ataxia progresses throughout life. Genomics. 2007;90(1):1–5. doi: 10.1016/j.ygeno.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 23.Armstrong JS, Khdour O, Hecht SM. Does oxidative stress contribute to the pathology of Friedreich’s ataxia? A radical question. The FASEB Journal. 2010;24(7):2152–2163. doi: 10.1096/fj.09-143222. [DOI] [PubMed] [Google Scholar]
- 24.Bayot A, Santos R, Camadro J, Rustin P. Friedreich’s ataxia: the vicious circle hypothesis revisited. BMC Medicine. 2011;9, article 112 doi: 10.1186/1741-7015-9-112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cho S, Lee MG, Yang JK, Lee JY, Song HK, Suh SW. Crystal structure of Escherichia coli CyaY protein reveals a previously unidentified fold for the evolutionarily conserved frataxin family. Proceedings of the National Academy of Sciences of the United States of America. 2000;97(16):8932–8937. doi: 10.1073/pnas.160270897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dhe-Paganon S, Shigeta R, Chi Y, Ristow M, Shoelson SE. Crystal structure of human frataxin. The Journal of Biological Chemistry. 2000;275(40):30753–30756. doi: 10.1074/jbc.C000407200. [DOI] [PubMed] [Google Scholar]
- 27.He Y, Alam SL, Proteasa SV, et al. Yeast frataxin solution structure, iron binding, and ferrochelatase interaction. Biochemistry. 2004;43(51):16254–16262. doi: 10.1021/bi0488193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Musco G, de Tommasi T, Stier G, et al. Assignment of the 1H, 15N, and 13C resonances of the C-terminal domain of frataxin, the protein responsible for Friedreich ataxia. Journal of Biomolecular NMR. 1999;15(1):87–88. doi: 10.1023/a:1008398832619. [DOI] [PubMed] [Google Scholar]
- 29.Nair M, Adinolfi S, Pastore C, Kelly G, Temussi P, Pastore A. Solution structure of the bacterial frataxin ortholog, CyaY: mapping the iron binding sites. Structure. 2004;12(11):2037–2048. doi: 10.1016/j.str.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 30.Prischi F, Konarev PV, Iannuzzi C, et al. Structural bases for the interaction of frataxin with the central components of iron-sulphur cluster assembly. Nature Communications. 2010;1(7, article 95) doi: 10.1038/ncomms1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Li H, Gakh O, Smith DY, IV, Isaya G. Oligomeric yeast frataxin drives assembly of core machinery for mitochondrial iron-sulfur cluster synthesis. The Journal of Biological Chemistry. 2009;284(33):21971–21980. doi: 10.1074/jbc.M109.011197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iannuzzi C, Adinolfi S, Howes BD, et al. The role of cyay in iron sulfur cluster assembly on the E. coli iscu scaffold protein. PLoS ONE. 2011;6(7) doi: 10.1371/journal.pone.0021992.e21992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Foury F, Pastore A, Trincal M. Acidic residues of yeast frataxin have an essential role in Fe-S cluster assembly. EMBO Reports. 2007;8(2):194–199. doi: 10.1038/sj.embor.7400881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Wang T, Craig EA. Binding of yeast frataxin to the scaffold for Fe-S cluster biogenesis, Isu. The Journal of Biological Chemistry. 2008;283(18):12674–12679. doi: 10.1074/jbc.M800399200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Correia AR, Wang T, Craig EA, Gomes CM. Iron-binding activity in yeast frataxin entails a trade off with stability in the α1/β1 acidic ridge region. Biochemical Journal. 2010;426(2):197–203. doi: 10.1042/BJ20091612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Cavadini P, Adamec J, Taroni F, Gakh O, Isaya G. Two-step processing of human frataxin by mitochondrial processing peptidase: precursor and intermediate forms are cleaved at different rates. The Journal of Biological Chemistry. 2000;275(52):41469–41475. doi: 10.1074/jbc.M006539200. [DOI] [PubMed] [Google Scholar]
- 37.Schmucker S, Argentini M, Carelle-Calmels N, Martelli A, Puccio H. The in vivo mitochondrial two-step maturation of human frataxin. Human Molecular Genetics. 2008;17(22):3521–3531. doi: 10.1093/hmg/ddn244. [DOI] [PubMed] [Google Scholar]
- 38.Condò I, Ventura N, Malisan F, Rufini A, Tomassini B, Testi R. In vivo maturation of human frataxin. Human Molecular Genetics. 2007;16(13):1534–1540. doi: 10.1093/hmg/ddm102. [DOI] [PubMed] [Google Scholar]
- 39.Gakh O, Bedekovics T, Duncan SF, Smith DY, IV, Berkholz DS, Isaya G. Normal and Friedreich ataxia cells express different isoforms of frataxin with complementary roles in iron-sulfur cluster assembly. The Journal of Biological Chemistry. 2010;285(49):38486–38501. doi: 10.1074/jbc.M110.145144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Seguin A, Bayot A, Dancis A, et al. Overexpression of the yeast frataxin homolog (Yfh1): contrasting effects on iron-sulfur cluster assembly, heme synthesis and resistance to oxidative stress. Mitochondrion. 2009;9(2):130–138. doi: 10.1016/j.mito.2009.01.007. [DOI] [PubMed] [Google Scholar]
- 41.Gakh O, Smith DY, IV, Isaya G. Assembly of the iron-binding protein frataxin in Saccharomyces cerevisiae responds to dynamic changes in mitochondrial iron influx and stress level. The Journal of Biological Chemistry. 2008;283(46):31500–31510. doi: 10.1074/jbc.M805415200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Correia AR, Ow SY, Wright PC, Gomes CM. The conserved Trp155 in human frataxin as a hotspot for oxidative stress related chemical modifications. Biochemical and Biophysical Research Communications. 2009;390(3):1007–1011. doi: 10.1016/j.bbrc.2009.10.095. [DOI] [PubMed] [Google Scholar]
- 43.Cossee M, Durr A, Schmitt M, et al. Friedreich's ataxia: point mutations and clinical presentation of compound heterozygotes. Annals of Neurology. 1999;45(2):200–206. doi: 10.1002/1531-8249(199902)45:2<200::aid-ana10>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 44.Correia AR, Adinolfi S, Pastore A, Gomes CM. Conformational stability of human frataxin and effect of Friedreich’s ataxia-related mutations on protein folding. Biochemical Journal. 2006;398(3):605–611. doi: 10.1042/BJ20060345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Correia AR, Pastore C, Adinolfi S, Pastore A, Gomes CM. Dynamics, stability and iron-binding activity of frataxin clinical mutants. FEBS Journal. 2008;275(14):3680–3690. doi: 10.1111/j.1742-4658.2008.06512.x. [DOI] [PubMed] [Google Scholar]
- 46.Calmels N, Schmucker S, Wattenhofer-Donzé M, et al. The first cellular models based on frataxin missense mutations that reproduce spontaneously the defects associated with Friedreich ataxia. PLoS ONE. 2009;4(7) doi: 10.1371/journal.pone.0006379.e6379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Li H, Gakh O, Smith DYT, Ranatunga WK, Isaya G. Missense mutations linked to Friedreich ataxia have different but synergistic effects on mitochondrial frataxin isoforms. The Journal of Biological Chemistry. 2013;288(6):4116–4127. doi: 10.1074/jbc.M112.435263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bayot A, Reichman S, Lebon S, et al. Cis-silencing of PIP5K1B evidenced in Friedreich's ataxia patient cells results in cytoskeleton anomalies. Human Molecular Genetics. 2013 doi: 10.1093/hmg/ddt144. [DOI] [PubMed] [Google Scholar]
- 49.Leal SS, Botelho HM, Gomes CM. Metal ions as modulators of protein conformation and misfolding in neurodegeneration. Coordination Chemistry Reviews. 2012;256(19-20):2253–2270. [Google Scholar]
- 50.Gomes CM. Protein misfolding in disease and small molecule therapies. Current Topics in Medicinal Chemistry. 2012;12(22):2460–2469. doi: 10.2174/1568026611212220002. [DOI] [PubMed] [Google Scholar]
- 51.Lefevre S, Brossas C, Auchère F, Boggetto N, Camadro JM, Santos R. Apn1 AP-endonuclease is essential for the repair of oxidatively damaged DNA bases in yeast frataxin-deficient cells. Human Molecular Genetics. 2012;21(18):4060–4072. doi: 10.1093/hmg/dds230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shan Y, Schoenfeld RA, Hayashi G, et al. Frataxin deficiency leads to defects in expression of antioxidants and Nrf2 expression in dorsal root ganglia of the Friedreich's ataxia YG8R mouse model. Antioxidants & Redox Signaling. 2013 doi: 10.1089/ars.2012.4537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Napoli E, Taroni F, Cortopassi GA. Frataxin, iron-sulfur clusters, heme, ROS, and aging. Antioxidants and Redox Signaling. 2006;8(3-4):506–516. doi: 10.1089/ars.2006.8.506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Irazusta V, Cabiscol E, Reverter-Branchat G, Ros J, Tamarit J. Manganese is the link between frataxin and iron-sulfur deficiency in the yeast model of Friedreich ataxia. The Journal of Biological Chemistry. 2006;281(18):12227–12232. doi: 10.1074/jbc.M511649200. [DOI] [PubMed] [Google Scholar]
- 55.Santos MM, Ohshima K, Pandolfo M. Frataxin deficiency enhances apoptosis in cells differentiating into neuroectoderm. Human Molecular Genetics. 2001;10(18):1935–1944. doi: 10.1093/hmg/10.18.1935. [DOI] [PubMed] [Google Scholar]
- 56.Anderson PR, Kirby K, Orr WC, Hilliker AJ, Phillips JP. Hydrogen peroxide scavenging rescues frataxin deficiency in a Drosophila model of Friedreich’s ataxia. Proceedings of the National Academy of Sciences of the United States of America. 2008;105(2):611–616. doi: 10.1073/pnas.0709691105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Paupe V, Dassa EP, Goncalves S, et al. Impaired nuclear Nrf2 translocation undermines the oxidative stress response in Friedreich ataxia. PLoS ONE. 2009;4(1) doi: 10.1371/journal.pone.0004253.e4253 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Lefevre S, Sliwa D, Auchre F, et al. The yeast metacaspase is implicated in oxidative stress response in frataxin-deficient cells. FEBS Letters. 2012;586(2):143–148. doi: 10.1016/j.febslet.2011.12.002. [DOI] [PubMed] [Google Scholar]
- 59.Santos R, Buisson N, Knight SAB, Dancis A, Camadro J, Lesuisse E. Candida albicans lacking the frataxin homologue: a relevant yeast model for studying the role of frataxin. Molecular Microbiology. 2004;54(2):507–519. doi: 10.1111/j.1365-2958.2004.04281.x. [DOI] [PubMed] [Google Scholar]
- 60.Vázquez-Manrique RP, González-Cabo P, Ros S, Aziz H, Baylis HA, Palau F. Reduction of Caenorhabditis elegans frataxin increases sensitivity to oxidative stress, reduces lifespan, and causes lethality in a mitochondrial complex II mutant. The FASEB Journal. 2006;20(1):172–174. doi: 10.1096/fj.05-4212fje. [DOI] [PubMed] [Google Scholar]
- 61.Llorens JV, Navarro JA, Martínez-Sebastían MJ, et al. Causative role of oxidative stress in a Drosophila model of Friedreich ataxia. The FASEB Journal. 2007;21(2):333–344. doi: 10.1096/fj.05-5709com. [DOI] [PubMed] [Google Scholar]
- 62.Runko AP, Griswold AJ, Min K. Overexpression of frataxin in the mitochondria increases resistance to oxidative stress and extends lifespan in Drosophila . FEBS Letters. 2008;582(5):715–719. doi: 10.1016/j.febslet.2008.01.046. [DOI] [PubMed] [Google Scholar]
- 63.Al-Mahdawi S, Pinto RM, Varshney D, et al. GAA repeat expansion mutation mouse models of Friedreich ataxia exhibit oxidative stress leading to progressive neuronal and cardiac pathology. Genomics. 2006;88(5):580–590. doi: 10.1016/j.ygeno.2006.06.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Tozzi G, Nuccetelli M, Lo Bello M, et al. Antioxidant enzymes in blood of patients with Friedreich’s ataxia. Archives of Disease in Childhood. 2002;86(5):376–379. doi: 10.1136/adc.86.5.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Jiralerspong S, Ge B, Hudson TJ, Pandolfo M. Manganese superoxide dismutase induction by iron is impaired in Friedreich ataxia cells. FEBS Letters. 2001;509(1):101–105. doi: 10.1016/s0014-5793(01)03140-4. [DOI] [PubMed] [Google Scholar]
- 66.Chantrel-Groussard K, Geromel V, Puccio H, et al. Disabled early recruitment of antioxidant defenses in Friedreich’s ataxia. Human Molecular Genetics. 2001;10(19):2061–2067. doi: 10.1093/hmg/10.19.2061. [DOI] [PubMed] [Google Scholar]
- 67.Tan G, Chen L, Lonnerdal B, Gellera C, Taroni FA, Cortopassi GA. Frataxin expression rescues mitochondrial dysfunctions in FRDA cells. Human Molecular Genetics. 2001;10(19):2099–2107. doi: 10.1093/hmg/10.19.2099. [DOI] [PubMed] [Google Scholar]
- 68.Wong A, Yang J, Cavadini P, et al. The Friedreich’s ataxia mutation confers cellular sensitivity to oxidant stress which is rescued by chelators of iron and calcium and inhibitors of apoptosis. Human Molecular Genetics. 1999;8(3):425–430. doi: 10.1093/hmg/8.3.425. [DOI] [PubMed] [Google Scholar]
- 69.Calabrese V, Lodi R, Tonon C, et al. Oxidative stress, mitochondrial dysfunction and cellular stress response in Friedreich’s ataxia. Journal of the Neurological Sciences. 2005;233(1-2):145–162. doi: 10.1016/j.jns.2005.03.012. [DOI] [PubMed] [Google Scholar]
- 70.Bulteau A, Dancis A, Gareil M, Montagne J, Camadro J, Lesuisse E. Oxidative stress and protease dysfunction in the yeast model of Friedreich ataxia. Free Radical Biology and Medicine. 2007;42(10):1561–1570. doi: 10.1016/j.freeradbiomed.2007.02.014. [DOI] [PubMed] [Google Scholar]
- 71.Gallogly MM, Mieyal JJ. Mechanisms of reversible protein glutathionylation in redox signaling and oxidative stress. Current Opinion in Pharmacology. 2007;7(4):381–391. doi: 10.1016/j.coph.2007.06.003. [DOI] [PubMed] [Google Scholar]
- 72.Piemonte F, Pastore A, Tozzi G, et al. Glutathione in blood of patients with Friedreich’s ataxia. European Journal of Clinical Investigation. 2001;31(11):1007–1011. doi: 10.1046/j.1365-2362.2001.00922.x. [DOI] [PubMed] [Google Scholar]
- 73.Auchère F, Santos R, Planamente S, Lesuisse E, Camadro J. Glutathione-dependent redox status of frataxin-deficient cells in a yeast model of Friedreich’s ataxia. Human Molecular Genetics. 2008;17(18):2790–2802. doi: 10.1093/hmg/ddn178. [DOI] [PubMed] [Google Scholar]
- 74.Pastore A, Tozzi G, Gaeta LM, et al. Actin glutathionylation increases in fibroblasts of patients with Friedreich’s ataxia: a potential role in the pathogenesis of the disease. The Journal of Biological Chemistry. 2003;278(43):42588–42595. doi: 10.1074/jbc.M301872200. [DOI] [PubMed] [Google Scholar]
- 75.Bulteau AL, Planamente S, Jornea L, et al. Changes in mitochondrial glutathione levels and protein thiol oxidation in Δyfh1 yeast cells and the lymphoblasts of patients with Friedreich’s ataxia. Biochimica et Biophysica Acta. 2012;1822(2):212–225. doi: 10.1016/j.bbadis.2011.11.003. [DOI] [PubMed] [Google Scholar]
- 76.Sparaco M, Gaeta LM, Santorelli FM, et al. Friedreich’s ataxia: oxidative stress and cytoskeletal abnormalities. Journal of the Neurological Sciences. 2009;287(1-2):111–118. doi: 10.1016/j.jns.2009.08.052. [DOI] [PubMed] [Google Scholar]
- 77.Nguyen T, Nioi P, Pickett CB. The Nrf2-antioxidant response element signaling pathway and its activation by oxidative stress. The Journal of Biological Chemistry. 2009;284(20):13291–13295. doi: 10.1074/jbc.R900010200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.D'Oria V, Petrini S, Travaglini L, et al. Frataxin deficiency leads to reduced expression and impaired translocation of NF-E2-related factor (Nrf2) in cultured motor neurons. International Journal of Molecular Sciences. 2013;10(4):7853–7865. doi: 10.3390/ijms14047853. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Philips T, Robberecht W. Neuroinflammation in amyotrophic lateral sclerosis: role of glial activation in motor neuron disease. The Lancet Neurology. 2011;10(3):253–263. doi: 10.1016/S1474-4422(11)70015-1. [DOI] [PubMed] [Google Scholar]
- 80.Lu C, Schoenfeld R, Shan Y, Tsai H, Hammock B, Cortopassi G. Frataxin deficiency induces Schwann cell inflammation and death. Biochimica et Biophysica Acta. 2009;1792(11):1052–1061. doi: 10.1016/j.bbadis.2009.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Newbern J, Birchmeier C. Nrg1/ErbB signaling networks in Schwann cell development and myelination. Seminars in Cell and Developmental Biology. 2010;21(9):922–928. doi: 10.1016/j.semcdb.2010.08.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Nave K, Trapp BD. Axon-glial signaling and the glial support of axon function. Annual Review of Neuroscience. 2008;31:535–561. doi: 10.1146/annurev.neuro.30.051606.094309. [DOI] [PubMed] [Google Scholar]
- 83.Morral JA, Davis AN, Qian J, Gelman BB, Koeppen AH. Pathology and pathogenesis of sensory neuropathy in Friedreich’s ataxia. Acta Neuropathologica. 2010;120(1):97–108. doi: 10.1007/s00401-010-0675-0. [DOI] [PubMed] [Google Scholar]
- 84.Cooke MS, Evans MD, Dizdaroglu M, Lunec J. Oxidative DNA damage: mechanisms, mutation, and disease. The FASEB Journal. 2003;17(10):1195–1214. doi: 10.1096/fj.02-0752rev. [DOI] [PubMed] [Google Scholar]
- 85.Svilar D, Goellner EM, Almeida KH, Sobol RW. Base excision repair and lesion-dependent subpathways for repair of oxidative DNA damage. Antioxidants and Redox Signaling. 2011;14(12):2491–2507. doi: 10.1089/ars.2010.3466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Gros L, Saparbaev MK, Laval J. Enzymology of the repair of free radicals-induced DNA damage. Oncogene. 2002;21(58):8905–8925. doi: 10.1038/sj.onc.1206005. [DOI] [PubMed] [Google Scholar]
- 87.Schulz JB, Dehmer T, Schöls L, et al. Oxidative stress in patients with Friedreich ataxia. Neurology. 2000;55(11):1719–1721. doi: 10.1212/wnl.55.11.1719. [DOI] [PubMed] [Google Scholar]
- 88.Di Prospero NA, Baker A, Jeffries N, Fischbeck KH. Neurological effects of high-dose idebenone in patients with Friedreich’s ataxia: a randomised, placebo-controlled trial. The Lancet Neurology. 2007;6(10):878–886. doi: 10.1016/S1474-4422(07)70220-X. [DOI] [PubMed] [Google Scholar]
- 89.Thierbach R, Drewes G, Fusser M, et al. The Friedreich’s ataxia protein frataxin modulates DNA base excision repair in prokaryotes and mammals. Biochemical Journal. 2010;432(1):165–172. doi: 10.1042/BJ20101116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Karthikeyan G, Lewis LK, Resnick MA. The mitochondrial protein frataxin prevents nuclear damage. Human Molecular Genetics. 2002;11(11):1351–1362. doi: 10.1093/hmg/11.11.1351. [DOI] [PubMed] [Google Scholar]
- 91.Karthikeyan G, Santos JH, Graziewicz MA, et al. Reduction in frataxin causes progressive accumulation of mitochondrial damage. Human Molecular Genetics. 2003;12(24):3331–3342. doi: 10.1093/hmg/ddg349. [DOI] [PubMed] [Google Scholar]
- 92.Haugen AC, Di Prospero NA, Parker JS, et al. Altered gene expression and DNA damage in peripheral blood cells from Friedreich’s ataxia patients: cellular model of pathology. PLoS Genetics. 2010;6(1) doi: 10.1371/journal.pgen.1000812.e1000812 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Wilson RB, Roof DM. Respiratory deficiency due to loss of mitochondrial dna in yeast lacking the frataxin homologue. Nature Genetics. 1997;16(4):352–357. doi: 10.1038/ng0897-352. [DOI] [PubMed] [Google Scholar]
- 94.Babcock M, de Silva D, Oaks R, et al. Regulation of mitochondrial iron accumulation by Yfh1p, a putative homolog of frataxin. Science. 1997;276(5319):1709–1712. doi: 10.1126/science.276.5319.1709. [DOI] [PubMed] [Google Scholar]
- 95.Foury F, Cazzalini O. Deletion of the yeast homologue of the human gene associated with Friedreich’s ataxia elicits iron accumulation in mitochondria. FEBS Letters. 1997;411(2-3):373–377. doi: 10.1016/s0014-5793(97)00734-5. [DOI] [PubMed] [Google Scholar]
- 96.Bradley JL, Blake JC, Chamberlain S, Thomas PK, Cooper JM, Schapira AHV. Clinical, biochemical and molecular genetic correlations in Friedreich’s ataxia. Human Molecular Genetics. 2000;9(2):275–282. doi: 10.1093/hmg/9.2.275. [DOI] [PubMed] [Google Scholar]
- 97.Houshmand M, Panahi MSS, Nafisi S, Soltanzadeh A, Alkandari FM. Identification and sizing of GAA trinucleotide repeat expansion, investigation for D-loop variations and mitochondrial deletions in Iranian patients with Friedreich’s ataxia. Mitochondrion. 2006;6(2):82–88. doi: 10.1016/j.mito.2006.01.005. [DOI] [PubMed] [Google Scholar]
- 98.Heidari MM, Houshmand M, Hosseinkhani S, Nafissi S, Scheiber-Mojdehkar B, Khatami M. A novel mitochondrial heteroplasmic C13806A point mutation associated with iranian Friedreich’s ataxia. Cellular and Molecular Neurobiology. 2009;29(2):225–233. doi: 10.1007/s10571-008-9315-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Evans HJ, Vijayalaxmi V, Pentland B, Newton MS. Mutagen hypersensitivity in Friedreich’s ataxia. Annals of Human Genetics. 1983;47(3):193–204. doi: 10.1111/j.1469-1809.1983.tb00988.x. [DOI] [PubMed] [Google Scholar]
- 100.Lewis PD, Corr JB, Arlett CF, Harcourt SA. Increased sensitivity to gamma irradiation of skin fibroblasts in Friedreich’s ataxia. The Lancet. 1979;2(8140):474–475. doi: 10.1016/s0140-6736(79)91530-7. [DOI] [PubMed] [Google Scholar]
- 101.Gimenez-Cassina A, Wade-Martins R, Gomez-Sebastian S, Corona J-C, Lim F, Diaz-Nido J. Infectious delivery and long-term persistence of transgene expression in the brain by a 135-kb iBAC-FXN genomic DNA expression vector. Gene Therapy. 2011;18(10):1015–1019. doi: 10.1038/gt.2011.45. [DOI] [PubMed] [Google Scholar]
- 102.Vyas PM, Tomamichel WJ, Pride PM, et al. A TAT-frataxin fusion protein increases lifespan and cardiac function in a conditional Friedreich’s ataxia mouse model. Human Molecular Genetics. 2012;21(6):1230–1247. doi: 10.1093/hmg/ddr554.ddr554 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Kearney M, Orrell RW, Fahey M, Pandolfo M. Antioxidants and other pharmacological treatments for Friedreich ataxia. Cochrane Database of Systematic Reviews. 2009;(4) doi: 10.1002/14651858.CD007791.pub2.CD007791 [DOI] [PubMed] [Google Scholar]
- 104.Lynch DR, Willi SM, Wilson RB, et al. A0001 in Friedreich ataxia: biochemical characterization and effects in a clinical trial. Movement Disorders. 2012;27(8):1026–1033. doi: 10.1002/mds.25058. [DOI] [PubMed] [Google Scholar]
- 105.Enns GM, Kinsman SL, Perlman SL, et al. Initial experience in the treatment of inherited mitochondrial disease with EPI-743. Molecular Genetics and Metabolism. 2012;105(1):91–102. doi: 10.1016/j.ymgme.2011.10.009. [DOI] [PubMed] [Google Scholar]
- 106.Valensi-Kurtz M, Lefler S, Cohen MA, et al. Enriched population of PNS neurons derived from human embryonic stem cells as a platform for studying peripheral neuropathies. PLoS ONE. 2010;5(2) doi: 10.1371/journal.pone.0009290.e9290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Liu J, Verma PJ, Evans-Galea MV, et al. Generation of induced pluripotent stem cell lines from Friedreich ataxia patients. Stem Cell Reviews and Reports. 2011;7(3):703–713. doi: 10.1007/s12015-010-9210-x. [DOI] [PubMed] [Google Scholar]
- 108.Ku S, Soragni E, Campau E, et al. Friedreich’s ataxia induced pluripotent stem cells model intergenerational GAATTC triplet repeat instability. Cell Stem Cell. 2010;7(5):631–637. doi: 10.1016/j.stem.2010.09.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Hick A, Wattenhofer-Donzé M, Chintawar S, et al. Neurons and cardiomyocytes derived from induced pluripotent stem cells as a model for mitochondrial defects in Friedreich's ataxia. Disease Models & Mechanisms. 2013;6(3):608–621. doi: 10.1242/dmm.010900. [DOI] [PMC free article] [PubMed] [Google Scholar]


