Abstract
The MMS19 gene of the yeast Saccharomyces cerevisiae encodes a polypeptide of unknown function which is required for both nucleotide excision repair (NER) and RNA polymerase II (RNAP II) transcription. Here we report the molecular cloning of human and mouse orthologs of the yeast MMS19 gene. Both human and Drosophila MMS19 cDNAs correct thermosensitive growth and sensitivity to killing by UV radiation in a yeast mutant deleted for the MMS19 gene, indicating functional conservation between the yeast and mammalian gene products. Alignment of the translated sequences of MMS19 from multiple eukaryotes, including mouse and human, revealed the presence of several conserved regions, including a HEAT repeat domain near the C-terminus. The presence of HEAT repeats, coupled with functional complementation of yeast mutant phenotypes by the orthologous protein from higher eukaryotes, suggests a role of Mms19 protein in the assembly of a multiprotein complex(es) required for NER and RNAP II transcription. Both the mouse and human genes are ubiquitously expressed as multiple transcripts, some of which appear to derive from alternative splicing. The ratio of different transcripts varies in several different tissue types.
INTRODUCTION
A genetic framework for elucidating the mechanism of nucleotide excision repair (NER) in eukaryotic cells was provided by the isolation and phenotypic characterization of mutant strains of the yeast Saccharomyces cerevisiae that are highly sensitive to UV radiation and to UV-mimetic chemicals but not to simple alkylating agents (reviewed in 1). Epistasis analyses involving many of these mutants established three distinct epistasis groups (reviewed in 1). Most members of the so-called RAD3 epistasis group were subsequently shown to encode polypeptides directly involved in the biochemical mechanism of NER in yeast (reviewed in 1). Among the multiple genes assigned to this epistasis group is one designated MMS19 (2). In contrast to other yeast mutants defective in NER, mms19 mutants are not only hypersensitive to UV radiation, but are additionally abnormally sensitive to the alkylating agent methylmethanesulfonate (MMS) (2,3). The mms19 mutation has been shown to compromise the incision step of NER (3–6).
The yeast MMS19 (yMMS19) gene has been cloned and characterized (5,6). Mms19 protein apparently does not participate in NER directly, since it is not required for this process in a reconstituted in vitro system (7). However, a strain deleted of the gene acquires a thermolabile defect in RNA polymerase II (RNAP II) transcription. This phenotype can be corrected in vitro by supplementing extracts with the basal transcription factor TFIIH, but not with Mms19 protein (5). It is well established that TFIIH is not only essential for RNAP II transcription, but is also indispensable for NER (8). Collectively, these results suggest that the yMMS19 gene has an influence on both RNAP II transcription and NER through an effect(s) on TFIIH. However, the nature of this effect is not understood nor is it clear why mms19 mutants are abnormally sensitive to alkylating agents such as MMS.
The majority of the proteins involved in NER in yeast are conserved in higher eukaryotes. Indeed, with the exception of the proteins encoded by the RAD7, RAD16 and MMS19 genes, human or mouse orthologs have been identified for every other known yeast NER gene (reviewed in 1). In the present studies we have cloned and sequenced human, mouse and Drosophila melanogaster orthologs of the yMMS19 gene and have mapped the human gene to chromosome 10q24. All the eukaryotic orthologs sequenced, as well as the orthologous S.cerevisiae, Schizosaccharomyces pombe and Arabidopsis thaliana translated sequences obtained from public databases, have a conserved HEAT repeat domain in the C-terminal region. We have also observed substantial correction of the phenotypes of UV radiation sensitivity and thermosensitivity for growth of a yeast mms19 deletion mutant following transfection of cells with plasmids that express the human or Drosophila cDNAs. Hence, our findings provide evidence for functional conservation of Mms19 protein between lower and higher eukaryotes. Finally, we provide evidence for complex processing of the human and mouse MMS19 primary transcripts, suggestive of complex regulation of expression of these genes. While these studies were in progress cloning of the human MMS19 (hMMS19) gene was independently reported (9). We have observed differences in the translated amino acid sequence of the hMMS19 gene from that previously reported. Additionally, the previously reported study failed to observe functional complementation of the UV radiation sensitivity and thermosensitivity of a yeast mms19 deletion mutant (9).
MATERIALS AND METHODS
Cloning the mouse and human MMS19 genes
The S.cerevisiae Mms19 protein was used to screen public databases using the TBLASTN algorithm (10). Two partially sequenced IMAGE clones [expressed sequence tag (EST) R89623 (clone 166947) derived from human adult brain and EST AA939567 (clone 1329989) derived from mouse thymus] were identified with homology to the 100–200 C-terminal amino acids of yeast Mms19 protein. These clones provided sequences for the design of primers. To obtain full-length human and mouse cDNAs a similar approach was followed. A human pEBS7 directional cDNA library derived from HeLa cells was screened by hybridization with a 702 bp PCR fragment amplified with primers 5′-CAGGACTCCTCAACAAGCAC-3′ and 5′-CCTGTGGTTTGTACGGCAGC-3′. Approximately 600 000 clones were plated and transferred to Hybond N+ membranes. A mouse testis 5′-STRETCH cDNA library (ML1020a; Clontech, Palo Alto, CA) was screened by hybridization with a 644 bp PCR fragment amplified with primers 5′-CAAGCCTGTGCTTTTACCAG-3′ and 5′-CAGCAGCACAGCTTCCTATTC-3′. Approximately one million recombinant bacteriophage plates were transferred to Hybond N+ membranes. Hybridization was carried out as previously described (11). Partial cDNA clones were obtained from both cDNA libraries. To obtain full-length human and mouse MMS19 cDNAs multiple rounds of 5′-RACE were performed according to the manufacturer’s suggested protocols (Gibco BRL, Rockville, MD) on human and mouse testis total RNA.
Databases and protein sequence analysis
Non-redundant public databases were iteratively searched using the gapped BLAST and PSI-BLAST programs as described (12,13). A cut-off of E < 0.05 was employed for inclusion of sequences in the position-specific weight matrices. Various sequences with homology to the hMMS19 ORF were identified. The most likely ORF for each of the MMS19 orthologs was deduced using a combination of BCM Genefinder, Genescan, Grail, ESTs and visual sequence analysis. Protein alignments were constructed with the CLUSTAL-X program (14). Sequences were partitioned into high complexity (predicted globular) and low complexity (predicted non-globular) using the SEG program (15). Protein secondary structure was predicted using the PHD program (16).
Chromosome mapping and fluorescence in situ hybridization (FISH)
PCR primers (5′-GCTTCAAGGACCAGCTGTGC and 5′-CCAAGACTGTCAGTGGG-3′) designed to produce a human-specific product of 96 bp from the 3′-end of the hMMS19 ORF were used to screen a human BAC library. Five clones were identified, designated 678A2, 678K9, 686A1, 628F13 and 737E11. Clone 737E11 was shown by PCR and sequence analysis to contain the complete MMS19 ORF. FISH was performed as described (17) with biotinylated 737E11 as the probe against normal male donor metaphase chromosomes from cells labeled with BrdUrd for the last 4.5 h of culture (18).
Northern blot analysis of MMS19 expression
Human multiple tissue northern blots I and II and a mouse multiple tissue northern blot (Clontech) containing 2 µg poly(A)+ RNA/lane were hybridized according to the manufacturer’s directions with labeled random primed probes corresponding to nucleotide positions 2358–3271 and 2729–3372, respectively. Probes corresponding to exons 3 and 8 of hMMS19 were also used. Membranes were washed to a final stringency of 0.1× SSC, 0.1% SDS at 65°C for 40 min.
Construction of plasmids for functional complementation
The vector pESC-TRP (Stratagene, La Jolla, CA) was used for expression of the human, Drosophila MMS19 (dMMS19) and yMMS19 genes. The yMMS19 gene was amplified from S.cerevisiae genomic DNA with primers 5′-GCGGCCGCGGGCCCGCCGGAACAATTGGCCTTAC-3′ and 5′-GAGCTCATGGTGATGGTGATGGTGATGGTGGTCGACCTCGAACGGGATTTGGCCTAATTC-3′. The dMMS19 coding region was amplified by RT–PCR from whole fly total RNA with primers (5′-CATGCGGCCGCGGGCCCGTGCCGCAATGACAACGCCCAC-3′ and 5′-CAGTGAGCTCTCAGTGATGGTGATGGTGGTGCCCGGGGTCGACATTCGGACTAGGCGACCGAC-3′) designed based on tentative exon identification (genomic clone AC007532). The hMMS19 gene was amplified by RT–PCR from testis total RNA with primers 5′-GCGGCCGCCGTGTTCGCGTTATGGCCG-3′ and 5′-GAGCTCATGGTGATGGTGATGGTGATGGTGCACTAGTAGGCTGCCAGGGCTCCCCAACAGAAACC-3′. High fidelity DNA polymerases were used for all amplifications. PCR products were cloned into pGEM-T (Promega, Madison, WI) and sequenced to confirm the absence of any mutations. Clones were then digested with ApaI and SalI (yMMS19) or with NotI and SpeI (hMMS19 and dMMS19) and inserts were subcloned into pESC-TRP.
Functional complementation
Yeast cells were grown to stationary phase at 28°C in supplemented minimal medium. Cells were harvested and washed in water. For complementation of methionine auxotrophy cells were spread on minimal medium plates supplemented with 2% galactose and the appropriate amino acids with or without methionine. Plates were incubated at 30 or 25°C and cell growth was assessed after 3–5 days. For complementation of thermosensitive growth cultures were incubated at 30 and 37°C. Cell growth was assessed by macroscopic analysis 4–6 days after plating and by quantification in a hemocytometer from liquid cultures. To determine sensitivity to UV radiation serial dilutions of cells were prepared in water and spread on supplemented minimal medium plates. Uracil and methionine were added to the plates to allow direct comparison with the wild-type strain. Plates were irradiated with increasing doses of UV light and incubated in the dark at 28°C. After 6–8 days colonies were counted and survival was calculated.
Yeast strains and media
The S.cerevisiae wild-type strain used for this study was W303-1B (MATa ade2-1 trp1-1 can1-100 his3-11,15 leu2-3,112 ura3-1). The isogenic mms19Δ mutant strain MGSC217 was described previously (6). The mms19Δ strain was transformed with the pESC-TRP constructs described above using standard protocols, to yield strains mms19Δ+pESC-TRP (MGSC217+empty vector), mms19Δ+yMMS19 (MGSC217+yMMS19), mms19Δ+hMMS19 (MGSC217+hMMS19) and mms19Δ+dMMS19 (MGSC217+dMMS19). All strains were maintained on selective YNB (0.67% yeast nitrogen base, 2% glucose, 2% bacto agar) supplemented appropriately for genetic markers.
RESULTS
cDNA and protein sequences of human, mouse and Drosophila MMS19 orthologs
Public databases were screened with the published amino acid sequence of the yMMS19 gene. Two ESTs derived from human adult brain (R89623) and from mouse thymus (AA939567) were identified with significant homology to the C-terminus of the yMMS19 ORF. To obtain full-length MMS19 cDNAs we screened human and mouse cDNA libraries with RT–PCR probes. The largest clone obtained from a human HeLa cell cDNA library was 3772 bp in length. This cDNA contains an ORF of 2616 bp which can encode a protein of 872 amino acids with a predicted molecular mass of 96 kDa. Alignment with the yeast Mms19 protein indicated amino acid sequence homology extending at least 100 amino acids upstream of the methionine start codon in this human transcript (data not shown), suggesting that this cDNA corresponds to an alternatively spliced form of the primary transcript. We therefore performed 5′-RACE from human testis total RNA using primers that map immediately 5′ to the first methionine codon in the cDNA. This confirmed the presence of alternatively spliced forms of the MMS19 gene (see below) and provided a full-length transcript. To verify these results we amplified the full-length ORF of the hMMS19 gene from human testis total RNA. A cDNA identical to the contiguous sequences previously aligned was obtained. The complete cDNA identified for the hMMS19 gene (GenBank accession no. AF319947) contains an ORF of 3090 nt which can encode a protein of 1030 amino acids (Fig. 1) with a predicted molecular mass of 113 kDa.
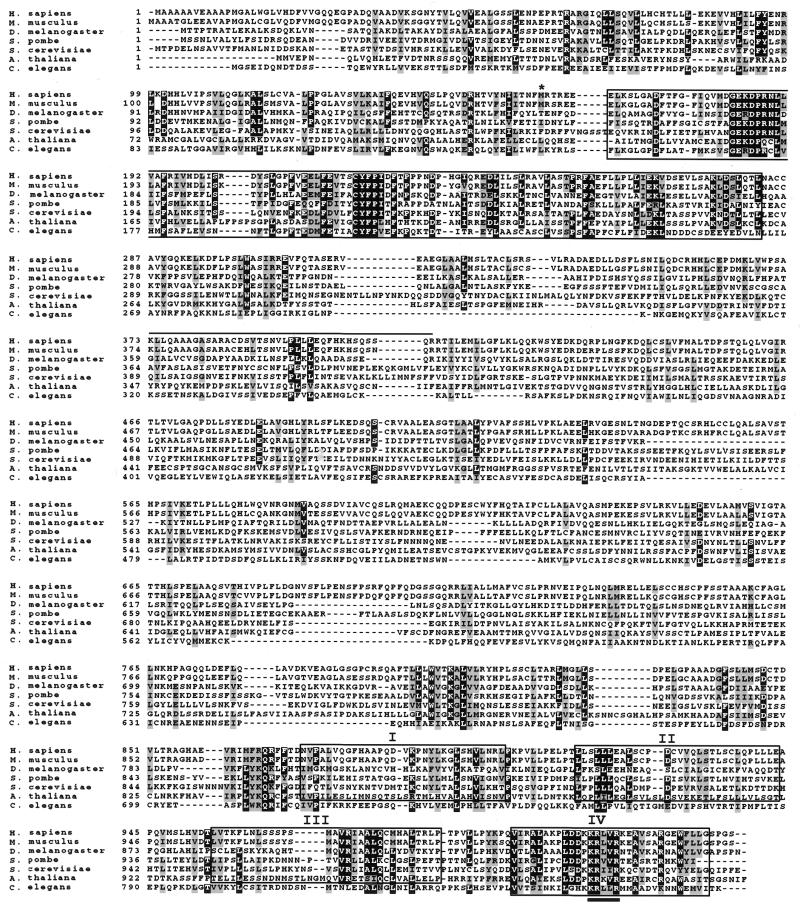
Figure 1.
Amino acid alignment of the Homo sapiens, Mus musculus, D.melanogaster, A.thaliana, C.elegans, S.pombe and S.cerevisiae Mms19 proteins. The alignment was generated using the CLUSTAL-X program. The highly conserved region of ∼120 amino acids at the N-terminus is boxed. Exon 8 (that can be alternatively spliced) corresponds to the first 43 amino acids of this region. The four HEAT repeats are also boxed and labeled I–IV. Only repeat IV of C.elegans is included in this box. The methionine start codon used with alternative splicing of exon 2b or 3 is indicated (*). The 39 amino acids that differ from the previously published MMS19 human protein (9) are overlined. Specifically they are the following: SCCRQLQVHLPVTLSPAMYCLYCWNSSTSTVAASGG. The conserved KRX[LV] sequence in HEAT repeat IV is underlined.
A mouse testis cDNA library yielded a single 977 bp clone in the Mms19 coding region. A full-length cDNA was obtained by multiple rounds of 5′-RACE from mouse testis total RNA. The longest mouse Mms19 cDNA identified (designated mMms19) is 3491 bp in length (GenBank accession no. AF319949) and predicts a polypeptide of 1031 amino acids (Fig. 1) with a molecular mass of 113 kDa. The dMMS19 gene was cloned from whole fly total RNA by RT–PCR. The gene comprises 2880 bp (GenBank accession no. AF319948) and can encode a protein of 958 amino acids (Fig. 1) with a calculated molecular mass of 107 kDa.
yMMS19 homologs from other eukaryotes, including S.pombe (CAB59878), Caenorhabitis elegans (AF067936) and A.thaliana (AB023039) were translated from genomic and partial cDNA clones obtained from public databases. Protein alignments including all the identified MMS19 orthologs confirmed extensive conservation of amino acid sequences (Fig. 1). The predicted amino acid sequences of the human and mouse Mms19 proteins are 90% identical and share 93% similarity. Both the human and mouse polypeptides are essentially the same size as that encoded by the yeast gene (1030, 1031 and 1032 amino acids, respectively). They also share a similar frequency of acidic (10.3% for human and mouse and 12.2% for yeast) and basic (11.5% for human and mouse and 12.1% for yeast) amino acids and a similar estimated pI of 5.92 (5.72 for yeast Mms19 protein). Overall the human and mouse polypeptides share 25% amino acid identity and 50% similarity with the orthologous yeast Mms19 protein. The predicted amino acid sequence for the human protein differs from that previously reported (9) over a stretch of 39 amino acids located between amino acid residues 373 and 412 (Fig. 1).
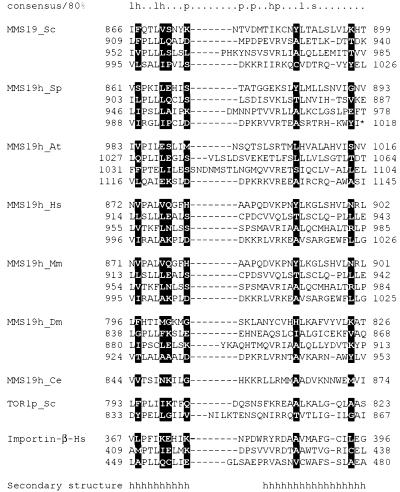
The Mms19 proteins are confidently predicted to possess predominantly α-helical structure (16), with a small α/β-domain potentially present in the middle part of the protein. Sequence analysis using the SEG program (15) suggests that the Mms19 sequences can be partitioned into two predicted globular regions. The longer N-terminal globular region spans amino acid positions 1–770, which includes a highly conserved region of ∼120 amino acids located between residues 167 and 285 of the yeast Mms19 protein. This region shares 42% amino acid sequence identity and 62% similarity with the mouse and human proteins (Fig. 1). A shorter C-terminal globular region spans amino acid positions ∼880–1032, whereas the intervening region is predicted to be non-globular. When the sequence of the C-terminal globular region was independently compared to the protein sequence database using the iterative PSI-BLAST program (12), moderate but statistically significant similarity was detected to a variety of proteins containing so-called HEAT repeats (19,20). Using an expect value of 0.05 as the cut-off for including sequences in the search profile the sequences of ∼500 HEAT repeat proteins were retrieved from the database in 10 search iterations without any obvious false positives. A more detailed analysis performed by searching the Mms19 sequences with a generic HEAT repeat profile (Fig. 2) showed the presence of four repeats (Figs 1 and 2). These comprise a tightly spaced cluster, an arrangement characteristic of HEAT repeat proteins (20). The four repeats are conserved in all available sequences of eukaryotic Mms19 orthologs, with the exception of the C.elegans sequence in which the HEAT repeat region is highly diverged and only the distal repeat is clearly conserved (Figs 1 and 2). Of particular note is the motif KRx[IV]R (alternative amino acids in brackets), which is conserved in the most distal repeat of the Mms19 orthologs (Fig. 1). This motif may function as a specific binding determinant.
Figure 2.
Multiple alignment of the HEAT repeats in the Mms19 orthologs Tor1p and β-importin. The alignment was constructed by parsing PSI-BLAST results, with the repeat boundaries derived from the crystal structure of β-importin (PDB code 1QGKA). The secondary structure underneath the alignment is from the 1QGKA structure: h indicates α-helix. The numbers show the positions of the first and last aligned residues in the corresponding protein sequences. The 80% consensus shows the following classes of amino acid residues: h, hydrophobic (ILVMFYWAC); l, aliphatic (ILV); p, polar (STDENQKRH); s, small (GASTVNAC); +, positively charged (KRH). The positions with the highest information content in the overall HEAT repeat alignment are highlighted in reverse shading.
Alternative splicing of the hMMS19 gene
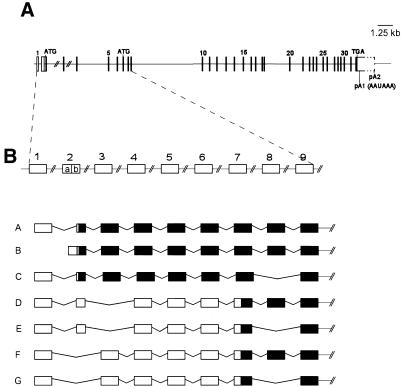
During cloning of the human and mouse cDNAs we identified different clones, presumably reflecting multiple transcripts. To determine whether these correspond to alternatively spliced products we established the exon/intron boundaries of the hMMS19 gene (Fig. 3A). The gene comprises at least 32 exons spanning >40 kb. The two highly conserved domains described above are encoded by exons 8–11 and exons 27–32, respectively. The most abundant hMMS19 cDNA (3530 bp) corresponds to ∼75% of all transcripts identified and includes all coding exons (2b-32) plus exon 1 in the 5′-UTR (Fig. 3B, transcript A). This cDNA can potentially encode a polypeptide of 1030 amino acids and utilizes the first polyadenylation signal in the 3′-UTR. Transcript B differs only in the 5′-UTR by the presence of exon 2a instead of exon 1 (Fig. 3B). The remaining cDNAs identified can encode polypeptides that are 43, 158 or 201 amino acids shorter than that encoded by transcripts A or B (1030 amino acids) (Fig. 3). Alternative splicing of exon 8 (Fig. 3B, transcripts E and F) is expected to delete part of the N-terminal conserved region in the human Mms19 protein. At least one of these transcripts (transcript D, Fig. 3B) uses the second polyadenylation site (pA2, Fig. 3A). Studies are in progress to evaluate the biological significance of these alternative transcripts. All detected transcripts include the region coding for the C-terminal HEAT repeats.
Figure 3.
(A) Schematic representation of the hMMS19 genomic structure. Horizontal lines represent introns. Rectangles/vertical lines correspond to exons. For orientation some exon numbers are displayed. The positions of the putative start codons ATG as well as the stop codon TGA are indicated. Coding sequences are represented as filled boxes. Untranslated regions are represented as open boxes. The first polyadenylation signal (pA1), at nucleotide position 3514, is the most frequently used. A downstream polyadenylation signal (pA2) is used in some transcripts. The complete length of exon 32 could not be determined due to the presence of Alu sequences. (B) Schematic representation of parts of the hMMS19 transcripts identified in normal tissues. Exons 10–32 were found in all transcripts and are not represented for simplification. Black boxes correspond to coding regions. Open boxes represent untranslated exons. Exon 1 or 2a (non-coding) is exclusively used at the 5′-UTR of the MMS19 gene. Transcripts A and B are shown as examples. Transcripts D and E were also identified with exon 2a instead of exon 1. Alternative splicing of exon 8 can apparently occur alone, as shown in transcript C, or in combination with alternative splicing of exon 2b or 3 (transcripts G and E). Both situations delete 43 amino acids in one of the two highly conserved domains of the Mms19 protein (see Fig. 1). Exons 2b and 3 can also be alternatively spliced, resulting in potentially even shorter polypeptides (transcripts D–G).
Human and mouse MMS19 gene expression patterns
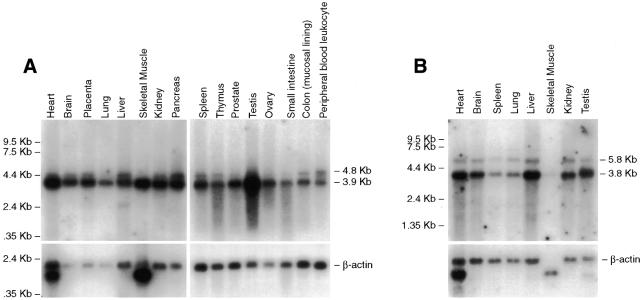
Northern blot analysis of human and mouse adult and fetal tissues revealed the presence of moderate to high steady-state levels of MMS19 mRNA in all tissues analyzed (Fig. 4A and B and data not shown). The most abundant transcript has an estimated size of 3.9 kb in human tissues (Fig. 4A) and 3.8 kb in mouse tissues (Fig. 4B). These presumably correspond to the most abundant cDNAs described above (3530 and 3491 bp for human and mouse, respectively). Both the human and mouse transcripts have consensus AAUAAA polyadenylation signals at positions 3514 and 3441, respectively, followed immediately by a poly(A) tail. Additional bands with estimated sizes of 4.8 and 5.8 kb in human and mouse tissues, respectively, were detected by northern blot analysis in most tissues examined (Fig. 4). Significant differences in the ratio of the two transcripts were observed in different tissues. For example, the 4.8 kb transcript is relatively abundant in human pancreas, but is almost undetectable in skeletal muscle (Fig. 4A). The 4.8 kb band presumably reflects transcripts with a 3′-UTR extending beyond the first polyadenylation signal, as represented in Figure 3A. This region of the gene is very rich in Alu repeat sequences, hence it was not possible to design a probe to confirm this presumption by northern blot analysis.
Figure 4.
Northern blot of mRNAs from various adult human (A) and mouse (B) tissues. Cloned and sequenced verified 3′-UTR human and mouse MMS19 products were used as probes (top). β-Actin is shown as an internal control (bottom). The marker track is shown on the left.
In an effort to correlate the transcripts detected by northern blot analysis with the putative alternative splice products at exons 3 and 8 we hybridized the human northern blots with probes derived from both exons. In all cases the results were indistinguishable from those obtained with a 3′-end probe (data not shown). We therefore conclude that both bands detected by northern blotting comprise a heterogeneous collection of alternatively spliced forms.
Chromosomal mapping of the hMMS19 gene
FISH with a BAC clone containing the entire hMMS19 ORF yielded a single site of hybridization at chromosomal band 10q24. This localization is consistent with hybrid mapping results for two STSs (A002G19 and Cda19h12) in the NCBI public database, which align with the 3′-UTR of the cloned hMMS19 gene.
Complementation of the yeast mms19Δ mutant by hMMS19 and dMMS19 cDNA
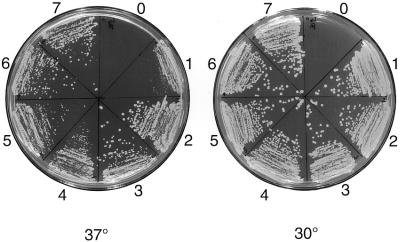
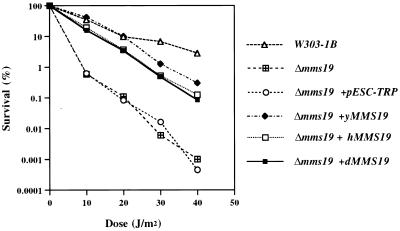
In view of the amino acid conservation between yeast Mms19 protein and that from higher eukaryotes, especially in the C- and N-terminal regions, we asked whether the MMS19 genes from higher organisms can functionally complement mutant phenotypes of the yeast mms19Δ strain, deleted of the entire MMS19 gene. We overexpressed both hMMS19 and dMMS19 cDNAs under control of the yeast GAL1 promoter in the yeast mms19 deletion mutant. As shown in Figure 5, both cDNAs corrected thermosensitivity for growth of the mms19 deletion mutant. Furthermore, whereas the doubling time of the yeast mutant is ∼19 h in liquid culture, at 37°C this doubling time was reduced to ∼3.5 h when the mutant strain was transformed with the human, Drosophila or yeast wild-type MMS19 genes. The full-length hMMS19 and dMMS19 genes also complemented the UV radiation sensitivity of the mms19 mutant (Fig. 6). However, these genes failed to complement the methionine auxotrophy of the yeast mms19 mutant (data not shown). As expected, the yMMS19 gene fully complemented the latter phenotype (data not shown).
Figure 5.
Rescue of thermosensitive growth of the mms19 deletion mutant. Strains are as follows: mms19Δ (0), mms19Δ+pESC-TRP (1), mms19Δ+yMMS19 (2 and 3), mms19Δ+dMMS19 (4 and 5), mms19Δ+hMMS19 (6 and 7). As positive controls strains were also grown at 30°C. In the absence of pESC-TRP the mms19Δ (0) strain is unable to grow.
Figure 6.
Functional complementation of the UV radiation sensitivity of the mms19Δ strain by overexpression of human (mms19Δ+hMMS19) or Drosophila (mms19Δ+dMMS19) MMS19 cDNAs. For comparison the survival curves are shown for mms19Δ transformed with pESC-TRP (empty expression vector) and with the yMMS19 ORF (expression vector with the wild-type yMMS19 gene). W303-1B is the isogenic wild-type strain.
DISCUSSION
We report here the identification of human, mouse and Drosophila orthologs of the S.cerevisiae MMS19 gene. Alignments of these as well as several other apparent orthologs with the yeast Mms19 protein show extensive amino acid sequence conservation as well as similarity in size. In particular, we have identified highly conserved HEAT repeat domains in the C-terminus of this family of proteins. HEAT repeats have been identified in a variety of cytoplasmic and nuclear eukaryotic proteins. The typical repeat unit consists of 30–34 amino acid residues and forms two α-helices separated by a short loop, as inferred from the crystal structure of human β-importin (21). Arrays of HEAT repeats form tandemly arranged bi-helical structures that appear to function as scaffolds for assembly of other subunits of the corresponding complexes (21–23). A common denominator for many of these proteins is their association with and involvement in the assembly of large, multisubunit protein complexes such as coated vesicles and microtubules. The recent demonstration of the presence of HEAT repeats in several chromatin-associated proteins, including subunits of cohesins and condensins and several transcription regulators, is particularly interesting (20).
Functional complementation of mutant phenotypes by NER proteins across species is the exception rather than the rule (24,25), presumably reflecting specific structural requirements for efficient assembly of multiprotein complexes. Hence, the observed complementation of thermosensitive growth and UV radiation sensitivity of the yeast mms19Δ mutant with the MMS19 genes from Drosophila and humans provides suggestive evidence for both structural and functional conservation of Mms19 protein.
While this manuscript was in preparation cloning and characterization of the hMMS19 gene was independently published (9). However, while the putative human polypeptide protein identified in our study is precisely the same size, it differs from the previously published sequence over a stretch of 39 amino acids in the central part of the protein. This difference does not appear to result from alternative splicing since we have characterized the genomic structure corresponding to the coding region. Furthermore, the mouse cDNA is 90% identical to our human cDNA clone and this identity includes the 39 amino acids that differ from the previously published translated sequence. We suggest that the previously published sequence (9) may have one or more mutations in the ORF and that the failure to observe correction of the yeast mms19Δ mutant phenotype may derive from such mutations. No GenBank accession number was provided to verify this. Alternatively, the gene previously reported might represent a slightly altered duplication of the yMMS19 gene. There is a precedent for duplication of a gene with an essential role in NER and RNAP II transcription. Specifically, p44 protein is a subunit of TFIIH that regulates the activity of XPD DNA helicase, another TFIIH subunit. In humans two closely linked p44 genes have been identified (p44t and p44c), which can encode proteins that differ by just three amino acids (26).
Using RT–PCR, molecular cloning and DNA sequencing we have demonstrated that the hMMS19 primary transcript undergoes extensive processing and uses two different polyadenylation sites. Extensive alternative splicing is not uncommon in genes that encode transcription factors and may play a critical role(s) in the regulation of developmental pathways and in cancer (27,28). Differences in the 5′-UTR have been implicated in regulating transcription efficiency (29) and different 3′-UTRs may be associated with differential stability or translatability of various mRNA products (reviewed in 30). Furthermore, different protein isoforms may regulate or interact with distinct sets of targets (31,32). For example, it was recently reported that splice variants of the Wilms’ tumor 1 gene (WT1) encode proteins that have opposite effects on tumorigenicity (33). It is possible that the hMMS19 gene evolved different protein isoforms with different functions.
The functional significance of the different MMS19 cDNAs cloned is supported by the identification of two major mRNAs in both human and mouse northern blots. We suggest that the larger transcript results from use of the second polyadenylation site and that this choice correlates with a different promoter. However, we were unable to precisely correlate the splice variants of MMS19 detected by RT–PCR and cloning with the transcripts detected by northern analysis. Previous studies (9) detected a single transcript at ∼4 kb in a mouse northern blot. It is likely that this corresponds to our transcript of ∼3.8 kb.
The hMMS19 gene maps to region 10q24 of the human genome, a region frequently rearranged in human cancer (34). However, no genetic disorders obviously associated with NER or transcription defects have been mapped to this region of the human genome. The precise function(s) of Mms19 protein remains elusive. It was recently demonstrated that cells from the hereditary human disease trichothiodystrophy (TTD), belonging to genetic complementation group A (TTD-A), have reduced levels of TFIIH activity (35). This observation led to the suggestion that the putative TTD-A protein normally determines the stability of the TFIIH complex. MMS19 is an attractive candidate for such a function. As mentioned earlier, RNAP II transcription in yeast mms19Δ extracts is inactivated by heat treatment (5). This thermolabile defect can be fully complemented by purified TFIIH, but not by purified yeast Mms19 protein, suggesting that Mms19 protein indeed influences the functional integrity of TFIIH (5). Consistent with this notion, the previously published form of human Mms19 protein was shown to physically interact with TFIIH through its XPD and XPB subunits (9). The observation of conserved HEAT repeats in the Mms19 proteins is consistent with a role of this protein in the assembly of TFIIH-containing complexes for NER and RNAP II transcription.
The genetic complementation group TTD-A is presently represented by a single patient (TTD1BR) which complemented the phenotypes of TTD cells with defects in TFIIH subunit XPB or XPD (36). We were unable to correct the NER defect of TTD1BR cells following microinjection of a construct containing full-length hMMS19 cDNA. However, we cannot exclude the possibility of a mutation in a regulatory region of MMS19 in TTD-A cells. Likewise, we cannot exclude differences in the relative abundance of specific MMS19 splice variants in the TTD-A disorder or in the several TTD-like syndromes previously described (37 and references therein). Experiments are in progress to evaluate these hypotheses.
Acknowledgments
ACKNOWLEDGEMENTS
We thank Dr Randy Legerski and Dr Jaap Brower for providing the HeLa cell cDNA pEBS7 library number 9 and the mms19 mutant deletion strain, respectively. We also thank Marzi Ranjbaran and Elizabeth Lutz for valuable technical assistance and our laboratory colleagues for valuable discussions. Studies in the E.C.F. laboratory are supported by grant CA44247 from the USPHS. M.S. acknowledges support from the Associazione Italiana per la Ricerca sul Cancro (AIRC).
DDBJ/EMBL/GenBank accession nos AF319947–AF319949
References
- 1.Friedberg E.C., Walker,G.C. and Siede,W. (1995) DNA Repair and Mutagenesis. American Society for Microbiology, Washington, DC.
- 2.Prakash L. and Prakash,S. (1977) Isolation and characterization of MMS-sensitive mutants of Saccharomyces cerevisiae. Genetics, 86, 33–55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Prakash L. and Prakash,S. (1979) Three additional genes involved in pyrimidine dimer removal in Saccharomyces cerevisiae: RAD7, RAD14 and MMS19. Mol. Gen. Genet., 176, 351–359. [DOI] [PubMed] [Google Scholar]
- 4.Miller R.D., Prakash,L. and Prakash,S. (1982) Genetic control of excision of Saccharomyces cerevisiae interstrand DNA cross-links induced by psoralen plus near-UV light. Mol. Cell. Biol., 2, 939–948. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lauder S., Bankmann,M., Guzder,S.N., Sung,P., Prakash,L. and Prakash,S. (1996) Dual requirement for the yeast MMS19 gene in DNA repair and RNA polymerase II transcription. Mol. Cell. Biol., 16, 6783–6793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lombaerts M., Tijsterman,M., Verhage,R.A. and Brouwer,J. (1997) Saccharomyces cerevisiae mms19 mutants are deficient in transcription-coupled and global nucleotide excision repair. Nucleic Acids Res., 25, 3974–3979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araujo S.J., Tirode,F., Coin,F., Pospiech,H., Syvaoja,J.E., Stucki,M., Hubscher,U., Egly,J.M. and Wood,R.D. (2000) Nucleotide excision repair of DNA with recombinant human proteins: definition of the minimal set of factors, active forms of TFIIH and modulation by CAK. Genes Dev., 14, 349–359. [PMC free article] [PubMed] [Google Scholar]
- 8.Svejstrup J.Q., Vichi,P. and Egly,J.M. (1996) The multiple roles of transcription/repair factor TFIIH. Trends Biochem. Sci., 21, 346–350. [PubMed] [Google Scholar]
- 9.Seroz T., Winkler,G.S., Auriol,J., Verhage,R.A., Vermeulen,W., Smit,B., Brouwer,J., Eker,A.P.M., Weeda,G., Egly,J.M. and Hoeijmakers,J.H.J. (2000) Cloning of a human homolog of the yeast nucleotide excision repair gene MMS19 and interaction with transcription repair factor TFIIH via the XPB and XPD helicases. Nucleic Acids Res., 28, 4506–4513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Altschul S.F., Gish,W., Miller,W., Myers,E.W. and Lipman,D.J. (1990) Basic local alignment search tool. J. Mol. Biol., 215, 403–410. [DOI] [PubMed] [Google Scholar]
- 11.Queimado L., Seruca,R., Costa-Pereira,A. and Castedo,S. (1995) Identification of two distinct regions of deletion at 6q in gastric carcinoma. Genes Chromosomes Cancer, 14, 28–34. [DOI] [PubMed] [Google Scholar]
- 12.Altschul S.F., Madden,T.L., Schaffer,A.A., Zhang,J., Zhang,Z., Miller,W. and Lipman,D.J. (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res., 25, 3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Altschul S.F. and Koonin,E.V. (1998) Iterated profile searches with PSI-BLAST—a tool for discovery in protein databases. Trends Biochem. Sci., 23, 444–447. [DOI] [PubMed] [Google Scholar]
- 14.Thompson J.D., Gibson,T.J., Plewniak,F., Jeanmougin,F. and Higgins,D.G. (1997) The CLUSTAL:X Windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res., 25, 4876–4882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wootton J.C. and Federhen,S. (1996) Analysis of compositionally biased regions in sequence databases. Methods Enzymol., 266, 554–571. [DOI] [PubMed] [Google Scholar]
- 16.Rost B. and Sander,C. (1994) Combining evolutionary information and neural networks to predict protein secondary structure. Proteins, 19, 55–72. [DOI] [PubMed] [Google Scholar]
- 17.Felsenstein J. (1996) Inferring phylogenies from protein sequences by parsimony, distance and likelihood methods. Methods Enzymol., 266, 418–427. [DOI] [PubMed] [Google Scholar]
- 18.Tonk V., Schneider,N.R., Delgado,M.R., Mao,J. and Schultz,R.A. (1996) Identification and molecular confirmation of a small chromosome 10q duplication [dir dup(10)(q24.2→q24.3)] inherited from a mother mosaic for the abnormality. Am. J. Med. Genet., 61, 16–20. [DOI] [PubMed] [Google Scholar]
- 19.Andrade M.A. and Bork,P. (1995) HEAT repeats in the Huntington’s disease protein [letter]. Nat. Genet., 11, 115–116. [DOI] [PubMed] [Google Scholar]
- 20.Neuwald A.F. and Hirano,T. (2000) HEAT repeats associated with condensins, cohesins and other complexes involved in chromosome-related functions. Genome Res., 10, 1445–1452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cingolani G., Petosa,C., Weis,K. and Muller,C.W. (1999) Structure of importin-β bound to the IBB domain of importin-α. Nature, 399, 221–229. [DOI] [PubMed] [Google Scholar]
- 22.Chook Y.M. and Blobel,G. (1999) Structure of the nuclear transport complex karyopherin-β2-Ran x GppNHp. Nature, 399, 230–237. [DOI] [PubMed] [Google Scholar]
- 23.Vetter I.R., Nowak,C., Nishimoto,T., Kuhlmann,J. and Wittinghofer,A. (1999) Structure of a Ran-binding domain complexed with Ran bound to a GTP analogue: implications for nuclear transport. Nature, 398, 39–46. [DOI] [PubMed] [Google Scholar]
- 24.Rodel C., Jupitz,T. and Schmidt,H. (1997) Complementation of the DNA repair-deficient swi10 mutant of fission yeast by the human ERCC1 gene. Nucleic Acids Res., 25, 2823–2827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sung P., Bailly,V., Weber,C., Thompson,L.H., Prakash,L. and Prakash,S. (1993) Human xeroderma pigmentosum group D gene encodes a DNA helicase. Nature, 365, 852–855. [DOI] [PubMed] [Google Scholar]
- 26.Burglen L., Seroz,T., Miniou,P., Lefebvre,S., Burlet,P., Munnich,A., Pequignot,E.V., Egly,J.M. and Melki,J. (1997) The gene encoding p44, a subunit of the transcription factor TFIIH, is involved in large-scale deletions associated with Werdnig-Hoffmann disease. Am. J. Hum. Genet., 60, 72–79. [PMC free article] [PubMed] [Google Scholar]
- 27.Lopez A.J. (1995) Developmental role of transcription factor isoforms generated by alternative splicing. Dev. Biol., 172, 396–411. [DOI] [PubMed] [Google Scholar]
- 28.MacDougall C., Harbison,D. and Bownes,M. (1995) The developmental consequences of alternate splicing in sex determination and differentiation in Drosophila. Dev. Biol., 172, 353–376. [DOI] [PubMed] [Google Scholar]
- 29.Gray N.K. and Wickens,M. (1998) Control of translation initiation in animals. Annu. Rev. Cell. Dev. Biol., 14, 399–458. [DOI] [PubMed] [Google Scholar]
- 30.Edwalds-Gilbert G., Veraldi,K.L. and Milcarek,C. (1997) Alternative poly(A) site selection in complex transcription units: means to an end? Nucleic Acids Res., 25, 2547–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.de Melker A.A. and Sonnenberg,A. (1999) Integrins: alternative splicing as a mechanism to regulate ligand binding and integrin signaling events. Bioessays, 21, 499–509. [DOI] [PubMed] [Google Scholar]
- 32.Hsu T., Gogos,J.A., Kirsh,S.A. and Kafatos,F.C. (1992) Multiple zinc finger forms resulting from developmentally regulated alternative splicing of a transcription factor gene. Science, 257, 1946–1950. [DOI] [PubMed] [Google Scholar]
- 33.Menke A.L., Riteco,N., van Ham,R.C., de Bruyne,C., de Rauscher,F.J., van der Eb,A.J. and Jochemsen,A.G. (1996) Wilms’ tumor 1 splice variants have opposite effects on the tumorigenicity of adenovirus-transformed baby-rat kidney cells. Oncogene, 12, 537–546. [PubMed] [Google Scholar]
- 34.Mitelman F., Mertens,F. and Johansson,B. (1997) A breakpoint map of recurrent chromosomal rearrangements in human neoplasia. Nat. Genet., 15, 417–474. [DOI] [PubMed] [Google Scholar]
- 35.Vermeulen W., Bergmann,E., Auriol,J., Rademakers,S., Frit,P., Appeldoorn,E., Hoeijmakers,J.H.J. and Egly,J.M. (2000) Sublimiting concentration of TFIIH transcription/DNA repair factor causes TTD-A trichothiodystrophy disorder. Nat. Genet., 26, 307–313. [DOI] [PubMed] [Google Scholar]
- 36.Stefanini M., Vermeulen,W., Weeda,G., Giliani,S., Nardo,T., Mezzina,M., Sarasin,A., Harper,J.I., Arlett,C.F., Hoeijmakers,J.H. et al. (1993) A new nucleotide-excision-repair gene associated with the disorder trichothiodystrophy. Am. J. Hum. Genet., 53, 817–821. [PMC free article] [PubMed] [Google Scholar]
- 37.de Boer J. and Hoeijmakers,J.H. (2000) Nucleotide excision repair and human syndromes. Carcinogenesis, 21, 453–460. [DOI] [PubMed] [Google Scholar]