Abstract
The interferon (IFN) response, induced as a side effect after transfection of nucleic acids into mammalian cells, is known but inadequately described. We followed the IFN response, the fate of cells, and the possible mechanisms leading to this response in NIH3T3 mouse fibroblasts after DNA nucleofection. The gateway destination vector, phGf, and its derivatives encoding toxic and non-toxic variants of the minor structural proteins of polyomaviruses, VP2 and VP3, were used. DNA vector sequences induced in cells the production of high levels of IFN and the upregulation of the IFN-inducible genes, Mx-1, STAT1, IRF1, and IRF7. The IFN response was not restricted to phGf-derived plasmids. In nucleofected cells, upregulation of the modified γ-histone 2A.X indicating DNA damage and inhibition of cell proliferation were also observed. Although 3T3 cells expressed the Toll-like receptor-9 (TLR9) and vectors used for nucleofection contained unmethylated CpGs, signaling leading to IFN induction was found to be TLR9 independent. However, the early activation of nuclear factor-kappa B suggested the participation of this transcription factor in IFN induction. Surprisingly, in contrast to nucleofection, transfection using a cationic polymer induced only a poor IFN response. Together, the results point to a strong side effect of nucleofection.
This article clearly provides data consistent with the induction of type I IFN in 3T3 cells following experimental introduction of CpG-containing DNA. They show that this process is independent of TLR9 but may involve mobilization of NF-κB.
Introduction
Transfection of mammalian cells with bacterial, viral, or synthetic nucleic acids has been acknowledged to induce type I interferon (IFN) responses. However, there has been little attention paid to these responses appearing as a side effect of cell transfection during studies of cellular responses. IFN induces tremendous effects on many cellular processes by activating different signaling cascades, such as Janus kinases-signal transducer and activator of transcription (JAK/STAT), mitogen-activated protein kinase (MAPK), or phosphatidylinositol 3-kinases/serine/threonine kinase Akt (PI3K/Akt) (reviewed in Pestka et al., 2004; Platanias, 2005; Stetson and Medzhitov, 2006). These pathways are involved, among other vital cellular processes, in the regulation of cell growth, proliferation, and apoptosis. In addition, IFN dramatically affects protein synthesis through activation of the protein kinase R (PKR). Activation of PKR results in the formation of an inactive complex of GDP-eIF2 (Guanosine diphosphate-eukaryotic initiation factor 2) that blocks translation (reviewed in García et al., 2006; Pitha and Kunzi, 2007; Pindel and Sadler, 2011).
For a long time, the only known mechanism used for immune cells to recognize nucleic acids was the sensing of unmethylated CpG motifs (cytosine next to guanine) by the endosomal membrane receptor, Toll-like receptor-9 (TLR9) (Krieg et al., 1995; Hemmi et al., 2000; Dalpke et al., 2006). Sensing of DNA by this receptor induces the production of type I IFN and other cytokines. Interestingly, it was also reported that non-immune cells are able to produce type I IFN in response to bacterial DNA and CpG motifs in a TLR9-dependent manner (Li et al., 2004; Platz et al., 2004; Di et al., 2009). Also, overexpression of TLR9 in non-immune cells reconstitutes CpG DNA responses (Bauer et al., 2001; Takeshita et al., 2001; Assaf et al., 2009). Recently, however, several cytoplasmic sensors that can recognize DNA by different mechanisms (CpG motif/TLR9 independent) have been reported for immune and non-immune cells: DNA-dependent activator of IFN-regulatory factor (DAI) (Takaoka et al., 2007), DEAH box polypeptide 9 (DHX9), DEAH box polypeptide 36 (DHX36) (Kim et al., 2010), DDX41 (Helicase) (Zhang et al., 2011b), absence in melanoma 2 (AIM2) (Hornung et al., 2009), IFN inducible protein 16 (IFI16) (Unterholzner et al., 2010), leucine-rich repeat containing protein (LRRF1P1l protein) (Yang et al., 2010), and the Ku70 protein (Zhang et al., 2011a). It is speculated that these sensors may recognize DNA in a sequence-independent manner. Nevertheless, the nature of the interaction between DNA and the cytoplasmic sensors is still unknown. With the exception of the AIM2 receptor, which induces caspase-1-dependent maturation of the pro-inflammatory cytokines, interleukin (IL)-1β and IL-18 (Hornung et al., 2009), the sensing of DNA by cytoplasmic sensors induces type I IFN production. The pathways of induction of the IFN response have been uncovered. In broad terms, six important signal molecules downstream of the receptors have been described as modulators of the response: the transcription factors, nuclear factor-kappa B (NF-κB) and IFN regulatory factor (IRF) 1, 3, 7, further, the protein kinase TANK-binding kinase 1 (TBK1), and the transmembrane ER resident protein, stimulator of IFN genes (STING) (Ishikawa et al., 2009 and reviewed in Keating et al., 2011; Sharma and Fitzgerald, 2011). Some of these proteins participate redundantly in different sensor signaling pathways of IFN induction.
The strength, and therefore, the outcome of the IFN response may depend on DNA properties (e.g., CpG content and methylation state), DNA delivery mechanism and cell type, as has been described in the context of immune system cells or even for other cells. In detail, murine dendritic cells produced inflammatory cytokines when stimulated with DNA containing CpG motifs from Escherichia coli or with naked plasmid DNA, but not when stimulated with calf thymus DNA (poor in CpG motifs). In addition, it was also found that the amount of secreted cytokines increased when the DNA was complexed with the cationic lipid-lipofectin (Yoshinaga et al., 2006). It was also demonstrated that peritoneal macrophages from mice launched strong immunoresponses to DNA/cationic liposome complexes in a CpG-dependent and independent manner (Yasuda et al., 2005). In a study using non-immune cells, it has been shown that among the transfection methods such as electroporation, lipofection by Fugene, or transfection by the cation polymer ExGen, the last one induced the highest levels of IFNβ. In addition, the authors of this study found that the IFN response was induced in a cell type-dependent manner (Rautsi et al., 2007). From the above studies, we can conclude that there may be specific ways of how the DNA is sensed, and an important part of this process may be dependent on the DNA entry into the cells and its transport toward the nucleus. For example, DNA transfected by lipofection or by cations engages the cellular endocytic pathways to reach the nucleus (Legendre and Szoka, 1992; Yasuda et al., 2005), while electroporation induces transient pores in the plasma membrane for DNA entry into the cells (Vasilkoski et al., 2006). Another recently developed method, nucleofection, combines electrical parameters with cell type-adapted solutions of unknown composition for delivery of DNA “straight into the cell nucleus” (Gresch et al., 2004). The influence of this particular method of transfection on IFN responses is currently unknown.
This study was preceded by a microarray analysis of mouse fibroblast cells transfected by nucleofection with mammalian expression vectors for transient expression of highly cytotoxic VP2 or VP3 structural proteins of the Mouse polyomavirus (MPyV). The study disclosed remarkable concomitant IFN and DNA damage response (DDR) and upregulation of the endosomal membrane receptor, TLR9, in cells transfected with a control empty plasmid. Here, we analyzed those responses, evaluated the role of TLR9 ligands (unmethylated CpG motifs) in the establishment of the IFN response and investigated the role of IRF3, and NF-κB transcription factors in the signaling for the IFN induction. As a model, we used the phGf gateway destination vector from Addgene and three of its derivatives carrying genes for VP2 or VP3 of MPyV and non-cytotoxic VP2 of the Merkel cell polyomavirus (MCPyV). For comparison, we used other vectors, pEGFP-N1 (Clontech) and two of its derivatives carrying the MPyV VP2 or VP3 genes and pcDNA3.1 (Invitrogen).
Materials and Methods
Cell lines, DNA, and transfections
Mouse fibroblasts NIH3T3 and mouse embryonic fibroblasts (MEF) were grown at 37°C in a humidified incubator with a 5% CO2 atmosphere, using DMEM (Dulbecco's modified Eagle's medium; Sigma-Aldrich) supplemented with 10% fetal bovine serum and 4 mM L-glutamine. Plasmid DNA was prepared using the EndoFree Maxi Kit (Qiagen), diluted in TE and stored at −20°C. Plasmid concentration and purity was determined by nanodrop. Transfections were performed by electroporation within a nucleofector™ device (program U-30 for 3T3 or A-23 for MEF), using Amaxa reagents (Amaxa Biosystems) or by cationic polymer by using TurboFect™ (Thermo Scientific) according to the manufacturer's instructions. Briefly, cells were seeded 24 h prior to nucleofection and 4×106 cells were transfected with 6 μg of plasmid DNA (∼1.0–1.5×1012 DNA molecules), except in the case of transfection with the small plasmid, phGfΔGΔR, when only 4 μg of DNA were used (1.2×1012 DNA molecules). For turbofection, 1×106 cells were seeded in 60-mm plates 24 h prior to transfection and then were transfected by 6 μg of DNA.
DNA constructs
The plasmids phGf, a Gateway® plasmid (# 22516), and their derivates ph2p (# 22521) and ph3p (# 22520) coding for variants of the MPyV minor proteins VP2 and VP3 were obtained from Addgene (Buck et al., 2006; Tolstov et al., 2009). We did modify the plasmids by deletion of the ORF coding for the EGF protein. Briefly, plasmids were double digested with StuI and ClaI, the overhangs were filled in by the Klenow fragment of DNA polymerase I and the plasmid DNA was circularized by ligation. The modified products were named phGfΔG, ph2pΔG, and ph3pΔG, respectively. In addition, for a control, we further modified the phGfΔG plasmid by deletion of region 2 (between lambda attachment sites [att sites]; Fig. 1A), which becomes substituted during gateway cloning. The resulting plasmid was named phGfΔGΔR. In detail, the phGfΔG plasmid was double digested with Nhe I and Nar I, the overhangs were filled in by the Klenow fragment of DNA polymerase I and the plasmid DNA was circularized by ligation. The phGf derivative plasmid ph2m (# 22518) carrying gene for the VP2 structural protein of the MCPyV was also obtained from Addgene (Tolstov et al., 2009). The vector pEGFP-N1 was obtained from Clontech. Its derivates, VP2-EGFP-N1 (carrying MPyV VP2 gene) or VP3-EGFP-N1 (carrying MPyV VP3 gene), had been previously prepared in our lab (Huerfano et al., 2010). The plasmid pcDNA3.1 was obtained from Invitrogen.
FIG. 1.
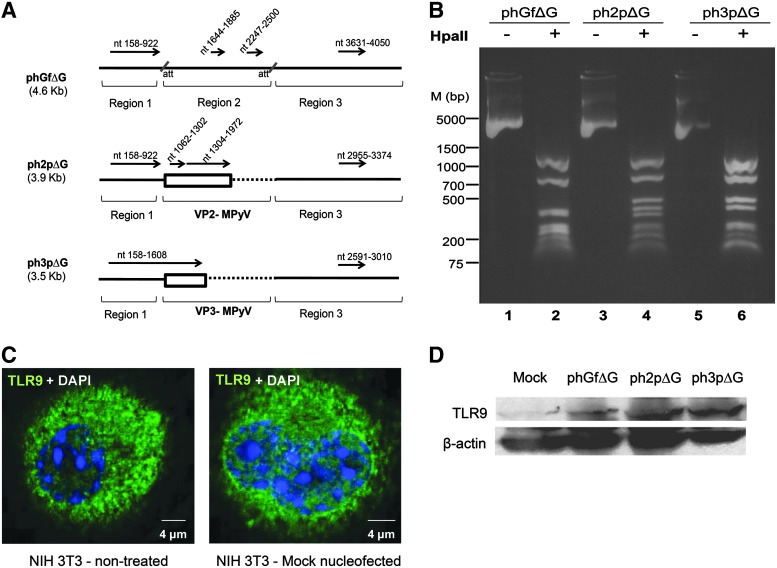
Unmethylated CpG motifs in the plasmids and presence of the Toll-like receptor-9 (TLR9) in the NIH3T3 cell line. (A) Analysis of the presence and distribution of CpG dinucleotides was performed by means of EMBOSSES CpGPlot. The CpG motifs of plasmid DNA are presented by black arrows, the position of the motifs within the plasmid are indicated on the top of the arrows in nucleotide (nt) numbers. The boxes in bold represent the sequences of VP2 and VP3 of Mouse polyomavirus (MPyV). Sequences of the phGfΔG plasmid were arbitrarily divided into three regions. Dotted lines in the expression vectors, ph2pΔ and ph3pΔG, indicate sequences deleted by cloning. (B) The methylation state of phGfΔG plasmid (lanes 1, 2), ph2pΔG plasmid (lanes 3, 4), or ph3pΔG plasmid (lanes 5, 6) was tested by Hpa II digestion. HpaII digested (lanes 2–4) and control non-digested (lanes 1, 3, 5) plasmids were resolved by 0.8% agarose electrophoresis. (C) Confocal section of NIH3T3 cells (untreated or mock-transfected cells) stained with antibody against the TLR9 receptor (green) and by DAPI. (D) Levels of the TLR9 protein in cells nucleofected with phGfΔG, ph2pΔG, or ph3pΔG plasmids and in mock-transfected cells (4 h post-transfection) were analyzed in Western blots using anti-TLR9 antibody. Antibody against β-actin was used as loading control.
CpG analysis
Sequences of the gateway plasmids used in the study were analyzed by using the EMBOSS, CpGPlot program (web-based program) available at EMBL-EBI (www.ebi.ac.uk). For the analysis, we used standard parameters (Window length 100, Obs/Exp CpG ratio 0.6, Min C+G 50%, and Min Length 200).
Plasmid DNA digestion and methylation
DNA was subjected to cleavage by restriction enzyme, HpaII (Thermo Scientific). Restriction digestion was confirmed by 0.8% agarose gel electrophoresis stained by Gel red nucleic acid stain™ (Biotium). In addition, when required, DNA was methylated using the CpG methyltransferase M.SssI (New England Biolabs), according to manufacturer's instructions.
Quantitative real-time polymerase chain reaction
Total RNA was extracted using the High Pure RNA Isolation Kit (Roche) according to the manufacturer's protocol. The RNA concentration was measured by a spectrophotometer, and the integrity was evaluated using an Agilent 2100 bioanalyzer (Agilent); reverse transcription was carried out with the iScriptcDNA Synthesis kit (Bio-Rad Laboratories) according to a producer's manual. cDNAs were amplified by polymerase chain reaction (PCR) using primers sets:
STAT1, 5′-GGAGGTGAACCTGACTTCCA-3′ and 5′-TCTGGTGCTTCCTTTGGTCT-3′; Mx-1, 5′-GGTCGGCTTCTGGTTTTGTA-3′ and 5′-GAACAGGTCCACTTCCTCCA-3′; IRF1, 5′-CCTGGGTCAGGACTTGGATA-3′ and 5′-TTCGGCTATCTTCCCTTCCT-3′; IRF7, 5′-GAAGACCCTGATCCTGGTGA-3′ and 5′-CCAGGTCCATGAGGAAGTGT-3′; and IFNβ, 5′-CCCTATGGAGATGACGGAGA-3′ and 5′-CTGTCTGCTGGTGGAGTTCA-3′.
Quantification of PCR products in real time was performed in a Light Cycler 480 II from Roche using the Light Cycler® 480 SYBR Green I Master kit, according to the manufacturer's protocol. Quantification of target gene expression was performed using Light Cycler 480 II software based on the relative quantification method, determining the concentration of target amplicons normalized to the reference β-actin gene.
IFNβ measurement
IFNβ was assessed in sample supernatants using the Verikine™ IFNβ kit, a sandwich enzyme-linked immunosorbent assay (ELISA) from PBL Biomedical Laboratories, according to the manufacturer's recommended protocol. In brief, after transfection, 1.2×106 cells were resuspended in 2.5 mL of DMEM supplemented with serum, seeded into 3-cm plates and incubated at 37°C in a humidified incubator with 5% CO2 atmosphere. Supernatants were collected at indicated times and ELISA was performed. Absorbance was measured by an ELISA reader (450 nm).
Evaluation of cytotoxicity
The release of LDH occurring upon cell lysis was quantified using a CytoTox 96 cytotoxicity assay kit (Promega), according to the manufacturer's instructions. Briefly, cells were transfected and then seeded on 24-well plates, and 2 h post-transfection, the medium was replaced by fresh medium to remove the cells that died due to the electrical pulse. Later, at indicated times, the medium from growing cells were collected to measure LDH. The total numbers of cells was obtained by treatment of cells with Triton X-100. Absorbance was measured by an ELISA reader (490 nm).
Proliferation assay
After transfection, the cells were resuspended in complete medium, seeded on 96-well tissue culture plates (confluence of ∼20%) and incubated at 37°C in a humidified incubator with 5% CO2 atmosphere. After 2 h, the medium was replaced by fresh medium and the cells were further incubated and, at selected times, we tested the ability of cells to cleave the tetrazolium salt to formazan as an indicator of an active metabolism in cells. Briefly, proliferation reagent, WST-1 (Roche) was suspended in medium without red phenol (ratio 1/10) and 100 μL of the suspension was added to cells. Cells were further incubated at 37 °C for 3 h and then the absorbance was measured by an ELISA reader (450 nm).
Western blot analysis
Cells were harvested at the indicated time points and washed with phosphate-buffered saline (PBS), then they were resuspended in ice-cold cell lysis buffer (10 mM Tris/HCl, pH 7.4, 1 mM EDTA, 150 mM NaCl, 1% Nonidet P-40, 1% sodium deoxycholate, and 0.1% SDS) supplemented with a protease inhibitor cocktail (Complete Mini EDTA free [Roche]). Cell lysis was carried out by incubating the cells for 20 min on ice. Cell debris was removed by centrifugation. The concentration of proteins was determined using a standard Bradford protein assay. Cellular proteins (50 μg) were applied to SDS/PAGE, blotted onto polyvinylidene difluoride membranes, immunostained with antibodies, and developed using an enhanced chemiluminiscence reagent (Pierce).
Immunofluorescence analysis
Cells were grown on coverslips and, at indicated times postnucleofection, cells were fixed, with 4% paraformaldehyde for 15 min and permeabilized with 0.5% Triton X-100 in PBS for 5 min. After washing in PBS, cells were incubated with 0.25% bovine serum albumin and 0.25% porcine skin gelatin in PBS for 30 min. Immunostaining with primary and secondary antibodies was carried out for 1 h and 30 min, respectively, with extensive washing with PBS after each incubation. Coverslips were mounted on droplets of glycerol with 4′, 6′-diamidino-2-phenylindole (DAPI). Images were obtained by using a Leica TCS SP2 confocal microscope.
Antibodies
The antibodies used were mouse monoclonal anti-phospho-Histone H2A.X (Ser 139) (Millipore); mouse monoclonal anti β-actin (Sigma-Aldrich); rabbit polyclonal anti-TLR9 (Abcam); rabbit polyclonal anti-IRF3 (Abcam); rabbit monoclonal anti-phospho IRF3 (Ser 396) (Cell Signaling Technology); rabbit polyclonal anti-NF-κB p65 (Santa Cruz); goat anti-mouse and goat anti-rabbit conjugated with peroxidase (Pierce); and goat anti-rabbit IgG conjugated with Alexa fluor 488 (Molecular Probes).
Results
Analysis of CpG distribution and methylation state of the plasmids
We have previously found that introduction of plasmid DNA by nucleofection into mouse fibroblasts induces upregulation of the endosomal receptor, TLR9 (our unpublished results). Since TLR9 recognizes unmethylated CpGs, we evaluated the quantity of the CpG motifs in the plasmids and their methylation state. For this study, we used the plasmid, phGf, a gateway-adapted destination plasmid and two of its derivatives, the expression vectors, ph2p and ph3p, that were previously prepared by insertion of the sequences encoding the minor structural proteins VP2 or VP3, of MPyV into phGf (Tolstov et al., 2009). The plasmids include an ORF that codes for the EGFP protein. We removed the EGFP sequences to exclude any cellular responses that could be launched by EGFP expression. The modified products were named phGfΔG (destination vector), ph2pΔG, and ph3pΔG (expression vectors). Table 1 summarizes the features of the plasmids. Before CpG analysis, we arbitrarily divided the sequence of phGfΔG into three regions (Fig. 1A) to facilitate the description of the CpG motifs. In expression vectors (ph2pΔG and ph3pΔG), the region 2 (between att sites) was lost during cloning by recombination and was replaced by a gene of interest, in this case by the MPyV VP2 or VP3 genes (Fig. 1A). We evaluated the presence of CpG motifs in the plasmids by using the cgplot program from the European Molecular Biology Laboratory-European Bioinformatics Institute (EMLB-EBI) commonly used to study vertebrate genomes to find promoters of housekeeping genes (Larsen et al., 1992). The program identifies regions of at least 200 nucleotides with a high content of GC (>50%) and observed/expected ratio of CpG greater than 0.6 (Gardiner-Garden and Frommer, 1987). The results of the analysis are summarized in Figure 1A. We found that the positive strand of PhGfΔG has four predicted islands, one long (∼700 bp) located in region 1, two short islands (∼200 bp) within region 2, and one medium size island (∼ 400 bp) in region 3. The expression plasmids coding for the VP2 and VP3 proteins retained the same long and middle size motifs of regions 1 and 3 as phGfΔG. In addition, the ph2pΔG plasmid has two islands in the VP2 sequence, a long one (669 bp) and a short one (241 bp); the ph3pΔG plasmid DNA has one island in the VP3 sequence that joins the long island of region 1, resulting in a very long island (∼1400 bp). For the complement strands of all plasmids used, similar regions were predicted as for the positive ones (Supplementary Table S1; Supplementary Data are available online at www.liebertpub.com/dna).
Table 1.
Plasmid Features and Its Expression Products
| Name | Size (bp) | Type of plasmid according to gateway system | Expression products |
|---|---|---|---|
| phGf | 5608 | Destination plasmid | EGFP |
| ph2p | 4932 | Expression vectors | EGFP and VP2 |
| ph3p | 4568 | EGFP and VP3 | |
| phGfΔG | 4601 | Destination plasmid | None |
| phGfΔGΔR | 2855 | Destination plasmid without region between att sites | None |
| ph2pΔG | 3925 | Expression vectors | VP2 |
| ph3pΔG | 3561 | VP3 | |
| pEGFP-N1 | 4728 | Non gateway | EGFP |
| pEGFP-VP2 | 5688 | Non gateway | EGFP-VP2 |
| pEGFP-VP3 | 5343 | Non gateway | EGFP-VP3 |
| pcDNA3.1 | 5428 | Non gateway | None |
Next, to follow the methylation state of some of the CpG sequences within the plasmids, we used the restriction enzyme, HpaII, which recognizes only unmethylated CpG within the sequence 5′…CCGG…3′ (Fig. 1B). The plasmid phGfΔG contains 23 repetitions of this sequence, while ph2pΔG or ph3pΔG contain 16 and 15, respectively (Supplementary Table S2). Some of the fragments produced by HpaII digestion have very similar length or are too small to be properly resolved by agarose gel electrophoresis. However, we were able to relate at least six bands in digest of each of the three plasmids to the sizes of the expected fragments (Supplementary Table S2). From this experiment, we can conclude that each plasmid contains at least some unmethylated CpG dinucleotides.
Evaluation of the presence of the TLR9 receptor in NIH3T3 fibroblasts
We hypothesized that the IFN response is induced after recognition of unmethylated CpGs by the TLR9 endosomal receptor. Supporting our hypothesis, three studies (Li et al., 2004; Platz et al., 2004; Di et al., 2009) reported responses to CpGs through TLR9 of nonimmune cells. Another two studies reported that both immune and nonimmune cells rapidly upregulate the production of the receptor after exposure to agonists of TLR9 (Bourke et al., 2003; Ewaschuk et al., 2007). Therefore, we followed the expression of the TLR9 in the 3T3 fibroblast cell line. First, we confirmed by indirect immunofluorescence that 3T3 fibroblasts produce TLR9 under standard growing conditions and that the production is not affected by mock nucleofection (Fig. 1C). In addition, we evaluated by western analysis the levels of TLR9 after nucleofection of the cells and found an upregulation of TLR9 after nucleofection with plasmid DNAs but not after mock nucleofection (Fig. 1D). Next, we characterized the type I IFN response rendered by introduction of plasmids into the mouse fibroblasts and evaluated the role of CpG motifs in this response.
Type I IFN response after plasmid DNA nucleofection
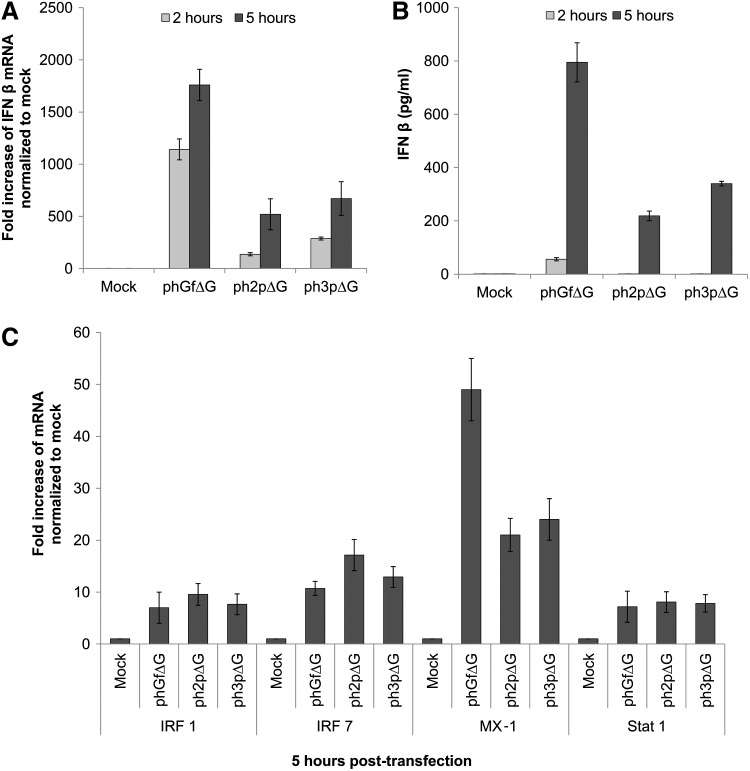
We first measured the levels of IFNβ mRNA by quantitative real-time-PCR (qRT-PCR) and secretion of IFNβ protein by ELISA (Fig. 2A, B, respectively). We detected high levels of IFNβ mRNA in cells transfected with phGfΔG and only moderate amounts of IFNβ mRNA in cells transfected with either ph2pΔG or ph3pΔG 2 h post-transfection. In agreement with this result, at this time post-transfection, we were able to detect IFNβ only in the growing medium of cells transfected with phGfΔG (the method we used for measuring IFN has a sensitivity of 15 pg/mL). However, later, 5 h post-transfection, a higher production of IFNβ mRNA correlated with high levels of IFNβ for all transfected cells. Nevertheless, the production of IFNβ was ∼2.5 times lower in cells transfected with either ph2pΔG or ph3pΔG in comparison with cells transfected with phGfΔG. To exclude any possible differences in the uptake of the plasmids, we determined the efficiency of transfection using the original phGf plasmid encoding EGFP as destination vector, or expression vectors (ph2pΔG or ph3pΔG) encoding the minor structural proteins of MPyV. We found that the efficiency of transfection was similar (∼25%–34%) for all three plasmids (Supplementary Table S3), indicating that the uptake of plasmids was also similar. All together, the above results show that region 2, which is present only in the phGfΔG vector, may contain sequences that positively affect vector recognition by DNA sensors. The levels of IFN produced in cells transfected with the phGfΔG plasmid were very high, comparable with those described, for example, in Rautsi et al. (2007) for potent inducers of IFN - mRNA from mouse muscles cells, or by in vitro transcribed RNA of vesicular stomatitis virus glycoprotein.
FIG. 2.
Interferon (IFN) type I response after nucleofection of mouse NIH3T3 fibroblasts with phGfΔG plasmid or its derivatives. Cells were transfected with phGfΔG, ph2pΔG, or ph3pΔG plasmids at the indicated times post-transfection. Cells were harvested for preparation of mRNAs (A, C) and growth medium supernatants for measurement of secreted IFN (B). (A) The level of IFNβ mRNA was measured by quantitative real-time polymerase chain reaction (qRT-PCR). (B) The production of IFNβ was analyzed by enzyme-linked immunosorbent assay (ELISA). (C) The levels of mRNA of genes regulated during IFN responses, IRF 1, 7, MX-1, and STAT 1, were measured by qRT-PCR. For all experiments (A–C), mock-transfected cells were included as a negative control. Data represent the mean values of three independent experiments.
The remarkably stronger IFN production induced by the phGfΔG vector in comparison with that induced by ph2pΔG or ph3pΔG raised a question of possible differences in subsequent steps of the IFN signaling that might result in a different expression of IFN-inducible genes. To answer this question, we quantified the mRNA levels of IFN-inducible genes, IRF1, IRF7, and genes activated during the establishment of the antiviral state, including Mx-1 and STAT1 (Fig. 2C). We found a remarkable but comparable induction of transcription of IRF1, IRF7, and STAT1 in cells transfected with either plasmid 5 h post-transfection. This indicates that in spite of the different levels of IFN produced, all three plasmids induce IFN signaling resulting in establishment of the antiviral state. However, supporting the very high IFNβ induction by the phGfΔG plasmid, markedly stronger Mx-1 expression was detected in cells transfected with this plasmid.
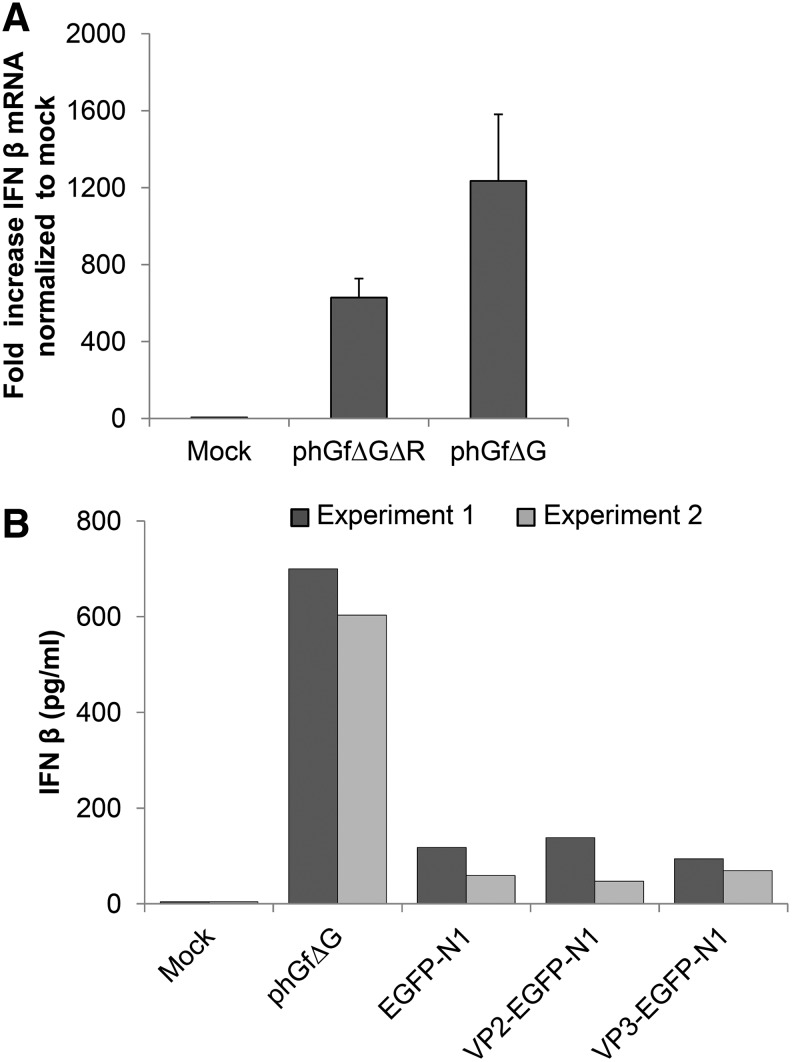
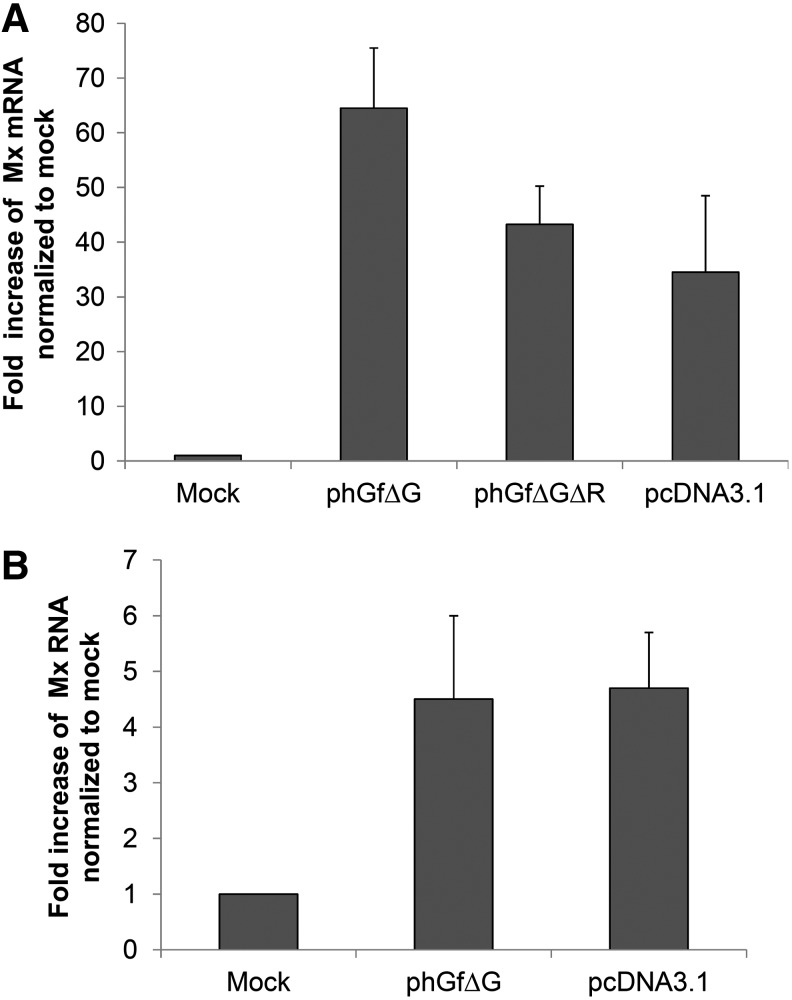
There is an important fact that should be considered. The destination vector, phGfΔG, induced a strong and very fast IFN response (within 2 h), while IFN response induced by expression vectors, ph2pΔG or ph3pΔG, was weaker and delayed (detected 5 h post-transfection). This makes us speculate about the nature of the IFN induction launched by expression vectors. There are three possibilities to explain this: (1) either DNA regions remaining in expression vectors from phGfΔG are less efficiently recognized by cells (e.g., due to the influence of different adjacent sequences) and therefore, the IFN response is delayed; (2) the induction is caused by the inserted nucleotide sequence of the minor proteins; and (3) the VP2 and VP3 gene products might be responsible for the delayed IFNβ production. To test the role of vector sequences in the IFN responses, we modified the destination vector phGfΔG by deleting the region between the att sites (see Fig. 1A), thus, generating the double modified destination vector phGfΔGΔR, which corresponds to phGfΔG vector sequences present in expression vectors ph2pΔG and ph3pΔG. Then, we nucleofected the cells by the double modified plasmid and quantified IFNβ mRNA levels. We found that levels of IFN induction are similar to those obtained after delivering the expression vectors by nucleofection (Fig. 3A). To test the contribution of gene or amino acid sequences of VP2 or VP3 to the IFN responses, we used the pEGFP-N1 vector and its derivatives, VP2-EGFP-N1 and VP3-EGFP-N1 carrying EGFP fusion genes for VP2 or VP3 (Huerfano et al., 2010). Features of these plasmids are shown in Table 1. A comparison of the IFN levels produced by transfection of pEGFP-N1 vector versus its derivatives, VP2-EGFP-N1 and VP3-EGFP-N1, is straightforward since the traditional method of cloning via restriction enzyme cleavage preserved all vector sequences after cloning. Efficiencies of transfection by using these plasmids were comparable (33%–39%; Supplementary Table S3), thus allowing us to compare responses induced by them. We measured the levels of IFNβ 5 h post-transfection (Fig. 3B) and found that cells transfected with pEGFP-N1, or with its derivatives, induced comparable amounts of IFNβ, indicating that neither VP2/VP3 gene sequences nor their products play a significant role in the response. From these experiments, we can conclude that similarly, in the case of ph2pΔG or ph3pΔG plasmids, the IFN response is induced by vector DNA sequences. An additional observation that can be extracted from the above data is that nucleofection of NIH3T3 cells induces an IFN response not only with phGfΔG-derived plasmids but also with the pEGFP-N1 plasmid. As pEGFP-N1 encodes EGFP, we cannot exclude the contribution of the EGFP protein to the response. Therefore, to test the induction of IFN responses by another unrelated plasmid, we investigated the antiviral state establishment (by quantification of Mx mRNA) after nucleofection by commonly used pcDNA3.1 (features of the plasmid are shown in Table 1). The results presented in Figure 4A show that nucleofection of the pcDNA3.1 plasmid also induced a remarkable upregulation of Mx gene expression. Thus, nucleofection of different plasmid DNAs into NIH3T3 apparently can induce IFN responses.
FIG. 3.
IFN induction after nucleofection of mouse fibroblasts with plasmid DNAs. (A) The double modified destination vector, phGfΔGΔR, was introduced into NIH3T3 cells by nucleofection and the levels of IFNβ mRNA at 5 h post-transfection were quantified by qRT-PCR. Data represent the values of three experiments. (B) NIH3T3 cells were nucleofected with VP2-EGFP-N1, VP3-EGFP-N1, or EGFP-N1, and at 5 h post-transfection, the supernatants of growing cells were used for detection of IFNβ levels by ELISA. Data represent the values of two independent experiments. For all experiments (A, B) mock-transfected cells were used as a negative and cells transfected with phGfΔG plasmid as a positive control.
FIG. 4.
Establishment of the antiviral state after nucleofection of NIH3T3 or mouse embryonic fibroblast (MEF) cells with plasmid DNAs. Mouse 3T3 fibroblast (A) or MEF cells (B) were nucleofected with the indicated plasmid and, 5 h post-transfection, the levels of Mx mRNA were quantified by qRT-PCR. Mock-transfected cells were used as a negative control.
We were interested whether dsDNA nucleofection induces IFN responses also in other cell lines. Therefore, we measured the levels of of Mx mRNA in primary MEF cells nucleofected with either pcDNA3.1 or phGfΔG plasmids. Quantitative PCR performed 5 h postnucleofection revealed that both plasmids induced upregulation of Mx gene expression (Fig. 4B). However, because of very low transfectability of these cells (only 5% of cells expressed EGFP after transfection with the phGf plasmid), we were not able to compare the IFN response of NIH3T3 and MEF cell lines. Nevertheless, we can conclude that, also MEF cells respond to nucleofection with dsDNA by induction of IFN responses.
DNA signaling and the cell fate
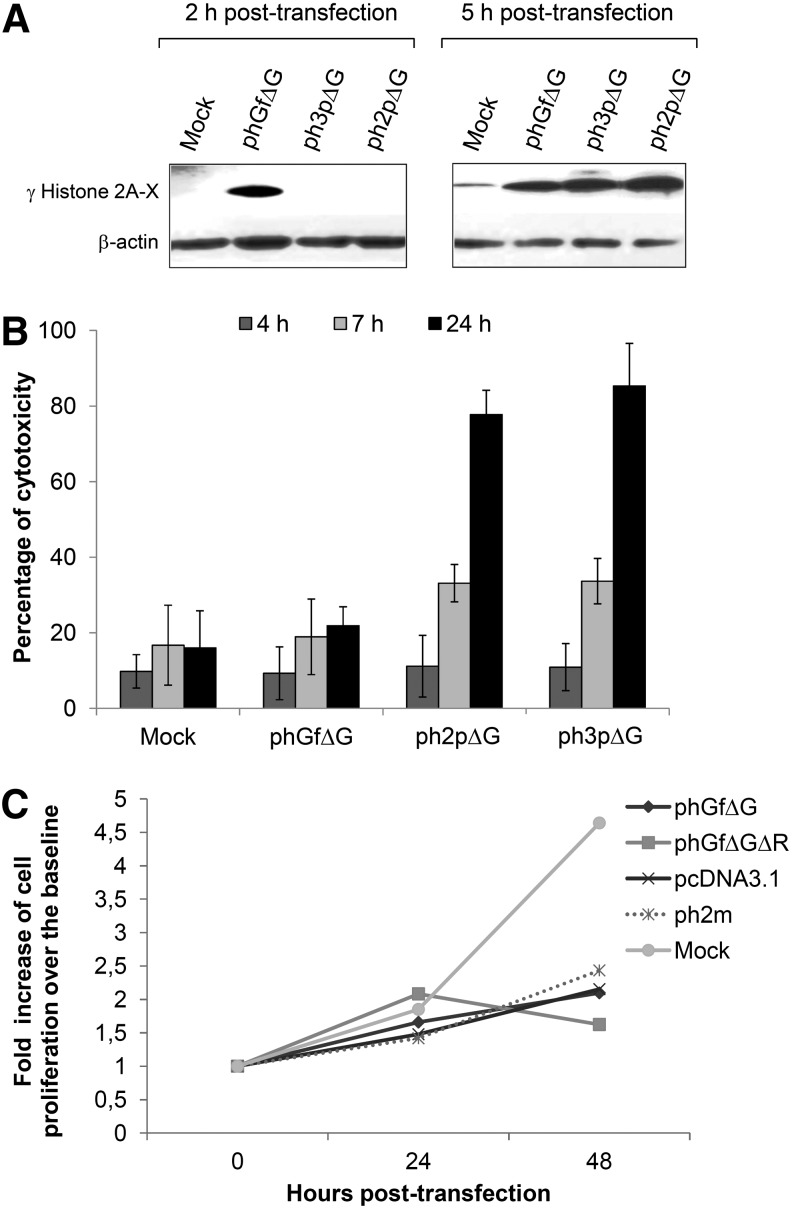
To follow possible DNA damage signaling induced by nucleofection with the gateway plasmids, we evaluated the presence of γ-histone 2A.X. This histone is a modified isoform of histone 2A.X, produced by phosphorylation of the serine in position 139 by ataxia telangiectasia mutated (ATM) or DNA-dependent protein kinase (DNA-P) or by other kinases in response to dsDNA breaks (Li et al., 2005). As soon as 2 h post-transfection, we observed an upregulation of γ-histone 2A.X in cells transfected with phGfΔG (Fig. 5A). The early sensing of DNA damage coincides with the early production of IFNβ (Fig. 2B). Interestingly, cells transfected with the expression vectors ph2pΔG or ph3pΔG exhibited DNA damage later (after 5 h post-transfection) at the time that coincided with the detection of IFNβ.
FIG. 5.
DNA damage signaling and fate of NIH3T3 cells after nucleofection with plasmid DNAs. Cells were nucleofected by Amaxa with phGfΔG, ph2pΔG, or ph3pΔG plasmids. At the indicated times post-transfection, cells or supernatants were collected. (A) Proteins of cell lysates were immunobloted for detection of γ-histone 2A.X. Detection of β-actin was performed as a control for sample loading. (B) The medium of the growing cells was collected to evaluate cytotoxicity by measuring LDH. Values are related to that of LDH release obtained by treatment with 9% Triton X-100 (=100%). Data represent the mean values obtained by measuring duplicates of three independent experiments. (C) Cell proliferation was followed by measuring bioreduction of tetrazolium salt, using an ELISA reader (450 nm). OD values were aligned to the baseline (0 time, OD value) and the resulting fold increase values were plotted. Three independent experiments were performed, and comparable results were obtained. For better graph clarity, the results of one representative experiment are presented. For all experiments (A–C), mock-transfected cells were used as a negative control.
As we detected γ-histone 2A.X upregulation sensing DNA damage in the cells nucleofected with the phGfΔG plasmid, we were interested in the fate of the transfected cells. As it has been described that prolonged IFNβ treatment induces cell death or inhibition of cell proliferation, we tested both cytotoxicity and the ability of the cells to proliferate. First, we followed cell death by measuring the release of lactate dehydrogenase as an indicator of membrane damage at different times post-transfection. Cells transfected with the phGfΔG plasmid did not undergo significant cell death when compared with the mock transfected control cells (Fig. 5B), indicating that, in spite of high IFN levels, there is no induction of cell death (within 24 h post-transfection). The cells transfected with the expression vectors coding for VP2 and VP3 died after 24 h as expected since the proteins were able to bind and perforate membranes and to induce cell death (Huerfano et al., 2010). Then, we followed the rate of cell division, after 24 and 48 h post-transfection. We could not compare the influence of the phGfΔG plasmid presence on cell proliferation with that of the MPyV VP2- and VP3-expressing vectors, ph2pΔG or ph3pΔG, for high toxicity of VP2 and VP3 proteins. Therefore, apart from phGfΔG, we included two additional plasmids in this experiment for comparison. One of them, the ph2m plasmid, is a derivative of phGf carrying the gene of the VP2 minor structural protein of the MCPyV. Unlike the MPyV structural proteins, VP2 of the MCPyV possess only weak (if any) cytotoxicity when measured up to 24 h postnucleofection (our unpublished results). The second plasmid, phGfΔGΔR, contains only phGfΔG sequences present in the expression vectors. We found that nucleofection of either of the plasmids, caused arrest of cell cycle progression (Fig. 5C). This indicates that nucleofection by the plasmids derived from the phGf gateway plasmid (but not mock nucleofection) induces inhibition of cell proliferation. In addition, we also evaluated the rate of proliferation of NIH3T3 cells after nucleofection with the pcDNA3.1 plasmid (Fig. 5C) and also found that transfection by this plasmid caused arrest of cell cycle progression.
Turbofect cationic transfection instead of nucleofection dramatically decreased IFNβ induction
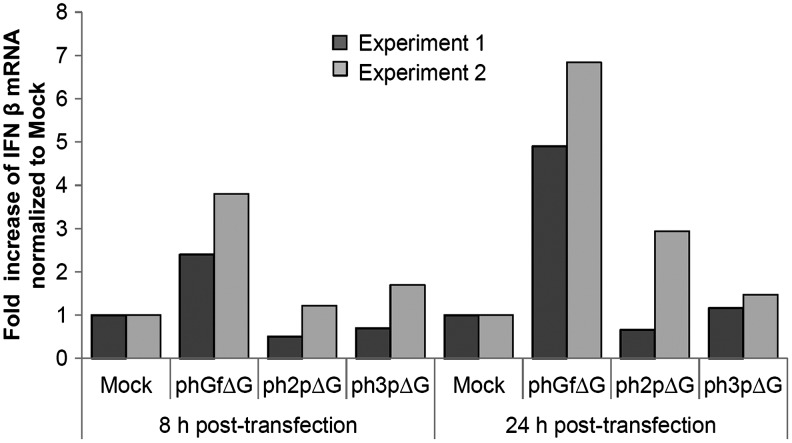
Transfection of naked DNA or wrapped by a cationic solution is known to induce differential IFN responses (Rautsi et al., 2007). Therefore, we evaluated the induction of IFN when transfecting cells by turbofect (cationic solution) with the phGfΔG destination vector or ph3pΔG and ph2pΔG expression vectors. First, we evaluated the efficiency of transfection as an indicator of DNA uptake using the destination vector, phGf carrying EGFP-coding sequences and expression vectors, ph2pΔG or ph3pΔG for expression of the MPyV VP2 or VP3 genes. We found that for all the plasmids, the amounts of cells expressing genes carried by vectors were similar (∼15%–18%, see Supplementary Table S3). Efficiency of transfection was lower (approximately one half) when compared to values reached by nucleofection. In agreement with our results, differences in efficiency of transfection by using different methods have been documented (Maurisse et al., 2010; Contreras et al., 2012). Next, we measured the levels of secreted IFNβ by ELISA. Surprisingly, we were not able to detect any IFN, indicating that IFN production by cells (if any) was under the detection limit of our system (15 pg/mL). Therefore, we proceeded by measuring IFNβ mRNA levels by RT-PCR at 8 and 24 h post-transfection. Figure 6 shows increased IFNβ transcription in cells transfected with all three plasmids when compared with mock-transfected cells. As in the nucleofection experiments, IFN transcription was the highest for cells transfected with the phGfΔG plasmid. However, the levels of IFN induction by all plasmids were dramatically decreased. While induction of IFN by phGfΔG plasmid was increased 1000-fold (in comparison with mock nucleofection), at the time 5 h-postnucleofection, only approximately sevenfold increase was detected 24 h postturbofection with the same plasmid (in comparison with mock turbofection). From all these results, it can be concluded that the method of DNA delivery into cells is crucial for the strength of the IFN response.
FIG. 6.
IFNβ induction in NIH3T3 cells transfected by Turbofect. Cells were transfected with phGfΔG, ph2pΔG, or ph3pΔG plasmids and at the indicated times post-transfection, mRNA was extracted. The levels of IFNβ mRNA were quantified by RT-PCR. Data of two independent experiments are presented. Mock-transfected cells were used as a negative control.
The mechanism of recognition of nucleofected DNA is TLR9 independent and involves NF-κB activation
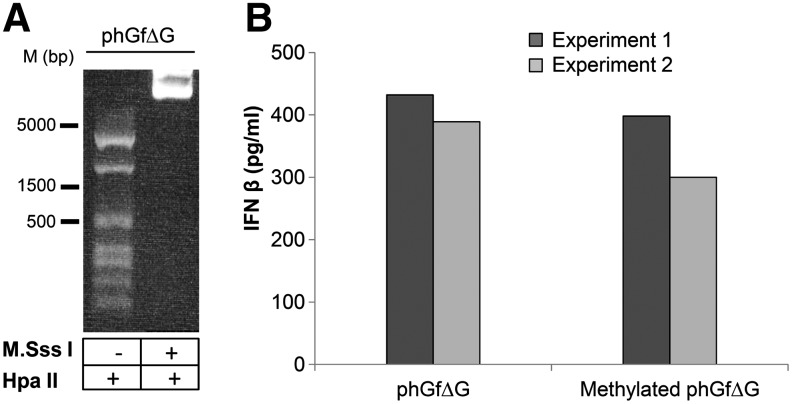
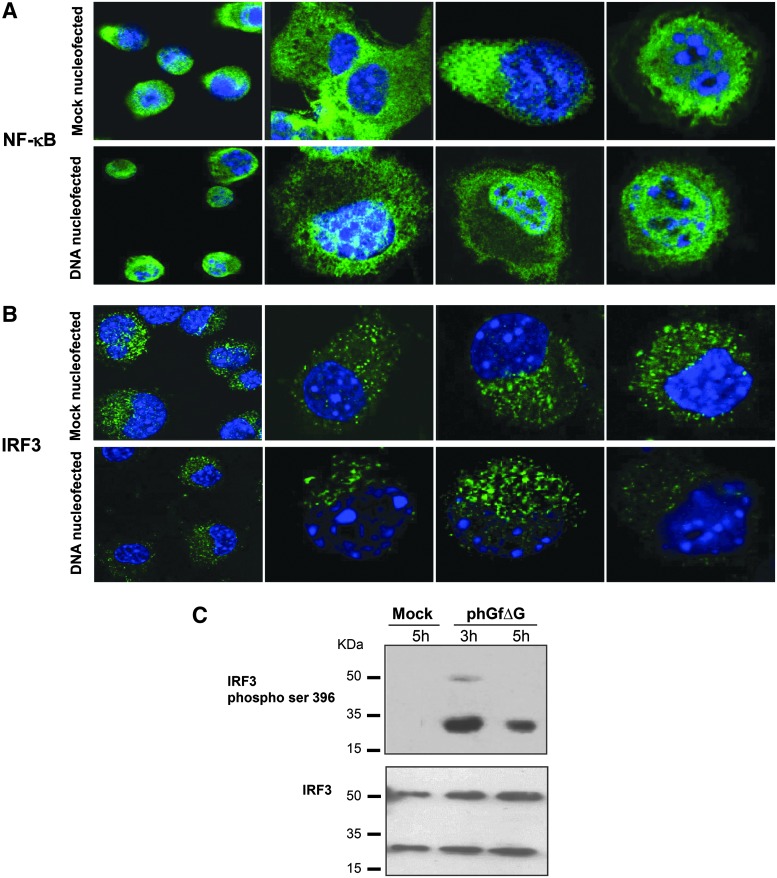
For evaluation of the contribution of the CpG motifs to the IFN responses, we methylated cytosine residues within the recognition sequence 5′…CG…3′ of the phGfΔG destination plasmid by the methyltransferase M.SssI. Methylation of the plasmid was confirmed by prevention of digestion by HpaII (Fig. 7A). Then, we transfected the cells with the unmethylated or methylated plasmids and, 5 h after nucleofection, measured the production of IFNβ by ELISA (Fig. 7B). We did not find any significant differences in the IFN levels in the cells transfected with methylated or unmethylated DNA. This indicates that there are apparently other features of the plasmid DNA rather than CpG motifs sensed by cells, and that, cytoplasmic DNA sensors (and not the endosomal TLR9 receptor), are responsible for sensing the DNA. To follow the possible pathways involved in IFN induction through DNA sensors, we investigated the role of IRF3, and NF-κB transcription factors in the signaling. IRF3 is ubiquitously expressed and activated by serine phosphorylation at its C-terminus, in the serine/threonine cluster site. Phosphorylation promotes dimerization of the protein. Dimers, translocate to the nucleus to bind specific DNA targets (Lin et al., 1998) Transcription factor, NF-κB, consists of homo- and heterodimers of NFκB1/p50 and RelA/p65 subunits. NF-κB dimers become activated by induced proteasomal degradation of the IkB inhibitor. Active NF-κB transcription factor subunits translocate to the nucleus and induce target gene expression (Baeuerle and Baltimore, 1988). We selected these two transcription factors because they were found to be activated by transfection of double-stranded oligonucleotides into mouse fibroblasts (Ferguson et al., 2012). In addition, IRF3 has been involved in signaling induced by at least four of nine described DNA sensors, while NF-κB has been involved alone or in conjunction with IRF3 in at least three of nine described sensors (Keating et al., 2011). We followed by confocal microscopy the translocation of NF-κB and IRF3 into the cell nucleus after nucleofection of cells with the phGfΔG plasmid. One hour post-transfection NF-κB exhibited marked nuclear localization in a significant subpopulation of cells (Fig. 8A) (∼12% of the whole cell population, where efficiency of nucleofection by phGf transfection was 25.5%), while only 4% of such cells were detected in mock-transfected cells. IRF3 was located almost exclusively in the cytoplasm at any of the times examined (40 min, 1, 3, and 5 h post-transfection) (Fig. 8B, shows representative pictures). We noticed that ∼2% of both nucleofected and mock-nucleofected cells exhibited a low level of IRF3 in the nucleus (see Fig. 8B picture 3, bottom panel). As IRF3 is known to have an important role in signaling through many DNA sensors, we decided to follow, in addition to nuclear translocation, serine phosphorylation of the protein at 1, 3, and 5 h post-transfection. Although we were not able to detect any phosphorylated IRF3 after 1 h (not shown) or 5 h post-transfection, very low signal of the phosphorylated protein was detected 3 h post-transfection (Fig. 8C, top). Intriguingly, from 3 h post-transfection, abundant levels of a product of low molecular weight recognized by the antibody against phosphorylated IRF3 were observed in plasmid-transfected but not in mock-transfected cells (Fig. 8C, top). The fact that the product was detected also by a polyclonal antibody recognizing IRF3 regardless of its phosporylation (in lysates of cells transfected with plasmid and in mock-transfected cells) (Fig. 8C, bottom) suggests that it might be a degradation product of IRF3. Together, the data suggest a role for NF-κB in IFN induction, while the role of IRF3 is not so clear. If there is a role of IRF3 in IFN induction, the activation seems to be dynamic and tightly regulated by cells. Thus, both nuclear translocation of IRF3 and its phosphorylation were difficult to follow.
FIG. 7.
Production of IFNβ by NIH3T3 cells after nucleofection with methylated or unmethylated phGfΔG plasmid DNA. (A) The phGfΔG plasmid was treated with the methyltransferase M. SssI to methylate the CpG motifs. Prevention of the digestion by HpaII indicated the methylation state of the plasmid. Untreated plasmid was used as a positive control of HpaII digestion. (B) Cells were transfected either with unmethylated or methylated plasmid, and the levels of IFNβ production were assayed by ELISA. The data represent the values of two independent experiments.
FIG. 8.
Activation analysis of transcription factors NfκB and IFN regulatory factor (IRF3). (A, B) Activation of the factors was followed 1 h postnucleofection of NIH3T3 cells with phGfΔG plasmid or after mock nucleofection. Selected confocal microscopy sections are presented. Cells were fixed and stained by (A) an antibody against nuclear factor-kappa (NF-κB) p-65 (green) and by DAPI (blue) or by (B) an IRF3-specific antibody (green) and DAPI (blue). (C) IRF3 phosphorylation in NIH3T3 cells nucleofected with phGfΔG or mock nucleofected was analyzed by Western blotting. Cells were lysed at the indicated times, separated by SDS electrophoresis, and blotted. Immunostaining was performed by phospho-IRF3 (Ser396) antibody, specific for the epitope containing phosphorylated serine at position 396. Polyclonal antibody against the IRF3 protein was used as a control.
Discussion
Transfection of cells by plasmid vectors is a widely used method to obtain efficient expression of desired genes for studying cellular responses to their expression or the behavior of their products. However, the insertion of DNA into the cells induces immunoresponses that could interfere with the result interpretation in an experimental setting. In this study, we documented a strong IFN response, DDR activation, and inhibition of cell proliferation induced by nucleofection of mouse fibroblasts with plasmid DNA. Interestingly, we did not find high levels of IFN when transfecting the same plasmids by the cationic polymer (Turbofect) into the cells. These results point to the role of a DNA entry mechanism as key for DNA sensor recognition. In detail, although the mechanism of DNA cell delivery during transfection is not completely understood it is known that DNA delivered by liposomes or cations enters the cells via endocytosis (Legendre and Szoka, 1992; Yasuda et al., 2005). Variations in the type of early endosomes that transport the DNA (fast maturing or static) or differences in the mechanism of DNA escape from endosomes may be responsible for the differences in DNA sensing and in the efficiency of nuclear delivery (review in Nguyen and Szoka, 2012).
Even less is known about electroporation. Until very recently, it was accepted that DNA may enter cells through transient membrane pores. In 2011, Rosazza et al. (2011) demonstrated that during DNA electroporation, actin polymerizes under the plasma membrane in places where DNA is aggregated. The authors postulated that there is formation of endocytic-like vesicles (Rosazza et al., 2011). Interestingly, nucleofection is a type of electroporation that combines electrical parameters with a buffer solution, which, by an unknown mechanism, delivers DNA straight into the cell nucleus (Gresch et al., 2004). We speculated that the “straight delivery” of DNA into the nucleus proposed as a mechanism for nucleofection, implies only a faster cytoplasmic DNA traffic, since, in our model, DNA cytoplasmic sensors were activated by nucleofected DNA. The way of trafficking of this DNA and the mechanism of its sensing by cell cytoplasmic sensors has to be assessed. Apparently, unlike cationic DNA delivery, nucleofection arouses DNA recognition by cytoplasmic DNA sensors. We found early postnucleofection translocation of NF-κB suggesting the role of this transcription factor in signaling, which leads to IFN production. The role of IRF3 in light of our results remains controversial. At first, lack of nuclear translocation of the protein strongly suggests that the factor is not signaling. The low levels of IRF3 in the nuclei of some cells (DNA or mock nucleofected) are consistent with the fact that the protein normally shuttles between the nucleus and the cytoplasm, and due to the presence of nuclear export signals in the protein, the cytoplasmic subpopulation is dominant (Lin et al., 1998). The presence of a small subpopulation of IRF3 phosphorylated at serine 396 in cells 3 h post-transfection with phGfΔG (Fig. 8C) may indicate some activation of the protein that, however, does not result in significant translocation of IRF3 but rather in its degradation as suggested by the appearance of low molecular weight product recognized by antibody specific for phosphorylated IRF3. The possibility that phosphorylation of IRF3 may represent a signal for subsequent degradation by the proteasome pathway was previously suggested (Lin et al., 1998). Further investigation will be required to identify the specific sensor responsible for dsDNA sensing in mouse NIH3T3 fibroblasts and the role of IRF3 in consequent signaling leading to IFN induction.
Although the plasmids used here were rich in unmethylated CpG motifs, and cells were overexpressing TLR9, we demonstrated that the motifs did not play an important role in foreign DNA recognition. The reason can be (1) in the method of transfection and/or (2) in the absence of recognition of sequences flanking the CpG motifs. Indeed, Yoshinaga et al. (2006) found that macrophages responded with higher production of inflammatory cytokines when stimulated by DNA containing CpGs complexed with cationic lipids rather than by naked DNA. In our hands, CpG motifs apparently did not contribute to the IFN signaling either when Amaxa nucleofection was used or during cation polymer delivery. Nevertheless, it is also possible that the sequences flanking CpG motifs in our vectors prevent receptor recognition. It has been described that differences in the levels of IFN response in humans and mice were caused by varying recognition of the CpG's flanking sequences by these two species (Weiner, 2000).
In addition, although we found some differences in IFN responses induced by Amaxa nucleofection with different plasmids, phGfΔG, phGfΔGΔR, or pcDNA3.1, we observed that the three plasmids have similar effects on cell proliferation. Consistent with our results, other side effects of nucleofection have been recently reported. Anderson et al. (2013) found that nucleofection induces transient Eif2α (eukaryotic initiation factor 2-α subunit) phosphorylation, which results in a general decrease in translation initiation events. Another group (Mello de Queiroz et al., 2012) reported a cell type-dependent increase in the metabolic rate of cells after nucleofection. Therefore, special consideration and additional controls must be taken into account when using transfection for studies of cell responses.
Taken together, all our data and previous reports suggest that the method of transfection, the nature of the DNA, and also transfected cell types determine the degree of the immunoresponse induced. Therefore, neither a specific method nor methylation of DNA can guarantee weak IFN responses and should be assessed for each specific experimental setting.
Concerning the particular set of plasmids used in this study, we should point out that when using the gateway system for cloning by replacement of a part of the vector sequences with genes of interest, the preferable control would be expression vectors with mutated ATG of inserted genes rather than the original destination vector to avoid misleading results.
We observed activation of the DDR pathway, which is a network that detects and repairs DNA breaks. How is the activation of this pathway in the context of transfection induced? There are two possible explanations: (1) transfected DNA is sensed as broken DNA and activates the response as suggested in Wake et al. (1984) or (2) DDR activation may be connected with IFN production. It has been described that the production of reactive oxygen species (ROS) induces DNA double-stranded breaks during the IFN response (Moiseeva et al., 2006). In fact, we detected high levels of ROS at 5 h postnucleofection (data not shown). In addition, it was recently described that macrophages exposed to γ-radiation concomitant with DDR activation undergo a type I IFN response indicating crosstalk between these two pathways (Mboko et al., 2012). However, how IFN is connected with DDR is not understood. The responses to transfection of DNA could be a good model for studying this process.
Supplementary Material
Acknowledgments
This work was generously supported by the Grant Agency of the Czech Republic GACR, no. P304/10/1511 (B.R.), by the project of the Ministry of Education, Youth and Sports of Czech Republic, MSM0021620858 (J.F.) and SVV-2013-267205 (S.H.).
Disclosure Statement
No competing financial interest exists.
References
- Anderson B.R. Karikó K. Weissman D. Nucleofection induces transient eIF2α phosphorylation by GCN2 and PERK. Gene Ther. 2013;20:136–142. doi: 10.1038/gt.2012.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Assaf A. Esteves H. Curnow S.J. Browning M.J. A threshold level of TLR9 mRNA predicts cellular responsiveness to CpG-ODN in haematological and non-haematological tumour cell lines. Cell Immunol. 2009;259:90–99. doi: 10.1016/j.cellimm.2009.06.003. [DOI] [PubMed] [Google Scholar]
- Baeuerle P. Baltimore D. IkB: a specific inhibitor of the NF-kB transcription factor. Science. 1988;242:540–546. doi: 10.1126/science.3140380. [DOI] [PubMed] [Google Scholar]
- Bauer S. Kirschning C.J. Häcker H. Redecke V. Hausmann S. Akira S. Wagner H. Lipford G.B. Human TLR9 confers responsiveness to bacterial DNA via species-specific CpG motif recognition. Proc Natl Acad Sci U S A. 2001;98:9237–9242. doi: 10.1073/pnas.161293498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bourke E. Bosisio D. Golay J. Polentarutti N. Mantovani A. The toll-like receptor repertoire of human B lymphocytes: inducible and selective expression of TLR9 and TLR10 in normal and transformed cells. Blood. 2003;102:956–963. doi: 10.1182/blood-2002-11-3355. [DOI] [PubMed] [Google Scholar]
- Buck C.B. Day P.M. Thompson C.D. Lubkowski J. Lu W. Lowy D.R. Schiller J.T. Human alpha-defensins block papillomavirus infection. Proc Natl Acad Sci U S A. 2006;103:1516–1521. doi: 10.1073/pnas.0508033103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Contreras J. Hsueh P.Y. Pei H. Hamm-Alvarez S.F. Use of nucleofection to efficiently transfect primary rabbit lacrimal gland acinar cells. Cytotechnology. 2012;64:149–156. doi: 10.1007/s10616-011-9404-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dalpke A. Frank J. Peter M. Heeg K. Activation of Toll-like receptor 9 by DNA from different bacterial species. Infect Immun. 2006;74:940–946. doi: 10.1128/IAI.74.2.940-946.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Di J.M. Pang J. Pu X.Y. Zhang Y. Liu X.P. Fang Y.Q. Ruan X.X. Gao X. Toll-like receptor 9 agonists promote IL-8 and TGF-beta1 production via activation of nuclear factor kappaB in PC-3 cells. Cancer Genet Cytogenet. 2009;192:60–67. doi: 10.1016/j.cancergencyto.2009.03.006. [DOI] [PubMed] [Google Scholar]
- Ewaschuk J.B. Backer J.L. Churchill T.A. Obermeier F. Krause D.O. Madsen K.L. Surface expression of Toll-like receptor 9 is upregulated on intestinal epithelial cells in response to pathogenic bacterial DNA. Infect Immun. 2007;75:2572–2579. doi: 10.1128/IAI.01662-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ferguson B.J. Mansur D.S. Peters N.E. Ren H. Smith G.L. DNA-PK is a DNA sensor for IRF-3-dependent innate immunity. Elife. 2012;1:e00047. doi: 10.7554/eLife.00047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- García M.A. Gil J. Ventoso I. Guerra S. Domingo E. Rivas C. Esteban M. Impact of protein kinase PKR in cell biology: from antiviral to antiproliferative action. Microbiol Mol Biol Rev. 2006;70:1032–1060. doi: 10.1128/MMBR.00027-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gardiner-Garden M. Frommer M. CpG islands in vertebrate genomes. J Mol Biol. 1987;196:261–282. doi: 10.1016/0022-2836(87)90689-9. [DOI] [PubMed] [Google Scholar]
- Gresch O. Engel F.B. Nesic D. Tran T.T. England H.M. Hickman E.S. Körner I. Gan L. Chen S. Castro-Obregon S. Hammermann R. Wolf J. Müller-Hartmann H. Nix M. Siebenkotten G. Kraus G. Lun K. New non-viral method for gene transfer into primary cells. Methods. 2004;33:151–163. doi: 10.1016/j.ymeth.2003.11.009. [DOI] [PubMed] [Google Scholar]
- Hemmi H. Takeuchi O. Kawai T. Kaisho T. Sato S. Sanjo H. Matsumoto M. Hoshino K. Wagner H. Takeda K. Akira S. A Toll-like receptor recognizes bacterial DNA. Nature. 2000;408:740–745. doi: 10.1038/35047123. [DOI] [PubMed] [Google Scholar]
- Hornung V. Ablasser A. Charrel-Dennis M. Bauernfeind F. Horvath G. Caffrey D.R. Latz E. Fitzgerald K. AAIM2 recognizes cytosolic dsDNA and forms a caspase-1-activating inflammasome with ASC. Nature. 2009;458:514–518. doi: 10.1038/nature07725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huerfano S. Zíla V. Boura E. Spanielová H. Stokrová J. Forstová J. Minor capsid proteins of mouse polyomavirus are inducers of apoptosis when produced individually but are only moderate contributors to cell death during the late phase of viral infection. FEBS J. 2010;277:1270–1283. doi: 10.1111/j.1742-4658.2010.07558.x. [DOI] [PubMed] [Google Scholar]
- Ishikawa H. Ma Z. Barber G.N. STING regulates intracellular DNA-mediated, type I interferon-dependent innate immunity. Nature. 2009;461:788–792. doi: 10.1038/nature08476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keating S.E. Baran M. Bowie A.G. Cytosolic DNA sensors regulating type I interferon induction. Trends Immunol. 2011;32:574–581. doi: 10.1016/j.it.2011.08.004. [DOI] [PubMed] [Google Scholar]
- Kim T. Pazhoor S. Bao M. Zhang Z. Hanabuchi S. Facchinetti V. Bover L. Plumas J. Chaperot L. Qin J. Liu Y.J. Aspartate-glutamate-alanine-histidine box motif (DEAH)/RNA helicase A helicases sense microbial DNA in human plasmacytoid dendritic cells. Proc Natl Acad Sci U S A. 2010;107:15181–15186. doi: 10.1073/pnas.1006539107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krieg A.M. Yi A.K. Matson S. Waldschmidt T.J. Bishop G.A. Teasdale R. Koretzky G.A. Klinman D.M. CpG motifs in bacterial DNA trigger direct B-cell activation. Nature. 1995;374:546–549. doi: 10.1038/374546a0. [DOI] [PubMed] [Google Scholar]
- Larsen F. Gundersen G. Lopez R. Prydz H. CpG islands as markers in the human genome. Genomics. 1992;13:1095–1107. doi: 10.1016/0888-7543(92)90024-m. [DOI] [PubMed] [Google Scholar]
- Legendre J.Y. Szoka F.C., Jr. Delivery of plasmid DNA into mammalian cell lines using pH-sensitive liposomes: comparison with cationic liposomes. Pharm Res. 1992;10:1235–1242. doi: 10.1023/a:1015836829670. [DOI] [PubMed] [Google Scholar]
- Li A. Eirín-López J.M. Ausió J. H2AX: tailoring histone H2A for chromatin-dependent genomic integrity. Biochem Cell Biol. 2005;83:505–515. doi: 10.1139/o05-114. [DOI] [PubMed] [Google Scholar]
- Li J. Ma Z. Tang Z.L. Stevens T. Pitt B. Li S. CpG DNA-mediated immune response in pulmonary endothelial cells. Am J Physiol Lung Cell Mol Physiol. 2004;287:L552–L558. doi: 10.1152/ajplung.00436.2003. [DOI] [PubMed] [Google Scholar]
- Lin R. Heylbroeck C. Pitha P.M. Hiscott J. Virus-dependent phosphorylation of the IRF-3 transcription factor regulates nuclear translocation, transactivation potential, and proteasome-mediated degradation. Mol Cell Biol. 1998;5:2986–2996. doi: 10.1128/mcb.18.5.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maurisse R. De Semir D. Emmamekhoo H. Bedayat B. Abdolmohammadi A. Parsi H. Gruenert D. Comparative transfection of DNA into primary and transformed mammalian cells from different lineages. BMC Biotechnol. 2010;10:9. doi: 10.1186/1472-6750-10-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mboko W.P. Mounce B.C. Wood B.M. Kulinski J.M. Corbett J.A. Tarakanova V.L. Coordinate regulation of DNA damage and type I interferon responses imposes an antiviral state that attenuates mouse gammaherpesvirus type 68 replication in primary macrophages. J Virol. 2012;86:6899–6912. doi: 10.1128/JVI.07119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mello de Queiroz F. Sánchez A. Agarwal J.R. Stühmer W. Pardo L.A. Nucleofectioninduces non-specific changes in the metabolic activity of transfected cells. Mol Biol Rep. 2012;39:2187–2194. doi: 10.1007/s11033-011-0967-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moiseeva O. Mallette F.A. Mukhopadhyay U.K. Moores A. Ferbeyre G. DNA damage signaling and p53-dependent Senescence after prologed interferon-β stimulation. Mol Bio Cell. 2006;17:1583–1592. doi: 10.1091/mbc.E05-09-0858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen J. Szoka F.C. Nucleic acid delivery: the missing pieces of the puzzle? Acc Chem Res. 2012;45:1153–1162. doi: 10.1021/ar3000162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pestka S. Krause C.D. Walter M.R. Interferons, interferon-like cytokines, and their receptors. Immunol Rev. 2004;202:8–32. doi: 10.1111/j.0105-2896.2004.00204.x. [DOI] [PubMed] [Google Scholar]
- Pindel A. Sadler A. The role of protein kinase R in the interferon response. J Interferon Cytokine Res. 2010;31:59–70. doi: 10.1089/jir.2010.0099. [DOI] [PubMed] [Google Scholar]
- Pitha P.M. Kunzi M.S. Type I interferon: the ever unfolding story. Curr Top Microbiol Immunol. 2007;316:41–70. doi: 10.1007/978-3-540-71329-6_4. [DOI] [PubMed] [Google Scholar]
- Platanias L.C. Mechanisms of type I and type II interferon mediated signalling. Nat Rev Immunol. 2005;5:375–386. doi: 10.1038/nri1604. [DOI] [PubMed] [Google Scholar]
- Platz J. Beisswenger C. Dalpke A. Koczulla R. Pinkenburg O. Vogelmeier C. Bals R. Microbial DNA induces a host defense reaction of human respiratory epithelial cells. J Immunol. 2004;173:1219. doi: 10.4049/jimmunol.173.2.1219. [DOI] [PubMed] [Google Scholar]
- Rautsi O. Lehmusvaara S. Salonen T. Häkkinen K. Sillanpää M. Hakkarainen T. Heikkinen S. Vähäkangas E. Ylä-Herttuala S. Hinkkanen A. Julkunen I. Wahlfors J. Pellinen R. Type I interferon response against viral and non-viral gene transfer in human tumor and primary cell lines. J Gene Med. 2007;9:122–135. doi: 10.1002/jgm.997. [DOI] [PubMed] [Google Scholar]
- Rosazza C. Escoffre J.M. Zumbusch A. Rols M.P. The actin cytoskeleton has an active role in the electrotransfer of plasmid DNA in mammalian cells. Mol Ther. 2011;19:913–921. doi: 10.1038/mt.2010.303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharma S. Fitzgerald K.A. Innate immune sensing of DNA. PLoS Pathog. 2011;7:e1001310. doi: 10.1371/journal.ppat.1001310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stetson D.B. Medzhitov R. Type I interferons in host defense. Immunity. 2006;25:373–381. doi: 10.1016/j.immuni.2006.08.007. [DOI] [PubMed] [Google Scholar]
- Takaoka A. Wang Z. Choi M.K. Yanai H. Negishi H. Ban T. Lu Y. Miyagishi M. Kodama T. Honda K. Ohba Y. Taniguchi T. DAI (DLM-1/ZBP1) is a cytosolic DNA sensor and an activator of innate immune response. Nature. 2007;448:501–505. doi: 10.1038/nature06013. [DOI] [PubMed] [Google Scholar]
- Takeshita F. Leifer C.A. Gursel I. Ishii K.J. Takeshita S. Gursel M. Klinman D.M. Cutting edge: role of Toll-like receptor 9 in CpG DNA-induced activation of human cells. J Immunol. 2001;167:3555–3558. doi: 10.4049/jimmunol.167.7.3555. [DOI] [PubMed] [Google Scholar]
- Tolstov Y.L. Pastrana D.V. Feng H. Becker J.C. Jenkins F.J. Moschos S. Chang Y. Buck C.B. Moore P.S. Human Merkel cell polyomavirus infection II. MCV is a common human infection that can be detected by conformational capsid epitope immunoassays. Int J Cancer. 2009;125:1250–1256. doi: 10.1002/ijc.24509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Unterholzner L. Keating S.E. Baran M. Horan K.A. Jensen S.B. Sharma S. Sirois C.M. Jin T. Latz E. Xiao T.S. Fitzgerald K.A. Paludan S.R. Bowie A.G. IFI16 is an innate immune sensor for intracellular DNA. Nat Immunol. 2010;11:997–1004. doi: 10.1038/ni.1932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasilkoski Z. Esser A.T. Gowrishankar T.R. Weaver J.C. Membrane electroporation: the absolute rate equation and nanosecond time scale pore creation. Phys Rev E Stat Nonlin Soft Matter Phys. 2006;74:21904–21916. doi: 10.1103/PhysRevE.74.021904. [DOI] [PubMed] [Google Scholar]
- Wake C.T. Gudewicz T. Porter T. White A. Wilson J.H. How damaged is the biologically active subpopulation of transfected DNA? Mol Cell Biol. 1984;3:387–398. doi: 10.1128/mcb.4.3.387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weiner G.J. The immunobiology and clinical potential of immunostimulatory CpG oligodeoxynucleotides. J. Leukocyte Biol. 2000;68:455–463. [PubMed] [Google Scholar]
- Yang P. An H. Liu X. Wen M. Zheng Y. Rui Y. Cao X. The cytosolic nucleic acid sensor LRRFIP1 mediates the production of type I interferon via a beta-catenin-dependent pathway. Nat Immunol. 2010;11:487–494. doi: 10.1038/ni.1876. [DOI] [PubMed] [Google Scholar]
- Yasuda K. Yamane I. Nishikawa M. Takakura Y. Macrophage activation by a DNA/cationic liposome complex requires endosomal acidification and TLR9-dependent and independent pathways. J Leukoc Biol. 2005;77:71–79. doi: 10.1189/jlb.0204089. [DOI] [PubMed] [Google Scholar]
- Yoshinaga T. Yasuda K. Ogawa Y. Nishikawa M. Takakura Y. DNA and its cationic lipid complexes induce CpG motif-dependent activation of murine dendritic cells. Immunology. 2006;120:295–302. doi: 10.1111/j.1365-2567.2006.02451.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang X. Brann T.W. Zhou M. Yang J. Oguariri R.M. Lidie K.B. Imamichi H. Huang D.W. Lempicki R.A. Baseler M.W. Veenstra T.D. Young H.A. Lane H.C. Imamichi T. Cutting edge: Ku70 is a novel cytosolic DNA sensor that induces type III rather than type I IFN. J Immunol. 2011a;186:4541–4545. doi: 10.4049/jimmunol.1003389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Z. Yuan B. Bao M. Lu N. Kim T. Liu Y.J. The helicase DDX41 senses intracellular DNA mediated by the adaptor STING in dendritic cells. Nat Immunol. 2011b;12:959–965. doi: 10.1038/ni.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.